Abstract
Eales’ disease is an idiopathic retinal vasculitis of the eye. The disease is predominantly characterized by recurrent vitreous hemorrhage. Interphotoreceptor retinol-binding protein 3 plays a significant role in the etiopathogenesis of this condition. It transports retinoids between the retinal pigment epithelium and the photoreceptors; hence, this protein is a potential target for docking studies. In silico data reveal that herbal molecules interact with regulatory domains of interphotoreceptor retinol-binding protein 3 (IRBP-3), resulting into significant docking score and also forms H-bond and several hydrophobic interactions between active residues of IRBP-3. These interactions between the active residues may lead to significant conformational change in that particular portion of the protein. This efficacy and suitability of ligand was determined on the basis of binding energy calculations. Ginkgolide showed minimum binding energy calculations among selected 10 other natural ligands. This fact of virtual screening for potential ligand can give new insights toward the therapeutic intonations and alterations toward the advances in treatment for Eales’ disease.
Keywords: Eales’ disease, IRBP-3, Docking, Hemorrhages, Venoocclusive, Genetic algorithm, Binding energy
Introduction
Eales’ disease is an idiopathic retinal vasculitis of the eye predominantly characterized by recurrent vitreous hemorrhage [1]. It eventually leads to painless loss of vision [2]. Most commonly, it affects healthy young adult males and is an important cause of preventable blindness in young adults [3]. Eales’ disease is distinctively characterized both by stage of inflammation as well as stage of proliferation [4].
It is well reported with the previous studies that interphotoreceptor retinol-binding protein 3 (IRBP-3) plays a significant role in the etiopathogenesis of this condition. IRBP-3 transports retinoids between the retinal pigment epithelium and the photoreceptors, a critical role in the visual process [5–8]. Thus, after analyzing the significant role of this selected protein, 3D protein model structures of this protein were generated through comparative modeling approaches. Prediction of putative functional site, another important finding, was determined by using computational approaches. Identification of these functional sites was another very significant outcome which was explored in earlier docking studies [9, 10].
Docking studies provide detailed view of drug–receptor interaction and have created new rational approach to drug designing where the structure of drug is designed based on its fit to three-dimensional structures of receptor site [11, 12]. Docking is frequently used to predict the binding orientation of any small molecule or drug candidates to their protein targets in order to predict the affinity and activity of the selected molecule with the target protein. Hence docking plays an important role in the area of pharmaceutical industry [13, 14].
Herbal medicine is still the mainstay of about 75–80 % of the world population, mainly in the developing countries, for primary health care because of better cultural acceptability, better compatibility with the human body, and lesser side effects [15]. The chemical constituents present in the phytochemical are a part of the physiological functions of living flora and hence they are believed to have better compatibility with the human body [16].
In this course of work, virtual screening was done by applying docking studies by AutoDock4.0, which gives the idea of most effective and physiologically suitable herbal molecule [17]. Virtual screening is the most important step for drug developmental studies [18, 19].
Methodology
Docking studies
Functionality of protein was determined by its 3D structure which are important for docking studies. 3D structure of interphotoreceptor retinol-binding protein 3 had already been predicted in previous course of work [8].
Docking is probably the best known of methods used to identify the fit between a receptor and a potential ligand. Predicting ligand–protein interactions is critical to success in many therapeutic research areas such as antibody modeling, elucidation of signal transduction pathways, and identification/optimization of peptide or protein inhibitors or activators for drug discovery.
The natural ligands selected through literature search for this study were docked into the modeled 3D structure of RBP using AutoDock4.0 for virtual screening. Gasteiger partial charges were added to the ligand atoms. Nonpolar hydrogen atoms were merged and rotable bonds were defined. Docking calculations were carried out on the protein models. Essential hydrogen atoms, Kollman charges, and solvation parameters were added with the aid of AutoDock tools. Affinity (grid) map of 60 × 60 × 60 angstrom grid points and spacing were generated using the auto grid program. AutoDock parameter that was set distance-dependent dielectric functions were used in the calculation of van der Waals and the electrostatic term, respectively. Docking simulations were performed using the Lamarckian genetic algorithm. Each docking experiment was derived from 10 different runs that were set to terminate after a maximum of 2,500,000 energy evaluations [20]. This process was carried out for all selected phytochemicals.
Result and discussion
In first duration of work, functionally important 3D structure of proteins was predicted along with the binding sites. In advance course of this work, these predicted binding sites were used for docking of selected herbal ligands with the protein RBP. Results are illustrated in Fig. 1. Figure 1 shows the interaction of RBP domain with alpha pinene, anethol, apigenin, aucubin, carotol, cinnamic aldehyde, coriandrol, ginkgolide, phyllantidine, and zivulgarin.
Fig. 1.
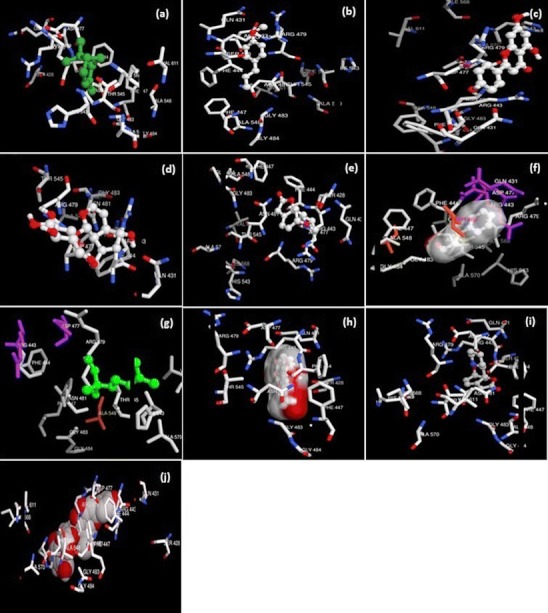
Drug receptor interaction. The docked complexes are a alpha pinene; b anethol; c apigenin; d aucubin; e carotol; f cinnamic aldehyde; g coriandrol; h ginkgolide; i phyllantidine; and j zivulgarin
Binding energy calculation results of drug receptor interaction, for different herbal compounds, are given in Table 1. Among the list of 60 selected herbal compounds or ligands, 10 was screened out on the basis of literature search, having major role in antiocular activity like inflammation, antioxidant, and anti-hemorrhages activity [21, 22]. All the ligands were docked deeply within the binding pocket region of RBP. As shown in Table 1, their AutoDock binding free energies (∆Gb, in kilocalories per moles) and inhibition constants were obtained. Among these 10 compounds, ginkgolide exhibited the lowest free energy −15.99 kcal/mol which is representative of the highest potential binding affinity with the binding site of RBP 3. A good docking interaction implies the prediction of ligand confirmation and orientation within targeted binding site and their lower interactions energies [23]. It can be well predicted from docking results that during this course ginkgolide was found to be most effective and strongly binding ligand with retinol-binding protein 3. This efficacy and suitability of ligand was determined on the basis of binding energy calculations. As ginkgolide is showing minimum binding energy calculations hence this was opted as the most suitable ligand against Eales’ disease for this selected protein ginkgolide could have good molecular interactions with the receptor IRBP-3 (Fig. 1; Table 1). Hence, the role of this compound in inhibiting IRBP-3activity is implied for consideration in Eales-related illness.
Table 1.
Result of drug receptor interaction
| Sample no. | Drug | Run | Binding energy ∆Gb (kcal/mol) | Intermolecular energy | Torsional energy | Internal energy | Histogram | Inhibition constant | Binding residues |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Alpha pinene | 3 | −6.24 | −6.24 | 0 | −0.10 | 10 | 26.88 | PRO409 |
| 2 | Anethol | 3 | −5.53 | −6.13 | 0.60 | −0.15 | 2 | 88.46 | GLN431 |
| 3 | Apigenin | 1 | −5.66 | −6.85 | 1.19 | −0.81 | 5 | 71.52 | TYR495 |
| 4 | Aucubin | 1 | −5.79 | −8.77 | 2.98 | −1.57 | 2 | 57.34 | SER428 |
| 5 | Carotol | 4 | −5.12 | −5.72 | 0.60 | −0.09 | 2 | 176.14 | PHE447 |
| 6 | Cinnamic aldehyde | 2 | −5.38 | −5.98 | 0.60 | −0.10 | 3 | 113.02 | ALA448 |
| 7 | Coriandrol | 8 | −5.53 | −7.02 | 1.49 | −0.26 | 6 | 88.67 | ARG479 |
| 8 | Ginkgolide | 8 | −15.99 | −16.88 | 0.89 | −0.19 | 7 | 1.92 | ASN487 |
| 9 | Phyllantidine | 5 | −7.38 | −7.38 | 0 | 0.00 | 2 | 3.89 | TYR495 |
| 10 | Zivulgarin | 9 | −5.74 | −10.51 | 4.77 | 192.63 | 3 | 62.18 | TRP465 |
Conclusion
The goal was to gain the details of interaction of various natural antiocular disorders agents (ligands) with IRBP-3 that has its role in Eales’ disease. The docking study revealed the important natural ligands involved in the interaction with IRBP-3. Analysis of the molecular interaction between IRBP-3 and natural ligands, based on binding energy, showed that this binding complex of selected ligands with IRBP-3 is reliable. Understanding ligand–protein interactions is important for the exploration of intracellular signaling pathways, modeling of protein complex structures, and for gaining acumens into various biochemical processes. Substantial study between human IRBP-3 and natural ligands was analyzed to recommend more and more proficient search for potential target molecule against Eales’ disease. Virtual screening for potential ligand can give new insights towards the therapeutic intonations and alterations towards the advances in treatment for Eales’ disease.
References
- 1.Sen A, Paine SK, Chowdhury IH, Mukherjee A, Chowdhury S, Mandal LK, et al. Assessment of gelatinase and tumour necrosis factor-a level in the vitreous and serum of patients with Eales disease: role of inflammation-mediated angiogenesis in the pathogenesis of Eales disease. Retina. 2011;31:1412–1420. doi: 10.1097/IAE.0b013e318203c199. [DOI] [PubMed] [Google Scholar]
- 2.Saxena S, Kumar D, Singh VK, Rajasingh J. Immunological studies in Eales disease: a review. Afro-Asian J Ophthalmol. 1995;13:19–22. [Google Scholar]
- 3.Therese KL, Deepa P, Therese J, Bagyalakshmi R, Biswas J, Madhavan HN. Association of mycobacteria with Eales’ disease. Indian J Med Res. 2007;126:56–62. [PubMed] [Google Scholar]
- 4.Das T, Pathengay A, Hussain N, Biswas J. Eales’ disease: diagnosis and management. Eye. 2010;24:472–482. doi: 10.1038/eye.2009.315. [DOI] [PubMed] [Google Scholar]
- 5.Saxena S, Pant AB, Khanna VK, Singh K, Shukla RK, Meyer CH, et al. Tumour necrosis factor-a-mediated severity of idiopathic retinal periphlebitis in young adults (Eales’ disease): implication for anti-TNF-a therapy. J Ocul Biol Dis Inf. 2010;3:35–38. doi: 10.1007/s12177-010-9053-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Danciger M, Kozak CA, Nickerson J, Redmond TM, Farber DB. Localization of the gene for interphotoreceptor retinoid-binding protein to mouse chromosome 14 near Np-1. Genomics. 1990;8:727–731. doi: 10.1016/0888-7543(90)90262-S. [DOI] [PubMed] [Google Scholar]
- 7.Saxena S, Rajasingh J, Biswas S, Kumar D, Shinohara T, Singh VK. Cellular immune response to retinal S-antigen and interphotoreceptor retinoid-binding protein fragments in Eales' disease patients. Pathobiology. 1999;67:39–44. doi: 10.1159/000028049. [DOI] [PubMed] [Google Scholar]
- 8.Tiwari A, Trivedi AC, Srivastava P, Pant AB, Saxena S. Comparative modeling of retinol-binding protein-3 and retinal S-antigen in Eales’ disease and prediction of their binding sites using computational methods. J Ocul Biol Dis Inf. 2010;3:88–91. doi: 10.1007/s12177-011-9060-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alasdair TR, Richard M. Q-Site Finder: an energy-based method for the prediction of protein– ligand binding sites. Bioinformatics. 2005;21:1908–1916. doi: 10.1093/bioinformatics/bti315. [DOI] [PubMed] [Google Scholar]
- 10.Srivastava P, Tiwari A, Trivedi AC. Computational prediction of 3D structure for the matrix protein 2 (BM2) of the influenza B virus. Int Med Technol Univ Med J. 2010;1:22–36. [Google Scholar]
- 11.Bothara KG, Patil AU, Saxena A. Importance of docking studies in drug design. Indian J Pharm Sci. 1998;60:333–337. [Google Scholar]
- 12.Claude CN. Guidebook on molecular modeling in drug design. Boston: Gulf Professional; 1996. [Google Scholar]
- 13.Guttula SV, Rao AA, Sridhar GR, Chakravarthy MS. Protein ligand interaction analysis an in silico potential drug target identification in diabetes mellitus and nephropathy. J Bioinforma Seq Anal. 2011;2:95–99. [Google Scholar]
- 14.Gaba M, Gaba P, Singh S, Gupta GD. An overview on molecular docking. Int J Drug Dev Res. 2010;2:219–231. [Google Scholar]
- 15.Kamboj VP. Herbal medicine. Curr Sci. 2000;78:35–39. [Google Scholar]
- 16.Sen S, Chakraborty R, Sridhar C, Reddy YSR, Biplab D. Free radicals, antioxidants, diseases and phytomedicines: current status and future prospect. Int J Pharm Sci Rev Res. 2010;3:91–100. [Google Scholar]
- 17.Huey R, Morris GM, Olson AJ, Goodsell DS. A semiempirical free energy force field with charge-based desolvation. J Comput Chem. 2007;28:1145–1152. doi: 10.1002/jcc.20634. [DOI] [PubMed] [Google Scholar]
- 18.Rester U. From virtuality to reality—virtual screening in lead discovery and lead optimization: a medicinal chemistry perspective. Curr Opin Drug Discov Dev. 2008;11:559–568. [PubMed] [Google Scholar]
- 19.Rollinger JM, Stuppner H, Langer T. Virtual screening for the discovery of bioactive natural products. Prog Drug Res. 2008;65:213–249. doi: 10.1007/978-3-7643-8117-2_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, et al. Automated docking using a Lamarckian genetic algorithm and empirical binding free energy function. J Comput Chem. 1998;19:1639–1662. doi: 10.1002/(SICI)1096-987X(19981115)19:14<1639::AID-JCC10>3.0.CO;2-B. [DOI] [Google Scholar]
- 21.Braquet P, Esanu A, Buisine E, Hosford D, Broquet C, Koltai M. Recent progress in ginkgolide research. Med Res Rev. 2006;11:295–355. doi: 10.1002/med.2610110303. [DOI] [PubMed] [Google Scholar]
- 22.Sandhu PS, Singh B, Gupta V, Bansal P, Kumar D. Potential herbs used in ocular diseases. Pharm Sci Res. 2011;3:1127–1140. [Google Scholar]
- 23.Camacho J, Vajda S. Protein docking along smooth association pathways. PNAS. 2011;98:1036–1041. doi: 10.1073/pnas.181147798. [DOI] [PMC free article] [PubMed] [Google Scholar]


