Abstract
The remediation of PCBs in soils and sediments remains a particularly difficult problem to solve. The possibility of in situ degradation by microorganisms has been pursued for many years since this approach has the potential to provide a cost-effective and environmentally sustainable alternative to dredging for treatment of PCB impacted sites. Because PCBs are hydrophobic and partition into organic material they accumulate in anoxic environments well poised to support anaerobic dechlorination of highly chlorinated commercial PCBs to congeners that are susceptible to complete aerobic degradation. Laboratory research over the past 25 years is now leading to new microbial technologies that could soon be tested for treatment of PCB impacted soils and sediments in the field.
Introduction
Polychlorinated biphenyls (PCBs) were manufactured as inert, stable, flame- and oxidation-resistant products for a variety of applications such as coolants and dielectric fluids in electrical equipment. Although their manufacture was banned in the U.S. in 1979 and subsequently worldwide in 2001, PCBs persist in the environment as a result of past disposal practices and accidents. Because PCBs are hydrophobic they partition preferentially to organic particles in the environment, which serve both as long-term reservoirs and as carriers that can distribute PCBs great distances from the original point source as a result of current and wind. Although sorbed PCBs resist migration into the water fraction, PCBs enter the food chain by ingestion and desorbtion in benthic microorganisms leading to eventual bioaccumulation and biomagnification of PCBs in organisms higher up in the food chain [1]. PCBs are listed as priority organic pollutants by the EPA (http://nlquery.epa.gov) due to the environmental impact and health risk that they pose and there has been a long search for cost-effective and environmentally sustainable methods such as bioremediation to treat them in situ.
Anaerobic Dechlorination
Discovery
Highly chlorinated PCBs common in many commercial Aroclors resist aerobic degradation until they are partially dechlorinated by anaerobic microbial dechlorination. The first evidence of anaerobic PCB dechlorination was based on changes in congener patterns observed downstream of a capacitor plant that released Aroclor 1242 into the Hudson River [2], which was attributed to microorganisms that could derive energy by using PCBs as electron acceptors; a process later termed dehalorespiration [3]. Quensen et al. [4] followed by others showed that microbial dechlorination of single PCB congeners and Aroclors could be reproduced in laboratory microcosms with PCB-impacted sediments from numerous sources ([5–6] and reviewed in [7–9]). Specific pathways and rates of PCB dechlorinating activity have been reported in freshwater, estuarine and most recently in marine sediments [10], and because they can vary greatly between sediments models have been developed recently to assist in predicting all potential dechlorination pathways for a specific site [11].
Identification and Growth in Culture of PCB Dechlorinating Bacteria
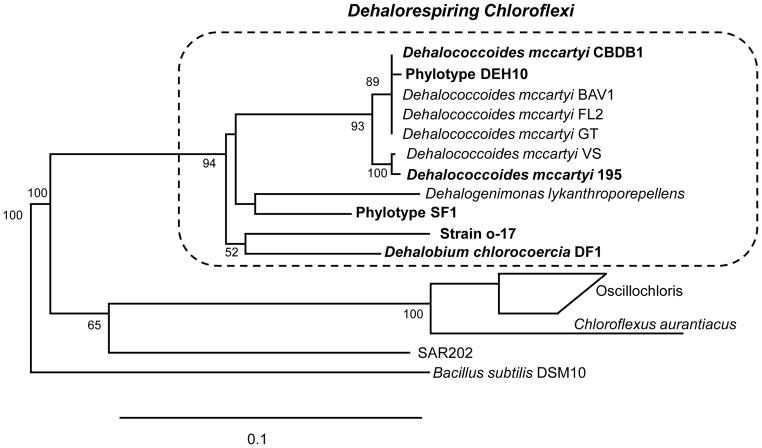
Identification of PCB dehalorespiring bacteria (Table 1 and Fig. 1) eluded investigators for a number of years because the microbes could only be grown in the presence of sediment or soil particles. Using a combination of selective enrichment in sediment microcosms and comparative sequence analysis of 16S rRNA genes after PCR amplification with universal primers, Holoman et al [12] first identified a phylotype within the Chloroflexi as the likely biocatalyst for PCB dechlorination. The identity of two PCB dechlorinating bacteria, strains o-17 and DF-1, were later confirmed in co-culture with Desulfovibrio spp. that were required for growth in a sediment-free medium [13–14]. These were the first reports of sustained anaerobic PCB dechlorination in the complete absence of sediment with PCBs serving as the sole electron acceptor and eventually led to the isolation of “Dehalobium chlorocoercia” DF-1 [15]. Dehalococcoides mccartyi strain 195 (previously D. ethenogenes [16]) has been shown to dechlorinate PCBs in the presence of chlorinated ethenes [17]. Later, D. mccartyi strain CBDB1 was demonstrated to dechlorinate a broad spectrum of PCBs in the absence of sediment [18]. Yoshida et al [19] reported reductive dechlorination of a tri- and tetra-chlorobiphenyl in a sediment-free consortium containing two phylotypes of Dehalobacter, but this activity has not yet been confirmed in pure culture. An alternative approach substituting silica powder for sediment has recently resulted in the sustainable growth of Aroclor-dechlorinating of microorganisms under sediment-free conditions [5,20]. A possible role of sediment in promoting reductive dechlorination could be to serve as a substrate for biofilm formation in close proximity to adsorbed hydrophobic PCBs. This conclusion is consistent with the formation of PCB degrading biofilms, described as “clay hutches”, on sandy clay soil contaminated with PCBs and the observation that biofilms form on the surface and eventually invade PCB droplets in water [21–22]. The ability to culture PCB dechlorinating bacteria in sediment -free medium was a critical achievement for eventual mass culturing of inoculum for bioaugmentation.
Table 1.
Dehalorespiring bacteria and phylotypes with confirmed PCB dechlorinating activities
| Strain or phylotype | Electron donor | Dechlorination activities | Culture status | Reference |
|---|---|---|---|---|
| “Dehalobium chlorocoercia” DF-1 | H2, formate | Double flanked meta/para | Isolate | Wu 2002 |
| Strain o-17 | Acetate | Flanked ortho/meta | Co-culture | Cutter 2001 |
| Phylotype DEH-10 | Unknowna | Double flanked meta/para Para flanked meta |
Sediment microcosm | Fagervold 2005, 2007 |
| Phylotype SF-1 | Unknowna | Double flanked meta Ortho flanked meta |
Sediment microcosm | Fagervold 2005, 2007 |
| Dehalococcoides sp. CBDB1 | Hydrogen | Double and single flanked para Double flanked meta |
Isolate | Adrian 2009 |
| Dehalococcoides mccartyi 195 | Hydrogen | Double flanked meta/para | Isolate | Fennell 2004 |
Grown with a mixture of acetate, propionate, butyrate
Specific activities of individual phylotypes not determined
Figure 1.
Phylogenetic tree showing the relationships between confirmed PCB dechlorinating bacteria and phylotypes (Bold) and other species within the dechlorinating Chloroflexi group based on comparative sequence analysis of 16S rRNA genes Bootstrap values over 50 are indicated at the branch points. The scale bar indicates 10 substitutions per 100 nucleotide positions.
In addition to a preference for solid substrates, some PCB dechlorinating bacteria require growth factors provided by other microorganisms. Dehalococcoides spp. often require acetate as a carbon source and cobalamin as a growth factor [16,23]. Growth and dechlorination by D. mccartyi 195 is also stimulated by an unidentified factor in sterile cell-free supernatant of Dehalococcoides enrichment cultures [16], but the addition of select amino acids will also stimulate growth and dechlorination rates [24]. D. chlorocoerciaDF-1 and strain o-17 require coculturing with a Desulfovibrio spp. or addition of sterile cell-free culture supernatant from pure Desulfovibrio spp., however the nature of this growth factor has also not been identified [15]. D. mccartyi 195 has been shown also to more rapidly grow and dechlorinate tetrachlorethene when grown in co-culture with D. vulgaris Hildenborough supplied with lactate as the sole carbon and energy source [25]. A sulfate reducer may supply growth factors such as amino acids and cobalamin (required for dehalogenases) as well as a slow release of electron donor to the dehalorespiring bacterium via inter-species hydrogen exchange.
Detection and In Situ Monitoring of PCB Dechlorinating Bacteria
PCB dechlorinating phylotypes are difficult to detect using universal 16S rRNA gene primers in nutrient-rich sediment because of their slow growth rates and low yields (<1%) relative to other indigenous species. Dehalococcoides sp. specific primers DHC1F/DHC1377R developed by Hendrickson et al [26] will detect PCB dechlorinating phylotypes within the Dehalococcoides spp. A complementary set of primers (Univ14F/Dehal1265R) developed by Watts et al. [27] will detect PCB dechlorinating phylotypes within the non-Dehalococcoides spp. including strain o-17 and D. chlorocoerciaDF-1 within the Chloroflexi. A group-specific 16S rRNA gene primer set, Chl348F and Dehal884R, will concurrently detect both Dehalococcoides spp. and o-17/DF-1-like PCB-dechlorinating species in soils and sediments [28]. A limitation of currently available 16S rRNA gene primer sets is that the assays are presumptive since they do not differentiate augmented from indigenous species, which might include both PCB dechlorinating and any non-dechlorinating phylotypes. Since 16S rRNA gene sequences will not differentiate strains within this clade, primers need to be designed for highly conserved protein or nucleotide encoding genes with unique sequence not detected in the indigenous background. Park et al [29] developed primers for known and putative reductive dehalogenase (rdh) gene homologs that differentiated two D. mccartyi-specific gene sequences from a background of indigenous Dehalococcoides phylotypes in sediment microcosms bioaugmented with D. mccartyi. The development of additional strain specific primer sets for quantitative monitoring of bioaugmentation inoculum by qPCR should be feasible with the availability of genome sequences for three of the known PCB dechlorinating bacteria D. mccartyi strains 195 and CBDB1, and D. chlorocoerocia, or by identifying putative dehalogenases from other PCB dechlorinators using degenerate rdh primers [30].
From Microcosm to Mesocosm to Field Trials
Biostimulation
Biostimulation of indigenous PCB dechlorinating bacteria has been achieved by halopriming with halogenated aromatic compounds. Halopriming may increase the biomass of the dehalogenating microbial catalysts, induce genes required for dechlorination, and possibly support dehalorespiration or cometabolism of additional PCB congeners. Bedard et al [31] first described the stimulation of weathered Aroclor 1260 dechlorination in sediments by addition of 2,5,3′,4′-tetrachlorobiphenyl and subsequently showed that the same could be achieved with bromated biphenyl congeners (PBBs) [32]. Although PBBs were more effective stimulants that could be completely dehalogenated [33], the deliberate addition of relatively high concentrations (0.6–1 PPM) of halogenated biphenyls into the environment would be subject to regulatory scrutiny. Halogenated benzoates and other halogenated aromatic compounds can also prime PCB dechlorination but they are not as effective as PBBs [34–36]. Most recently, Park et al. expanded the list of haloprimers to include the fungicide pentachloronitrobenzene, which was demonstrated to stimulate more dechlorination of weathered PCBs than tetrachlorobenzene [29,36].
Biostimulation has also been observed after addition of a slow release electron donor. Addition of Feo as a source of cathodic hydrogen stimulated the microbial dechlorination of selected PCB congeners in microcosms containing PCB-impacted Baltimore Harbor sediment [37] and of Aroclor 1254 in a marine sediment [38]. Periodic addition of Feo was observed to stimulate the indigenous population of Dehalococcoides in a microcosm study with PCB impacted sediments from Lake Hartwell, New Bedfod Harbor and Rosanna Marsh [39]. The low levels of hydrogen released by periodic replenishment with Feo provided Dehalococcoides a greater competitive advantage over other hydrogen utilizers such as methanogens and sulfate reducing bacteria, but the effect of Feo on PCB dechlorination activity in the microcosms was not reported. In contrast Feo did not stimulate reductive dechlorination of PCBs in microcosms containing sediment from the Raisin River in Michigan unless they were bioaugmented with an actively dechlorinating culture, which suggests that biostimulation will not be effective in sites that lack a viable indigenous population of PCB dechlorinating bacteria [40]. Although biostimulation with Feo has the potential to be an effective cost-effective treatment for in situ treatment of PCBs, with or without bioaugmentation, the effect of Feo on dechlorination of weathered Aroclor-impacted sediments remains to be tested.
Electrochemical techniques have been used in the past to treat pollutants in groundwater or sediment [41]. The use of carbon cloth electrodes to supply electron donor and acceptor directly to microbes was recently demonstrated to stimulate the dechlorination of tetrachlorobenzene [42]. The method enables one to control the redox, hydrogen and oxygen supply to microorganism within electrochemically reactive sediment caps. Applying an electric current to sediment microcosms, Chun et al [43] recently demonstrated the removal of up to 60% (by mass) of weathered PCBs from Fox River sediment. This result was dependent on the action of anaerobic and aerobic microbes when voltage exceeded 2.2V and H2 and O2 were generated. However, degradation was most apparent in the absence of electrolytic O2 generation with 1.5V applied, suggesting an expanded role for anaerobes in the degradation of the PCBs.
Bioaugmentation
Another potential approach for in situ treatment of PCBs is bioaugmentation with dehalogenating microorganisms. Bedard et al. [44] observed in an enrichment culture that a critical mass of cells was required before reductive dechlorination of spiked Aroclor 1260 was detected and proposed that low indigenous numbers of dehalorespiring bacteria explains why substantial attenuation of PCBs is rarely observed in the environment. However, there have been very few studies to date describing anaerobic bioaugmentation with PCB dechlorinating isolates to stimulate in situ treatment of Aroclor-impacted sediments. May et al. [15] showed that bioaugmentation with DF-1 stimulated the reductive dechlorination of weathered Aroclor 1260 (4.6 ppm) in contaminated soil microcosms, and Krumins [36] reported that the addition of D. mccartyi and pentachloronitrobenzene stimulated the dechlorination of weathered Aroclors 1248, 1254, and 1260 (2.1 ppm) in sediment microcosms. More recently Payne et al [45] demonstrated 56% reduction (by mass) of total penta- and higher chlorinated PCBs in open mesocosms containing weathered Aroclor 1260 (1.3 ppm) after bioaugmentation with D. chlorocoercia DF1, which was sustained within the indigenous microbial population after 120 days. These combined studies provide the most convincing evidence to date that using bioaugmentation for in situ treatment of weathered PCBs is potentially feasible.
Coupling Anaerobic PCB Dechlorination with Aerobic Degradation
Extensive dechlorination of Aroclor 1260 has been observed by the complementary activities of three member consortia in sediment microcosms [46] and with an individual isolate, D. mccartyi CBDB1, in sediment-free culture [18]. As early as 1995 it was recognized that the anaerobic dechlorination of more highly chlorinated congeners followed by the aerobic degradation of those dechlorination products was occurring in the environment [47], and this was suggested to be a potential treatment strategy for PCB impacted sediment. Several investigators have demonstrated that sequentially treating PCB impacted sediment in an anaerobic PCB dehalorespiring enrichment followed by transfer in an aerobic culture containing B. xenovorans LB400 effectively degraded Aroclors by as much a 70% [48–49]. However, all sequential anaerobic-aerobic studies to date have been conducted in closed microcosms and do not represent in situ conditions. One current limitation of this approach is that Aroclors contain varying percentages of congeners with tri- and tetra ortho CBs that are recalcitrant to aerobic degradation. Since reductive dechlorination of ortho-chlorines has been reported infrequently in the environment [32], in situ treatment of a PCB impacted site might require bioaugmentation with an ortho-dechlorinating microorganism in order to prevent a build up of recalcitrant ortho-PCBs. Fagervold [50] reported that addition of the strain o-17 in co-culture with other PCB dechlorinating microorganisms reduced the accumulation of ortho-CBs in sediment microcosms. Sequential bioaugmentation by anaerobic dechlorination with a consortium containing strain o-17 and aerobic degradation with recombinant strains such as Burkholderia xenovorans LB400 (ohb), which effectively grows on and mineralizes ortho substituted PCBs [51], has the potential to lead to more complete degradation of Aroclors.
In situ treatment of PCBs – from laboratory to field
In situ treatment will require sufficient scale-up of biomass to bioaugment large areas of impacted sediment. Payne et al [45] showed that approximately 105 cells g−1 (wet wt) sediment provided a sufficient critical mass of cells to effectively stimulate dechlorination of weathered Aroclor 1260. Based on this cell density and the assumption that bioamendment applied to the top cm will be distributed deeper into the sediment by bioturbation, one km2 of PCB impacted sediment would require 1015 cells grown in a culture volume of 10,000 l and maximum cell density of 108 cells ml−1. Although large-scale culturing of Dehalococcoides sp. grown on chloroethenes has been reported in volumes up to 3,200 l [52], bioaugmentation of dechlorinating species grown at large scale with PCBs would restrict their distribution in the environment. Thus far the only electron acceptors known to support growth of PCB dechlorinating bacteria are halogenated aliphatic or aromatic compounds that are also considered persistent organic pollutants. Unless a non-toxic electron acceptor is identified, methods need to be developed for one that can be readily removed from the cells. Miller et al [53] reported that D. chlorocoerocia pre-grown with tetrachloroethene showed no significant lag in growth when transferred to 2,3,4,5-tetrachlorobiphenyl, which suggests that residual volatile substrates such as chlorinated ethenes could be sparged from cultures prior to harvesting. Alternatively, substituting more readily used electron acceptors such as PPBs might be a viable approach for application in the field.[Bedard 1998]
A suitable means for deploying PCB dechlorinating bacteria in the field is also required. Unlike more soluble organohalides such as chloroethenes, which can be bioaugmented by pumping microorganisms and nutrients into groundwater, PCBs are hydrophobic and tend to become immobilized by adsorption to soil and sediment particles. Effective bioaugmentation of PCB impacted soils and sediments will require a method for inoculating sediment either by direct injection or deployment on solid particles. Dehalogenating microorganisms enriched in microbial granules have been proposed as a mean for deployment in sediments [54–55]. Payne et al [45] recently showed that bioaugmentation of sediments contaminated with weathered Aroclor 1260 was equally effective either by direct injection or on GAC particles. Organic particles such as clay or GAC would strongly sorb PCBs in an aqueous environment and provide substrate for biofilm formation in close proximity to the hydrophobic PCBs [21–22]. The ability to use a solid substrate such as clay or GAC particles for inoculation of cells offers a possible solution for dispersing cells in the field.
Conclusion
Currently the predominant treatment option for PCBs in sediments is dredging followed by stabilization by dewatering and landfilling, but this approach is environmentally disruptive and unsustainable. Passive capping limits exposure of PCBs to the food chain, but since PCBs remain in the environment a potential long-term risk due to gradual or acute disruption of the cap remains. Development of a tractable microbial in situ treatment system would provide a cost-effective, and environmentally sustainable alternative to dredging by reducing the health risks associated with sediment disruption, reducing overall energy use, effectively negating the requirement for extensive waste management and obviating the requirement for substantial habitat restoration. Over the years, several anaerobic bacteria with a broad range of PCB dechlorinating activity have been described and show great potential to be coupled with aerobic PCB degrading bacteria. Novel means of supplying electron donor to the dechlorinators and electron acceptors, methods to mass culture and harvest PCB dechlorinators, design of molecular tools for monitoring the fate of inocula, and approaches for field deployment are currently under development. While much remains to be done to develop methods to advance degradation further, may of the critical components are in place to begin field trials and optimize this biotechnology for effective in situ treatment in PCB-impacted environments (Fig. 2).
Figure 2.
Proposed scheme for in situ treatment of PCB impacted sediments using biaugmentation.
Highlights.
Microbial catalysts with different PCB dechlorinating activities are cultured
Biostimulation and bioaugmentation has been successful in the laboratory
Molecular tools for monitoring dechlorinating bacteria in situ are available
Methods to deploy these catalysts in the field are currently under development
Acknowledgments
This work was supported by U.S. Department of Defense, Environmental Security Technology Certification Program (ER-201215) and the National Institute of Environmental Health Science Superfund Research Program (5R01ES-016197-02). We thank Dr. R. Payne for assistance with figure preparation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Recommended Reading
•• of outstanding interest
• of special interest
- 1.Kelly BC, Ikonomou MG, Blair JD, Morin AE, Gobas FAPC. Food web-specific biomagnification of persistent organic pollutants. Science. 2007;317:236–239. doi: 10.1126/science.1138275. [DOI] [PubMed] [Google Scholar]
- 2.Brown JJF, Wagner RE, Bedard DL, Brennan MJ, Carnahan JC, May RJ. PCB transformations in upper Hudson sediments. Northeastern Environ Sci. 1984;3:167–179. [Google Scholar]
- 3.Holliger C, Hahn D, Harmsen H, Ludwig W, Schumacher W, Tindall B, Vazquez F, Weiss N, Zehnder AJB. Dehalobacter restrictus gen. nov. and sp. nov. a strictly anaerobic bacterium that reductively dechlorinates tetra- and trichloroethene in an anaerobic respiration. Arch Microbiol. 1998;169:313–321. doi: 10.1007/s002030050577. [DOI] [PubMed] [Google Scholar]
- 4.Quensen JF, Tiedje JM, Boyd SA. Reductive dechlorination of polychlorinated biphenyls by anaerobic microorganisms from sediments. Science. 1988;242:752–754. doi: 10.1126/science.242.4879.752. [DOI] [PubMed] [Google Scholar]
- 5.Dudkova V, Demnerova K, Bedard DL. Sediment-free anaerobic microbial enrichments with novel dechlorinating activity against highly chlorinated commercial PCBs. J Chem Technol Biotechnol. 2012;87:1254–1262. [Google Scholar]
- 6.Ho C, Liu S. Effect of coplanar PCB concentration on dechlorinating microbial communities and dechlorination in estuarine sediments. Chemosphere. 2011;82:48–55. doi: 10.1016/j.chemosphere.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 7.Field JA, Cervantes FJ, Lettinga G. Role of quinones in the biodegradation of priority pollutants: a review. Water Sci Tech. 2000;42:215–223. [Google Scholar]
- 8.Bedard DL. A case study for microbial biodegradation: anaerobic bacterial reductive dechlorination of polychlorinated biphenyls—from sediment to defined medium. Annu Rev Microbiol. 2008;62:253–270. doi: 10.1146/annurev.micro.62.081307.162733. [DOI] [PubMed] [Google Scholar]
- 9.Beyer A, Biziuk M. Environmental Fate and Global Distribution of Polychlorinated Biphenyls. Reviews Environ Contam Toxicol. 2009;201:137–158. doi: 10.1007/978-1-4419-0032-6_5. [DOI] [PubMed] [Google Scholar]
- 10.Zanaroli G, Balloi A, Negroni A, Borruso L, Daffonchio D, Fava F. A Chloroflexi bacterium dechlorinates polychlorinated biphenyls in marine sediments under in situ-like biogeochemical conditions. Journal of Hazardous Materials. 2012;209–210:449–457. doi: 10.1016/j.jhazmat.2012.01.042. [DOI] [PubMed] [Google Scholar]
- 11.Hughs AS, Van Briesen JM, Small MJ. Identification of structural properties associated with polychlorinated biphenyl dechlorination processes. Environ Sci Technol. 2010;44:2842–2848. doi: 10.1021/es902109w. [DOI] [PubMed] [Google Scholar]
- 12.Pulliam Holoman TR, Elberson MA, Cutter LA, May HD, Sowers KR. Characterization of a defined 2,3,5,6-tetrachlorobiphenyl-ortho-dechlorinating microbial community by comparative sequence analysis of genes coding for 16S rRNA. Appl Environ Microbiol. 1998;64:3359–3367. doi: 10.1128/aem.64.9.3359-3367.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cutter L, Sowers KR, May HD. Microbial dechlorination of 2,3,5,6-tetrachlorobiphenyl under anaerobic conditions in the absence of soil or sediment. Appl Environ Microbiol. 1998;64:2966–2969. doi: 10.1128/aem.64.8.2966-2969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Q, Sowers KR, May HD. Establishment of a polychlorinated biphenyl-dechlorinating microbial consortium, specific for doubly flanked chlorines in a defined, sediment-free medium. Appl Environ Microbiol. 2000;66:49–53. doi: 10.1128/aem.66.1.49-53.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.May HD, Miller GS, Kjellerup BV, Sowers KR. Dehalorespiration with polychlorinated biphenyls by an anaerobic ultramicrobacterium. Appl Environ Microbiol. 2008;74:2089–2094. doi: 10.1128/AEM.01450-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Löffler FE, Yan J, Ritalahti KM, Adrian L, Edwards EA, Konstantinidis KT, Müller JA, Fullerton H, Zinder SH, Spormann AM. Dehalococcoides mccartyi gen. nov., sp. nov., obligate organohalide-respiring anaerobic bacteria, relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidetes classis nov., within the phylum Chloroflexi. Int J Syst Evol Microbiol. 2012 Epub ahead of print. [Google Scholar]
- 17.Fennell DE, Nijenhuis I, Wilson SF, Zinder SH, Häggblom MM. Dehalococcoides ethenogenes strain 195 reductively dechlorinates diverse chlorinated aromatic pollutants. Environ Sci Technol. 2004;38:2075–2081. doi: 10.1021/es034989b. [DOI] [PubMed] [Google Scholar]
- 18••.Adrian L, Dudkova V, Demnerova K, Bedard DL. “Dehalococcoides” sp. strain CBDB1 extensively dechlorinates the commercial polychlorinated biphenyl mixture Aroclor 1260. Appl Environ Microbiol. 2009;75:4516–4524. doi: 10.1128/AEM.00102-09. Dehalococcoides mccartyi strain CBDB1 is capable of dechlorinating a very broad range of PCB congeners and will extensively dechlorinate Aroclor 1248 and 1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshida N, Ye L, Baba D, Katayama A. Reductive dechlorination of polychlorinated biphenyl and dibenzo-p-dioxins in an enrichments culture containing Dehalobacter species. Microbes Environ. 2009;24:343–346. doi: 10.1264/jsme2.me09132. [DOI] [PubMed] [Google Scholar]
- 20.Bedard DL, Bailey JJ, Reiss BL, Jerzak GVS. Development and Characterization of Stable Sediment-Free Anaerobic Bacterial Enrichment Cultures That Dechlorinate Aroclor 1260. Appl Environ Microbiol. 2006;72:2460–2470. doi: 10.1128/AEM.72.4.2460-2470.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lunsdorf H, Erb RW, Abraham W-R, Timmis KN. “Clay Hutches”: a novel interaction between bacteria and clay minerals. Environ Microbiol. 2000;2:161–168. doi: 10.1046/j.1462-2920.2000.00086.x. [DOI] [PubMed] [Google Scholar]
- 22.Macedo AJ, Kuhlicke U, Neu TR, Timmis KN, Abraham WR. Three stages of a biofilm community developing at the liquid-liquid interface between polychlorinated biphenyls and water. Appl Environ Microbiol. 2005;71:7301–7309. doi: 10.1128/AEM.71.11.7301-7309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He J, Holmes V, Lee P, Alvarez-Cohen L. Influence of vitamin B12 and co-cultures on the growthof Dehalococcoides isolates in defined medium. Appl Environ Microbiol. 2007;73:2847–2853. doi: 10.1128/AEM.02574-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhuang W-Q, Yi S, Feng X, Zinder S, Tang Y, Alvarez-Cohen L. Selective utilization of exogenous amino acids by Dehalococcoides ethenogenes strain 195 and its effects on growth and dechlorination activity. Appl Environ Microbiol. 2011;77:7797–7803. doi: 10.1128/AEM.05676-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25•.Men Y, Feil H, VerBerkmoes N, Shah M, Johnson DR, Lee P, West K, Zinder S, Andersen G, Alvarez-Cohen L. Sustainable syntrophic growth of Dehalococcoides ethenogenes strain 195 with Desulfovibrio vulgaris Hildenborough and Methanobacterium congolense: global transcriptomic and proteomic analyses. ISME Journal. 2011;6:410–421. doi: 10.1038/ismej.2011.111. This report clarifies at the cellular and transcriptome level the complex growth requirements of Dehalococcoides sp. particularly when grown in association with a Desulfovibrio sp., a genus commonly associated with dechlorinating bacteria including those that attack PCBs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hendrickson ER, Payne JA, Young RM, Starr MG, Perry MP, Fahnestock S, Ellis DE, Ebersole RC. Molecular analysis of Dehalococcoides 16S ribosomal DNA from chloroethene-contaminated sites throughout North America and Europe. Appl Environ Microbiol. 2002;68:485–495. doi: 10.1128/AEM.68.2.485-495.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watts JEM, Fagervold SK, Sowers KR, May HD. A PCR based specific assay reveals a population of bacteria within the Chloroflexi associated with the reductive dehalogenation of polychlorinated biphenyls. Microbiology. 2005;151:2039–2046. doi: 10.1099/mic.0.27819-0. [DOI] [PubMed] [Google Scholar]
- 28.Fagervold SK, Watts JEM, May HD, Sowers KR. Sequential reductive dechlorination of meta-chlorinated polychlorinated biphenyl congeners in sediment microcosms by two different Chloroflexi phylotypes. Appl Environ Microbiol. 2005;71:8085–8090. doi: 10.1128/AEM.71.12.8085-8090.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park J-W, Krumins V, Kjellerup BV, Fennell DE, Rodenburg LA, Sowers KR, Kerkhof LJ, Häggblom MM. The effect of co-substrate activation on indigenous and bioaugmented PCB dechlorinating bacterial communities in sediment microcosms. Appl Microbiol Biotechnol. 2011;89:2005–2017. doi: 10.1007/s00253-010-2958-8. [DOI] [PubMed] [Google Scholar]
- 30.Krajmalnik-Brown R, Hölscher T, Thompson IN, Saunders M, Ritalahti KM, Löffler FE. Multiple nonidentical reductive-dehalogenase-homologous genes are common in Dehalococcoides. Appl Environ Microbiol. 2004;70:5290–5297. doi: 10.1128/AEM.70.9.5290-5297.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bedard DL, Bunnell SC, Smullen LA. Stimulation of microbial para-dechlorination of polychlorinated biphenyls that have persisted in housatonic river sediment for decades. Environ Sci Technol. 1996;30:687–694. [Google Scholar]
- 32.Bedard DL. Polychlorinated Biphenyls in Aquatic Sediments: Environmental Fate and Outlook for Biological Treatment. In: Häggblom M, Bossert I, editors. In Dehalogenation: Microbial Processes and Environmental Applications. Kluwer Press; 2003. pp. 443–465. [Google Scholar]
- 33.Bedard DL, VanDort H, Deweerd KA. Brominated biphenyls prime extensive microbial reductive dehalogenation of Aroclor 1260 in Housatonic River sediment. Appl Environ Microbiol. 1998;64:1786–1795. doi: 10.1128/aem.64.5.1786-1795.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deweerd KA, Bedard DL. Use of halogenated benzoates and other halogenated aromatic compounds to stimulate the microbial dechlorination of PCBs. Environ Sci Technol. 1999;33:2057–2063. [Google Scholar]
- 35.Cho YC, Ostrofsky EB, Sokol RC, Frohnhoefer RC, Rhee GY. Enhancement of microbial PCB dechlorination by chlorobenzoates, chlorophenols and chlorobenzenes. FEMS Microb Ecol. 2002;42:51–58. doi: 10.1111/j.1574-6941.2002.tb00994.x. [DOI] [PubMed] [Google Scholar]
- 36.Krumins V, Park JW, Son EK, Rodenburg LA, Kerkhof LJ, Haggblom MM, Fennell DE. PCB dechlorination enhancement in Anacostia River sediment microcosms. Wat Res. 2009;43:4549–4558. doi: 10.1016/j.watres.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 37.Rysavy JP, Yan T, Novak PJ. Enrichment of anaerobic polychlorinated biphenyl dechlorinators from sediment with iron as a hydrogen source. Water Res. 2005;39:569–578. doi: 10.1016/j.watres.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 38.Zanaroli G, Negroni A, Vignola M, Nuzzo A, Shub H-Y, Favaa F. Enhancement of microbial reductive dechlorination of polychlorinated biphenyls (PCBs) in a marine sediment by nanoscale zerovalent iron (NZVI) particles. J Chem Technol Biotechnol. 2012;87:1246–1253. [Google Scholar]
- 39.Varadhan AS, hodadoust AP, Brenner RC. Effect of biostimulation on the microbial community in PCB-contaminated sediments through periodic amendment of sediment with iron. J Ind Microbiol Biotechnol. 2011;38:1691–1707. doi: 10.1007/s10295-011-0959-y. [DOI] [PubMed] [Google Scholar]
- 40.Winchell LJ, Novak PJ. Enhancing polychlorinated biphenyl dechlorination in fresh water sediment with biostimulation and bioaugmentation. Chemosphere. 2008;71:176–182. doi: 10.1016/j.chemosphere.2007.10.021. [DOI] [PubMed] [Google Scholar]
- 41.Ho S, Sheridan P, Athmer C, Heitkamp M, Brackin J, Weber D, Brodsky P. Integrated in situ soil remediation technology: the Lasagna process. Environ Sci Technol. 1995;29:2528–2534. doi: 10.1021/es00010a011. [DOI] [PubMed] [Google Scholar]
- 42•.Sun M, Yan F, Zhang R, Reible D, Lowry G, Gregory K. Redox Control and Hydrogen Production in Sediment Caps Using Carbon Cloth Electrodes. Environ Sci Technol. 2010;44:8209–8215. doi: 10.1021/es101003j. Biodegradation within capped sediments may be limited and here an innovative electrochemical approach to create reactive sediment caps was proven with tetrachlorobenzene. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43•.Chun CL, Payne RB, Sowers KR, May HD. Electrical Stimulation of Microbial PCB Degradation in Sediment. Water Research. 2012 doi: 10.1016/j.watres.2012.09.038. In press. High total PCB mass was removed by an innovative bioelectrochemical approach applied to weathered PCBs in sediment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bedard DL, Ritalahti KM, Loffler FE. The Dehalococcoides population in sediment-free mixed cultures metabolically dechlorinates the commercial polychlorinated biphenyl mixture Aroclor 1260. Appl Environ Microbiol. 2007;73:2513–2521. doi: 10.1128/AEM.02909-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45••.Payne RB, Chun C, May HD, Sowers KR. Enhanced reductive dechlorination of polychlorinated biphenyl impacted sediment by bioaugmentation with a dehalorespiring bacterium. Env Sci Technol. 2011;45:8772–8779. doi: 10.1021/es201553c. Significant dechlorination of Aroclor impacted sediments after bioaugmentation in open mesocosms and inoculum remained viable after 120 days. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fagervold SK, May HD, Sowers KR. Microbial reductive dechlorination of Aroclor 1260 in Baltimore Harbor sediment microcosms is catalyzed by three phylotypes within the phylum Chloroflexi. Appl Environ Microbiol. 2007;73:3009–3018. doi: 10.1128/AEM.02958-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abramowicz DA. Aerobic and anaerobic PCB biodegradation in the environment. Environmental Health Perspectives. 1995;103:97–99. doi: 10.1289/ehp.95103s497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Evans BS, Dudley CA, Klasson KT. Sequential anaerobic-aerobic biodegradation of PCBs in soil slurry microcosms. Appl Biochem Biotechnol. 1996;57–58:885–894. doi: 10.1007/BF02941769. [DOI] [PubMed] [Google Scholar]
- 49.Master ER, Lai VW, Kuipers B, Cullen WR, Mohn WW. Sequential anaerobic-aerobic treatment of soil contaminated with weathered Aroclor 1260. Environ Sci Technol. 2002;36:100–103. doi: 10.1021/es001930l. [DOI] [PubMed] [Google Scholar]
- 50•.Fagervold SK, Watts JEM, May HD, Sowers KR. Effects of bioaugmentation on indigenous PCB dechlorinating activity in sediment microcosms. Wat Res. 2011;45:3899–3907. doi: 10.1016/j.watres.2011.04.048. Aroclor dechlorination pathways can be redirected by bioaugmentation with different dehalorespiring bacteria. [DOI] [PubMed] [Google Scholar]
- 51.Rodrigues JLM, Kachel CA, Aiello MR, Quensen JF, Maltseva OV, Tsoi TV, Tiedje JM. Degradation of Aroclor 1242 Dechlorination Products in Sediments by Burkholderia xenovorans LB400(ohb) and Rhodococcus sp. Strain RHA1(fcb) Appl Environ Microbiol. 2006;72:2476–2482. doi: 10.1128/AEM.72.4.2476-2482.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vainberg S, Condee CW, Steffan RJ. Large-scale production of bacterial consortia for remediaiton of chlorinated solvent-contaminated groundwater. J Ind Microbiol. 2009;36:1189–1197. doi: 10.1007/s10295-009-0600-5. [DOI] [PubMed] [Google Scholar]
- 53.Miller GS, Milliken CE, Sowers KR, May HD. Reductive dechlorination of tetrachloroethene to trans-dichloroethene and cis-dichloroethene by PCB-dechlorinating bacterium DF-1. Environ Sci Technol. 2005;39:2631–2635. doi: 10.1021/es048849t. [DOI] [PubMed] [Google Scholar]
- 54.Nollet H, Verstraete W. Development of microbial granules for PCB dechlorination. Commun Agric Appl Biol Sci. 2003;68:185–189. [PubMed] [Google Scholar]
- 55.Natarajan MR, Nye J, Wu W-M, Wang H, Jain MK. Reductive dechlorination of PCB contaminated Raisin River sediments by anaerobic microbial granules. Biotechnol Bioeng. 1997;55:181–190. doi: 10.1002/(SICI)1097-0290(19970705)55:1<182::AID-BIT19>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]