Abstract
MicroRNAs (miRNAs) have emerged as critical regulators of gene expression through translational inhibition and RNA decay, and have been implicated in the regulation of cellular differentiation, proliferation, angiogenesis, and apoptosis. In this study, we use global bioinformatics analysis of miRNA and mRNA microarrays to predict novel miRNA-mRNA interactions in human embryonic stem cells (hESCs) and induced pluripotent stem cells (iPSCs). In particular, we demonstrate a regulatory feedback loop between the miR-302 cluster and two transcription factors, NR2F2 and OCT4. Our data show high expression of miR-302 and OCT4 in pluripotent cells, while NR2F2 is expressed exclusively in differentiated cells. Target analysis predicts that NR2F2 is a direct target of the miR-302, which we experimentally confirm by reporter luciferase assays and real-time PCR. We also demonstrate that NR2F2 directly inhibits the activity of the OCT4 promoter and thus diminishes the positive feedback loop between OCT4 and the miR-302. Importantly, higher reprogramming efficiencies were obtained when we reprogrammed human adipose-derived stem cells (hASCs) into iPSCs using four factors (KLF4, C-MYC, OCT4, and SOX2) plus miR-302 (this reprogramming cocktail is hereafter referred to as “KMOS3”) when compared to using four factors (“KMOS”). Furthermore, shRNA knockdown of NR2F2 mimics the over-expression of miR-302 by also enhancing reprogramming efficiency. Interestingly, we were unable to generate iPSCs from miR-302a/b/c/d alone, which is in contrast to previous publications that have reported that miR-302 by itself can reprogram human skin cancer cells and human hair follicle cells. Taken together, these findings demonstrate that miR-302 inhibits NR2F2 and promotes pluripotency through indirect positive regulation of OCT4. This feedback loop represents an important new mechanism for understanding and inducing pluripotency in somatic cells.
Keywords: microRNA, reprogramming, induced pluripotent stem cells, OCT4
INTRODUCTION
Embryonic stem cells (ESCs) are derived from the inner cell mass of mammalian blastocyst stage embryos and are characterized by two main properties: self-renewal and pluripotency. Self-renewal confers indefinite ESC proliferation in vitro, while pluripotency provides nearly unlimited differentiation potential. Induced pluripotent stem cells (iPSCs) provide a revolutionary method for deriving pluripotent cells artificially, and are generated from somatic cells by forced over-expression of defined combinations of transcription factors that include OCT4, SOX2, KLF4, and C-MYC 1-3, or OCT4, SOX2, NANOG, and LIN28 4, 5. iPSCs are a promising cell source for regenerative biology, and appear to be similar to ESCs in terms of proliferation and differentiation capacity 1, 4. iPSCs therefore hold great promise for not only basic stem cell biology but also regenerative medicine, which aims to differentiate patient-specific iPSC lines into any cell lineage (e.g., neurons, cardiomyocytes, etc.) for a wide range of therapeutic applications 6-10.
A core set of transcription factors that includes OCT4, SOX2, and NANOG has previously been shown to participate in the regulation of pluripotency and self-renewal in ESCs and iPSCs 11. Rapidly being added to this list of factors are microRNAs (miRNAs), which are post-transcriptional regulatory factors 12. For example, Dicer-/- embryos, which lack miRNA processing, display early embryo lethality 13. Similarly, embryos deficient in Dgcr8 (another essential protein for microRNA biogenesis) will arrest in early development, though this phenotype appears to be less severely affected than the Dicer-/- embryos 14. Both Dicer-/- and Dgcr8-/- mESCs show impairment in cell proliferation and differentiation 14, 15. More specifically, a distinct set of miRNAs and miRNA clusters has been shown to be highly expressed in ESCs, including let-7, miR-302/367 cluster, miR-17-92, and miR-371-373 3, 16, 17. Some of these “embryonic” miRNAs are believed to regulate differentiation directly by targeting key regulation factors 18. In mammals, the miR-302/367 cluster includes miR-302a/b/c/d and miR-367, which are located in the first intron of the LARP7 gene on human chromosome 4. Various aspects of miR-302 have been investigated, including its target genes and potential role in embryonic self-renewal and pluripotency, as well as its reprogramming potency 19-23. However, the specific mechanism(s) by which miR-302 regulates pluripotency remains to be elucidated.
In this study, we predict that the mRNA targets of miRNAs expressed in undifferentiated pluripotent stem cells would be unique to hESCs and iPSCs, and focus on a regulatory loop between miR-302 and Nuclear receptor subfamily 2, group F, member 2 (NR2F2) that appears to affect OCT4 expression in human cells. Furthermore, we show increased generation of bona fide iPSC lines when miR-302 is added to the standard reprogramming factor cocktail, but this effect is diminished when NR2F2 is included in the cocktail. We also show that shRNA against NR2F2 boosts the reprogramming efficiency in a manner similar to miR-302 over-expression. These findings demonstrate that the interference of NR2F2 expression by miR-302 (or shRNA) is an important new mechanism for increasing the efficiency of deriving iPSCs.
MATERIALS AND METHODS
Cell culture and maintenance of hESCs and iPSCs
IMR90 (ATCC), human adipose-derived stem cells (hASCs), adult primary fibroblast, and HeLa (ATCC) were maintained with DMEM containing 10% FBS, Glutamax-I, 4.5 g/L glucose, 110 mg/L sodium pyruvate, 50 U/mL penicillin, and 50 μg/mL streptomycin at 37 °C, 95% air, and 5% CO2 in a humidified incubator. HUVEC was maintained with EGM2 medium (LONZA). hESCs (H7) and hiPSCs were maintained on Matrigel-coated (BD Biosciences) culture dishes with mTeSR-1 medium (STEMCELL Technologies).
Derivation of human adipose stem cells (hASCs)
hASCs were collected from adipose tissue of a female undergoing elective lipoaspiration, in accordance with Stanford University human IRB guidelines. After washing the tissues in serial dilutions of dilute Betadine and PBS, they were subsequently digested with an equal volume of 0.075% type II collagenase in HBSS in 37°C water bath with agitation at 125 rpm for 30 min. After inactivation of collagenase with serum, the stromal vascular fraction was pelleted via centrifugation at 1200g for 5 min. The cell pellet was re-suspended and filtered through a 100 μm cell strainer, and the collected cells were seeded on 10-cm dishes for further expansion. Note that the phenotype and other characteristics of hASCs have been previously described 24.
Reprogramming of hASCs to iPSCs
The cells were seeded in 6-well plates and transduced with individual lentiviruses carrying human OCT4, SOX2, KLF4, or C-MYC, with or without the miR-302a/b/c/d (System Biosciences). On day 5 after transduction, 50,000 cells were transferred onto mouse embryonic fibroblast (MEF) feeder cells. On the next day, the culture medium was switched from hASC growth medium to hESC cell growth medium containing DMEM/F12, 20% knockout serum replacement, 0.1 mM nonessential amino acids, 2 mM l-glutamine, 0.1 mM β-mercaptoethanol, and 4 ng/ml rhFGF-2. Typically, hESC-like colonies were observed around day 12. At day 21, the hESC-like colonies were stained with TRA-1-60 and AKP for calculation of reprogramming efficiency (100% × the number of triple positive colonies / 50,000), or placed onto Matrigel-coated plates with feeder-free maintenance conditions for further analysis.
Microarray data analysis: (A) MicroRNA microarray normalization and data analysis
We used our previous data to determine the miRNA profiles of all three cell types: IMR90 fibroblast, iPSCs (derived from IMR90 fibroblast), and hESCs 3. Using Sanger miRBase Version 10.0 miRNA expression microarrays (LC Sciences, Houston, TX), we analyzed 697 unique miRNAs across biological duplicates of each cell type. Data adjustment includes data filtering, Log2 transformation, and gene centering, and normalization. Gene centering and normalization transform the Log2 values using the mean and the standard deviation of individual genes across all samples. miRNAs with fold change > = 2.0 are considered significantly expressed in iPSCs and hESCs compared to those in IMR90 fibroblast. (B)
Gene expression data analysis
Gene expression data from IMR90 fibroblast, iPSCs (derived from IMR90 fibroblast), and hESCs were obtained with the Agilent 4×44 K whole human genome microarray platform. The adjusted data were subjected to analysis of variance (ANOVA), incorporating the Benjamini Hochberg FDR multiple testing correction, with a significance level of P-value <0.05 to get the differentially expressed genes between different groups. Probe sets were further filtered on the basis of a fold-change cut off of 2.0.
Bioinformatics analysi
MicroRNA target prediction was performed using TargetScan v6.2 25, Microcosm v5.0 26, and miRanda 27, followed by expression correlation between miRNA-mRNA pairs. Matinspector (http://www.genomatix.de) and GeneGo (http://www.genego.com/metacore.php) were used for searching the transcription factor binding sites as well as analyzing the miRNA-mRNA network, respectively. MatInspector is a software tool that utilizes a large library of matrix descriptions for transcription factor binding sites (TFBS) to locate matches in DNA sequences. By introducing a matrix family concept, optimized thresholds, and comparative analysis, the program produces concise results that avoid redundant and false-positive matches. It assigns a quality rating to matches and thus allows quality-based filtering and selection of matches. GeneGo’s MetaCore™ is an integrated “knowledge-based” platform for pathway analysis of OMICs data and gene lists. MetaCore™ is based on a proprietary manually curated database of human protein-protein, protein-DNA, and protein compound interactions, as well as metabolic and signaling pathways for human, mouse, and rat, supported by proprietary ontologies and controlled vocabulary.
Luciferase assay
For luciferase reporter, the 3′-UTR segment of human NR2F2 was amplified by PCR from human cDNA and inserted into the pGL3 control vector (Promega) immediately downstream from the stop codon of Firefly luciferase. The mutant structures were generated with QuikChange® Lightning Site-Directed Mutagenesis Kit (Stratagene) according to manufacturer’s instruction. In 3’UTR assay, human HeLa cells (ATCC) were pre-plated in 24-well tissue culture plates at a concentration of 5 × 104 cells per well. On the next day, the cells were transfected with 40 ng of pRL-TK and 200 ng of UTR reporters, as well as miRNA precursors (Ambion), miR-302a/b/c/d plasmid or miR-367 plasmid. All the transfections were performed with Lipofectamine 2000 (Invitrogen) in Opti-MEM (Invitrogen). For promoter activity assay, the promoter segments of OCT4 and miR-302 cluster were amplified by PCR from human genome DNA and inserted into the pGL3 basic vector (Promega). Electroporation on hESC was performed by Amaxa® Human Stem Cell Nucleofector® Kit II according to manufacturer’s manual. Cell lysates were harvested at 24 and/or 48 hours post-transfection. The firefly and Renilla luciferase activities were measured by using Dual-Luciferase Reporter System (Promega).
In vitro differentiation
Spontaneous differentiation of hASC-iPSCs by forming embryoid body (EB) suspensions was performed in hESC medium without rhFGF-2. Cytokine-induced differentiation was performed with Activin A and BMP4 (R&D Systems), which were added to cell cultures at 20 ng/ml/day in hESC medium without rhFGF-2 for 7 days. (A) For endothelial differentiation, EBs were formed in hESC medium without rhFGF-2 on day 0. On day 1, the medium was supplemented with 20 ng/ml Activin A and 20 ng/ml BMP4; on day 3, the medium was supplemented with 10 ng/ml rhFGF-2 (R&D Systems), 20 ng/ml Activin A, and 20 ng/ml BMP4; on day 5, the EBs were transferred onto Matrigel-coated dishes and the medium was supplemented with 20 ng/ml BMP4, 10 ng/ml rhFGF-2, and 20 ng/ml VEGF-A (R&D Systems); on day 8, the medium was supplemented with 10 ng/ml rhFGF-2 and 20 ng/ml VEGF-A. On day 15, the EBs were harvested by Accutase (Sigma) and sorted by CD31 for purifying endothelial cells. (B) For cardiac differentiation, we employed chemically defined differentiation methods modified from previous reports 28, 29. Briefly, when the iPSC colonies reached the 95% confluence on day 0, the cells were supplemented with a basal medium comprised of RPMI 1640 medium (Invitrogen), 2% B27 supplement minus Insulin (Invitrogen) and 1% penicillin/streptomycin, as well as CHIR99021, a selective inhibitor of glycogen synthase kinase 3β (GSK3β) which activates the canonical Wnt signaling pathway. On day 1, the medium was replaced with basal medium without CHIR99021. On day 3, IWR-1, a Wnt antagonist, was added to basal medium for another 2 days. On day 5, the medium was replaced with basal medium. On day 7, the cells were supplemented with the medium comprised of RPMI 1640 (Invitrogen), 2% B27 supplement, and 1% penicillin/streptomycin. Afterwards, the medium was exchanged every 2 days. (C) For neural stem cell differentiation, the EBs were formed in Neurobasal medium (Invitrogen) supplemented with B-27 (minus vitamin A), 10 ng/ml EGF, and 10ng/ml rhFGF-2 for 7 days. Afterwards, the EBs were dissociated into single cells for further immunostaining.
Fluorescence-activated cell sorting (FACS)
FACS was carried out using a BD Aria II (BD Biosciences) at the Stanford Shared FACS Facility, and data were analyzed by FlowJo software (Tree Star Inc). PE mouse anti-human CD31 (BD Biosciences) was used for endothelial cell specific direct staining. The isotype-matched antibodies were used for background fluorescence.
Bisulfite pyrosequencing
Genomic DNA was subjected to sodium bisulfite treatment using the EZ DNA Methylation-Direct™ Kit according to the manufacturer’s instruction (Zymo Research, Orange, CA). Approximately 25 ng bisulfite-treated DNA was amplified by PCR with NANOG specific primers. The amplified product was cloned into pGEM-T Easy vector (Promega). For each cell type, 10 clones were sequenced. The sequencing results were further analyzed by BiQ analyzer software.
Statistical analysis
Unless otherwise noted, non–microarray data are presented as mean ± S.D. Data were compared using standard or repeated measures, using ANOVA where appropriate. Pairwise comparisons were performed using a two-tailed Student’s t-test. For all data, differences were considered significant for P < 0.05.
RESULTS
Bioinformatic analysis of miRNA and mRNA interactions in pluripotent cells
Using miRNA array data from our previous publication 3 (results summarized in Table S1) and also newly generated mRNA array data, we performed target prediction analysis (see Experiment Procedures for TargetScan, miRanda and Microcosm methods) and miRNA-mRNA gene expression correlation measurements in pluripotent cells vs. IMR90 fetal fibroblasts. Our bioinformatic algorithm is shown in Figure S1. A set of differentially expressed miRNAs was predicted to be significantly correlated (p<0.01) to 23 pluripotency-related genes in both iPSCs and hESCs (Table S2). Several crucial stem cell transcriptional factors are predicted to be miRNA targets in our analysis. For example, TBX3, a self-renewal regulator in embryonic stem cells 30, appears to be a target of miR-106a, 106b, 17, 20a, 25, 92a, 92b, and 93. We were particularly interested in the interaction between the miR-302 cluster, which is up-regulated in pluripotent cells, and NR2F2, which is up-regulated in somatic cells (Figure 1A) and contains miR-302 binding sites in its 3’UTR. Note that because miR-302a, b, c, and d share the same target genes, we selected miR-302b as a surrogate for the miR-302 in the majority of our analyses and subsequent molecular studies. Real-time PCR confirmed that miR-302b and OCT4 were highly expressed in pluripotent cells, while NR2F2 was highly expressed in a variety of somatic cells (Figure 1B). We also measured NR2F2 expression during spontaneous differentiation (Figure 1C) and during cytokine-induced differentiation (Figure 1D). In both differentiation conditions, the levels of both OCT4 and miR-302b decreased as hESCs began to differentiate; conversely, the level of NR2F2 increased. Overall, these expression results demonstrate an inverse correlation between OCT4/miR-302 and NR2F2.
Figure 1. Differential expression of OCT4, miR-302, and NR2F2.
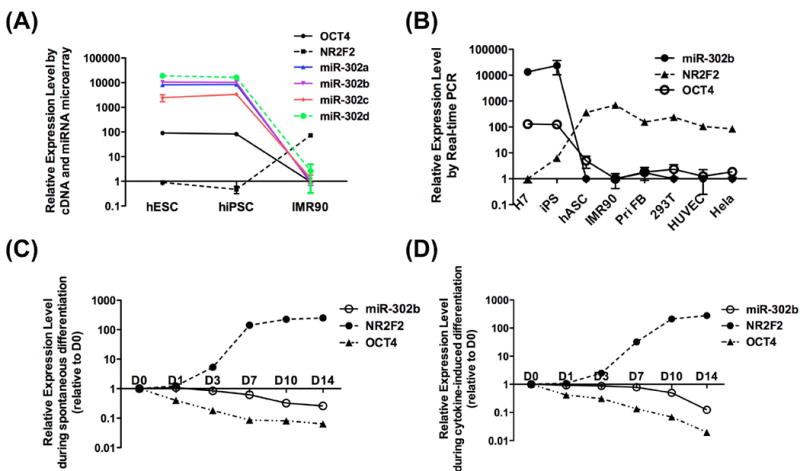
(A) microarray data show that OCT4 and miR-302abcd are highly expressed in pluripotent cells but not in IMR90 fibroblasts. The opposite pattern of expression is seen for NR2F2. (B) Real-time PCR confirms that OCT4 and miR-302b are highly expressed in pluripotent cells (H7 and iPSCs), whereas NR2F2 is highly expressed in differentiated cells such as hASC, IMR90, primary fibroblast, 293T, HUVEC, and HeLa cells. During spontaneous differentiation (C) and cytokine-induced differentiation (D), the expression of OCT4 and miR-302b gradually decreased in hESCs. Conversely, the expression of NR2F2 increased.
MiR-302 directly suppresses the transcription factor NR2F2
To investigate whether NR2F2 can be directly targeted by miR-302, we constructed a firefly luciferase reporter vector carrying the wild-type 3’UTR of NR2F2 (“T-WT”), which contains two predicted wild-type binding sites for miR-302. For negative controls, we created three mutant reporters: (1) T-MT1 reporter with the first 6-bp mismatched seed region, (2) T-MT2 reporter with the second 6-bp mismatched seed region, and (3) T-MT1,2 reporter with both mismatched seed regions (Figure 2A and 2B). Similar luciferase reporter systems have been successfully used by the authors to confirm miRNA-target gene interactions 31-33. The luciferase reporter T-WT was co-transfected with the precursor mimic miR-302b into HeLa cells. Compared to all three negative control luciferase vectors, the wild-type T-WT luciferase activity showed the largest decrease in activity after introduction of the miR-302b precursor mimic (Figure 2C). Mutations in either of the miR-302 binding sites resulted in a modest reduction of luciferase activity, and mutations at both sites negated the inhibition completely (Figure 2C). We did not observe significant differences between T-MT1 and T-MT2, indicating that these two binding sites may play similar roles in the binding activity between the miR-302 and its target. After replacing the miR-302b mimic with a lentiviral vector expressing miR-302a/b/c/d (Lv-302), we obtained similar results as with the miR-302b mimic (Figure S2A). Because miR-367 lies within the 302/367 cluster, we also wondered whether miR-367 might also regulate NR2F2. We found no direct relationship between miR-367 and NR2F2 based on our in silico predictions, which was confirmed in vitro using a luciferase assay of miR-367 against T-WT (Figure S2B). We also investigated whether the miR-302b mimic could inhibit endogenous NR2F2 expression in HeLa cells. Compared to control, miR-302b inhibited endogenous NR2F2 mRNA expression by almost 50% (Figure 2D).
Figure 2. MiR-302 suppresses NR2F2.
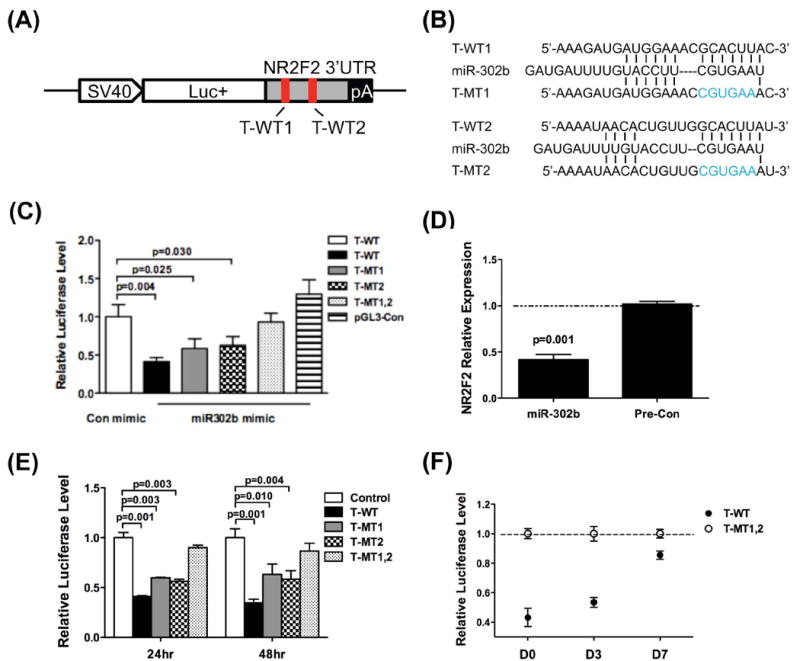
(A) Schematic diagram of the 3’UTR segment of NR2F2 used in the pGL3 control vector (red bars = sites of mutated sequences). (B) miR-302b binding sites and mutant sequences. (C) In HeLa cells, the T-WT luciferase activity was significantly decreased by the miR-302b precursor. However, miR-302b precursor did not repress the vector with double mutations (T-MT1,2), and showed diminished inhibition in vectors with single mutations (T-MT1 and T-MT2). (D) miR-302b precursor inhibits the endogenous expression of NR2F2 in HeLa cells. (E) The activity of T-WT reporter is inhibited in hESCs. Vectors with binding site mutations relieve this suppression. Control = blank luciferase reporter without 3’UTR. (F) During differentiation, the repression of reporters decreased gradually due to the decreasing expression of miR-302.
To investigate whether endogenous miR-302 directly targets the NR2F2 3’UTR in hESCs, the 3’UTR luciferase reporter constructs were transfected into H7 hESCs by electroporation (blank luciferase reporter without 3’UTR was used as control). Note that we validated our electroporation technique by transfecting a GFP reporter vector into H7 cells and achieved more than 80% transfection efficiency (Figure S3). After 24 hours of transfection, we observed a significant repression of the wild-type luciferase reporter (T-WT) in comparison to control reporter (Figure 2E). Similar to our findings in HeLa cells, the two binding site mutations (T-MT1 or T-MT2) resulted in reduced inhibitory activity, and the double mutation (T-MT1,2) exhibited very little inhibitory activity by endogenous miR-302. The effect persisted after 48 hours post-transfection (Figure 2E), suggesting that NR2F2 is directly regulated by endogenous miR-302 repression.
We next determined if the repression of NR2F2 by miR-302 is mitigated during hESC differentiation. In this experiment, the ratio of the NR2F2 double mutant to wild-type luciferase reporter indicates the degree of inhibition by endogenous miR-302 18. Upon culturing hESCs in ultra-low attachment dishes in basic fibroblast growth factor (bFGF) free medium, the inhibition decreased gradually as differentiation progressed from day 0 to day 7 (Figure 2F), which we attribute to decreasing expression of endogenous miR-302 during differentiation (Figures 1C and 1D). Taken together, our results demonstrate that miR-302 does indeed inhibit NR2F2 expression through direct binding of the two regulatory sites found in the 3’UTR of NR2F2, and that this regulatory relationship may directly affect pluripotency and differentiation.
The OCT4, NR2F2 and miR-302 circuit
Several groups have reported that OCT4 binds to the promoter of the miR-302 cluster and increases its expression 22, 34, and that NR2F2 inhibits OCT4 through direct binding of its promoter 35, 36. To study these regulatory relationships in the context of our new data showing that miR-302 also targets NR2F2, we employed a pGL3-Basic plasmid to engineer two new luciferase reporter constructs: pOCT4-Luc (partial promoter region of OCT4 driving firefly luciferase) and p302-Luc (partial promoter region of miR-302 cluster driving firefly luciferase) (Figure 3A). pOCT4-Luc or control reporter (pGL3-Basic) were transfected into HeLa cells along with a reference reporter (pRL-TK). Afterwards, the cells were transduced by a lentivirus carrying miR-302a/b/c/d (Lv-302) or GFP (Lv-GFP) as control. Compared to Lv-GFP, Lv-302 significantly increased the reporter activity of pOCT4-Luc (Figure 3B). By contrast, the control reporter pGL3-Basic was not affected by Lv-302 transduction.
Figure 3. Luciferase assays of OCT4 and miR-302 promoter activity.
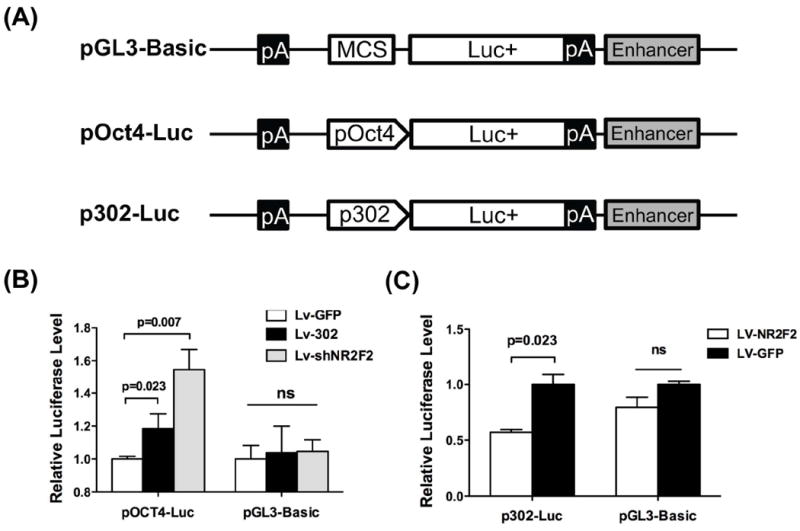
(A) Schematic diagram for constructing promoter segments of OCT4 and miR-302 cluster into pGL3 basic vectors. (B) MiR-302a/b/c/d (Lv-302) and shRNA against NR2F2 (Lv-shNR2F2) increase the activity of OCT4 promoter reporter (pOCT4-Luc) compared to control reporter (pGL3-Basic) in HeLa cells. GFP control (Lv-GFP) lentiviral transduction did not cause increase in activity in either pOCT4-Luc or control, as expected. Scale bar=50 μm. (C) Exogenous expression of NR2F2 (Lv-NR2F2) inhibits the activity of miR-302 cluster promoter reporter in H7 cells relative to control Lv-GFP. “ns” represents non-statistical significance.
To determine whether NR2F2 may be a key mediator in the regulatory loop between miR-302 and OCT4, we next transduced HeLa cells with a lentivirus expressing shRNA against NR2F2 (Lv-shNR2F2) in place of Lv-302. Note that transduction of the shRNA construct against NR2F2 led to approximately 50% knockdown of endogenous NR2F2 (Figure S4). We found that Lv-shNR2F2 increased pOCT4-Luc reporter activity (Figure 3B), confirming previous studies showing that NR2F2 silences OCT4 by directly binding to its promoter 35. We next transfected the promoter reporter (p302-Luc) into hESCs along with a lentivirus expressing NR2F2 (Lv-NR2F2) or Lv-GFP (control). Compared to Lv-GFP, Lv-NR2F2 significantly repressed the reporter activity of p302-Luc, whereas Lv-NR2F2 did not affect reporter activity of pGL3-Basic (Figure 3C), indicating the inhibition of OCT4 by NR2F2 led to the suppression of reporter activity of p302-Luc. Taken together, these results suggest that there exists a regulatory circuit in which miR-302, NR2F2, and OCT4 are the important players: (a) OCT4 upregulates miR-302, (b) miR-302 suppresses NR2F2, and (c) NR2F2 suppresses OCT4.
MiR-302 enhances reprogramming by suppressing NR2F2
Because endogenous miR-302 promotes endogenous OCT4 expression, we asked whether miR-302a/b/c/d could enhance reprogramming similar to recent studies 19, 37. We have previously reported that human adipose stem cells (hASCs) can be efficiently reprogrammed into bona fide iPSCs using OCT4, SOX2, KLF4, and C-MYC 38, and we therefore opted to use hASCs in our experiments. Interestingly, we were unable to generate iPSCs from miR-302a/b/c/d alone, which is in contrast to previous publications that have reported that miR-302 by itself can reprogram human skin cancer cells and human hair follicle cells 21, 39. Because we also could not successfully reprogram hASCs when OCT4 was removed from the cocktail (KMS3), we believe that OCT4 is essential for reprogramming and that it cannot be replaced by miR-302 (Figure 4A). Although we were unable to reprogram hASCs using miR-302 alone, we did find that miR-302 improved the reprogramming efficiency by 4-fold (0.143 ±0.017% vs 0.035±0.012%; p=0.007), when used in combination with the four transcription factors (Figure 4A). All bona fide iPSC colonies were alkaline phosphatase (AKP) and TRA-1-60 positive and displayed hESC-like morphology (Figure 4B and S6). Of note, expression of GFP (a marker used in our vectors) was silenced during the reprogramming process, which suggests that the reprogramming vectors were silenced and endogenous pluripotency programs turned on (Figure 4B). Overall, our results demonstrate that although miR-302 can improve the reprogramming efficiency, it cannot replace OCT4.
Figure 4. The miR-302 enhances reprogramming.
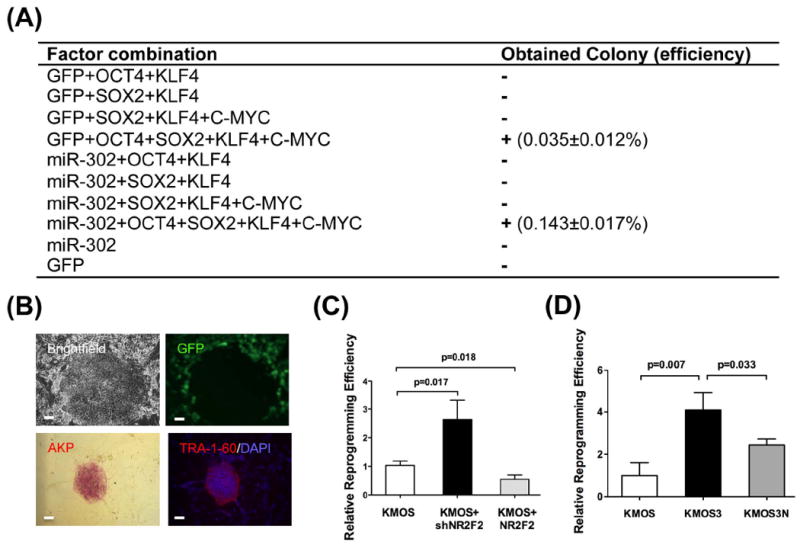
(A) Reprogramming experiments using different combinations of factors and microRNAs. ‘+’ = successful reprogramming, ‘−’ = not successful. Scale bar=50 μm. (B) Bona fide iPSC colonies showed AKP+, TRA-1-60+ and hESC-like morphology. Note that the GFP expression was silenced during the reprogramming process. Scale bar=50 μm. (C) Improved reprogramming efficiency with NR2F2 knockdown. Compared to standard KMOS reprogramming, the addition of shRNA against NR2F2 increases the reprogramming efficiency. Conversely, over-expression of NR2F2 inhibited reprogramming. (D) The ~4-fold increase in reprogramming efficiency caused by miR-302 is partially reduced with the addition of NR2F2 to the reprogramming factor cocktail. This finding confirms that NR2F2 is a key gene target in the reprogramming process.
In our 3’UTR luciferase data, we showed that miR-302 directly represses NR2F2. We hypothesized that NR2F2 knockdown might improve the reprogramming efficiency in a manner similar to miR-302 over-expression. After hASCs were transduced with a lentivirus expressing shRNA against NR2F2 (Lv-shNR2F2), we observed a significant improvement in reprogramming efficiency compared to KMOS (Figure 4C). Next we tested whether the converse is true, wherein over-expression of NR2F2 might impair reprogramming. Indeed, the up-regulation of NR2F2 by lentivirus significantly inhibits the reprogramming efficiency compared to KMOS (Figure 4D). Taken together, our data demonstrate that inhibition of NR2F2 by miR-302 or shRNA knockdown is an important mechanism for reprogramming.
To demonstrate that NR2F2 is indeed an important target of miR-302 during the reprogramming process, we added lentivirus expressing NR2F2 to our KMOS3 cocktail. The addition of NR2F2 resulted in a partial loss of the increased reprogramming efficiency that is caused by miR-302 (Figure 4D). However, NR2F2 did not completely inhibit the increase in reprogramming that we have observed in our experiments with miR-302, suggesting that miR-302 improves reprogramming via targeting other genes in addition to NR2F2 21, 37, 40, 41.
Confirmation of pluripotency in KMOS3-derived iPSCs
While our iPSCs derived using KMOS and KMOS3 appeared morphologically identical, we wanted to confirm that no genotypic or phenotypic changes occurred as a direct result of adding miR-302 to the reprogramming cocktail. All iPSC lines were positive for pluripotent markers (Figure 5A) and showed similar OCT4 protein expression by Western blot (Figure 5B). Whole genome expression microarrays, hierarchical clustering (Figure 5C) and principal component analysis (Figure 5D) demonstrated that all iPSC lines were distinct from hASCs but very similar to hESCs. Bisulfite gene sequencing revealed that the promoter region of the pluripotency factor NANOG was demethylated at similar levels in KMOS3-iPSC and hESC lines (Figure 5E). Karyotype analysis revealed no abnormalities (Figure 5F), and a teratoma formation assay of KMOS3-iPSCs yielded all three germ layers (Figure S7). We next differentiated KMOS- and KMOS3-iPSCs into endothelial cells, cardiomyocytes, and neural stem cells using published protocols 23, 29. Endothelial cells derived from both lines expressed similar levels of both CD31 and CD144 markers and showed no obvious differences in tube-like formation and LDL uptake (Figure S8). After 14 days of cardiac differentiation, spontaneously contracting cell layers from both iPSC lines were observed under light microscopy (Movie S1 and S2). Immunostaining of dissociated cardiomyocytes showed that these cells expressed similar levels of the cardiac specific markers TNNT2 and MEF2C (Figure S9). Finally, immunostaining of differentiated neural stem cells showed similar levels of NESTIN and SOX2 in both iPSC lines (Figure S10), which are known neural markers 42, 43. In conclusion, we detected no significant genotypic or phenotypic differences between KMOS3- and KMOS-iPSCs.
Figure 5. Characterization of iPSC lines.
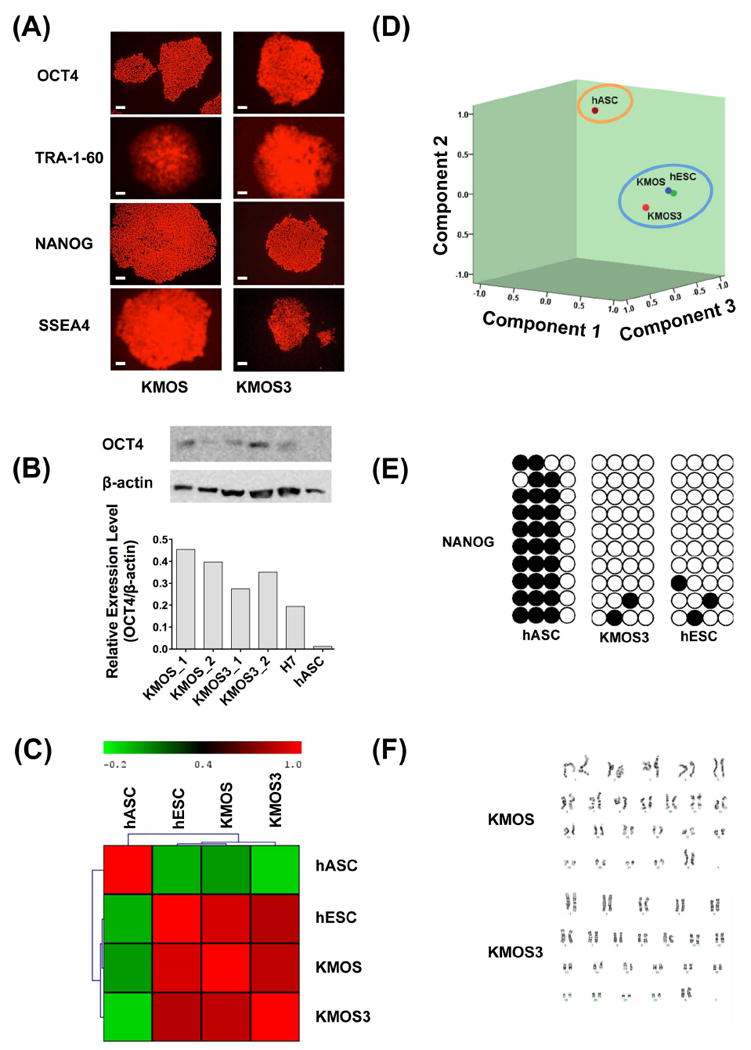
(A) Immunofluorescence staining shows that iPSC lines express pluripotent markers OCT4, NANOG, TRA-1-60, and SSEA4. (B) Western blot shows that the protein levels of OCT4 are expressed at comparable levels in KMOS- and KMOS3-derived iPSCs. Scale bar=50 μm. (C) Hierarchical clustering of mRNA microarray data demonstrates that all iPSC lines are distinct from hASCs but similar to hESCs. (D) Principal component analysis (PCA) indicates that iPSCs and hESCs cluster together. (E) NANOG promoter methylation is similar between KMOS3-iPSCs and hESCs. (F) Both iPSC lines have normal karyotypes. KMOS = KLF4, cMYC, OCT4, and SOX2, and KMOS3 = all factors plus miR-302
DISCUSSION
Understanding the regulatory core circuitry of self-renewal and differentiation in ESCs and iPSCs is critical to our understanding of pluripotency. Many transcription factors that take part in the regulation of pluripotency have already been characterized, including OCT4, SOX2, NANOG, TCF3, and many others 12, 44-46. Likewise, some miRNAs have been discovered that may also play a role in pluripotency 47, including let-7 48, miR-302/367 cluster 22, miR-17-92 49, miR-371-373 16, miR-145 18, and others. Nucleotides 2-7 in the mature forms of miRNAs represents the key region for target mRNA recognition, which leads to miRNA-mRNA interactions within RISC and degradation and/or translational inhibition. In general, these small molecules achieve their effects by binding the 3’UTR of hundreds of target genes, resulting in diminished translation of entire gene programs. For example, in human fibroblasts, miR-302b and miR-372 target several genes that are involved in cell cycle, epithelial-mesenchymal transition, epigenetic regulation and vesicular transport, including TGFBR2 and RHOC 37. Target recognition is therefore critical for the elucidation of miRNA function under physiological and pathological conditions 50-53.
Our 3’UTR luciferase experiments with miR-302, OCT4, and NR2F2 can be integrated into previously described transcription factor interactions to create a broader pluripotency network (Figure S11). To briefly summarize this network, the miR-302/367 cluster is expressed at high levels in pluripotent cells 3, and its promoter is directly bound by many ESC-specific transcription factors such as OCT4 that can increase transcription 22, 54. Previous reports have demonstrated that miR-302 indirectly regulates a number of downstream targets including the transcription factors B-MYB 55, 56 and E2F2 56, 57, which are up-regulated in pluripotent cells. B-MYB regulates the expression of both OCT4 and SOX2 and is a critical regulator of cell cycle progression, whereas E2F2 regulates B-MYB to maintain the pluripotent state. REX1 and NANOG are direct targets of OCT4 56, and NANOG itself is a transcriptional regulator of REX-1 in pluripotent cells 58. Previous studies suggest that NR2F2 can silence the OCT4 promoter through direct binding 35, and we now report that NR2F2 is post-transcriptionally repressed by miR-302.
NR2F2, also called Chicken Ovalbumin Upstream Promoter Transcription Factor II (COUP-TFII), is an orphan member of the steroid/thyroid hormone receptor superfamily. NR2F2 is involved in the control of development, cellular differentiation, angiogenesis, growth, and metabolism 59, 60. In the human trophoblast, NR2F2 plays an important role by transactivating TFAP2A expression 61. In the development of mouse right presomitic mesoderm, NR2F2 controls asymmetric retinoic acid signaling, suggesting that it has a role in controlling somatic symmetry 62. NR2F2 (as well as NR2F1) is transiently expressed in the ventricular zone of the early embryonic central nervous system, temporally specifying neural progenitor cells, which greatly contributes to the development and advancement of the central nervous system 63. In mouse embryonic development and tumorigenesis in adults, NR2F2 can modulate lymphatic vessel development through direct regulation of neuropilin-2 64. NR2F2 also regulates PROX1 in lymphatic endothelial cells during differentiation and maintenance of lymphatic vessels 65.
To these known roles in development, our data demonstrate that NR2F2 may also act as a critical molecular switch for diminishing pluripotency and priming the cell for differentiation. Specifically, NR2F2 appears to mediate the interaction between miR-302 and OCT4 by acting as a target of miR-302, which leads to reduced inhibition of OCT4. Conversely, in differentiated cells, the low expression of the miR-302 does not suppress NR2F2 protein production, thus allowing NR2F2 to bind to the promoter of OCT4 and inhibit its transcription (Figure S12). Corroborating previous reports 22, 34, we also note that OCT4 can promote the expression of miR-302 cluster by binding its promoter region. Clearly, the overall picture of pluripotency and its core regulatory factors is only just emerging, and future studies undoubtedly will identify even more novel interactions and molecules that also play a role in this network. However, our current understanding of the triumvirate of miR-302, NR2F2, and OCT4 already allows us to hypothesize how each might affect the reprogramming of somatic cells into iPSCs, which we investigated next.
At present, iPSC reprogramming remains a highly inefficient process, and new strategies to boost the efficiency of iPSCs generation are urgently needed. Recent papers have indicated that the over-expression of the miR-302/367 cluster and addition of valproic acid (VPA) can rapidly and efficiently reprogram mouse and human fibroblasts into iPSCs without exogenous transcription factors 66. These authors claimed that miR-367 plays a critical role in the process of miR-302/367-mediated reprogramming, and can activate the expression of OCT4. A second study has demonstrated that an inducible miR-302 can reprogram human hair follicle cells (hHFCs) to iPSCs 21. A third group has found that mature miRNAs, including miR-200c, the miR-302s, and miR-369, can also reprogram mouse and human cells 19. In our study, we were unable to reprogram somatic cells using miR-302 alone, which we attribute to differences in methodology (e.g., the absence of VPA), differences in starting somatic cell type, and/or lack of other key miRNAs. In addition, anecdotal experience from various independent labs suggests that the reprogramming efficiency of iPSCs using miRNAs is not as high as previously reported. However, further studies are needed to determine the cause(s) of this discrepancy.
Although our results show that miR-302 improves the reprogramming efficiency via targeting NR2F2, we found that addition of NR2F2 to the KMOS3 cocktail did not completely inhibit this boost in efficiency. Clearly the function of miRNAs are complex, and a single miRNA can target hundreds of genes 67. Recent studies have shown that miR-302 also targets epigenetic regulators (AOF1/2, MECP1-p66, MECP2 and MBD2) 21, cell cycle regulators (Cyclin D1/D2, CDK2, BMI-1, PTEN) 40, 68, 69, TGF-β regulators (Lefty1/2, TGFBR2) 23, 37, 41, and BMP inhibitors (DAZAP2, SLAIN1, and TOB2) 40. Therefore, NR2F2 is likely not the only gene that is targeted during the reprogramming process, and these other target genes may play complementary roles during reprogramming. However, our data does support the conclusion that NR2F2 is a critical target of miR-302 for reprogramming, and is likely one of the key players in this complex process.
In summary, we were able to achieve higher reprogramming efficiencies when using miR-302 in conjunction with the standard cocktail of OCT4, SOX2, KLF4, and c-MYC. We therefore believe that direct inhibition of NR2F2 by miR-302 is a new and important mechanism for understanding pluripotency and for enhancing iPSC derivation. Future studies should elucidate even more complex interactions and molecules that together govern pluripotency. To this end, we believe that the interaction of miR-302 and NR2F2 will be a major component within this broader network, and this interaction can be manipulated in order to enhance the derivation of iPSCs.
Supplementary Material
Figure S1. Algorithm for correlating miRNAs with their target mRNAs in pluripotent cells.
Figure S2. (A) The miR-302 inhibits NR2F2 3’UTR reporter activity. The lentiviral miR-302a/b/c/d (Lv-302) suppresses the activity of T-WT reporter. However, mutations in either binding site reduce this suppression. The reporter with double 3’UTR mutations showed luciferase activity similar to control (pGL3-Con). (B) The miR-367 can not inhibit NR2F2 3’UTR reporter activity.
Figure S3. Electroporation of GFP reporter into H7 hESCs. The hESCs were electroporated with GFP reporter using the Amaxa® Human Stem Cell Nucleofector® Kit II. With electroporation, typical transfection efficiency is estimated at 80%. Scale bar=50 μm.
Figure S4. Knockdown of NR2F2. The shRNA against NR2F2 led to approximately 50% knockdown of endogenous NR2F2 in HeLa cells.
Figure S5. The expression level of both miR-302b and NR2F2 upon the reprogramming initiation. (A) The reprogramming with the addition of Lv-shNR2F2 or Lv-NR2F2. (B) The reprogramming with Lv-302 and/or Lv-NR2F2.
Figure S6. Typical colonies used for the evaluation of reprogramming efficiency. These colonies were AKP+, TRA-1-60+, and had hESC-like morphology. Note that colonies that lacked TRA-1-60 and/or did not have hESC-like morphology were discarded from our analysis. Scale bar=50 μm.
Figure S7. Representative hematoxylin and eosin staining of a solid encapsulated teratoma derived from typical iPSC lines implanted into the sub-renal capsule of SCID mice. Histological analysis of tissue demonstrated presence of glandular structures (endoderm), cartilage (mesoderm), and neural rosettes (ectoderm). Scale bar=250 μm.
Figure S8. Characterization of KMOS and KMOS3-iPSC-derived endothelial cells (ECs). The differentiated and purified endothelial cells expressed typical EC markers such as CD31 and CD144. Furthermore, these endothelial cells showed tube formation on Matrigel and low-density lipoprotein uptake, indicating that they are functional endothelial cells. Scale bar=50 μm.
Figure S9. Characterization of KMOS- and KMOS3-iPSC-derived cardiomyocytes. The differentiated cardiomyocytes expressed cardiac-specific TNNT2 and transcription factor MEF2C. Scale bar=50 μm.
Figure S10. Characterization of iPSC-derived neural stem cells (NSCs). The differentiated NSCs expressed NESTIN and transcription factor SOX2. Scale bar=50 μm.
Figure S11. Schematic diagram showing predicted miR-302b induced regulatory circuit in pluripotent cells.
Figure S12. OCT4, miR-302, and NR2F2 regulatory models in pluripotent cells and non-pluripotent cells based on experimental results from our study. Blue line represents clustering in pluripotent cells; black line represents clustering in differentiated cells.
Movie S1. After 14 days of cardiac differentiation of KMOS iPSCs, spontaneously contracting cell layers are observed under light microscopy.
Movie S2. After 14 days of cardiac differentiation of KMOS3 iPSCs, spontaneously contracting cell layers are observed under light microscopy.
Acknowledgments
This research is supported in part by Burroughs Wellcome Foundation, NIH DP2OD004437, R01 HL113006, RC1 AG036142 (J.C.W.), and AHA Postdoctoral fellowship 10POST3730079 (S.H.).
Footnotes
Author contribution: Shijun Hu: Conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing; Kitchener D. Wilson: Conception and design, collection and/or assembly of data, data analysis and interpretation; Zhumur Ghosh: Conception and design, collection and/or assembly of data, data analysis and interpretation; Leng Han: Collection and/or assembly of data, data analysis and interpretation; Yongming Wang: data analysis and interpretation; Feng Lan: collection and/or assembly of data, data analysis and interpretation; Katherine J. Ransohoff: collection and/or assembly of data; Joseph C. Wu: Conception and design, data analysis and interpretation, financial support
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
None
References
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Wilson KD, Venkatasubrahmanyam S, Jia F, et al. MicroRNA profiling of human-induced pluripotent stem cells. Stem Cells Dev. 2009;18:749–758. doi: 10.1089/scd.2008.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 5.Jia F, Wilson KD, Sun N, et al. A nonviral minicircle vector for deriving human iPS cells. Nat Methods. 2010;7:197–199. doi: 10.1038/nmeth.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han JW, Yoon YS. Induced pluripotent stem cells: emerging techniques for nuclear reprogramming. Antioxid Redox Signal. 2011;15:1799–1820. doi: 10.1089/ars.2010.3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alcon A, Cagavi Bozkulak E, Qyang Y. Regenerating functional heart tissue for myocardial repair. Cell Mol Life Sci. 2012 doi: 10.1007/s00018-012-0942-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plath K, Lowry WE. Progress in understanding reprogramming to the induced pluripotent state. Nat Rev Genet. 2011;12:253–265. doi: 10.1038/nrg2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robinton DA, Daley GQ. The promise of induced pluripotent stem cells in research and therapy. Nature. 2012;481:295–305. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tiscornia G, Vivas EL, Belmonte JC. Diseases in a dish: modeling human genetic disorders using induced pluripotent cells. Nat Med. 2011;17:1570–1576. doi: 10.1038/nm.2504. [DOI] [PubMed] [Google Scholar]
- 11.Boyer LA, Lee TI, Cole MF, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marson A, Levine SS, Cole MF, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernstein E, Kim SY, Carmell MA, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Medvid R, Melton C, et al. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murchison EP, Partridge JF, Tam OH, et al. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci U S A. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stadler B, Ivanovska I, Mehta K, et al. Characterization of microRNAs involved in embryonic stem cell states. Stem Cells Dev. 2010;19:935–950. doi: 10.1089/scd.2009.0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ren J, Jin P, Wang E, et al. MicroRNA and gene expression patterns in the differentiation of human embryonic stem cells. J Transl Med. 2009;7:20. doi: 10.1186/1479-5876-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu N, Papagiannakopoulos T, Pan G, et al. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 19.Miyoshi N, Ishii H, Nagano H, et al. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell. 2011;8:633–638. doi: 10.1016/j.stem.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 20.Rosa A, Brivanlou AH. A regulatory circuitry comprised of miR-302 and the transcription factors OCT4 and NR2F2 regulates human embryonic stem cell differentiation. EMBO J. 2011;30:237–248. doi: 10.1038/emboj.2010.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin SL, Chang DC, Lin CH, et al. Regulation of somatic cell reprogramming through inducible mir-302 expression. Nucleic Acids Res. 2011;39:1054–1065. doi: 10.1093/nar/gkq850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Card DA, Hebbar PB, Li L, et al. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol Cell Biol. 2008;28:6426–6438. doi: 10.1128/MCB.00359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barroso-Deljesus A, Lucena-Aguilar G, Sanchez L, et al. The Nodal inhibitor Lefty is negatively modulated by the microRNA miR-302 in human embryonic stem cells. FASEB J. 2011 doi: 10.1096/fj.10-172221. [DOI] [PubMed] [Google Scholar]
- 24.Bunnell BA, Flaat M, Gagliardi C, et al. Adipose-derived stem cells: isolation, expansion and differentiation. Methods. 2008;45:115–120. doi: 10.1016/j.ymeth.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis BP, Shih IH, Jones-Rhoades MW, et al. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 26.Griffiths-Jones S, Saini HK, van Dongen S, et al. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 28.Hudson J, Titmarsh D, Hidalgo A, et al. Primitive Cardiac Cells from Human Embryonic Stem Cells. Stem Cells Dev. 2011 doi: 10.1089/scd.2011.0254. [DOI] [PubMed] [Google Scholar]
- 29.Gonzalez R, Lee JW, Schultz PG. Stepwise chemically induced cardiomyocyte specification of human embryonic stem cells. Angew Chem Int Ed Engl. 2011;50:11181–11185. doi: 10.1002/anie.201103909. [DOI] [PubMed] [Google Scholar]
- 30.Han J, Yuan P, Yang H, et al. Tbx3 improves the germ-line competency of induced pluripotent stem cells. Nature. 2010;463:1096–1100. doi: 10.1038/nature08735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu SJ, Ren G, Liu JL, et al. MicroRNA expression and regulation in mouse uterus during embryo implantation. J Biol Chem. 2008;283:23473–23484. doi: 10.1074/jbc.M800406200. [DOI] [PubMed] [Google Scholar]
- 32.Hu S, Huang M, Li Z, et al. MicroRNA-210 as a novel therapy for treatment of ischemic heart disease. Circulation. 2010;122:S124–131. doi: 10.1161/CIRCULATIONAHA.109.928424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu S, Huang M, Nguyen PK, et al. Novel microRNA prosurvival cocktail for improving engraftment and function of cardiac progenitor cell transplantation. Circulation. 2011;124:S27–34. doi: 10.1161/CIRCULATIONAHA.111.017954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu H, Deng S, Zhao Z, et al. Oct4 regulates the miR-302 cluster in P19 mouse embryonic carcinoma cells. Mol Biol Rep. 2011;38:2155–2160. doi: 10.1007/s11033-010-0343-4. [DOI] [PubMed] [Google Scholar]
- 35.Ben-Shushan E, Sharir H, Pikarsky E, et al. A dynamic balance between ARP-1/COUP-TFII, EAR-3/COUP-TFI, and retinoic acid receptor:retinoid X receptor heterodimers regulates Oct-3/4 expression in embryonal carcinoma cells. Mol Cell Biol. 1995;15:1034–1048. doi: 10.1128/mcb.15.2.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schoorlemmer J, van Puijenbroek A, van Den Eijnden M, et al. Characterization of a negative retinoic acid response element in the murine Oct4 promoter. Mol Cell Biol. 1994;14:1122–1136. doi: 10.1128/mcb.14.2.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subramanyam D, Lamouille S, Judson RL, et al. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol. 2011 doi: 10.1038/nbt.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun N, Panetta NJ, Gupta DM, et al. Feeder-free derivation of induced pluripotent stem cells from adult human adipose stem cells. Proc Natl Acad Sci U S A. 2009;106:15720–15725. doi: 10.1073/pnas.0908450106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin SL, Chang DC, Chang-Lin S, et al. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA. 2008;14:2115–2124. doi: 10.1261/rna.1162708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lipchina I, Elkabetz Y, Hafner M, et al. Genome-wide identification of microRNA targets in human ES cells reveals a role for miR-302 in modulating BMP response. Genes Dev. 2011;25:2173–2186. doi: 10.1101/gad.17221311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosa A, Spagnoli FM, Brivanlou AH. The miR-430/427/302 family controls mesendodermal fate specification via species-specific target selection. Dev Cell. 2009;16:517–527. doi: 10.1016/j.devcel.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 42.Pollard SM, Conti L, Sun Y, et al. Adherent neural stem (NS) cells from fetal and adult forebrain. Cereb Cortex. 2006;16(Suppl 1):i112–120. doi: 10.1093/cercor/bhj167. [DOI] [PubMed] [Google Scholar]
- 43.Conti L, Pollard SM, Gorba T, et al. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 2005;3:e283. doi: 10.1371/journal.pbio.0030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Z, Oron E, Nelson B, et al. Distinct Lineage Specification Roles for NANOG, OCT4, and SOX2 in Human Embryonic Stem Cells. Cell Stem Cell. 2012;10:440–454. doi: 10.1016/j.stem.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 45.Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ivanova N, Dobrin R, Lu R, et al. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- 47.Mallanna SK, Rizzino A. Emerging roles of microRNAs in the control of embryonic stem cells and the generation of induced pluripotent stem cells. Dev Biol. 2010;344:16–25. doi: 10.1016/j.ydbio.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Foshay KM, Gallicano GI. miR-17 family miRNAs are expressed during early mammalian development and regulate stem cell differentiation. Dev Biol. 2009;326:431–443. doi: 10.1016/j.ydbio.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 50.van Rooij E. Introduction to the series on microRNAs in the cardiovascular system. Circ Res. 2012;110:481–482. doi: 10.1161/CIRCRESAHA.111.257311. [DOI] [PubMed] [Google Scholar]
- 51.Zhu H, Fan GC. Role of microRNAs in the reperfused myocardium towards post-infarct remodelling. Cardiovasc Res. 2012;94:284–292. doi: 10.1093/cvr/cvr291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev. 2011;91:827–887. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- 53.Ambros V. MicroRNAs and developmental timing. Curr Opin Genet Dev. 2011;21:511–517. doi: 10.1016/j.gde.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barroso-delJesus A, Romero-Lopez C, Lucena-Aguilar G, et al. Embryonic stem cell-specific miR302-367 cluster: human gene structure and functional characterization of its core promoter. Mol Cell Biol. 2008;28:6609–6619. doi: 10.1128/MCB.00398-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tarasov KV, Tarasova YS, Tam WL, et al. B-MYB is essential for normal cell cycle progression and chromosomal stability of embryonic stem cells. PLoS One. 2008;3:e2478. doi: 10.1371/journal.pone.0002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boheler KR. Stem cell pluripotency: a cellular trait that depends on transcription factors, chromatin state and a checkpoint deficient cell cycle. J Cell Physiol. 2009;221:10–17. doi: 10.1002/jcp.21866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu W, Giangrande PH, Nevins JR. E2Fs link the control of G1/S and G2/M transcription. EMBO J. 2004;23:4615–4626. doi: 10.1038/sj.emboj.7600459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shi W, Wang H, Pan G, et al. Regulation of the pluripotency marker Rex-1 by Nanog and Sox2. J Biol Chem. 2006;281:23319–23325. doi: 10.1074/jbc.M601811200. [DOI] [PubMed] [Google Scholar]
- 59.Pereira FA, Tsai MJ, Tsai SY. COUP-TF orphan nuclear receptors in development and differentiation. Cell Mol Life Sci. 2000;57:1388–1398. doi: 10.1007/PL00000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martin LJ, Tremblay JJ. Nuclear receptors in Leydig cell gene expression and function. Biol Reprod. 2010;83:3–14. doi: 10.1095/biolreprod.110.083824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hubert MA, Sherritt SL, Bachurski CJ, et al. Involvement of transcription factor NR2F2 in human trophoblast differentiation. PLoS One. 2010;5:e9417. doi: 10.1371/journal.pone.0009417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vilhais-Neto GC, Maruhashi M, Smith KT, et al. Rere controls retinoic acid signaling and somite bilateral symmetry. Nature. 2010;463:953–957. doi: 10.1038/nature08763. [DOI] [PubMed] [Google Scholar]
- 63.Naka H, Nakamura S, Shimazaki T, et al. Requirement for COUP-TFI and II in the temporal specification of neural stem cells in CNS development. Nat Neurosci. 2008;11:1014–1023. doi: 10.1038/nn.2168. [DOI] [PubMed] [Google Scholar]
- 64.Lin FJ, Chen X, Qin J, et al. Direct transcriptional regulation of neuropilin-2 by COUP-TFII modulates multiple steps in murine lymphatic vessel development. J Clin Invest. 2010;120:1694–1707. doi: 10.1172/JCI40101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamazaki T, Yoshimatsu Y, Morishita Y, et al. COUP-TFII regulates the functions of Prox1 in lymphatic endothelial cells through direct interaction. Genes Cells. 2009;14:425–434. doi: 10.1111/j.1365-2443.2008.01279.x. [DOI] [PubMed] [Google Scholar]
- 66.Anokye-Danso F, Trivedi CM, Juhr D, et al. Highly Efficient miRNA-Mediated Reprogramming of Mouse and Human Somatic Cells to Pluripotency. Cell Stem Cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin SL, Chang DC, Ying SY, et al. MicroRNA miR-302 inhibits the tumorigenecity of human pluripotent stem cells by coordinate suppression of the CDK2 and CDK4/6 cell cycle pathways. Cancer Res. 2010;70:9473–9482. doi: 10.1158/0008-5472.CAN-10-2746. [DOI] [PubMed] [Google Scholar]
- 69.Lee NS, Kim JS, Cho WJ, et al. miR-302b maintains “stemness” of human embryonal carcinoma cells by post-transcriptional regulation of Cyclin D2 expression. Biochem Biophys Res Commun. 2008;377:434–440. doi: 10.1016/j.bbrc.2008.09.159. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Algorithm for correlating miRNAs with their target mRNAs in pluripotent cells.
Figure S2. (A) The miR-302 inhibits NR2F2 3’UTR reporter activity. The lentiviral miR-302a/b/c/d (Lv-302) suppresses the activity of T-WT reporter. However, mutations in either binding site reduce this suppression. The reporter with double 3’UTR mutations showed luciferase activity similar to control (pGL3-Con). (B) The miR-367 can not inhibit NR2F2 3’UTR reporter activity.
Figure S3. Electroporation of GFP reporter into H7 hESCs. The hESCs were electroporated with GFP reporter using the Amaxa® Human Stem Cell Nucleofector® Kit II. With electroporation, typical transfection efficiency is estimated at 80%. Scale bar=50 μm.
Figure S4. Knockdown of NR2F2. The shRNA against NR2F2 led to approximately 50% knockdown of endogenous NR2F2 in HeLa cells.
Figure S5. The expression level of both miR-302b and NR2F2 upon the reprogramming initiation. (A) The reprogramming with the addition of Lv-shNR2F2 or Lv-NR2F2. (B) The reprogramming with Lv-302 and/or Lv-NR2F2.
Figure S6. Typical colonies used for the evaluation of reprogramming efficiency. These colonies were AKP+, TRA-1-60+, and had hESC-like morphology. Note that colonies that lacked TRA-1-60 and/or did not have hESC-like morphology were discarded from our analysis. Scale bar=50 μm.
Figure S7. Representative hematoxylin and eosin staining of a solid encapsulated teratoma derived from typical iPSC lines implanted into the sub-renal capsule of SCID mice. Histological analysis of tissue demonstrated presence of glandular structures (endoderm), cartilage (mesoderm), and neural rosettes (ectoderm). Scale bar=250 μm.
Figure S8. Characterization of KMOS and KMOS3-iPSC-derived endothelial cells (ECs). The differentiated and purified endothelial cells expressed typical EC markers such as CD31 and CD144. Furthermore, these endothelial cells showed tube formation on Matrigel and low-density lipoprotein uptake, indicating that they are functional endothelial cells. Scale bar=50 μm.
Figure S9. Characterization of KMOS- and KMOS3-iPSC-derived cardiomyocytes. The differentiated cardiomyocytes expressed cardiac-specific TNNT2 and transcription factor MEF2C. Scale bar=50 μm.
Figure S10. Characterization of iPSC-derived neural stem cells (NSCs). The differentiated NSCs expressed NESTIN and transcription factor SOX2. Scale bar=50 μm.
Figure S11. Schematic diagram showing predicted miR-302b induced regulatory circuit in pluripotent cells.
Figure S12. OCT4, miR-302, and NR2F2 regulatory models in pluripotent cells and non-pluripotent cells based on experimental results from our study. Blue line represents clustering in pluripotent cells; black line represents clustering in differentiated cells.
Movie S1. After 14 days of cardiac differentiation of KMOS iPSCs, spontaneously contracting cell layers are observed under light microscopy.
Movie S2. After 14 days of cardiac differentiation of KMOS3 iPSCs, spontaneously contracting cell layers are observed under light microscopy.


