Abstract
Historically it has been virtually impossible to experimentally determine the contribution of residual protein entropy to fundamental protein activities such as the binding of ligands. Recent progress has illuminated the possibility of employing NMR relaxation methods to quantitatively determine the role of changes in conformational entropy in molecular recognition by proteins. The method rests on using fast internal protein dynamics as a proxy. Initial results reveal a large and variable role for conformational entropy in the binding of ligands by proteins. Such a role for conformational entropy in molecular recognition has significant implications for enzymology, signal transduction, allosteric regulation and the development of protein-directed pharmaceuticals.
Introduction
The nature of the folded state of proteins defines their potential for engaging in chemical phenomena in a biologically productive way. Indeed, the many forces that govern the stability of the native state are complex and continue to be difficult to unravel and dissect through experiment, simulation or theory. This is particularly true for two principal components defining the physical character of the native state ensemble: the residual conformational (configurational) entropy and the entropy of solvent, which often couched in discussions of the hydrophobic effect. Over the past decade, the plasticity of proteins has moved to center stage as ever more powerful experimental and analytical technologies emerge. Indeed, it is now clear that the residual conformational entropy of protein is larger than perhaps captured by common representations of the energy landscapes of proteins. The presence of such large reservoirs of entropy in the native states of proteins raises the important question of whether it is coupled to the energetics of protein functions that exist beyond those involved in the folding and stability of protein molecules. Here we will focus on the role of conformational entropy in perhaps the simplest of functions exhibited by the native state of proteins: the binding of ligands and the use of the attendant free energy to guide biological action. The formation of protein complexes involves a complicated manifold of interactions that are diverse and complex. This complexity is reflected in the difficulty of computing the energetics of interactions involving proteins using molecular structure alone [1].
Molecular recognition by proteins is fundamental to almost every biological process, particularly those protein-ligand complexes underlying enzymatic catalysis. A variety of mechanistic frameworks have been constructed to describe the physical basis for specific high affinity interactions between protein molecules and their ligands. They have ranged from the simple “lock and key” interaction model [2] to the “induced fit” [3] and the “conformational selection” models [4*]. Each of these derives from an increasing recognition of the plasticity and energetics of the ensemble of structures that proteins occupy and the potential role for travelling across this complex energy landscape in protein function. Here we highlight recent advances in the experimental characterization of protein conformational entropy using a “dynamical proxy” derived from NMR relaxation phenomena.
Motion as a Proxy for Conformation Entropy
Expression of the total binding free energy emphasizes that the entropy of binding is comprised of contributions from the protein, the ligand and the solvent:
| (1) |
The free energy of binding (ΔGbind) and the enthalpy of binding (ΔHbind) can in favorable cases be directly measured using isothermal titration calorimetry and the associated total binding entropy (ΔSbind) obtained by arithmetic. Unfortunately, as equation (1) indicates, the molecular origins of these thermodynamic parameters remain obscure. Views of associations involving proteins have largely been seen through the lens of structure, which helps reveal the details of the interactions governing the enthalpy. In distinct contrast, the origin of the binding entropy remain difficult to grasp as it inherently involves a manifold of states that the protein, ligand and solvent can occupy, each having its own probability for existence. Historically, the contributions by solvent entropy to binding thermodynamics have often dominated discussions and are usually framed in terms of the hydrophobic effect [5]. Hydrophobic solvation by water remains the subject of extensive analysis [6].
In principle, the entropic contributions of a structured protein to the binding of a ligand (ΔSprotein) includes both changes in its internal conformational (configurational) entropy (ΔSconf), as well as changes in rotational and translational entropy (ΔSRT) [1]. Equation 1 emphasizes that the measurement of the entropy of binding does not resolve contributions from protein conformational entropy. Arguments from fundamental considerations [7] and early observations from simulation (e.g. [8] [9]) and experiment (e.g. [10,11]) indicated that proteins fluctuate about a structure closely similar to that observed by crystallography and that these fluctuations could reflect significant residual conformational entropy. Yet it is only recently that experimental methods and strategies have been created to quantitatively assess this and related ideas.
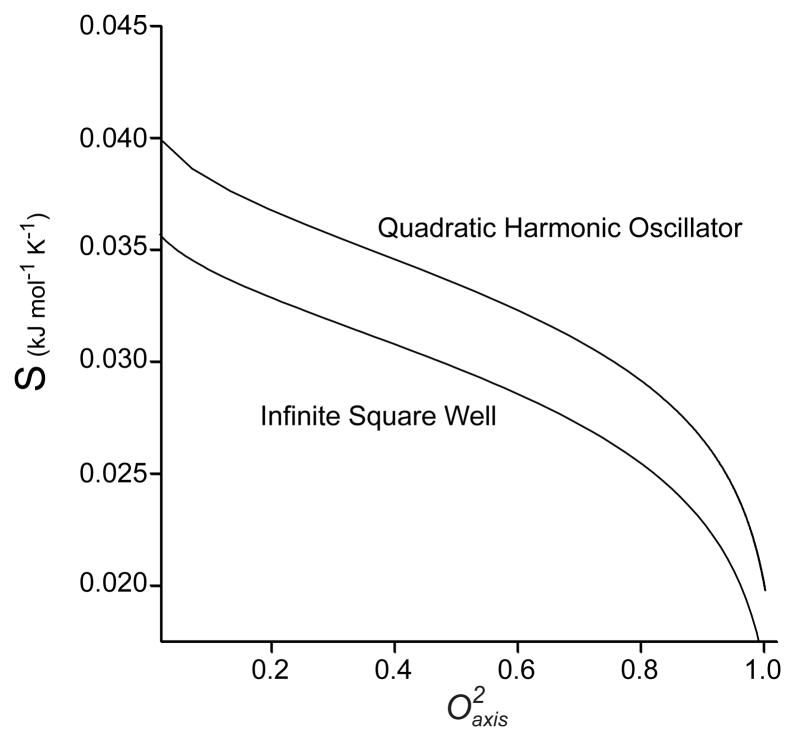
How to experimentally measure ΔSconf is not obvious. During the past two decades there has been a significant effort to develop solution NMR methods that serve to provide measures of motion between different microscopic structural states [12,13]. Solution NMR spectroscopy has emerged as the most powerful experimental technique for accessing protein motion in a site-resolved comprehensive manner. Motion expressed on the sub-nanosecond timescale corresponds to significant entropy [14] and NMR relaxation methods are particularly well suited for its characterization [12]. Slower motions, such as those detected by dispersion-type NMR relaxation experiments [15], are generally interpreted to involve large components of the protein interconverting between a very small number of states and would, to a first approximation, not contribute significantly to the conformational entropy. A popular and very robust way of capturing the essential character of the fast motions that gives rise to NMR relaxation phenomena is the so-called “model free approach” of Lipari and Szabo [16]. Roughly speaking, the spatial disorder of the NMR probe is captured in the squared generalized order parameter (O2). The order parameter by definition ranges from zero to one, corresponding to complete isotropic disorder and complete rigidity of the NMR interaction vector within the molecular frame, respectively. It is this motional parameter that offers the potential to provide access to statements about thermodynamics in general [17] and conformational entropy in particular [18,19]. This leads directly to the idea of using measures of motion as a proxy for conformational entropy. This is illustrated quantitatively in Figure 1 where two simple models of the potential energy function governing the motion of an NMR relaxation “spy” are parametrically related to the corresponding entropy. From these types of calculations one learns that obtaining absolute entropies is model-dependent and likely not accessible. However, differences in measures of motion (i.e. ΔO2) seem more useful as the slopes of parametric relationships such as those shown in Figure 1 are very similar for intermediate order parameters [12].
Figure 1.
Parametric relationships between two simple potentials, an infinite square well and a quadratic simple harmonic oscillator, and the corresponding Lipari-Szabo squared generalized order parameter.
The role of protein conformational entropy in ligand binding
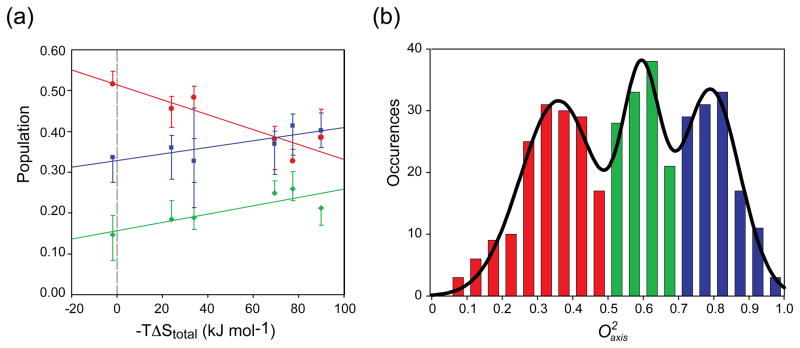
Some time ago it was appreciated that NMR relaxation studies indicated that methyl-bearing amino acids exhibit three types of motion in globular protein [12]. These types or classes of motion often (but not always) segregate into three distinct distributions of order parameters describing motion of the methyl symmetry axis ( ). The so-called “J-class” corresponds to side chain having significant sub-nanosecond rotameric interconversion that leads to averaging of the J-coupling constant across the associated torsion angle. The “ω-class” corresponds to side chain methyl groups showing highly restricted motion reminiscent of the backbone. The “α-class” arises from large amplitude motion within a single rotameric well and does not involve significant (i.e. experimentally detectable) rotameric interconversion. The detailed structural and energetic contexts giving rise to these classes remain unknown [12]. Nevertheless, the variation of these distributions upon ligand binding provides a powerful and model-independent insight into the potential importance of conformational entropy in molecular recognition by proteins. The initial example was the high affinity binding of calcium-activated calmodulin to peptides corresponding to the calmodulin-binding domains of enzymes regulated by this interaction. The three-class distribution is remarkably distinct in these calmodulin complexes (Figure 2a). What is most interesting is that the relative populations of the three classes are modulated by the binding of the target domains and is surprisingly linearly correlated with the overall entropy of binding (Figure 2b). There is no physical law requiring such a relationship but its existence indicates that nature knows about the conformational entropy that the motion represents and it has been employed in the adjustment of the overall free energy of binding. To a first approximation, a linear relationship between the binding entropy and its components would naturally arise during evolution driven by selection based on the free energy of association. Interestingly, the population of the J-class, and the entropy that it represents, is anti-correlated the populations of the α and ω classes. Using a model-dependent proxy of the conformational entropy, one can also estimate the apparent contribution of conformational entropy to the binding of the target domains. A linear relationship between the apparent change in conformational entropy and the total binding entropy is recapitulated [20]. Unfortunately, this model-dependent approach has many issues. These include questions about the validity of the potential energy model used, the fact that only methyl-bearing side chains are considered, the effects of correlated motion are completely ignored and so on [12,20]. Thus tremendous uncertainty is present. Recently this has been overcome by subsuming the microscopic details and taking an empirical approach to directly calibrate the “entropy meter” represented by changes in dynamics [21**].
Figure 2.
(a) Summed distribution of the amplitude of methyl-bearing side chain motion of calmodulin in complex with six different target domains. The solid lines represent fitted 3-Gaussian distributions centred on values of 0.35 (J-class, red), 0.58 (α-class, green) and axis 0.78 (ω-class, blue). (b) Correlation of motional class with the total entropy of binding. The change in population of the J, α and ω classes upon formation of each calmodulin complex with the corresponding binding entropy and have fitted linear correlation coefficients (R2) of −0.83, +0.74 and +0.70, respectively. Error bars reflect the variation of the population of each motional class that results from an increase or decrease in the measured values by two standard deviations. See [20] for more details.
Calibration of an “entropy meter”
The idea rests on the fundamental assumptions that methyl-bearing side chains are numerous enough to provide good coverage and are sufficiently coupled to neighboring side chains to report on the local disorder (entropy). This leads to an apparently simple relationship between what can be measured (protein motion) or confidently calculated (solvent entropy) and what is desired (conformational entropy):
| (2) |
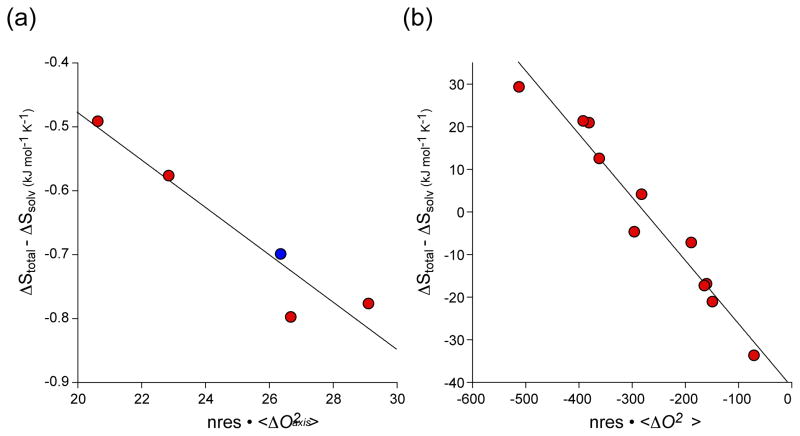
where ΔStot, ΔSsol, ΔSconf, ΔSRT and ΔSother are the changes in total system entropy, solvent entropy, conformational entropy, rotational-translational entropy and undocumented entropy, which is mostly solvent entropy from ion pair dissociation and solvation. By postulate, ΔSconf is linearly related to the residue-weighted change in the dynamics of the target domain and the protein (e.g. calmodulin) upon binding. Linearity is strongly supported by the simple simulations illustrated in Fig. 1 and elsewhere [22]. “m” is the desired empirical scaling factor relating changes in motion to changes in conformation entropy. Implementation in the context of the calmodulin complexes is somewhat involved because of uncertainty about the nature of the free ligand peptide (necessary to calculate the change in solvent entropy) and the details of translation-rotational entropy changes, which was assumed to be constant across the complexes. A mutant far from the interaction surface but having different binding thermodynamics was used to buttress the initial data set derived from the wild-type complexes. A remarkably linear calibration line is obtained (Figure 3a) [21**]. Thus the calmodulin example strongly suggests that quantitative measures of conformational entropy can be obtained using a dynamical proxy provided by NMR relaxation.
Figure 3.
Calibration of the dynamical proxy for protein conformational entropy. (a) The binding ofcalmodulin -binding domains to calcium saturated calmodulin [21**]. Order parameters derived from deuterium methyl relaxation in calmodulin and the target domains. Red symbols represent wild-type calmodulin binding to four different target domains. The blue symbol corresponds to a complex involving the E84K mutant of calmodulin. The data givea linear regression statistic R of 0.95and a slope (m) of−0.037 ± 0.007 kJ K−1 mol res−1that allows for empirical calibration of the conversion of changes in side-chain dynamics to a quantitative estimate of changes in conformational entropy. (b) The binding of DNA to activated CAP [23**]. The wild-type and mutant CAP proteins ligated with cAMP or cGMP were studied. Order parameters were derived from cross-correlated 13C-1H relaxation in CAP. The data givea linear regression statistic R of 0.98 and aslope (m) of −0.15 ± 0.01 kJ K−1mol res−1.
A second, and perhaps more convincing, demonstration of this approach has recently appeared. Using the catabolite activator protein (CAP), Tzeng & Kalodimos used an impressive collection of mutant proteins to undertake an empirical analysis of the thermodynamic origins of high affinity binding of DNA to the cAMP-activated protein [23**]. The advantage of the CAP system is that the same binding ligand (DNA) is used, the structure does not change upon binding and an array of structurally benign mutations remote from the binding interface were available. This eliminated most uncertainty in the variation of ΔSRT, ΔSsolv and ΔSother, which may have hampered the analysis of the calmodulin complexes described above. The resulting empirical calibration for CAP is most impressive (Figure 3b). Interestingly, the “m” values for calmodulin and CAP are quite different, which suggests that there may not be a universal scaling and that each protein studied in this way must be calibrated separately. This remains to be resolved. It was also found that the CAP variants have markedly different affinities for DNA, despite the CAP2-DNA-binding interfaces being essentially identical in the various complexes. Importantly, it was seen that the conformational entropy changes can inhibit (disfavor) the binding of CAP variants that are structurally poised for optimal DNA binding or can stimulate (favor) the binding activity of CAP variants that only transiently populate the DNA-binding-domain active state. Indeed, a study of the association of the carbohydrate to galectin-3 by Akke and coworkers indicated that affinity is driven by a favorable change in protein conformational entropy [24]. Collectively, these examples show how changes in fast internal dynamics (conformational entropy) and slow internal dynamics (energetically excited conformational states) can regulate binding activity in a way that cannot be predicted on the basis of the protein’s predominant ground-state structure. A highly complementary crystallographic approach is also emerging. Taking advantage of the considerable redundancy in diffraction data, Alber and coworkers have developed an analytical strategy for recovering minor rotameric states [25], which has revealed the richness suggested by the dynamical studies emphasized here [26*]. Clearly, conformational entropy can no longer be ignored when considering the origins of the free energy governing molecular recognition by proteins.
The role of protein conformational entropy in more sophisticated functions
In addition to providing a template for catalytic chemistry, it is the ability of proteins to move the free energy of binding of one ligand to modulate the free energy of subsequent binding that arguably makes proteins so special in biology. The pioneering models of Monod–Wyman–Changeux [27] and Koshland–Nemethy–Filmer [28], which rest on two-state transitions of individual protein subunits within an oligomer, have recently been supplemented by a more general representation that incorporates the protein ensemble [29*,30]. This view reveals a number of previously unrecognized possible mechanisms for allosteric regulation. This model emphasizes the potential for a significant role for conformational entropy in allosteric regulation as speculated some time ago [31]. Indeed, given the confirmation that measurable changes in side chain motion correspond to significant changes in conformational entropy, a number of examples of consistent with Cooper’s hypothesis have been revealed. These include the aforementioned CAP [32] and a surprising allosteric response in a putatively non-allosteric PDZ domain [33], for example. The use of motion (conformational entropy) to transmit allosteric free energy through a protein implies a limited degree of side chain coupling. On the one hand, neighboring side chains need to be coupled to some extent to move the “signal” through the protein. However, on the other hand, the motion must have limited coupling in order to provide directionality for the signal in the context of a pathways-type model for allosteric regulation. Experimental delineation of coupling of internal protein motion is difficult. A recent pressure-perturbation illuminated regions of variable coupling of fast internal motion in the small protein ubiquitin [34*]. When combined with temperature- [35,36] and mutational-perturbation [37*–39], these types of approaches should reveal the dynamical structure of proteins and will potentially provide a direct test of emerging theoretical treatments of this issue [40,41].
Protein hydration and protein motion
The interaction of proteins with ligands necessarily involves the modification of the interaction of water with the binding surface [42,43]. In many cases, water is simply excluded while in others the interface remains wet. In one interesting case, it has been shown using high-resolution crystallography that the interaction of water in the unliganded state of galetcin-3 mimics that seen in the protein’s interaction with the hydroxyl groups of the bound carbohydrate ligand [44]. This is a particularly good example of the likely importance of specific protein-water interactions in molecular recognition and emphasizes the need to understand the dynamical coupling between the protein and water. In this regard, there has been a flurry of technical developments of methods for site-resolved probes of protein hydration dynamics and motional coupling [45,46*,47*,48*,49*–51]. Generally, it is found that the interaction of water with the surface of proteins is quite variable with respect to residence time and local dynamics. Interestingly, a novel solution NMR approach, based on the favorable properties of proteins encapsulated within the protective water core of a reverse micelle, suggested a clustering of dynamics of hydration water on the surface ubiquitin [51]. Furthermore, less mobile water is centered on regions of the protein surface that are directly involved in protein-protein interactions. This argues that ubiquitin has evolved to maximize the hydrophobic effect for surfaces involved in molecular recognition. The generality of this observation remains to be explored.
Future Challenges
The recent demonstrations that an empirically calibrated “entropy meter” based on a dynamical proxy seems to provide self-consistent and robust estimates of the change in conformational entropy of a protein upon a change in functional state is promising. These initial efforts are arguably crude and one anticipates that further refinements will be forth coming both in the type of experimental data that is obtained and the manner in which it is interpreted. In this vein, it is important to note that the changes in fast internal protein motion upon ligand binding are relatively small and remain a considerable challenge for simulation. Notable improvements have recently been made in the ability of molecular dynamics simulations to capture motion on the backbone [52,53] and side chains [54*]. However, these types of simulations are not yet capable of recapitulating the small experimentally observed dynamical changes that generally accompany a change in the functional state of a protein. There are, of course, statistical thermodynamic approaches, such as the enhanced Monte Carlo sampling algorithms that have been used with some success to understand the calmodulin system [55,56]. The availability of the types of experimental studies described here will undoubtedly provide strong reference sets for improvements to computational approaches to this most important topic.
Highlights.
Internal protein motion can be used as a proxy for conformational entropy
Fast motion interconverts states with significant conformational entropy
Fast internal motion can be characterized in a site resolved way using solution NMR relaxation phenomena
Changes in motion can be converted to changes in conformational entropy
Conformational entropy can contribute significantly to ligand binding by proteins
Acknowledgments
We thank Professor Kim Sharp for continuing discussions. We gratefully acknowledge support from the National Institutes of Health (GM102447).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Recommended Reading
- 1*.Zhou HX, Gilson MK. Theory of free energy and entropy in noncovalent binding. Chem Rev. 2009;109:4092–4107. doi: 10.1021/cr800551w. An excellent recent review of the issues underlying a comprehensive theoretical understanding of ligand binding. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fischer E. Einfluss der confiruation auf die wirkung der enzyme. Ber Dt Chem Ges. 1894;27:2895–2993. [Google Scholar]
- 3.Koshland DE. Application of a theory of enzyme specificity to protein synthesis. Proc Natl Acad Sci USA. 1958;44:98–104. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4*.Hammes GG, Chang YC, Oas TG. Conformational selection or induced fit: A flux description of reaction mechanism. Proc Natl Acad Sci U S A. 2009;106:13737–13741. doi: 10.1073/pnas.0907195106. Clear description of the two kinetic models for ligand association and how to distinguish them. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanford C. The hydrophobic effect and the organization of living matter. Science. 1978;200:1012–1018. doi: 10.1126/science.653353. [DOI] [PubMed] [Google Scholar]
- 6.Chandler D. Interfaces and the driving force of hydrophobic assembly. Nature. 2005;437:640–647. doi: 10.1038/nature04162. [DOI] [PubMed] [Google Scholar]
- 7.Cooper A. Thermodynamic fluctuations in protein molecules. Proc Natl Acad Sci U S A. 1976;73:2740–2741. doi: 10.1073/pnas.73.8.2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levitt M, Warshel A. Computer simulation of protein folding. Nature. 1975;253:694–698. doi: 10.1038/253694a0. [DOI] [PubMed] [Google Scholar]
- 9.McCammon JA, Gelin BR, Karplus M. Dynamics of folded proteins. Nature. 1977;267:585–590. doi: 10.1038/267585a0. [DOI] [PubMed] [Google Scholar]
- 10.Frauenfelder H, Petsko GA, Tsernoglou D. Temperature-dependent x-ray diffraction as a probe of protein strutural dynamics. Nature. 1979;280:558–563. doi: 10.1038/280558a0. [DOI] [PubMed] [Google Scholar]
- 11.Gurd FR, Rothgeb TM. Motions in proteins. Adv Protein Chem. 1979;33:73–165. doi: 10.1016/s0065-3233(08)60459-3. [DOI] [PubMed] [Google Scholar]
- 12.Igumenova TI, Frederick KK, Wand AJ. Characterization of the fast dynamics of protein amino acid side chains using NMR relaxation in solution. Chem Rev. 2006;106:1672–1699. doi: 10.1021/cr040422h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng JW. Exposing the moving parts of proteins with NMR spectroscopy. J Phys Chem Lett. 2012;3:1039–1051. doi: 10.1021/jz3002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karplus M, Ichiye T, Pettitt BM. Configurational entropy of native proteins. Biophys J. 1987;52:1083–1085. doi: 10.1016/S0006-3495(87)83303-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mittermaier A, Kay LE. Review - new tools provide new insights in NMR studies of protein dynamics. Science. 2006;312:224–228. doi: 10.1126/science.1124964. [DOI] [PubMed] [Google Scholar]
- 16.Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic- resonance relaxation in macromolecules. 1. Theory and range of validity. J Am Chem Soc. 1982;104:4546–4559. [Google Scholar]
- 17.Akke M, Bruschweiler R, Palmer AG. NMR order parameters and free-energy – an analytical approach and its application to cooperative Ca2+ binding by calbindin-d(9k) J Am Chem Soc. 1993;115:9832–9833. [Google Scholar]
- 18.Li Z, Raychaudhuri S, Wand AJ. Insights into the local residual entropy of proteins provided by NMR relaxation. Protein Sci. 1996;5:2647–2650. doi: 10.1002/pro.5560051228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang D, Kay LE. Contributions to conformational entropy arising from bond vector fluctuations measured from NMR-derived order parameters. Application to protein folding. J Mol Biol. 1996;263:369–382. doi: 10.1006/jmbi.1996.0581. [DOI] [PubMed] [Google Scholar]
- 20.Frederick KK, Marlow MS, Valentine KG, Wand AJ. Conformational entropy in molecular recognition by proteins. Nature. 2007;448:325–329. doi: 10.1038/nature05959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21**.Marlow MS, Dogan J, Frederick KK, Valentine KG, Wand AJ. The role of conformational entropy in molecular recognition by calmodulin. Nat Chem Biol. 2010;6:352–358. doi: 10.1038/nchembio.347. Introduction of the idea of empirically calibrating the dynamical proxy for conformational entropy and it use to show its importance in molecular recognition by calmodulin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee AL, Sharp KA, Kranz JK, Song XJ, Wand AJ. Temperature dependence of the internal dynamics of a calmodulin-peptide complex. Biochemistry. 2002;41:13814–13825. doi: 10.1021/bi026380d. [DOI] [PubMed] [Google Scholar]
- 23**.Tzeng S–R, Kalodimos C. Protein activity regulation by conformational entropy. Nature. 2012;488:236–240. doi: 10.1038/nature11271. Superb illustration of the range of contributions that slow and fast motions can have in the energetics of molecular recognition. Includes the second use of the “entropy meter” approach. [DOI] [PubMed] [Google Scholar]
- 24*.Diehl C, Engstrom O, Delaine T, Hakansson M, Genheden S, Modig K, Leffler H, Ryde U, Nilsson UJ, Akke M. Protein flexibility and conformational entropy in ligand design targeting the carbohydrate recognition domain of galectin-3. J Am Chem Soc. 2010;132:14577–14589. doi: 10.1021/ja105852y. A case where conformational entropy drives the high affinity association of a protein with a small molecule ligand. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lang PT, Ng HL, Fraser JS, Corn JE, Echols N, Sales M, Holton JM, Alber T. Automated electron-density sampling reveals widespread conformational polymorphism in proteins. Protein Sci. 2010;19:1420–1431. doi: 10.1002/pro.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26*.Fraser JS, van den Bedem H, Samelson AJ, Lang PT, Holton JM, Echols N, Alber T. Accessing protein conformational ensembles using room-temperature x-ray crystallography. Proc Natl Acad Sci U S A. 2011;108:16247–16252. doi: 10.1073/pnas.1111325108. Illustration of the power of the cystallographic analysis described in [25] to delineate minor conformations and illuminate the corresponding conformational entropy. Highly complementary to the NMR relaxation methods emphasized here. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monod J, Wyman J, Changeaux JP. On the nature of allosteric transitions: A plausible model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 28.Koshland DE, Nemethy G, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5:365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- 29*.Hilser VJ, Thompson EB. Intrinsic disorder as a mechanism to optimize allosteric coupling in proteins. Proc Natl Acad Sci U S A. 2007;104:8311–8315. doi: 10.1073/pnas.0700329104. Introduction of an allosteric paradigm in the context of the protein ensemble that highlights a previously unrecognized mechanism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wrabl JO, Gu J, Liu T, Schrank TP, Whitten ST, Hilser VJ. The role of protein conformational fluctuations in allostery, function, and evolution. Biophys Chem. 2011;159:129–141. doi: 10.1016/j.bpc.2011.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cooper A, Dryden DTF. Allostery without conformational change - a plausible model. Eur Biophys J Biophys Lett. 1984;11:103–109. doi: 10.1007/BF00276625. [DOI] [PubMed] [Google Scholar]
- 32.Tzeng SR, Kalodimos CG. Dynamic activation of an allosteric regulatory protein. Nature. 2009;462:368–372. doi: 10.1038/nature08560. [DOI] [PubMed] [Google Scholar]
- 33.Petit CM, Zhang J, Sapienza PJ, Fuentes EJ, Lee AL. Hidden dynamic allostery in a PDZ domain. Proc Natl Acad Sci U S A. 2009;106:18249–18254. doi: 10.1073/pnas.0904492106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34*.Fu Y, Kasinath V, Moorman VR, Nucci NV, Hilser VJ, Wand AJ. Coupled motion in proteins revealed by pressure perturbation. J Am Chem Soc. 2012:8543–8550. doi: 10.1021/ja3004655. A novel strategy for identifying regions of coupled motion in proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee AL, Wand AJ. Microscopic origins of entropy, heat capacity and the glass transition in proteins. Nature. 2001;411:501–504. doi: 10.1038/35078119. [DOI] [PubMed] [Google Scholar]
- 36.Song XJ, Flynn PF, Sharp KA, Wand AJ. Temperature dependence of fast dynamics in proteins. Biophys J. 2007;92:L43–45. doi: 10.1529/biophysj.106.102160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37*.Mauldin RV, Lee AL. Nuclear magnetic resonance study of the role of M42 in the solution dynamics of Escherichia coli dihydrofolate reductase. Biochemistry. 2010;49:1606–1615. doi: 10.1021/bi901798g. An excellent illustration of the potential intracies of dynamic perturbation of proteins by the binding of a drug. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mauldin RV, Carroll MJ, Lee AL. Dynamic dysfunction in dihydrofolate reductase results from antifolate drug binding. Modulation of dynamics within a strctural state. Structure. 2009;17:386–394. doi: 10.1016/j.str.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fuentes EJ, Gilmore SA, Mauldin RV, Lee AL. Evaluation of energetic and dynamic coupling networks in a pdz domain protein. J Mol Biol. 2006;364:337–351. doi: 10.1016/j.jmb.2006.08.076. [DOI] [PubMed] [Google Scholar]
- 40.Halabi N, Rivoire O, Leibler S, Ranganathan R. Protein sectors: Evolutionary units of three-dimensional structure. Cell. 2009;138:774–786. doi: 10.1016/j.cell.2009.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reynolds KA, McLaughlin RN, Ranganathan R. Hot spots for allosteric regulation on protein surfaces. Cell. 2011;147:1564–1575. doi: 10.1016/j.cell.2011.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levy Y, Onuchic JN. Water mediation in protein folding and molecular recognition. Ann Rev Biophys Biomol Struct. 2006;35:389–415. doi: 10.1146/annurev.biophys.35.040405.102134. [DOI] [PubMed] [Google Scholar]
- 43.Hummer G. Molecular binding under water’s influence. Nat Chem. 2010;2:906–907. doi: 10.1038/nchem.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saraboji K, Hakansson M, Genheden S, Diehl C, Qvist J, Weininger U, Nilsson UJ, Leffler H, Ryde U, Akke M, et al. The carbohydrate-binding site in galectin-3 is preorganized to recognize a sugarlike framework of oxygens: Ultra-high-resolution structures and water dynamics. Biochemistry. 2012;51:296–306. doi: 10.1021/bi201459p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qiu WH, Kao YT, Zhang LY, Yang Y, Wang LJ, Stites WE, Zhong DP, Zewail AH. Protein surface hydration mapped by site-specific mutations. Proc Natl Acad Sci U S A. 2006;103:13979–13984. doi: 10.1073/pnas.0606235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46*.Zhang LY, Yang Y, Kao YT, Wang LJ, Zhong DP. Protein hydration dynamics and molecular mechanism of coupled water-protein fluctuations. J Am Chem Soc. 2009;131:10677–10691. doi: 10.1021/ja902918p. An excellent illustration of the use of a fluorescence spectroscopy strategy to map protein hydration. [DOI] [PubMed] [Google Scholar]
- 47*.Nucci NV, Pometun MS, Wand AJ. Site-resolved measurement of water-protein interactions by solution NMR. Nat Struct Mol Biol. 2011;18:245–249. doi: 10.1038/nsmb.1955. Utilizes the favorable properties of proteins encapsulated to enable site-resolved studies of protein hydration by solution NMR methods. See also [51] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48*.Armstrong BD, Han SG. Overhauser dynamic nuclear polarization to study local water dynamics. J Am Chem Soc. 2009;131:4641–4647. doi: 10.1021/ja809259q. A novel approach utilizing the liquid DNP effect for the site-resolved characterization of protein-water interactions. [DOI] [PubMed] [Google Scholar]
- 49*.King JT, Arthur EJ, Brooks CL, Kubarych KJ. Site-specific hydration dynamics of globular proteins and the role of constrained water in solvent exchange with amphiphilic cosolvents. J Phys Chem B. 2012;116:5604–5611. doi: 10.1021/jp300835k. An exciting new IR approach to illuminating solvent and protein fluctuations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ortony JH, Cheng CY, Franck JM, Kausik R, Pavlova A, Hunt J, Han SI. Probing the hydration water diffusion of macromolecular surfaces and interfaces. New J Phys. 2011;13 [Google Scholar]
- 51.Nucci NV, Pometun MS, Wand AJ. Mapping the hydration dynamics of ubiquitin. J Am Chem Soc. 2011;133:12326–12329. doi: 10.1021/ja202033k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li DW, Bruschweiler R. NMR-based protein potentials. Angew Chem Intl Ed. 2010;49:6778–6780. doi: 10.1002/anie.201001898. [DOI] [PubMed] [Google Scholar]
- 53.Lindorff-Larsen K, Maragakis P, Piana S, Eastwood MP, Dror RO, Shaw DE. Systematic validation of protein force fields against experimental data. PLoS One. 2012;7 doi: 10.1371/journal.pone.0032131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54*.Scouras AD, Daggett V. The dynameomics rotamer library. Amino acid side chain conformations and dynamics from comprehensive molecular dynamics simulations in water. Protein Sci. 2011;20:341–352. doi: 10.1002/pro.565. Presents a significant step forward in the accuracy of methyl side chain order parameters predicted from molecular dynamics simulations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dubay KH, Bothma JP, Geissler PL. Long-range intra-protein communication can be transmitted by correlated side-chain fluctuations alone. PLoS Comput Biol. 2011;7:e1002168. doi: 10.1371/journal.pcbi.1002168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DuBay KH, Geissler PL. Calculation of proteins’ total side-chain torsional entropy and its influence on protein-ligand interactions. J Mol Biol. 2009;391:484–497. doi: 10.1016/j.jmb.2009.05.068. [DOI] [PubMed] [Google Scholar]