Abstract
ATP-dependent initiation factors help process replication origins and coordinate replisome assembly to control the onset of DNA synthesis. Although the specific properties and regulatory mechanisms of initiator proteins can vary greatly between different organisms, certain nucleotide-binding elements and assembly patterns appear preserved not only within the three domains of cellular life (bacteria, archaea, and eukaryotes), but also with certain classes of double-stranded DNA viruses. Structural studies of replication initiation proteins, both as higher-order oligomers and in complex with cognate DNA substrates, are revealing how an evolutionarily-related ATPase fold can support different modes of macromolecular assembly and function. Comparative studies between initiation systems in turn provide clues as to how duplex origin regions may be melted during initiation events.
Introduction
The faithful copying of genetic material depends on the precise spatial and temporal control of chromosome replication. In cells, replication onset is regulated by dedicated ATP-dependent initiation factors, which associate with cis-encoded loci (origins) to define start positions for strand synthesis (reviewed in [1]). Once engaged, cellular initiation complexes both unwind origin DNA and recruit requisite catalytic and scaffolding factors needed to construct a functional replisome [2,3]. Although the essential components of many model replication initiation systems largely have been identified, the detailed biophysical mechanisms by which these factors operate are still poorly understood. This deficiency has hindered our knowledge of how initiation programs are executed in an error-free manner to enable cell proliferation and preserve genetic integrity.
Broadly speaking, initiation complexes must carry out at least three key tasks, including origin recognition, origin melting, and the placement of ring-shaped DNA unwinding enzymes (helicases) onto DNA (Table 1). For cellular organisms, these activities often are served by distinct factors, termed initiators and helicase-loaders, respectively. However, in some instances, the replicative helicase can distinguish between origin and non-origin regions, or facilitate origin unwinding itself. Once loaded, replicative helicases help recruit strand synthesis machineries such as primases, polymerases, and polymerase clamp-loader complexes to complete replisome formation [2,4].
Table 1.
A subset of cellular and viral initiation factors.
| Replication Factor | Bacteria | Simian virus 40 | Papillomavirus | Eukaryotes | Archaea |
|---|---|---|---|---|---|
| Initiator | DnaA | L Tag | E1 | ORC | Orc1/Cdc6 |
| Helicase | DnaB | L Tag | E1 | MCM2-7 | MCM |
| Helicase Loader | DnaC | L Tag | E2 | Cdc6/Cdt1 | Orc1/Cdc6? |
A sizable number of eukaryotic and prokaryotic replication initiation factors, as well as those of papilloma and polyoma double-stranded DNA viruses, are predicated on the AAA+ (ATPases Associated with various cellular Activities) family of nucleotidyl hydrolases (Fig 1) [5]. This structural conservation has suggested that certain mechanistic properties of these proteins may overlap. Consistent with this idea, ATP can modulate the DNA binding and/or melting activities of initiation factors (e.g., see [6–11]), as well as promote the stable formation of higher-order initiator complexes through canonical AAA+-type interactions (Fig 1c, d) [12–18]. However, a paucity of structural models for key nucleotide- and DNA-bound intermediates has limited our understanding of how initiation proteins covert a latent chromosomal origin into an active replication fork. In this review, we detail recent findings that have started to reveal the means by which AAA+ type enzymes required for initiation stretch and untwist DNA to drive replication origin opening, a committing step toward copying the genome.
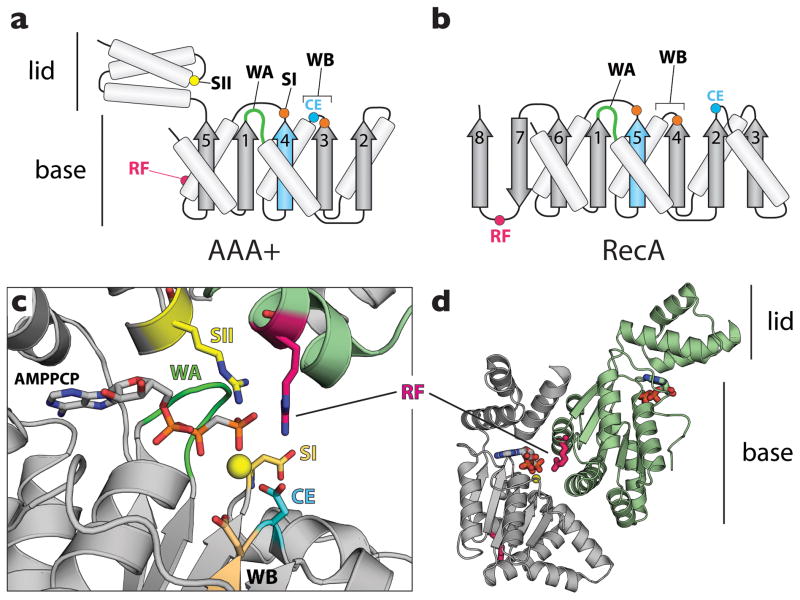
Figure 1.
ASCE ATPase architecture. AAA+ enzymes share a common structural core with RecA-type ATPases, together forming the Additional Strand Catalytic glutamate (E) (ASCE) supergroup of P-loop NTPases [79]. (a) Topology diagram for a AAA+ ATPase. AAA+ proteins frequently possess a nucleotide-binding pocket formed by two subdomains, a base αβ α-nucleotide-binding fold and a C-terminal α-helical lid domain. (b) Topology diagram for a RecA ATPase. In panels a and b, the additional strand and catalytic glutamate are highlighted in cyan. (c) Signature sequence motifs of AAA+ proteins. WA – Walker A (dark green), a glycine-rich element that forms a distinctive loop for cradling the phosphates of ATP. WB – Walker B (cyan and orange), a motif that contains acidic residues that interact with an associated magnesium ion (yellow sphere). SI/SII – sensor I (orange) and sensor II (yellow) elements, which reside near and can interact with the γ-phosphate of ATP. (d) Assembly patterns in AAA+ ATPase oligomers. A typical AAA+ dimer is shown (DnaA in this instance; PDB ID: 3R8F [9]). A conserved arginine residue – the arginine finger (RF, magenta) – extends from one AAA+ subunit (light green) to contact the γ-phosphate of ATP bound to a neighboring protomer (gray).
Initiation pathways in different organisms
Bacteria
In bacteria, replication typically begins with the recognition of a single chromosomal origin, oriC, by the initiator protein DnaA (reviewed in [19]). Although oriCs of different species vary significantly in size and organization [20], all contain a series of strong and weak DnaA binding sites that lie adjacent to an AT-rich DNA Unwinding Element (DUE) [21,22]. During the first steps of initiation, a subset of DnaA molecules initially engages duplex sites within oriC using a Helix-Turn-Helix (HTH) type of DNA binding domain that resides C-terminal to the initiator’s core AAA+ fold [23–25]. As initiation proceeds (and in the presence of ATP), additional DnaA protomers associate with initiators anchored at the origin to assemble into a large nucleoprotein complex that opens the DUE (Fig 2a) [7,16,18,21,22,26,27]. Following origin opening, the DnaA proteins of many bacterial species collaborate with an auxiliary helicase loader (DnaC in E. coli) to recruit and deposit two hexameric DnaB helicases onto the melted origin (reviewed in [3]).
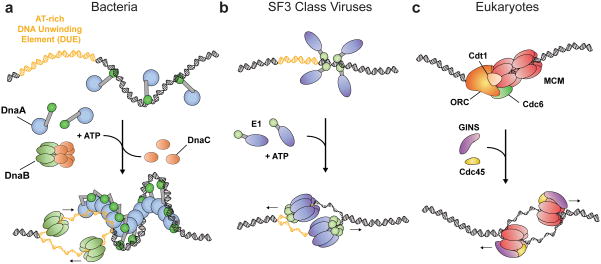
Figure 2.
Replication initiation mechanisms in cells and papilloma/polyomaviridae. (a) Bacteria. DnaA monomers are displayed as a blue oval (AAA+ domain) connected to a green oval (DNA binding domain). (Top)DnaA monomers associate with oriC. (Bottom) DnaA homo-oligomerizes in the presence of ATP, melting the DUE (yellow) and aiding the loading of the DnaB helicase (light green) with DnaC (orange). (b) SF3 viruses. Initiators are displayed as a purple oval (AAA+ domain) connected to a light green oval (DNA binding domain). (Top) monomers associate with the origin. (Bottom) Viral initiators melt the origin in the presence of ATP (yellow – DUE), eventually assembling into two hexameric helicases. (c) Eukaryotes. (Top) Activation of ORC by ATP stimulates formation of the pre-replicative complex (pre-RC), composed of ORC (orange), an MCM2-7 double-hexamer (red ovals), and the loader/chaperone proteins Cdc6 (green) and Cdt1 (tan). (Bottom) After loading, Cdc45 (yellow) and the GINS (purple) complex stimulate duplex melting by MCM2-7, giving rise to two hexameric helicases.
Viruses
The AAA+ ATPase initiators of polyoma and papilloma viruses comprise a different branch of the AAA+ lineage (the superfamily-3 (SF3) clade) [28], and are distinctive in that they act both as an initiator and as the replicative helicase (reviewed in [29]). As with DnaA, initiators such as the SV40 Large T-antigen (LTag) and papillomavirus E1 protein first associate with a viral origin using a dedicated duplex-DNA binding domain [30,31]. These early-binding initiators then recruit additional copies of themselves to the origin, ultimately forming two LTag or E1 hexamers around melted origin DNA strands in an ATP-dependent manner (Fig 2b) [32,33]. Although LTag is principally responsible for origin firing in SV40 [6], chaperoning of the papillomavirus E1 protein onto DNA is aided by a partner protein known as E2 [34,35]. Dissociation of E2 in turn allows for the assembly of a metastable, double-trimer E1 intermediate that facilitates origin melting [32].
Eukaryotes
In eukaryotes, initiation is coordinated by a six-subunit assembly known as the origin recognition complex (ORC) [36,37]. Phylogenetic and electron microscopy studies have shown that ORC forms a crescent-shaped particle predicated on a central core of as many as five AAA+ subunits (Orc1-Orc5) [28,38,39]. ORC localizes to duplex replication origins through the use of both specialized DNA-binding domains and interactions with partner proteins or nucleosomes(reviewed in [2]). Once bound to an origin, ORC helps recruit and load the heterohexameric Mini-Chromosome Maintenance (MCM2-7) helicase onto DNA (Fig 2c). ATP is required for MCM2-7 loading by ORC [11,40,41], as are two additional proteins – Cdc6 and Cdt1 [14,42–45]; together with ORC, these factors collectively form a discrete particle known as the pre-replicative complex (Pre-RC) (Table 1) [46]. Like ORC, MCM2-7 and Cdc6 retain a central AAA+ domain. However, only Cdc6 belongs to the same initiator clade as Orc1-Orc5 (and DnaA) [28]; MCM2-7 instead falls within a different “pre-sensor II (PSII)” group, so named for the presence of an extra α-helix that precedes the sensor II motif of the helicase’s ATPase fold [28].
Following pre-RC formation, ATP-turnover by ORC and Cdc6 triggers MCM release [43], resetting the initiation complex and allowing for additional MCM2-7 loading events prior to replication [10]. Notably, MCM2-7 complexes are first loaded as inactive double hexamers that encircle double-stranded DNA [47,48]. Activation requires specific phosphorylation events [49,50], as well as the association of the accessory proteins GINS and Cdc45, giving rise to a complex known as the CMG [51–55]. The MCM2-7 double hexamer appears to dissociate either during or after formation of the CMG [56], which itself has recently been shown to encircle only one strand during DNA unwinding [57].
Archaea
Archaeal organisms possess homologs of eukaryotic replication initiation factors (Table 1) (reviewed in [58]). Although archaeal initiators are most closely related to eukaryotic Orc1 and Cdc6 [28], they share a common organization with DnaA, bearing an initiator-type AAA+ domain fused to a C-terminal, duplex-DNA binding domain [59–61]. Co-crystal structures have shown that the Orc1 DNA binding domain, which comprises a winged-helix (WH) fold, can deform cognate origin sites upon engagement; interestingly, however, Orc1 AAA+ domains also can bind to and distort DNA directly [62,63]. At present, archaea appear to use a hexameric MCM-type helicase to unwind template DNA for replication fork progression [58]. How archaeal Orc1 proteins help load MCMs onto origins, and how ATP supports Orc1 function, remains enigmatic [58].
Structural insights into origin firing in bacteria
All replication initiation machineries that act on duplex DNA must overcome certain common physical challenges. Foremost among these is the separation of paired DNA strands to create a replication bubble that exposes template DNA. AAA+ domains are integral for DNA opening by cellular and SF3 initiation systems, although how these elements promote melting is still ill-defined.
A molecular picture of how AAA+ ATPases aid origin opening is starting to emerge, particularly for the bacterial initiator DnaA. ATP binding has been shown to accompany a conformational change in DnaA [64], facilitating the formation of a right-handed, spiral filament both in vitro and in vivo [65,66]. Helix formation, which is unusual for AAA+ proteins [67], appears to be supported by the presence of an α-helical insertion within the core AAA+ fold of DnaA that nudges adjacent subunits out of plane from each other (Fig. 3a) [64]. This so-termed Initiator Specific Motif (ISM) is present in all cellular initiators and serves to distinguish this group of proteins from other AAA+ ATPases [28,62]. The ISM, together with a pair of neighboring helices (α5–α6), also comprisesa cryptic single-stranded DNA-binding site within DnaAthat is activatedupon self-assembly of the initiator in the presence of ATP and is critical for origin melting [66,68].
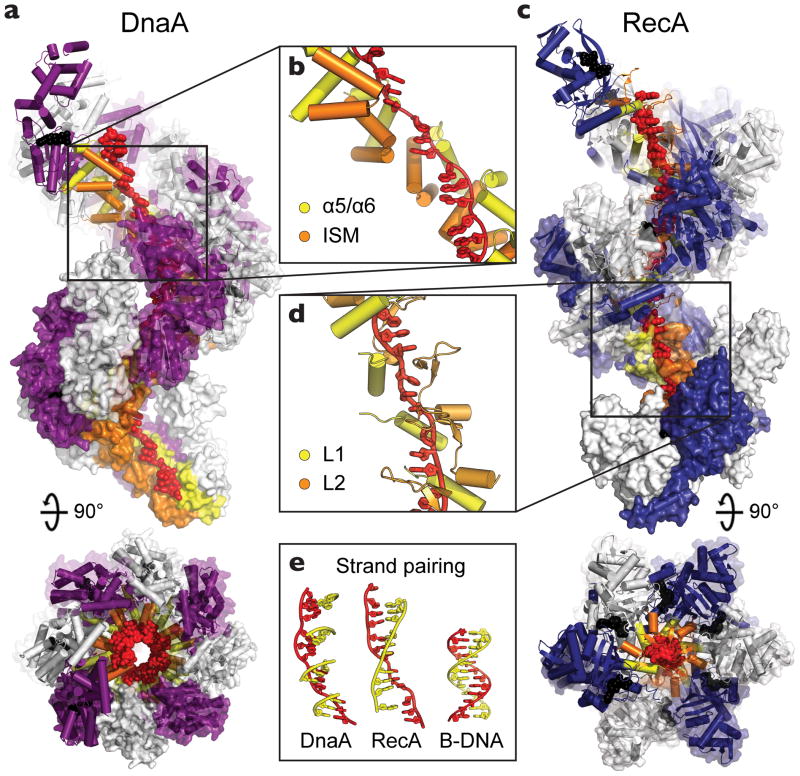
Figure 3.
DnaA binds single-stranded DNA in a manner similar to RecA. (a) Side and top views of oligomerized DnaA (PDB ID: 3R8F), showing twelve DnaA subunits, each colored white or purple, complexed with DNA (red). AMPPCP and Mg2+ are shown as black spheres and the DNA engagement elements, helices α5/α6 and the ISM, are colored yellow and orange, respectively. (b) Close-up of the DnaA oligomer bound to the backbone of DNA. (c) Side and top views of a RecA filament, constructed by aligning four copies of a RecA pentamer model (PDB ID: 3CMW [69]). Twelve RecA subunits, each colored white or blue, are shown complexed with DNA (red). ADP•AlF4 and Mg2+ are shown as black spheres and the DNA engagement elements, loops L1 and L2, are colored yellow and orange, respectively. (d) Close-up of the RecA oligomer bound to the backbone of DNA. Both DnaA and RecA bind DNA in an extended conformation with large gaps between trinucleotide base stacks. (e) (left) Cartoon model showing how complementary base triplets (yellow) would pair (in a B-DNA manner) with single-stranded-DNA bound to DnaA (red). Formation of a continuous base-paired strand is prevented by the orientation of successive DnaA-bound triplets. (middle) Same DNA view, but as seen in RecA, which orients triplets to allow pairing of an extended complementary strand for promoting duplex formation and strand exchange (PDB ID: 3CMX [69]). (right) B-DNA.
A recent crystal structure of DnaA bound to single-stranded DNA shows that upon forming a helical oligomer, the initiator can grip and extend the phosphodiester backbone by ~50% compared to B-form DNA (Fig 3a, b) [9]. An unanticipated feature of this behavior is its striking similarity to the homologous recombination protein, RecA (Fig 3c, d) [69]. Although the cellular function of RecA – catalyzing DNA strand-exchange to support DNA repair [70] – is fundamentally different from that of DnaA, both proteins are based on an evolutionarily-shared ATPase fold belonging to the ASCE superfamily of P-loop NTPases (Fig 1) [28]. This congruence extends to the mechanism of substrate engagement by both proteins, which is manifest by the binding of three nucleotides of DNA to each protomer, concomitant with the introduction of small gaps between tri-nucleotide segments that stretch the nucleic acid substrate (Fig 3c, d). The DNAs bound by RecA and DnaA differ, however, in the relative displacement of their trinucleotide elements: in RecA, successive trinucleotides are oriented in a smoothly-spiraled arrangement to permit the contiguous pairing of a complementary strand [69], whereas in DnaA these segments are offset in a manner that impedes the formation of a continuous duplex (Fig 3e). These similarities and differences suggest that both RecA and DnaA initially extend DNA to promote duplex separation, but that the initiator further reconfigures the substrate to prevent re-pairing and to stabilize an open replication bubble [9].
The disposition of the single-stranded DNA bound to an ATP-charged DnaA oligomer suggests that the complex seen crystallographically likely corresponds to a state after DnaA has unwound DNA [9]. But how does DnaA melt oriC in the first place? Clues derive not only from the single-stranded DNA-bound structure of DnaA, but also from biochemical studies showing that DnaA can actively catalyze the disruption of short, model duplexes [9]. This strand separation activity depends on the single-stranded DNA-binding site within DnaA’s AAA+ domain, as well as on the active, ATP-dependent assembly of the initiator, indicating that neighboring ATPase regions can directly engage double-stranded DNA and collaborate to pry open the duplex. While there is currently no structural picture of how the DnaA AAA+ domain might engage a double-stranded substrate, consideration of related initiation systems offers some valuable insights. For example, the ISMs of archaeal Orc1 proteins have been observed to bind and distort double-stranded origin DNA directly [62,63]. Viral SF3-class initiators similarly can bind duplexes through their AAA+ fold, and further are able to melt these substrates before enveloping one of the two DNA strands [8,71]. Comparisons of bacterial, archaeal/eukaryl, and papilloma/polyomavirus initiators shows that all of these factors engage the backbone of target DNAs using the same face of their core AAA+ ATPase folds [9] (Fig 4a–d). Moreover, this site and mode of binding further extends to distantly-related polymerase clamp-loaders [72], and appears to be used by MCMs as well (Fig 4e, f). Although the direction of the bound strand can run either 5′→3′ or 3′→5′ through the nucleic-acid binding site of a particular ATPase (Fig 4g), the overall similarities in how these disparate proteins engage client substrates is consistent with a prospective ability of the DnaA AAA+ domain to bind both single and double-stranded DNAs.
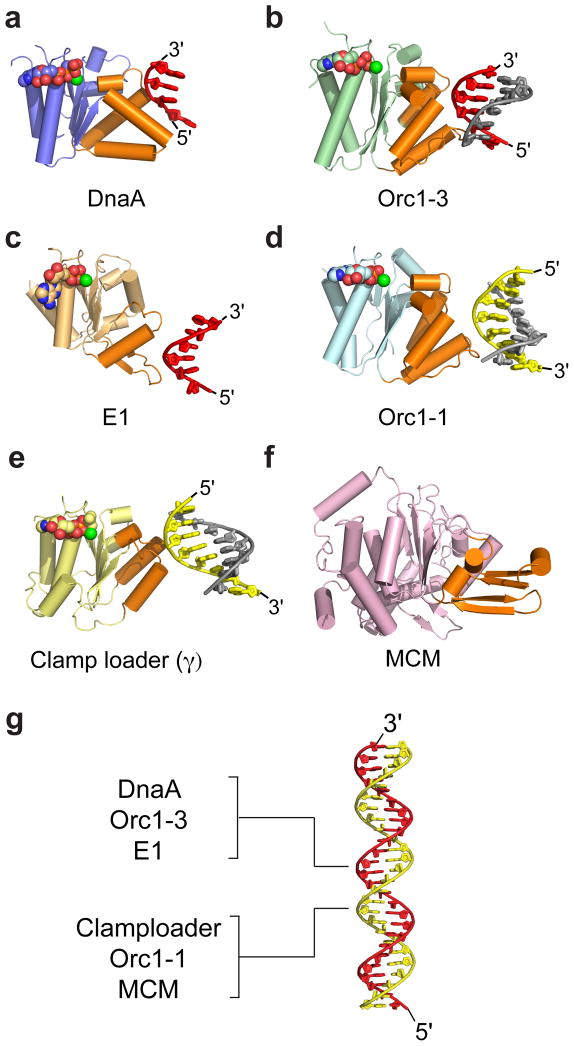
Figure 4.
DNA recognition by replicative AAA+ proteins. Nucleic acid substrates are all engaged using structural elements that reside on the same face of the AAA+ domain (orange). (a) Bacterial initiator DnaA (AAA+ domain, blue) bound to single-stranded DNA [9]. (b) Archaeal initiator Orc1-3 (AAA+ domain, green) bound to duplex DNA (PDB ID: 2QBY [62]). (c) Viral initiator E1 (AAA+ domain, light orange) bound to single-stranded DNA (PDB ID: 2GXA [71]). (d) Archaeal initiator Orc1-1 (AAA+ domain, cyan) bound to duplex DNA (PDB ID: 2QBY). (e) Bacterial clamp-loader subunit γ (AAA+ domain, yellow) bound to primer-template DNA (PDB ID: 3GLF [72]). (f) Archaeal Mcm (AAA+ domain, pink) (PDB ID: 3F9V [80]); although a DNA-complex has yet to emerge for this enzyme, mutagenesis studies have implicated the highlighted regions in binding substrate and helicase activity [81,82]. (g) Duplex DNA (red/yellow) with the strand bound by various AAA+ proteins indicated. For panels a–f, DNA is shown as red/grey or yellow/gray cartoons. Nucleotide co-factors are represented as spheres colored by atom.
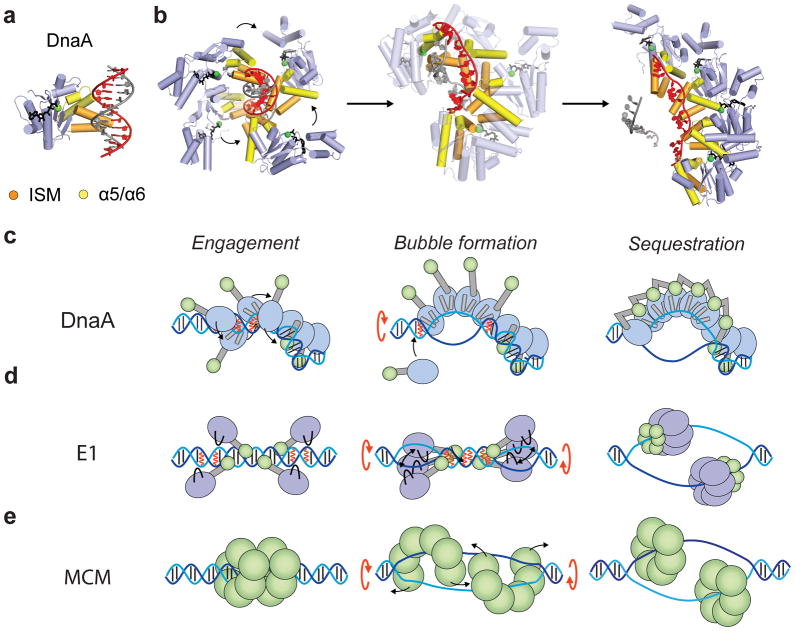
Using this logic, a possible sequence of molecular events that underlie origin opening in bacteria can be described as follows. In this scheme, several DnaA monomers first associate with duplex sites adjacent to the DUE using their C-terminal HTH DNA-binding domains (Fig 2a) [23–25]. Following binding, the AAA+ domains of adjacent DnaA molecules co-assemble into a helical oligomer [64,65], forming an organizing complex that wraps duplex oriC elements into a solenoidal supercoil [16,64], but that precedes DUE melting [26]. Next, the AAA+ domains of several DnaA protomers collectively engage one strand of a duplex DUE region. A model for this pre-melting state can be generated by aligning individual, ssDNA-bound DnaA protomers onto successive trinucleotide segments of a double-stranded DNA fragment (Fig 5a, b); the resultant arrangement is locally similar to the manner in which Orc1 AAA+ domains bind to duplex DNA (Fig 5c) [62,63], and further places the nucleotide-binding face of each protomer in close proximity to the arginine finger of a partner subunit (Fig 5b). As additional DnaA protomers bind to the DUE, the additive contributions of their effects on the structure of the duplex destabilize the AT-rich (and labile) region, opening up a bubble that can be sequestered by the stable association of adjacent ATPase domains (Fig 5c). It is at present uncertain how single DnaA protomers are initially recruited to the DUE. Biochemical studies have found evidence for the existence of several types of DnaA oriC complexes [16,27,66]; in this vein, the organizing complex bound to the flanking duplex oriC sites could act as a nucleation center to support the formation of a dedicated melting subcomplex (Fig. 5c). Once initiation is complete, ATP hydrolysis would promote the disassembly of DnaA oligomers [73,74], freeing DNA strands for copying by replicative polymerases.
Figure 5.
Models for origin melting. (a) Alignment of a single-stranded, DNA-bound DnaA monomer onto one strand of duplex DNA (red) results in an arrangement similar to that seen for Orc1 when bound to a double-stranded substrate (cf. Fig 4c). (b) Possible mechanism for DNA opening by DnaA. Four DnaA molecules are shown bound to consecutive trinucleotide segments along one strand of the duplex DUE. DnaA binding destabilizes this metastable region, promoting bubble formation and sequestration of a single DUE strand by a DnaA oligomer. The intermediate state (middle) corresponds to a linear interpolation between the initial and final states (Yale Morph Server) [83]. Bound nucleotide and Mg2+ are represented as black sticks and green spheres, respectively. (c) Stages of origin opening by DnaA. Each DnaA monomer is displayed as a blue oval (AAA+ domain) connected to a green oval (DBD). DNA binding elements (ISM and α5/α6) are displayed as gray cylinders. (d) Stages of origin opening by E1. Each E1 monomer is displayed as a purple oval (AAA+ domain) connected to a green oval (DBD). DNA binding elements are displayed as black loops. (e) Stages of duplex DNA melting by MCM. MCM monomers are represented as green ovals. In panels c–e the two strands of duplex DNA are displayed in blue and cyan with black lines for bases. Red lines represent regions where duplex DNA is destabilized. Black arrows refer to protein movements and red arrows to DNA untwisting as a bubble is formed.
Origin melting by other AAA+ ATPases
The nucleic-acid binding strategy employed by DnaA may have parallels with other initiation systems, albeit with some notable distinctions as well. For example, like the bacterial initiator, papillomavirus E1 and SV40 LTag both undergo a monomer/multimer transition, forming higher-order complexes that engage and deform/underwind a duplex AT-rich region [16,18,21,22,26,29,32]. In E1, DNA melting depends on a β-hairpin element [8,29], which emanates from the same face of SF3 AAA+ fold as the ISM of DnaA and Orc1 (Fig. 4). In contrast to DnaA, however, the expansion of two early-stage bubbles appears to require multiple rounds of ATP turnover by two trimeric E1 subcomplexes, which untwists DNA to open the origin (Fig 5d) [8]. After melting, additional E1 protomers assemble with the trimers [8], and eventually fully encircle single-stranded DNA within two topologically-closed rings [71], as opposed to adopting the extended RecA-like spiral.
How origins are opened by eukaryotic initiation factors is less well-resolved. Nevertheless, there are hints that this process may exhibit an intriguing blend of similarities and differences compared to bacterial and viral SF3-type initiation systems. For example, based on phylogenetic signatures, the AAA+ subunits of ORC and Cdc6 likely all contain an ISM [62,63]. ORC, however, has not yet been observed to melt duplexes, either prior to or concomitant with helicase loading [11,47,48]. In this regard, it is notable that the local architecture of ORC-type ISMs differs from that of DnaA (at least in archaeal Orc1 orthologs), forming not a “V”-shaped helical wedge, but rather a parallel, helix-loop-helix motif (Fig 4a, b). Because the DnaA ISM appears at least partly responsible for pushing oligomerized initiator protomers into a spiral and extending single-stranded DNA [9,64], this structural difference raises the possibility that assembled ORC subunits may adopt a more planar configuration that allows adjoining ISMs to engage duplex DNA, but not to stretch and melt it.
If ORC does not melt origin DNA directly, the most likely candidate for such a function would seem to be MCM2-7. However, while MCM double hexamers loaded by ORC do encircle double-stranded DNA, they are not immediately competent to unwind the duplex [47,48]. This finding, along with three other observations a need for phosphorylation, the stable binding of Cdc45•GINs to activate MCM2-7 [47,75], and the apparent encirclement of single DNA strands by the resultant CMG [57] – suggests that origin melting could occur as a consequence of MCM2-7 isomerization between double and single-hexamer states [57,76,77]. Were MCM2-7 to prove capable of catalyzing the initial origin melting event (in addition to its demonstrated role as a replicative helicase), the heterohexamer’s AAA+ domains would likely drive the process, potentially emulating viral SF3 enzymes by engaging one strand of duplex DNA, restructuring the nucleic acid substrate through ATP-dependent conformational changes, and eventually sequestering a single DNA strand within a heterohexameric ring (Fig 5e) [57,76,77]. How isomerization might take place alongside DNA opening is not clear, but could rely on an ATP-dependent untwisting approach analogous to that proposed for E1. Alternatively, a wide variety of structural forms have been observed for eukaryotic and archaeal MCMs, including an intriguing helical state [78], which we note displays right-handed chirality and pitch similar to that used by DnaA to promote DNA unpairing. Additional studies will be needed to resolve these issues.
Conclusions
Although many structural and mechanistic aspects of replication initiation are still poorly understood, trends pertaining to how duplex origin segments are melted are starting to emerge. Cellular and SF3-type initiation factors are predicated on an evolutionarily-related AAA+ ATPase fold that assembles into higher-order oligomers through the formation of a canonical, bipartite catalytic center. Despite bearing numerous distinguishing structural augmentations, initiation proteins display a striking similarity in how they engage nucleic acid targets, in particular the use of a common face of the AAA+ region to bind to the backbone of a single DNA strand (even when that substrate is paired within a duplex). However, architectural differences between these substrate-binding surfaces also correlate with distinct structural states formed upon oligomerization (e.g., spirals vs. closed rings), and in turn appear to underpin unique functional traits. Hence, the AAA+ domains of eukaryotic and archaeal initiators assist with origin recognition and the loading of a hexameric helicase (which itself may melt DNA); SF3 viral initiators by contrast both bind and open origin regions, and then further interconvert into hexameric helicases that processively unwind DNA. Bacterial DnaA exhibits a blend of these properties, with an ability to engage and melt oriC and to aid helicase loading. Future work will undoubtedly provide more surprises regarding how cellular and viral systems have adapted and exploited a common ancestral ATPase fold to accommodate specialized roles for supporting the initiation of DNA replication.
Highlights.
AAA+ ATPase proteins in the initiation of replication engage DNA in a similar manner.
ATP-dependent assembly stretches and untwists DNA to drive replication origin opening.
Distinct spiral and ring-shaped assemblies conduct initiation in different systems.
Acknowledgments
This work was supported by a Human Frontier Science Program fellowship (to KED) and the NIGMS (to JMB, GM071747).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Duderstadt KE, Berger JM. AAA+ ATPases in the initiation of DNA replication. Critical reviews in biochemistry and molecular biology. 2008;43:163–187. doi: 10.1080/10409230802058296. [DOI] [PubMed] [Google Scholar]
- 2.Stillman B. Origin recognition and the chromosome cycle. FEBS letters. 2005;579:877–884. doi: 10.1016/j.febslet.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 3.Kaguni JM. DnaA: controlling the initiation of bacterial DNA replication and more. Annual Review of Microbiology. 2006;60:351–375. doi: 10.1146/annurev.micro.60.080805.142111. [DOI] [PubMed] [Google Scholar]
- 4.Johnson A, O’Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annual Review of Biochemistry. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- 5.Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome research. 1999;9:27–43. [PubMed] [Google Scholar]
- 6.Borowiec JA, Hurwitz J. Localized melting and structural changes in the SV40 origin of replication induced by T-antigen. The EMBO journal. 1988;7:3149–3158. doi: 10.1002/j.1460-2075.1988.tb03182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sekimizu K, Bramhill D, Kornberg A. ATP activates dnaA protein in initiating replication of plasmids bearing the origin of the E. coli chromosome. Cell. 1987;50:259–265. doi: 10.1016/0092-8674(87)90221-2. [DOI] [PubMed] [Google Scholar]
- ••8.Schuck S, Stenlund A. Mechanistic analysis of local ori melting and helicase assembly by the papillomavirus E1 protein. Mol Cell. 2011;43:776–787. doi: 10.1016/j.molcel.2011.06.026. An extensive study of origin melting by E1 using time-dependent permangante reactivity and assembly assays with a library of mutant E1 proteins reveals distinct stages of the melting process catalyzed by a double-trimer intermediate. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••9.Duderstadt KE, Chuang K, Berger JM. DNA stretching by bacterial initiators promotes replication origin opening. Nature. 2011;478:209–213. doi: 10.1038/nature10455. A compilation of structural and biochemical studies showing that nucleotide-assembled DnaA forms a spiral oligomer that binds, melts, and stretches ssDNA in a manner similar to RecA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowers JL, Randell JC, Chen S, Bell SP. ATP hydrolysis by ORC catalyzes reiterative Mcm2-7 assembly at a defined origin of replication. Molecular cell. 2004;16:967–978. doi: 10.1016/j.molcel.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 11.Klemm RD, Austin RJ, Bell SP. Coordinate binding of ATP and origin DNA regulates the ATPase activity of the origin recognition complex. Cell. 1997;88:493–502. doi: 10.1016/s0092-8674(00)81889-9. [DOI] [PubMed] [Google Scholar]
- 12.Ranjan A, Gossen M. A structural role for ATP in the formation and stability of the human origin recognition complex. Proc Natl Acad Sci U S A. 2006;103:4864–4869. doi: 10.1073/pnas.0510305103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siddiqui K, Stillman B. ATP-dependent assembly of the human origin recognition complex. J Biol Chem. 2007;282:32370–32383. doi: 10.1074/jbc.M705905200. [DOI] [PubMed] [Google Scholar]
- 14.Perkins G, Diffley JF. Nucleotide-dependent prereplicative complex assembly by Cdc6p, a homolog of eukaryotic and prokaryotic clamp-loaders. Molecular cell. 1998;2:23–32. doi: 10.1016/s1097-2765(00)80110-0. [DOI] [PubMed] [Google Scholar]
- 15.Funnell BE, Baker TA, Kornberg A. In vitro assembly of a prepriming complex at the origin of the Escherichia coli chromosome. The Journal of biological chemistry. 1987;262:10327–10334. [PubMed] [Google Scholar]
- 16.Crooke E, Thresher R, Hwang DS, Griffith J, Kornberg A. Replicatively active complexes of DnaA protein and the Escherichia coli chromosomal origin observed in the electron microscope. Journal of Molecular Biology. 1993;233:16–24. doi: 10.1006/jmbi.1993.1481. [DOI] [PubMed] [Google Scholar]
- 17.Kawakami H, Keyamura K, Katayama T. Formation of an ATP-DnaA-specific initiation complex requires DnaA Arginine 285, a conserved motif in the AAA+ protein family. The Journal of biological chemistry. 2005;280:27420–27430. doi: 10.1074/jbc.M502764200. [DOI] [PubMed] [Google Scholar]
- 18.Margulies C, Kaguni JM. Ordered and sequential binding of DnaA protein to oriC, the chromosomal origin of Escherichia coli. The Journal of biological chemistry. 1996;271:17035–17040. doi: 10.1074/jbc.271.29.17035. [DOI] [PubMed] [Google Scholar]
- 19.Katayama T, Ozaki S, Keyamura K, Fujimitsu K. Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nature reviews Microbiology. 2010;8:163–170. doi: 10.1038/nrmicro2314. [DOI] [PubMed] [Google Scholar]
- 20.Mackiewicz P, Zakrzewska-Czerwinska J, Zawilak A, Dudek MR, Cebrat S. Where does bacterial replication start? Rules for predicting the oriC region. Nucleic acids research. 2004;32:3781–3791. doi: 10.1093/nar/gkh699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kowalski D, Eddy MJ. The DNA unwinding element: a novel, cis-acting component that facilitates opening of the Escherichia coli replication origin. The EMBO journal. 1989;8:4335–4344. doi: 10.1002/j.1460-2075.1989.tb08620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bramhill D, Kornberg A. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell. 1988;52:743–755. doi: 10.1016/0092-8674(88)90412-6. [DOI] [PubMed] [Google Scholar]
- 23.Roth A, Messer W. The DNA binding domain of the initiator protein DnaA. The EMBO journal. 1995;14:2106–2111. doi: 10.1002/j.1460-2075.1995.tb07202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sutton MD, Kaguni JM. Threonine 435 of Escherichia coli DnaA protein confers sequence-specific DNA binding activity. J Biol Chem. 1997;272:23017–23024. doi: 10.1074/jbc.272.37.23017. [DOI] [PubMed] [Google Scholar]
- 25.Fujikawa N, Kurumizaka H, Nureki O, Terada T, Shirouzu M, Katayama T, Yokoyama S. Structural basis of replication origin recognition by the DnaA protein. Nucleic acids research. 2003;31:2077–2086. doi: 10.1093/nar/gkg309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller DT, Grimwade JE, Betteridge T, Rozgaja T, Torgue JJ, Leonard AC. Bacterial origin recognition complexes direct assembly of higher-order DnaA oligomeric structures. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18479–18484. doi: 10.1073/pnas.0909472106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •27.Ozaki S, Katayama T. Highly organized DnaA-oriC complexes recruit the single-stranded DNA for replication initiation. Nucleic Acids Res. 2012;40:1648–1665. doi: 10.1093/nar/gkr832. This paper, together with several other recent papers from the same group, dissects the importance of various types of DnaA binding sites in E. coli oriC, and proposes a model for origin opening in which DNA looping allows a preformed DnaA oligomer to engage and process the DUE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. Journal of structural biology. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 29.Hickman AB, Dyda F. Binding and unwinding: SF3 viral helicases. Current opinion in structural biology. 2005;15:77–85. doi: 10.1016/j.sbi.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 30.Meinke G, Phelan P, Moine S, Bochkareva E, Bochkarev A, Bullock PA, Bohm A. The crystal structure of the SV40 T-antigen origin binding domain in complex with DNA. PLoS biology. 2007;5:e23. doi: 10.1371/journal.pbio.0050023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Enemark EJ, Stenlund A, Joshua-Tor L. Crystal structures of two intermediates in the assembly of the papillomavirus replication initiation complex. The EMBO journal. 2002;21:1487–1496. doi: 10.1093/emboj/21.6.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schuck S, Stenlund A. Assembly of a double hexameric helicase. Molecular cell. 2005;20:377–389. doi: 10.1016/j.molcel.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 33.Valle M, Gruss C, Halmer L, Carazo JM, Donate LE. Large T-antigen double hexamers imaged at the simian virus 40 origin of replication. Molecular and cellular biology. 2000;20:34–41. doi: 10.1128/mcb.20.1.34-41.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohr IJ, Clark R, Sun S, Androphy EJ, MacPherson P, Botchan MR. Targeting the E1 replication protein to the papillomavirus origin of replication by complex formation with the E2 transactivator. Science (New York, NY) 1990;250:1694–1699. doi: 10.1126/science.2176744. [DOI] [PubMed] [Google Scholar]
- 35.Sedman J, Stenlund A. Co-operative interaction between the initiator E1 and the transcriptional activator E2 is required for replicator specific DNA replication of bovine papillomavirus in vivo and in vitro. The EMBO journal. 1995;14:6218–6228. doi: 10.1002/j.1460-2075.1995.tb00312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 37.Diffley JF, Cocker JH. Protein-DNA interactions at a yeast replication origin. Nature. 1992;357:169–172. doi: 10.1038/357169a0. [DOI] [PubMed] [Google Scholar]
- •38.Sun J, Kawakami H, Zech J, Speck C, Stillman B, Li H. Cdc6-induced conformational changes in ORC bound to origin DNA revealed by cryo-electron microscopy. Structure. 2012;20:534–544. doi: 10.1016/j.str.2012.01.011. A single-particle cryo-EM structure of Saccharomyces cerevisiae ORC, Cdc6 and ARS1 origin DNA in the presence of ATPγS shows that Cdc6 binding changes the conformation of ORC, leading to the bending and wrapping of origin DNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clarey MG, Erzberger JP, Grob P, Leschziner AE, Berger JM, Nogales E, Botchan M. Nucleotide-dependent conformational changes in the DnaA-like core of the origin recognition complex. Nature structural & molecular biology. 2006;13:684–690. doi: 10.1038/nsmb1121. [DOI] [PubMed] [Google Scholar]
- 40.Klemm RD, Bell SP. ATP bound to the origin recognition complex is important for preRC formation. Proc Natl Acad Sci U S A. 2001;98:8361–8367. doi: 10.1073/pnas.131006898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee DG, Makhov AM, Klemm RD, Griffith JD, Bell SP. Regulation of origin recognition complex conformation and ATPase activity: differential effects of single-stranded and double-stranded DNA binding. The EMBO journal. 2000;19:4774–4782. doi: 10.1093/emboj/19.17.4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seki T, Diffley JF. Stepwise assembly of initiation proteins at budding yeast replication origins in vitro. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:14115–14120. doi: 10.1073/pnas.97.26.14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Randell JC, Bowers JL, Rodriguez HK, Bell SP. Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2-7 helicase. Molecular cell. 2006;21:29–39. doi: 10.1016/j.molcel.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 44.Hofmann JF, Beach D. cdt1 is an essential target of the Cdc10/Sct1 transcription factor: requirement for DNA replication and inhibition of mitosis. The EMBO journal. 1994;13:425–434. doi: 10.1002/j.1460-2075.1994.tb06277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nishitani H, Lygerou Z, Nishimoto T, Nurse P. The Cdt1 protein is required to license DNA for replication in fission yeast. Nature. 2000;404:625–628. doi: 10.1038/35007110. [DOI] [PubMed] [Google Scholar]
- 46.Diffley JF, Cocker JH, Dowell SJ, Rowley A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell. 1994;78:303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- 47.Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, Diffley JF. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139:719–730. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, Li H, Stillman B, Speck C. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:20240–20245. doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Francis LI, Randell JC, Takara TJ, Uchima L, Bell SP. Incorporation into the prereplicative complex activates the Mcm2-7 helicase for Cdc7-Dbf4 phosphorylation. Genes Dev. 2009;23:643–654. doi: 10.1101/gad.1759609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheu YJ, Stillman B. Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Molecular cell. 2006;24:101–113. doi: 10.1016/j.molcel.2006.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kanemaki M, Sanchez-Diaz A, Gambus A, Labib K. Functional proteomic identification of DNA replication proteins by induced proteolysis in vivo. Nature. 2003;423:720–724. doi: 10.1038/nature01692. [DOI] [PubMed] [Google Scholar]
- 52.Kubota Y, Takase Y, Komori Y, Hashimoto Y, Arata T, Kamimura Y, Araki H, Takisawa H. A novel ring-like complex of Xenopus proteins essential for the initiation of DNA replication. Genes & development. 2003;17:1141–1152. doi: 10.1101/gad.1070003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takayama Y, Kamimura Y, Okawa M, Muramatsu S, Sugino A, Araki H. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes & development. 2003;17:1153–1165. doi: 10.1101/gad.1065903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gambus A. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nature cell biology. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- 55.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •56.Yardimci H, Loveland AB, Habuchi S, van Oijen AM, Walter JC. Uncoupling of sister replisomes during eukaryotic DNA replication. Mol Cell. 2010;40:834–840. doi: 10.1016/j.molcel.2010.11.027. Single-molecule fluorescence imaging in Xenopus replication assays provide evidence that MCM2-7 double hexamers decouple from each other as initiation transitions to elongation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •57.Fu YV, Yardimci H, Long DT, Ho TV, Guainazzi A, Bermudez VP, Hurwitz J, van Oijen A, Scharer OD, Walter JC. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell. 2011;146:931–941. doi: 10.1016/j.cell.2011.07.045. By presenting replisomes with strand-specific roadblocks in Xenopus egg extracts, the authors show that the eukaryotic DNA helicase, CMG, translocates on single-stranded-DNA. This finding, together with prior work establishing an MCM2-7 double hexamer as the ORC-depedent loading intermediate [47,48], suggests that origin melting is catalyzed by the helicase itself. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barry ER, Bell SD. DNA replication in the archaea. Microbiology and molecular biology reviews: MMBR. 2006;70:876–887. doi: 10.1128/MMBR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu J, Smith CL, DeRyckere D, DeAngelis K, Martin GS, Berger JM. Structure and function of Cdc6/Cdc18: implications for origin recognition and checkpoint control. Molecular cell. 2000;6:637–648. doi: 10.1016/s1097-2765(00)00062-9. [DOI] [PubMed] [Google Scholar]
- 60.Erzberger JP, Pirruccello MM, Berger JM. The structure of bacterial DnaA: implications for general mechanisms underlying DNA replication initiation. The EMBO journal. 2002;21:4763–4773. doi: 10.1093/emboj/cdf496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Singleton MR, Morales R, Grainge I, Cook N, Isupov MN, Wigley DB. Conformational changes induced by nucleotide binding in Cdc6/ORC from Aeropyrum pernix. Journal of Molecular Biology. 2004;343:547–557. doi: 10.1016/j.jmb.2004.08.044. [DOI] [PubMed] [Google Scholar]
- 62.Dueber EL, Corn JE, Bell SD, Berger JM. Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex. Science (New York, NY) 2007;317:1210–1213. doi: 10.1126/science.1143690. [DOI] [PubMed] [Google Scholar]
- 63.Gaudier M, Schuwirth BS, Westcott SL, Wigley DB. Structural basis of DNA replication origin recognition by an ORC protein. Science (New York, NY) 2007;317:1213–1216. doi: 10.1126/science.1143664. [DOI] [PubMed] [Google Scholar]
- 64.Erzberger JP, Mott ML, Berger JM. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nature structural & molecular biology. 2006;13:676–683. doi: 10.1038/nsmb1115. [DOI] [PubMed] [Google Scholar]
- ••65.Scholefield G, Errington J, Murray H. Soj/ParA stalls DNA replication by inhibiting helix formation of the initiator protein DnaA. EMBO J. 2012;31:1542–1555. doi: 10.1038/emboj.2012.6. Cysteine-specific crosslinking is used to demonstrate that DnaA forms helical oligomers in vivo, and that the size of the assembly is regulated by the Soj protein though a direct interaction with the DnaA AAA+ domain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Duderstadt KE, Mott ML, Crisona NJ, Chuang K, Yang H, Berger JM. Origin remodeling and opening in bacteria rely on distinct assembly states of the DnaA initiator. The Journal of biological chemistry. 2010;285:28229–28239. doi: 10.1074/jbc.M110.147975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ogura T, Wilkinson AJ. AAA+ superfamily ATPases: common structure--diverse function. Genes to cells: devoted to molecular & cellular mechanisms. 2001;6:575–597. doi: 10.1046/j.1365-2443.2001.00447.x. [DOI] [PubMed] [Google Scholar]
- 68.Ozaki S, Kawakami H, Nakamura K, Fujikawa N, Kagawa W, Park SY, Yokoyama S, Kurumizaka H, Katayama T. A Common Mechanism for the ATP-DnaA-dependent Formation of Open Complexes at the Replication Origin. The Journal of biological chemistry. 2008;283:8351–8362. doi: 10.1074/jbc.M708684200. [DOI] [PubMed] [Google Scholar]
- 69.Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–484. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- 70.Cox MM. Motoring along with the bacterial RecA protein. Nature reviews Molecular cell biology. 2007;8:127–138. doi: 10.1038/nrm2099. [DOI] [PubMed] [Google Scholar]
- 71.Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 72.Simonetta KR, Kazmirski SL, Goedken ER, Cantor AJ, Kelch BA, McNally R, Seyedin SN, Makino DL, O’Donnell M, Kuriyan J. The mechanism of ATP-dependent primer-template recognition by a clamp loader complex. Cell. 2009;137:659–671. doi: 10.1016/j.cell.2009.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kurokawa K, Nishida S, Emoto A, Sekimizu K, Katayama T. Replication cycle-coordinated change of the adenine nucleotide-bound forms of DnaA protein in Escherichia coli. The EMBO journal. 1999;18:6642–6652. doi: 10.1093/emboj/18.23.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Katayama T, Kubota T, Kurokawa K, Crooke E, Sekimizu K. The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E. coli chromosomal replicase. Cell. 1998;94:61–71. doi: 10.1016/s0092-8674(00)81222-2. [DOI] [PubMed] [Google Scholar]
- •75.Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Molecular cell. 2010;37:247–258. doi: 10.1016/j.molcel.2009.12.030. Biochemical studies dissect the network of interactions within the Drosophila CMG complex to show that Cdc45 and GINS enhance the ATPase, DNA binding and unwinding activities of Mcm2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •76.Costa A, Ilves I, Tamberg N, Petojevic T, Nogales E, Botchan MR, Berger JM. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nature structural & molecular biology. 2011 doi: 10.1038/nsmb.2004. EM reconstructions of Mcm2-7 and the full CMG complex from Drosophila melanogaster suggest a structural mechanism by which the eukaryotic helicase may isomerize during origin loading and melting. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Remus D, Diffley JF. Eukaryotic DNA replication control: lock and load, then fire. Current opinion in cell biology. 2009;21:771–777. doi: 10.1016/j.ceb.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 78.Chen YJ, Yu X, Kasiviswanathan R, Shin JH, Kelman Z, Egelman EH. Structural polymorphism of Methanothermobacter thermautotrophicus MCM. Journal of Molecular Biology. 2005;346:389–394. doi: 10.1016/j.jmb.2004.11.076. [DOI] [PubMed] [Google Scholar]
- 79.Leipe DD, Koonin EV, Aravind L. Evolution and classification of P-loop kinases and related proteins. Journal of Molecular Biology. 2003;333:781–815. doi: 10.1016/j.jmb.2003.08.040. [DOI] [PubMed] [Google Scholar]
- 80.Brewster AS, Wang G, Yu X, Greenleaf WB, Carazo JM, Tjajadi M, Klein MG, Chen XS. Crystal structure of a near-full-length archaeal MCM: functional insights for an AAA+ hexameric helicase. Proc Natl Acad Sci U S A. 2008;105:20191–20196. doi: 10.1073/pnas.0808037105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jenkinson ER, Chong JP. Minichromosome maintenance helicase activity is controlled by N- and C-terminal motifs and requires the ATPase domain helix-2 insert. Proc Natl Acad Sci U S A. 2006;103:7613–7618. doi: 10.1073/pnas.0509297103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McGeoch AT, Trakselis MA, Laskey RA, Bell SD. Organization of the archaeal MCM complex on DNA and implications for the helicase mechanism. Nat Struct Mol Biol. 2005;12:756–762. doi: 10.1038/nsmb974. [DOI] [PubMed] [Google Scholar]
- 83.Krebs WG, Gerstein M. The morph server: a standardized system for analyzing and visualizing macromolecular motions in a database framework. Nucleic Acids Res. 2000;28:1665–1675. doi: 10.1093/nar/28.8.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]