Abstract
Poly(ADP-ribose) polymerase 1 (PARP-1) regulates gene transcription, cell death signaling, and DNA repair through production of the posttranslational modification poly(ADP-ribose). During the cellular response to genotoxic stress PARP-1 rapidly associates with DNA damage, which robustly stimulates poly(ADP-ribose) production over a low basal level of PARP-1 activity. DNA damage-dependent PARP-1 activity is central to understanding PARP-1 biological function, but structural insights into the mechanisms underlying this mode of regulation have remained elusive, in part due to the highly modular six-domain architecture of PARP-1. Recent structural studies have illustrated how PARP-1 uses specialized zinc fingers to detect DNA breaks through sequence-independent interaction with exposed nucleotide bases, a common feature of damaged and abnormal DNA structures. The mechanism of coupling DNA damage detection to elevated poly(ADP-ribose) production has been elucidated based on a crystal structure of the essential domains of PARP-1 in complex with a DNA strand break. The multiple domains of PARP-1 collapse onto damaged DNA, forming a network of interdomain contacts that introduce destabilizing alterations in the catalytic domain leading to an enhanced rate of poly(ADP-ribose) production.
Introduction
Poly(ADP-ribose) is a reversible posttranslational modification synthesized from NAD+ by poly(ADP-ribose) polymerases, or PARPs, most commonly in response to cellular stress signals such as DNA damage. The most abundant PARP enzyme in cells, PARP-1 creates long and branched poly(ADP-ribose) covalently attached onto target proteins involved in gene transcription, DNA damage repair, and cell death signaling (reviewed in [1]). Due to PARP-1 involvement in DNA damage repair, inhibitors of PARP-1 are being actively pursued for the treatment of cancer (reviewed in [2]). The primary target for PARP-1 mediated poly(ADP-ribosyl)ation is PARP-1 itself, an activity termed automodification. Several other nuclear targets are modified by PARP-1 (reviewed in [3]); however the mechanism for PARP-1 substrate selection is not well understood. PARP-1 catalytic activity is chiefly regulated through its interaction with DNA damage. Binding to DNA strand breaks elevates the PAR synthesis activity of PARP-1 over 500-fold above basal levels of DNA-independent activity. DNA-dependent PARP-1 automodification is an immediate and robust cellular response to DNA damage that contributes to the recruitment of DNA repair and chromatin remodeling factors to sites of DNA breaks [4-7]. Recent structural studies highlighted in this article have begun to establish the physical basis for PARP-1 structure-specific detection of DNA breaks, and the mechanism for DNA damage-dependent activation of PARP-1 catalytic activity.
PARP-1 “beads-on-a-string” architecture
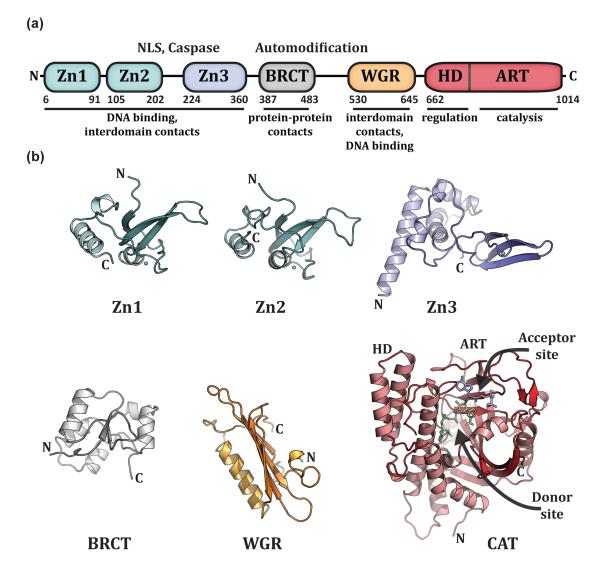
PARP-1 has a modular architecture with DNA binding, catalytic, and regulatory functions distributed among six independently folded domains (Figure 1a). Structures of each of the individual domains of PARP-1 in the absence of DNA have been determined (Figure 1b) [8-12](RIKEN,Structural Genomics Consortium). Located at the PARP-1 N-terminus, two homologous zinc finger domains, Zn1 and Zn2, recognize particular DNA structures, rather than specific DNA sequences [8,13-16]. A third zinc finger domain, Zn3, has a structure and function that is distinct from that of the Zn1 and Zn2 domains [9,10]. In addition to the zinc finger domains, human PARP-1 has a BRCT (BRCA1 C–terminus) fold that is located within the central region of PARP-1 that bears the major sites of automodification. A WGR domain is located adjacent to the catalytic domain; it is named after a well-conserved region of the amino acid sequence, Trp-Gly-Arg (WGR in single-letter code). The catalytic domain (CAT) of PARP-1 is composed of two subdomains, HD and ART. The ART subdomain is conserved in other ADP-ribosyl transferases (ARTs) of the ARTD class [17-19], and includes the amino acids involved in binding NAD+ and performing catalysis [12,20,21]. The active site of the CAT carries out three distinct activities: the initial ADP-ribose attachment to amino acid side chains, the elongation of the polymer with additional ADP-ribose modules, and the introduction of branches in the polymer chain. The helical subdomain (HD) of the CAT is composed of six α-helices with connecting linkers of variable sizes [12]. The majority of PARP-1 structural analysis has focused on the CAT domain, and there is a growing collection of structures of the CAT domain bound to PARP inhibitors [12,20,22-29], thus defining features of the catalytic active site and pointing to new avenues for drug design. There are also apo and inhibitor-bound structures determined for the catalytic domains of related PARP family members [30-38], which will help to understand the variation of catalytic activities [i.e. mono-versus poly(ADP-ribose) production], and will guide the design of more potent and specific inhibitors.
Figure 1. PARP-1 has a highly modular, “beads-on-a-string” architecture.
(a). Schematic representation of human PARP-1 domains. A bipartite nuclear localization signal (NLS) and a caspase 3 cleavage site (Caspase) are located between the Zn2 and Zn3 domains. A BRCA C-terminus (BRCT) fold is located within the region of PARP-1 that is primarily targeted for automodification. The catatlyic domain is composed of an alpha-helical subdomain (HD) and an ADP-ribosyl transferase subdomain (ART). (b). Crystal and/or NMR structures have been determined for each of the PARP-1 domains in the absence of DNA. Shown are the NMR structures of the homologous Zn1 and Zn2 domains (PDB code 2dmj and 2cs2, respectively [64], see also [8]), the NMR structure of the Zn3 domain (PDB code 2jvn, see also reference [9] for crystal structure), the NMR structure of the BRCT fold (PDB code 2cok [64], see also reference [11]), the NMR structure of the WGR domain (PDB code 2cr9 [64]), and the crystal structure of the catalytic domain (PDB code 1a26 [12]). Catalytic subdomains are labeled (HD and ART). The catalytic domain structure was determined in the presence of an NAD+ analogue that has defined the “acceptor site” for poly(ADP-ribose) formation. NAD+ is modeled in the “donor site” based on the related structure of diphthetia toxin [19]. All structure depictions were made using PYMOL (www.pymol.org).
How DNA damage regulates PARP-1 catalytic activity has been a long-standing question that is central to our understanding of PARP-1 biological functions. The CAT domain alone is capable of attaching poly(ADP-ribose) onto target proteins at a low basal level of activity that is independent of interaction with DNA [39]. The Zn1, Zn3, and WGR domains are required in combination with CAT for the robust DNA damage-dependent activity that is a hallmark of PARP-1 function. The highly modular, beads-on-a-string architecture of PARP-1 has complicated structural analysis and made it difficult to envision how the multiple domains of PARP-1 collectively function to stimulate catalytic activity in the presence of DNA damage. Recent structural studies have shed light on how PARP-1 binds to breaks in the DNA [8,13,40,41]. Moreover, the crystal structure of PARP-1 essential domains on DNA has revealed how the multiple domains of PARP-1 assemble on DNA breaks to create a network of interdomain communication that ultimately leads to destabilization and activation of the CAT domain [41].
DNA damage detection by PARP-like zinc fingers
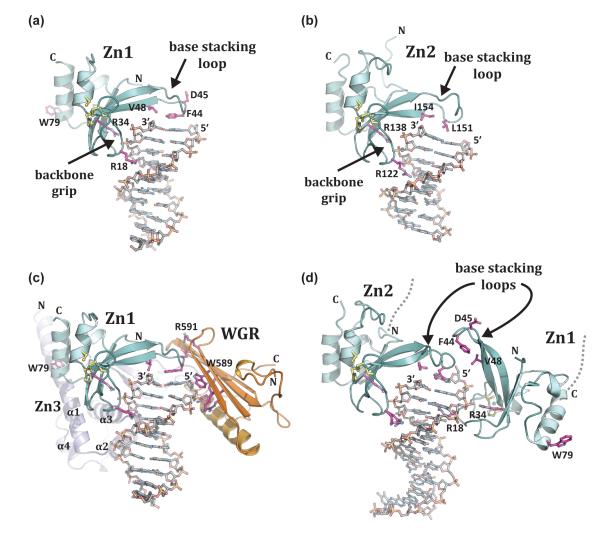
The structure of a PARP-like DNA binding zinc finger was first determined for the homologous N-terminal zinc finger domain of DNA ligase III [42], where it serves a nick sensing function [43,44]. The structural basis for DNA strand break detection has recently come from crystal structures of the human PARP-1 zinc fingers in complex with different models of DNA damage. The individual zinc fingers of human PARP-1, Zn1 and Zn2, were each crystallized in complex with DNA double strand breaks [13]. These structures define the features of damaged DNA that are recognized by PARP-1. Most notably, the interaction is mediated through contacts with exposed nucleotide bases using a loop connecting two β-strands, termed the “base stacking loop” due to its use of hydrophobic residues to stack onto the DNA break (Figure 2a, b). A second mode of interaction is formed between a continuous segment of the DNA phosphate backbone and a region of the zinc finger termed the “backbone grip.” Structure-based mutagenesis has confirmed the importance of both features to the overall binding affinity of the Zn1 and Zn2 domains [8,13]. Thus, the signature features of damaged DNA recognized by PARP-like zinc fingers are nucleotide bases available for interaction and a continuous region of phosphate backbone, both common features of damaged and abnormal DNA structures.
Figure 2. PARP-1 recognizes exposed nucleotide bases as a signature of DNA damage.
(a). Crystal structure of human Zn1 domain in complex with a DNA double strand break (PDB code 3od8 [13]). Together with panel b, these structures illustrate the features of damage DNA that are recognized by PARP-1 zinc finger domains: a continuous phosphate backbone engaged by conserved Arg residues on the “backbone grip”, and exposed nucleotide bases engaged by hydrophobic residues on the “base stacking loop”. D45 and W79 are key Zn1 residues involved in contacts with other PARP-1 domains. (b). Crystal structure of human Zn2 in complex with a DNA double strand break (PDB code 3odc [13]). (c). The Zn1, Zn3, and WGR domains collectively assemble on a DNA double strand break, with each domain forming specific protein-DNA, and protein-protein contacts with adjacent domains [41] (see also Figure 3a). Zn1 has the same orientation as in panel a. Residues critical for PARP-1 DNA-dependent activity are labeled: WGR residues W589 and R591, and Zn1 residue W79. (d). Crystal structure of the tandem Zn1–Zn2 domains of PARP-1 bound to a DNA double strand break with a single 5’ nucleotide overhang [40]. Zn2 forms DNA contacts similar to that seen for the isolated domain bound to DNA in panel b (the same conserved residues are shown). Zn1 binds to DNA with opposite polarity to that seen in panel a, with Arg18 positioned in the major groove rather than the minor groove, and the “base stacking loop” forming protein-protein contacts rather than protein-DNA contacts.
A recent structure of human PARP-1 domains in complex with a DNA double-strand break demonstrated a PARP-1 interface with DNA damage that is distributed over three domains [41], providing new insights into the DNA binding activities of PARP-1. The Zn1, Zn3, and WGR domains collectively form a continuous interface with DNA that traverses the terminus of the double-strand break and extends over roughly seven base pairs of DNA (Figure 2c). PARP-1 mainly contacts the ribose-phosphate backbone of the DNA mediating sequence-independent interactions, as expected for a repair protein that binds to a variety of damaged DNA structures independent of DNA sequence context. Zn1 makes the most extensive interactions with the DNA double-strand break, and contacts DNA using the backbone grip and the base stacking loop as observed in the crystal structure of the individual Zn1 domain bound to DNA [13]. The N-terminal α-helical region of Zn3 domain binds to the DNA on the same side as Zn1, extending the Zn1 interaction with DNA toward the continuous duplex region, and away from the break. WGR domain binds to the 5’-terminus of the break, with the DNA backbone held between the central β-sheet of WGR and an α-helix that runs parallel to the β-sheet. WGR contacts with DNA extend the contacts made by the Zn1 base stacking loop, which rests on the nucleotide bases at the end of the DNA. Mutations that target DNA contact residues affect DNA damage-dependent PARP-1 catalytic activity, indicating that the observed contacts are functionally important.
A recent structure of the tandem Zn1–Zn2 domains was determined in complex with DNA damage represented by a double-strand break with a single 5’ nucleotide overhang [40]. In this structure, Zn2 engages DNA as seen in the individual Zn2–DNA complex (Figure 2, compare Zn2 in panel b and d), with the base stacking loop engaging the nucleotide bases at the end of the DNA duplex and conserved Arg residues bracing the phosphate backbone. Unexpectedly, Zn1 binds to DNA with an opposite polarity to that seen in other PARP-1 zinc finger structures (Figure 2, compare Zn1 in panel a and d). The backbone grip uses the same conserved Arg 34 residue to bind to the phosphate backbone, but the entire domain is flipped such that Arg 18 is directed into the DNA major groove rather than the minor groove (Figure 2d). Furthermore, the base stacking loop of Zn1 does not engage DNA, but rather forms contacts with the base stacking loop of Zn2 (Figure 2d). It was not possible to model the linker connecting the two zinc fingers; however, the termini of the two zinc fingers engaged on one DNA end are not close enough in the structure to be physically linked. A model was therefore proposed in which the two zinc fingers originate from two different polypeptides, and the observed interaction between Zn1 and Zn2 is responsible for assembling two PARP-1 molecules onto DNA. The positioning of Zn1 and Zn2 on DNA in this complex is not compatible with the Zn1–Zn3–WGR complex bound to a similar DNA structure (Figure 2, compare panel c and d), primarily because Zn1 will not be able to form key contacts with WGR and Zn3 (see W79 and D45 in Figure 2d). Further experimentation is necessary to establish whether the observed conformation of Zn1 and Zn2 is consistent with the activation of PARP-1, and whether the observed interface between Zn1 and Zn2 is substantial enough to drive homodimerization of PARP-1 in solution.
DNA damage-induced PARP-1 interdomain communication
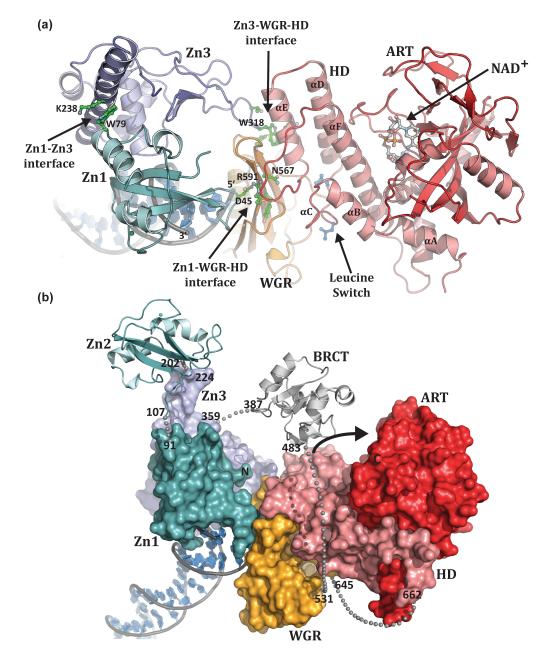
The crystal structure of PARP-1 essential domains (Zn1, Zn3, WGR-CAT) bound to a DNA double-strand break has provided the first views of how the multiple domains of PARP-1 assemble on DNA to form the active enzyme (Figure 3a) [41]. This study has taken advantage of the fact that the Zn2 and BRCT domains are not required for DNA-dependent activity on double-strand breaks [13,41,45] and that PARP-1 activity can be reconstituted by mixing the isolated Zn1, Zn3 and WGR-CAT fragments with DNA [41]. The PARP-1/DNA structure reveals that a single copy of each PARP-1 domain assembles into a compact complex on the DNA break (Figure 3a, b). The WGR domain occupies a central location in the complex, making contacts with all the other domains (Zn1, Zn3 and CAT) and the DNA, thus establishing an important functional role for this conserved domain.
Figure 3. PARP-1 domains collectively assemble on DNA damage.
(a). Human PARP-1 domains Zn1, Zn3, and WGR–CAT were crystallized in complex with a DNA double-strand break (PDB code 4dqy [41]). The PARP-1/DNA complex illustrates how DNA damage detection is coupled to structural transitions in the catalytic domain that elevate poly(ADP-ribosyl)ation activity [41]. Three essential interdomain contact regions form upon PARP-1 interaction with DNA: Zn1–WGR–HD, Zn3–WGR–HD, and Zn1–Zn3. Mutations that target has captured interdomain contacts that are relevant to PARP-1 regulation. Interdomain communication with the HD displaces conserved Leu residues from the hydrophobic interior of the HD, leading to destabilization of the CAT that correlates with an elevation in PARP-1 catalytic activity. (b). A model for the approximate positioning of the Zn2 and BRCT domains within the PARP-1/DNA complex. Zn1, Zn3, and WGR-CAT are shown as surfaces, labeled, and colored as in Figure 1a. The Zn2 and BRCT domains are drawn in schematic representation. Their positioning is based on the relative location of the termini of adjacent domains in the structure. The numbering and location of linker residues are shown. The arrow indicates the location of the PARP-1 automodification region near the catalytic active site.
The PARP-1/DNA structure demonstrates that multiple interdomain contacts are formed upon PARP-1 binding to DNA, and it demonstrates how interaction with DNA damage is vital to the formation of three key interacting regions between PARP-1 domains: Zn1–WGR–HD, Zn3– WGR–HD, and Zn1–Zn3 (Figure 3a). In the Zn1–WGR–HD interface, Zn1 uses one side of the base stacking loop to contact the DNA double-strand break, while the other side contacts the WGR near a turn between two β strands. A critical contact at the Zn1–WGR interface is a salt bridge formed between Asp45 of Zn1 and Arg591 of WGR. Arg591 also contacts the HD in a region connecting αE and αF (Figure 3a). WGR thus provides a bridge between the Zn1 DNA damage interface and the catalytic domain. WGR residues Asn567 and Tyr569 make additional contributions to the WGR–HD interface.
The Zn1 base stacking loop interaction with DNA damage is a critical factor that regulates PARP-1 DNA-dependent activity [13], and it appears to be a pivotal aspect of PARP-1 domain assembly on DNA damage. NMR structures of human Zn1 indicate that the base stacking loop has a flexible conformation and a high degree of mobility in the absence of DNA (PDB code 2dmj and [8]). The interaction with DNA damage likely serves to stabilize this region of the Zn1 structure into a single conformation and represents a conformational transition that triggers PARP-1 domain assembly and activation in response to DNA damage.
At the Zn3–WGR–HD interface, an extended loop of the Zn3 zinc ribbon fold contacts the WGR and HD. Zn3 residue Trp318 occupies a central location at this interface (Figure 3a). Trp318 is engulfed in a pocket formed by HD and WGR, stacked between WGR residue Lys633 and HD residue Arg735. At the Zn1–Zn3 interface, the N-terminal helical region of Zn3 rests against the Zn1 domain, near their points of contact with the DNA (Figure 3a). The Zn1–Zn3 interaction is primarily formed by residues on the first α-helix of Zn3, and a turn before the C-terminal α-helix of Zn1. The major contacts are formed between Zn3 residue Trp246 and Zn1 residue Arg78. Similarly, Zn1 residue Trp79 interacts with Zn3 residue Lys238 (Figure 3a). Several residues involved in interdomain contacts in the PARP-1/DNA crystal structure were targeted by site-directed mutagenesis in full-length PARP-1. All mutants showed moderate to severe reduction in DNA-dependent PARP-1 activity confirming the importance of the three domain interfaces observed in the PARP-1/DNA structure [41]. The importance of interdomain communication to PARP-1 activity suggests that these contacts represent new targets for PARP-1 inhibitor design, with the advantage that these interfaces are specific to PARP-1 and thus avoid PARP family cross reactivity with inhibitors that target the NAD+ binding pocket.
Coupling DNA damage detection to catalytic activity
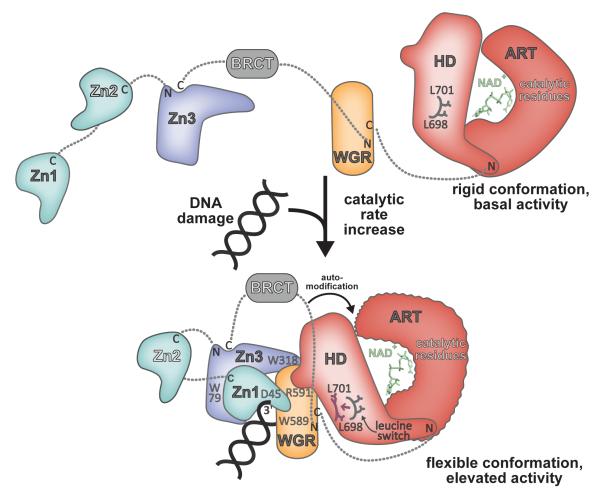
Upon binding to a DNA break, PARP-1 domains collapse together and establish interdomain contacts that collectively distort the structure of HD when compared to HD structures determined for isolated CAT domains [41]. The most prominent change in HD structure occurs near the interface with WGR, in the region surrounding αC (Figure 3a). In CAT structures determined in the absence of DNA and regulatory domains, αC forms an interface with αF and αB and contributes Leu698 and Leu701 to the hydrophobic interior of the HD [12]. In the PARP-1 complex with DNA, αC is remodeled to no longer resemble a helix, and the entire region moves away from αF and αB and toward the WGR interface, displacing conserved residues Leu698 and Leu701 from the hydrophobic core of the HD (Figure 4). The structure thus suggests that the DNA damage-induced conformation of PARP-1 acts to distort and destabilize the HD, and that HD distortion underlies PARP-1 activation. Site-directed mutants of HD residues facing the interior of the HD hydrophobic core show a substantial increase in DNA independent activity that correlates with a decrease in thermal stability [41], indicating that these mutations mimic the effect of DNA binding on PARP-1 activity. In support of this model, PARP-1 binding to DNA decreases thermal stability of full-length PARP-1, and the decrease in thermal stability depends on the CAT domain and the ability of PARP-1 to form interdomain contacts observed in the structure of PARP-1 in complex with DNA [41].
Figure 4. Model for DNA damage-dependent activation of PARP-1.
In the absence of DNA damage, PARP-1 domains exist in an extended, “beads-on-a-string” conformation. The HD serves as a modulator of PARP-1 activity, holding the ART in a rigid conformation. Upon detecting DNA damage the Zn1, Zn3, and WGR domains collapse together, forming a network of interdomain contacts that perturb the structure of the HD, displacing a “leucine switch” that decreases the stability of the catalytic domain and increases the catalytic activity. A more flexible, dynamic ART conformation is more efficient to perform the multi-step synthesis of poly(ADP-ribose). The collapsed conformation positions the automodication region adjacent to the catalytic domain, providing substrate specificity and contributing to an enhanced rate of poly(ADP-ribose) production.
The decrease in thermal stability observed for PARP-1 in the presence of DNA is likely to be a consequence of the disruption of the HD hydrophobic interior by the PARP-1 regulatory domains following binding to the break. One hypothesis is that the decrease in thermal stability of the HD leads to an increase in protein dynamics in the ART and consequently an increase in poly(ADP-ribosyl)ation activity. Indeed, the folds of the HD and ART subdomains are tightly linked, since αA of the HD contributes directly to the fold of the ART, suggesting that their stabilities and dynamics could be linked. Poly(ADP-ribosyl)ation is a multi-step enzymatic reaction involving repeated cycles of binding to NAD+ substrate, NAD+ hydrolysis and attachment of the ADP-ribose to the target residue, and displacement of the ADP-ribose from the donor to the acceptor site. Therefore an increase in protein dynamics of the ART could help increase the rate of some of these steps and consequently increase the catalytic rate of the poly(ADP-ribosyl)ation reaction. The stimulation of the poly(ADP-ribosyl)ation activity by DNA is not expected to modify the ability of PARP-1 to bind to the substrate NAD+ since DNA binding does not alter PARP-1 affinity for NAD+, but rather increases the rate of the reaction [46,47]. Consistently, crystal structures of the isolated PARP-1 CAT have shown an active site open and accessible for binding to various inhibitors that occupy the NAD+ binding site. The increase in PARP-1 activity in the presence of DNA is also unlikely to be related to access of target protein substrate to the catalytic site. Indeed, molecular dynamic calculations that have positioned an extended histone peptide tail into a rigid catalytic site have shown that the CAT can accommodate a protein substrate in the absence of DNA [18].
In addition to the structural changes that increase PARP-1 activity, it is likely that the positioning of the region of automodification in close proximity to the catalytic domain will contribute to PARP-1 DNA damage-dependent activity by increasing exposure to protein substrate. The positioning of the automodification region can also explain the strong preference for PARP-1 to attach polymers to itself, rather than heteromodification of other molecules.
Remaining Questions and Challenges
The modular domain architecture has challenged structural analysis of PARP-1, but the divide and conquer approach has provided key insights into PARP-1 mechanism of action that are consistent with biochemical analysis of the full-length protein. Nonetheless, a structure of full-length PARP-1 bound to DNA damage will build on our understanding of how the domains of PARP-1 assemble and communicate. For example, the BRCT domain is not strictly required for DNA damage-dependent activity, but its positioning within the PARP-1/DNA complex will have important implications for how PARP-1 recruits partner proteins for heteromodification, and how the automodification reaction might proceed. It will also be critical to understand how PARP-1 engages a variety of damaged DNA structures. Thus far, PARP-1 has been captured in complex with damaged DNA structures that represent a double strand break with only one end available for binding. PARP-1 is activated by a variety of DNA structures, perhaps most importantly single strand breaks and gaps in DNA that are commonly found in repair pathways associated with PARP-1 function. These types of DNA damage leave two ends available for binding, and it will be important to understand how PARP-1 parses its DNA binding domains to engage these substrates, and how this might influence PARP-1 interdomain communication necessary for activation. An even more difficult challenge will be to understand how PARP-1 engages unbroken, but abnormal DNA structures like that found in nucleosomal linker DNA, where PARP-1 binds to influence chromatin structure and gene transcription [48].
Are there multiple ways that PARP-1 assembles for activation? A growing number of biophysical studies have observed monomeric interactions between PARP-1 and DNA damage [49,8,13], including the Zn1–Zn2 fragment interaction with single strand breaks, and the full-length protein with a double strand break [41]. However, PARP-1 has measurable in vitro activity when domains are “mixed-and-matched” from separate polypeptides [9,10,13,40,45,47], suggesting there is some fluidity in how the domains can assemble. The Zn1–Zn2 crystal structure on a double strand break (Figure 2d) infers a dimer assembly, which presents a testable model for an alternative mode of PARP-1 activation. Importantly, the interdomain contacts observed in the monomeric PARP-1/DNA complex (Figure 3) are essential for activation on a variety of damaged DNA structures (MF Langelier et al., unpublished), so this assembly of PARP-1 domains is most likely to represent a consistent feature of all DNA-dependent mechanisms.
The complex of PARP-1 domains bound to DNA illustrates a damage-dependent intramolecular mechanism for rapidly elevating the low basal level of PARP-1 activity through perturbations to the HD (Figure 4). PARP-1 activity is also regulated through other signals besides DNA damage, such as posttranslational modifications [50,51] and interaction with nucleosomes [48] and histones [52]. It will be interesting to investigate whether these activating signals also act through perturbations to the HD structure. PARP-2 and PARP-3 CAT domains also contain an HD [35,38], but their overall domain organizations are quite different from PARP-1. Thus, it will also be interesting to see how the HD of PARP-2 and PARP-3 will be regulated, and whether they ultimately use a similar destabilization mechanism.
Conclusion
The structural biology of poly(ADP-ribose) signaling has made important advances over the past several years. In addition to the structures highlighted in this review, we have recently seen the first structures of poly(ADP-ribose) glycohydrolase (PARG) enzymes that reverse the modification [53-55], protein modules that recognize the modification (WWE [56,57] and PBZ [58-61]), catalytic domains from additional PARP family enzymes [30,32,34], and the regulatory ankyrin domains of PARP-5, or Tankyrase, in complex with target protein peptides [62,63]. Collectively, these structural insights have moved forward our understanding of the biology and regulation of this interesting posttranslational modification, and pave the way for exciting new discoveries.
Highlights.
**
PARP-1 zinc finger domains recognize exposed nucleotide bases as a DNA damage signal
**
The modular domain architecture of PARP-1 collapses onto DNA damage as a monomer
**
Interdomain communication is critical to PARP-1 activation in response to DNA damage
**
A destabilized PARP-1 catalytic domain has increased poly(ADP-ribosyl)ation activity
Acknowledgements
Research in the Pascal laboratory is supported by the NIH (GM087282) and the American Cancer Society (RSG0918301DMC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell. 2010;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci. 2008;13:3046–3082. doi: 10.2741/2909. [DOI] [PubMed] [Google Scholar]
- 4.Haince JF, McDonald D, Rodrigue A, Dery U, Masson JY, Hendzel MJ, Poirier GG. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J Biol Chem. 2008;283:1197–1208. doi: 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]
- 5.Gottschalk AJ, Timinszky G, Kong SE, Jin J, Cai Y, Swanson SK, Washburn MP, Florens L, Ladurner AG, Conaway JW, et al. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc Natl Acad Sci U S A. 2009;106:13770–13774. doi: 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Timinszky G, Till S, Hassa PO, Hothorn M, Kustatscher G, Nijmeijer B, Colombelli J, Altmeyer M, Stelzer EH, Scheffzek K, et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat Struct Mol Biol. 2009;16:923–929. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- 7.Ahel D, Horejsi Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP, et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8 *.Eustermann S, Videler H, Yang JC, Cole PT, Gruszka D, Veprintsev D, Neuhaus D. The DNA-binding domain of human PARP-1 interacts with DNA single-strand breaks as a monomer through its second zinc finger. J Mol Biol. 2011;407:149–170. doi: 10.1016/j.jmb.2011.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9 *.Langelier MF, Servent KM, Rogers EE, Pascal JM. A Third Zinc-binding Domain of Human Poly(ADP-ribose) Polymerase-1 Coordinates DNA-dependent Enzyme Activation. J Biol Chem. 2008;283:4105–4114. doi: 10.1074/jbc.M708558200. [DOI] [PubMed] [Google Scholar]
- 10 *.Tao Z, Gao P, Hoffman DW, Liu HW. Domain C of Human Poly(ADP-ribose) Polymerase-1 Is Important for Enzyme Activity and Contains a Novel Zinc-Ribbon Motif. Biochemistry. 2008;47:5804–13. doi: 10.1021/bi800018a. [DOI] [PubMed] [Google Scholar]
- 11.Loeffler PA, Cuneo MJ, Mueller GA, DeRose EF, Gabel SA, London RE. Structural studies of the PARP-1 BRCT domain. BMC Struct Biol. 2011;11:37. doi: 10.1186/1472-6807-11-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruf A, Mennissier de Murcia J, de Murcia G, Schulz GE. Structure of the catalytic fragment of poly(AD-ribose) polymerase from chicken. Proc Natl Acad Sci U S A. 1996;93:7481–7485. doi: 10.1073/pnas.93.15.7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13 *.Langelier MF, Planck JL, Roy S, Pascal JM. Crystal structures of poly(ADP-ribose) polymerase-1 (PARP-1) zinc fingers bound to DNA: structural and functional insights into DNA-dependent PARP-1 activity. J Biol Chem. 2011;286:10690–10701. doi: 10.1074/jbc.M110.202507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lonskaya I, Potaman VN, Shlyakhtenko LS, Oussatcheva EA, Lyubchenko YL, Soldatenkov VA. Regulation of poly(ADP-ribose) polymerase-1 by DNA structure-specific binding. J Biol Chem. 2005;280:17076–17083. doi: 10.1074/jbc.M413483200. [DOI] [PubMed] [Google Scholar]
- 15.Pion E, Bombarda E, Stiegler P, Ullmann GM, Mely Y, de Murcia G, Gerard D. Poly(ADP-ribose) polymerase-1 dimerizes at a 5’ recessed DNA end in vitro: a fluorescence study. Biochemistry. 2003;42:12409–12417. doi: 10.1021/bi0344432. [DOI] [PubMed] [Google Scholar]
- 16.D’Silva I, Pelletier JD, Lagueux J, D’Amours D, Chaudhry MA, Weinfeld M, Lees-Miller SP, Poirier GG. Relative affinities of poly(ADP-ribose) polymerase and DNA-dependent protein kinase for DNA strand interruptions. Biochim Biophys Acta. 1999;1430:119–126. doi: 10.1016/s0167-4838(98)00278-7. [DOI] [PubMed] [Google Scholar]
- 17.Otto H, Reche PA, Bazan F, Dittmar K, Haag F, Koch-Nolte F. In silico characterization of the family of PARP-like poly(ADP-ribosyl)transferases (pARTs) BMC Genomics. 2005;6:139. doi: 10.1186/1471-2164-6-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Messner S, Altmeyer M, Zhao H, Pozivil A, Roschitzki B, Gehrig P, Rutishauser D, Huang D, Caflisch A, Hottiger MO. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res. 2010;38:6350–6362. doi: 10.1093/nar/gkq463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bell CE, Eisenberg D. Crystal structure of diphtheria toxin bound to nicotinamide adenine dinucleotide. Biochemistry. 1996;35:1137–1149. doi: 10.1021/bi9520848. [DOI] [PubMed] [Google Scholar]
- 20.Ruf A, de Murcia G, Schulz GE. Inhibitor and NAD+ binding to poly(ADP-ribose) polymerase as derived from crystal structures and homology modeling. Biochemistry. 1998;37:3893–3900. doi: 10.1021/bi972383s. [DOI] [PubMed] [Google Scholar]
- 21.Rolli V, Ruf A, Augustin A, Schulz GE, Ménissier-de Murcia J, de Murcia G. From DNA damage and stress signalling to cell death: Poly ADP-ribosylation reactions. Oxford University Press; New York: 2000. Poly(ADP-ribose) polymerase: Structure and function; pp. 35–79. [Google Scholar]
- 22.Ruf A, Rolli V, de Murcia G, Schulz GE. The mechanism of the elongation and branching reaction of poly(ADP-ribose) polymerase as derived from crystal structures and mutagenesis. J Mol Biol. 1998;278:57–65. doi: 10.1006/jmbi.1998.1673. [DOI] [PubMed] [Google Scholar]
- 23.Kinoshita T, Nakanishi I, Warizaya M, Iwashita A, Kido Y, Hattori K, Fujii T. Inhibitor-induced structural change of the active site of human poly(ADP-ribose) polymerase. FEBS Lett. 2004;556:43–46. doi: 10.1016/s0014-5793(03)01362-0. [DOI] [PubMed] [Google Scholar]
- 24.Gandhi VB, Luo Y, Liu X, Shi Y, Klinghofer V, Johnson EF, Park C, Giranda VL, Penning TD, Zhu GD. Discovery and SAR of substituted 3-oxoisoindoline-4-carboxamides as potent inhibitors of poly(ADP-ribose) polymerase (PARP) for the treatment of cancer. Bioorg Med Chem Lett. 2010;20:1023–1026. doi: 10.1016/j.bmcl.2009.12.042. [DOI] [PubMed] [Google Scholar]
- 25.Miyashiro J, Woods KW, Park CH, Liu X, Shi Y, Johnson EF, Bouska JJ, Olson AM, Luo Y, Fry EH, et al. Synthesis and SAR of novel tricyclic quinoxalinone inhibitors of poly(ADP-ribose)polymerase-1 (PARP-1) Bioorg Med Chem Lett. 2009;19:4050–4054. doi: 10.1016/j.bmcl.2009.06.016. [DOI] [PubMed] [Google Scholar]
- 26.Iwashita A, Hattori K, Yamamoto H, Ishida J, Kido Y, Kamijo K, Murano K, Miyake H, Kinoshita T, Warizaya M, et al. Discovery of quinazolinone and quinoxaline derivatives as potent and selective poly(ADP-ribose) polymerase-1/2 inhibitors. FEBS Lett. 2005;579:1389–1393. doi: 10.1016/j.febslet.2005.01.036. [DOI] [PubMed] [Google Scholar]
- 27.Hattori K, Kido Y, Yamamoto H, Ishida J, Kamijo K, Murano K, Ohkubo M, Kinoshita T, Iwashita A, Mihara K, et al. Rational approaches to discovery of orally active and brain-penetrable quinazolinone inhibitors of poly(ADP-ribose)polymerase. J Med Chem. 2004;47:4151–4154. doi: 10.1021/jm0499256. [DOI] [PubMed] [Google Scholar]
- 28.Penning TD. Small-molecule PARP modulators--current status and future therapeutic potential. Curr Opin Drug Discov Devel. 2010;13:577–586. [PubMed] [Google Scholar]
- 29.Penning TD, Zhu GD, Gong J, Thomas S, Gandhi VB, Liu X, Shi Y, Klinghofer V, Johnson EF, Park CH, et al. Optimization of phenyl-substituted benzimidazole carboxamide poly(ADP-ribose) polymerase inhibitors: identification of (S)-2-(2-fluoro-4-(pyrrolidin-2-yl)phenyl)-1H-benzimidazole-4-carboxamide (A-966492), a highly potent and efficacious inhibitor. J Med Chem. 2010;53:3142–3153. doi: 10.1021/jm901775y. [DOI] [PubMed] [Google Scholar]
- 30.Wahlberg E, Karlberg T, Kouznetsova E, Markova N, Macchiarulo A, Thorsell AG, Pol E, Frostell A, Ekblad T, Oncu D, et al. Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nat Biotechnol. 2012;30:283–288. doi: 10.1038/nbt.2121. [DOI] [PubMed] [Google Scholar]
- 31.Karlberg T, Hammarstrom M, Schutz P, Svensson L, Schuler H. Crystal structure of the catalytic domain of human PARP2 in complex with PARP inhibitor ABT-888. Biochemistry. 2010;49:1056–1058. doi: 10.1021/bi902079y. [DOI] [PubMed] [Google Scholar]
- 32.Karlberg T, Thorsell AG, Kallas A, Schuler H. Crystal structure of human ADP-ribose transferase ARTD15/PARP16 reveals a novel putative regulatory domain. J Biol Chem. 2012;287:24077–24081. doi: 10.1074/jbc.M112.379289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karlberg T, Markova N, Johansson I, Hammarstrom M, Schutz P, Weigelt J, Schuler H. Structural basis for the interaction between tankyrase-2 and a potent Wnt-signaling inhibitor. J Med Chem. 2010;53:5352–5355. doi: 10.1021/jm100249w. [DOI] [PubMed] [Google Scholar]
- 34.Andersson CD, Karlberg T, Ekblad T, Lindgren AE, Thorsell AG, Spjut S, Uciechowska U, Niemiec MS, Wittung-Stafshede P, Weigelt J, et al. Discovery of ligands for ADP-ribosyltransferases via docking-based virtual screening. J Med Chem. 2012;55:7706–7718. doi: 10.1021/jm300746d. [DOI] [PubMed] [Google Scholar]
- 35.Lehtio L, Jemth AS, Collins R, Loseva O, Johansson A, Markova N, Hammarstrom M, Flores A, Holmberg-Schiavone L, Weigelt J, et al. Structural basis for inhibitor specificity in human poly(ADP-ribose) polymerase-3. J Med Chem. 2009;52:3108–3111. doi: 10.1021/jm900052j. [DOI] [PubMed] [Google Scholar]
- 36.Kirby CA, Cheung A, Fazal A, Shultz MD, Stams T. Structure of human tankyrase 1 in complex with small-molecule inhibitors PJ34 and XAV939. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012;68:115–118. doi: 10.1107/S1744309111051219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gunaydin H, Gu Y, Huang X. Novel binding mode of a potent and selective tankyrase inhibitor. PLoS One. 2012;7:e33740. doi: 10.1371/journal.pone.0033740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oliver AW, Ame JC, Roe SM, Good V, de Murcia G, Pearl LH. Crystal structure of the catalytic fragment of murine poly(ADP-ribose) polymerase-2. Nucleic Acids Res. 2004;32:456–464. doi: 10.1093/nar/gkh215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simonin F, Hofferer L, Panzeter PL, Muller S, de Murcia G, Althaus FR. The carboxyl-terminal domain of human poly(ADP-ribose) polymerase. Overproduction in Escherichia coli, large scale purification, and characterization. J Biol Chem. 1993;268:13454–13461. [PubMed] [Google Scholar]
- 40 *.Ali AA, Timinszky G, Arribas-Bosacoma R, Kozlowski M, Hassa PO, Hassler M, Ladurner AG, Pearl LH, Oliver AW. The zinc-finger domains of PARP1 cooperate to recognize DNA strand breaks. Nat Struct Mol Biol. 2012;19:685–692. doi: 10.1038/nsmb.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41 **.Langelier MF, Planck JL, Roy S, Pascal JM. Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science. 2012;336:728–732. doi: 10.1126/science.1216338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kulczyk AW, Yang JC, Neuhaus D. Solution structure and DNA binding of the zinc-finger domain from DNA ligase IIIalpha. J Mol Biol. 2004;341:723–738. doi: 10.1016/j.jmb.2004.06.035. [DOI] [PubMed] [Google Scholar]
- 43.Mackey ZB, Niedergang C, Murcia JM, Leppard J, Au K, Chen J, de Murcia G, Tomkinson AE. DNA ligase III is recruited to DNA strand breaks by a zinc finger motif homologous to that of poly(ADP-ribose) polymerase. Identification of two functionally distinct DNA binding regions within DNA ligase III. J Biol Chem. 1999;274:21679–21687. doi: 10.1074/jbc.274.31.21679. [DOI] [PubMed] [Google Scholar]
- 44.Cotner-Gohara E, Kim IK, Tomkinson AE, Ellenberger T. Two DNA-binding and nick recognition modules in human DNA ligase III. J Biol Chem. 2008;283:10764–10772. doi: 10.1074/jbc.M708175200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altmeyer M, Messner S, Hassa PO, Fey M, Hottiger MO. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res. 2009;37:3723–3738. doi: 10.1093/nar/gkp229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clark NJ, Kramer M, Muthurajan UM, Luger K. Alternative modes of binding of poly(ADP-ribose) polymerase 1 to free DNA and nucleosomes. J Biol Chem. 2012;287:32430–32439. doi: 10.1074/jbc.M112.397067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langelier MF, Ruhl DD, Planck JL, Kraus WL, Pascal JM. The Zn3 domain of human poly(ADP-ribose) polymerase-1 (PARP-1) functions in both DNA-dependent poly(ADP-ribose) synthesis activity and chromatin compaction. J Biol Chem. 2010;285:18877–18887. doi: 10.1074/jbc.M110.105668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim MY, Mauro S, Gevry N, Lis JT, Kraus WL. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell. 2004;119:803–814. doi: 10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 49.Lilyestrom W, van der Woerd MJ, Clark N, Luger K. Structural and biophysical studies of human PARP-1 in complex with damaged DNA. J Mol Biol. 2010;395:983–994. doi: 10.1016/j.jmb.2009.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kauppinen TM, Chan WY, Suh SW, Wiggins AK, Huang EJ, Swanson RA. Direct phosphorylation and regulation of poly(ADP-ribose) polymerase-1 by extracellular signal-regulated kinases 1/2. Proc Natl Acad Sci U S A. 2006;103:7136–7141. doi: 10.1073/pnas.0508606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gagne JP, Moreel X, Gagne P, Labelle Y, Droit A, Chevalier-Pare M, Bourassa S, McDonald D, Hendzel MJ, Prigent C, et al. Proteomic investigation of phosphorylation sites in poly(ADP-ribose) polymerase-1 and poly(ADP-ribose) glycohydrolase. J Proteome Res. 2009;8:1014–1029. doi: 10.1021/pr800810n. [DOI] [PubMed] [Google Scholar]
- 52.Pinnola A, Naumova N, Shah M, Tulin AV. Nucleosomal core histones mediate dynamic regulation of poly (ADP-Ribose) polymerase 1 protein binding to chromatin and induction of its enzymatic activity. J Biol Chem. 2007;282:32511–32519. doi: 10.1074/jbc.M705989200. [DOI] [PubMed] [Google Scholar]
- 53 **.Slade D, Dunstan MS, Barkauskaite E, Weston R, Lafite P, Dixon N, Ahel M, Leys D, Ahel I. The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature. 2011;477:616–620. doi: 10.1038/nature10404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54 *.Dunstan MS, Barkauskaite E, Lafite P, Knezevic CE, Brassington A, Ahel M, Hergenrother PJ, Leys D, Ahel I. Structure and mechanism of a canonical poly(ADP-ribose) glycohydrolase. Nat Commun. 2012;3:878. doi: 10.1038/ncomms1889. [DOI] [PubMed] [Google Scholar]
- 55 *.Kim IK, Kiefer JR, Ho CM, Stegeman RA, Classen S, Tainer JA, Ellenberger T. Structure of mammalian poly(ADP-ribose) glycohydrolase reveals a flexible tyrosine clasp as a substrate-binding element. Nat Struct Mol Biol. 2012;19:653–656. doi: 10.1038/nsmb.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Z, Michaud GA, Cheng Z, Zhang Y, Hinds TR, Fan E, Cong F, Xu W. Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination. Genes Dev. 2012;26:235–240. doi: 10.1101/gad.182618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He F, Tsuda K, Takahashi M, Kuwasako K, Terada T, Shirouzu M, Watanabe S, Kigawa T, Kobayashi N, Guntert P, et al. Structural insight into the interaction of ADP-ribose with the PARP WWE domains. FEBS Lett. 2012;586:3858–3864. doi: 10.1016/j.febslet.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 58.Eustermann S, Brockmann C, Mehrotra PV, Yang JC, Loakes D, West SC, Ahel I, Neuhaus D. Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose) Nat Struct Mol Biol. 2010;17:241–243. doi: 10.1038/nsmb.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li GY, McCulloch RD, Fenton AL, Cheung M, Meng L, Ikura M, Koch CA. Structure and identification of ADP-ribose recognition motifs of APLF and role in the DNA damage response. Proc Natl Acad Sci U S A. 2010;107:9129–9134. doi: 10.1073/pnas.1000556107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Isogai S, Kanno S, Ariyoshi M, Tochio H, Ito Y, Yasui A, Shirakawa M. Solution structure of a zinc-finger domain that binds to poly-ADP-ribose. Genes Cells. 2010;15:101–110. doi: 10.1111/j.1365-2443.2009.01369.x. [DOI] [PubMed] [Google Scholar]
- 61.Oberoi J, Richards MW, Crumpler S, Brown N, Blagg J, Bayliss R. Structural basis of poly(ADP-ribose) recognition by the multizinc binding domain of checkpoint with forkhead-associated and RING Domains (CHFR) J Biol Chem. 2010;285:39348–39358. doi: 10.1074/jbc.M110.159855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62 **.Guettler S, LaRose J, Petsalaki E, Gish G, Scotter A, Pawson T, Rottapel R, Sicheri F. Structural basis and sequence rules for substrate recognition by Tankyrase explain the basis for cherubism disease. Cell. 2011;147:1340–1354. doi: 10.1016/j.cell.2011.10.046. [DOI] [PubMed] [Google Scholar]
- 63 **.Morrone S, Cheng Z, Moon RT, Cong F, Xu W. Crystal structure of a Tankyrase-Axin complex and its implications for Axin turnover and Tankyrase substrate recruitment. Proc Natl Acad Sci U S A. 2012;109:1500–1505. doi: 10.1073/pnas.1116618109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Riken Structural Genomics/Proteomics Initiative. www.rsgi.riken.go.jp/rsgi_e.