Abstract
Severe mitochondria deficiency leads to a number of devastating degenerative disorders, yet, mild mitochondrial dysfunction in different species, including the nematode Caenorhabditis elegans, can have pro-longevity effects. This apparent paradox indicates that cellular adaptation to partial mitochondrial stress can induce beneficial responses, but how this is achieved is largely unknown. Complete absence of frataxin, the mitochondrial protein defective in patients with Friedreich's ataxia, is lethal in C. elegans, while its partial deficiency extends animal lifespan in a p53 dependent manner.
In this paper we provide further insight into frataxin control of C. elegans longevity by showing that a substantial reduction of frataxin protein expression is required to extend lifespan, affect sensory neurons functionality, remodel lipid metabolism and trigger autophagy. We find that Beclin and p53 genes are required to induce autophagy and concurrently reduce lipid storages and extend animal lifespan in response to frataxin suppression. Reciprocally, frataxin expression modulates autophagy in the absence of p53. Human Friedreich ataxia-derived lymphoblasts also display increased autophagy, indicating an evolutionarily conserved response to reduced frataxin expression.
In sum, we demonstrate a causal connection between induction of autophagy and lifespan extension following reduced frataxin expression, thus providing the rationale for investigating autophagy in the pathogenesis and treatment of Friedreich's ataxia and possibly other human mitochondria-associated disorders.
Keywords: Aging, Nematode, Mitochondria, Frataxin, p53/cep-1, Autophagy, Fat
Highlights
► Substantial fratxin protein decrease is required to extend lifespan in C. elegans ► Substantial fratxin protein decrease is required to trigger autophagy ► Life-extension correlates with reduced fat content and antioxidants induction ► Lifespan and fat content are inversely modulated by p53-regulated autophagy ► Autophagy induction is an evolutionary conserved response to frataxin deficiency
1. Introduction
Mutations in genes that directly or indirectly affect the functionality of the mitochondrial respiratory chain (MRC) lead to a variety of devastating disorders in humans (Wallace, 2005). Friedreich's ataxia (FRDA), the most frequently inherited recessive ataxia, is one such disease and it is ascribed to severe deficiency of frataxin, a nuclear-encoded mitochondrial protein involved in the biogenesis of iron–sulfur cluster (ISC) containing proteins (Campuzano et al., 1996; Puccio et al., 2001). Residual levels of frataxin are critical for survival and inversely correlate with disease onset, progression and severity (McDaniel et al., 2001). Symptoms only appear when levels of frataxin are severely decreased, and non-pathological levels of frataxin deficiency are associated with alterations in gene expression profiles (Haugen et al., 2010; Huang et al., 2009). These observations suggest that animals attempt to cope with partial frataxin deficiency by inducing adaptive responses, which, if characterized, may reveal novel therapeutic strategies to prevent or postpone the established disease in humans.
In the nematode Caenorhabditis elegans (C. elegans), complete knock-out of the frataxin ortholog (frh-1) or severe deficiency of other nuclear-encoded MRC proteins leads to pathological phenotypes such as arrested development or short lifespan (Rea et al., 2007; Ventura and Rea, 2007). On the other hand, partial suppression of the same genes, including frh-1 (Ventura et al., 2005), consistent with the induction of beneficial adaptive responses under these conditions, translates into life extension (Ni and Lee, 2010). Nevertheless, after these initial findings, contrasting results were published on lifespan regulation following frh-1 silencing in C. elegans (Vazquez-Manrique et al., 2006; Zarse et al., 2007). Although some explanations (Ventura et al., 2006) could be evoked to reconcile the opposite lifespan outcomes observed in response to frataxin suppression, these conflicting data still await compelling experimental clarification, which are required to gain insight into the biology of aging and to help establish an appropriate C. elegans model to study Friedreich's ataxia.
The extended longevity of animals with reduced expression of genes directly or indirectly involved in regulating MRC functionality is associated with the induction of different stress responses and it is regulated among others by the p53 C. elegans homolog cep-1 (Torgovnick et al., 2010; Ventura et al., 2009). P53 integrates several intrinsic and extrinsic stress signals to modulate intracellular responses and its activation is impaired in frataxin-deficient mammalian cells (Guccini et al., 2011; Palomo et al., 2011). Besides its classical function in response to DNA damage, p53 controls mitochondrial energy metabolism, antioxidant defenses, and autophagy (Maddocks and Vousden, 2011). Macroautophagy (hereafter referred to as autophagy) is a fundamental housekeeping program responsible for recycling of cellular components and necessary for tissue homeostasis; it is required for normal growth, development, proper energy metabolism and aging, and can be stimulated in response to a variety of different stressors, such as oxidative stress, energy deprivation, hypoxia, and mitochondria or DNA damage (Kroemer et al., 2010). The autophagic process is finely regulated by several proteins, which cooperate to coordinate the nucleation, elongation and degradation of autophagosomes in the lysosomes (Klionsky et al., 2010), and are very well conserved between humans and nematodes. Excessive or hindered activation of this finely regulated program can lead to cell death and has been associated with many diseases in humans, including neurodegenerative disorders (Levine and Kroemer, 2008).
Here we show that a substantial amount of frataxin protein expression must be reduced to trigger autophagy, in turn extending lifespan and reducing lipid content in C. elegans. Interestingly, we found that a reciprocal interplay exists in regulating autophagy between levels of frataxin expression and p53/cep-1. Collectively, we demonstrate a causal connection between induction of autophagy and lifespan extension following reduced frataxin expression, and provide a rationale for investigating autophagy in the pathogenesis and treatment of FRDA and possibly other human mitochondrial-associated disorders (HMAD).
2. Materials and methods
2.1. Nematode strains and maintenance
We employed standard nematode culture conditions (Stiernagle, 2006). All strains were maintained at 20 °C on Nematode Growth Media agar supplemented with Escherichia coli (OP50 or transformed HT115). The complete list of strains utilized in this work can be found in Supplemental Information (SI).
2.2. RNAi feeding
The different dsRNA constructs against frh-1 (I, II, III, IV) were generated using pL440 vector through standard molecular cloning techniques and utilized to transform HT115(DE3) E. coli for RNAi feeding. Unless otherwise indicated, all the experiments in this study were carried out in animals fed for three consecutive generations with bacteria expressing frh-1IV dsRNA, which targets the entire frh-1 CDS. See SI.
2.3. Lifespan and statistical analysis
Survival curves and statistical analyses were carried out as previously described (Ventura et al., 2009). Data from survival assays are summarized in Supplementary Tables. Figures display survival curves of pooled populations utilized for statistical analysis.
2.4. Quantitative RT-PCR
Quantification of mRNA in frh-1 RNAi-treated animals relative to vector-only treated animals was undertaken from three different biological samples. cDNA from each biological sample was run in triplicate, using specific primer pairs (available upon request). Different methodologies were utilized for different subset of genes, to design primers and to quantify gene expression. See SI. Regardless, fold change in gene expression in frh-1 (RNAi) animals was considered significantly different from empty-vector treated animals when a two-tailed Student's t-test resulted in p < 0.05.
2.5. Western blot analysis
Protocols for western blot analysis in both nematodes and mammalian cells are described in SI. Generation of a specific antibody against C. elegans FRH-1 is carefully described in SI. Briefly, Frataxin cDNA was utilized to synthesize recombinant frataxin protein (GeneScript) and to immunize Balb/c mice. Somatic cell hybrids were prepared using the mouse myeloma cells lines NS-1 and hybridoma supernatants were screened for immunoreactivity to recombinant frataxin. Positive hybridoma cell lines were cloned and one MAb was selected and designated 1D2.
2.6. Autophagy quantification
2.6.1. Nematodes
The level of autophagy (autophagosomal accumulation) was assessed by quantifying the number of GFP positive foci in embryos and in seam cells of L3 larvae, using the translational reporter strain GFP::LGG-1. See SI.
2.6.2. Mammalian cells
The basal level of autophagy (LC3 processing) in mammalian cells was assessed by western blot quantification of LC3II accumulation normalized to the level of tubulin in the same cellular extract. Autophagic flux was similarly assessed by LC3II quantification after treating cells for the indicated time with the lysosome inhibitor NH4Cl.
2.7. Lipid and lysosomes content quantification
Lysosome related organelles (LROs) were quantified by vital Nile Red staining, while fat content was quantified by fixing animals and staining them with Oil Red O. See SI.
2.8. Quantification of ATP, reactive oxygen species and GSSG/GSH content
ATP and ROS measurements were carried out as carefully described in SI. The ratio between oxidized and reduced pools of glutathione was assessed through the Prpl7::Grx1-roGFP2 strain. All assays were carried out in one-day old adults in, at least, triplicate. See SI.
2.9. Behavioral assays
Behavioral assays were carried out in one-day old adults in, at least, triplicate, each time by two independent operators. See SI.
Pumping rate was calculated as the number of pharyngeal pumps per minute in 15 animals. Defecation was scored by averaging the time between two consecutive pBoc contractions in 15 animals. Locomotion was assessed by counting the changes in the body bend at the mid-body point per minute of 10 animals placed on solid agar plates with no bacteria. Mechanosensation was calculated by touching 10 animals with an eyelash alternating 6 touches on the head (backward movement) and 6 touches on the tail (forward movement). Chemotaxis was calculated as carefully described in SI. Briefly, a population of 80–100 age-synchronized animals was spotted in the centre of the testing plate and the number of worms at attractant and control was counted every 15 minutes for two hours to calculate the Chemotaxis Index (CI). CI = (A − B) / (A + B + C), where A is the number of worms at attractant, B is the number of worms at control and C is the number of animals which did not reach any of the two spots at the end of the two hours.
2.10. Statistical analysis
Unless otherwise specified all statistical analysis were calculated with the unpaired Student's t-test and considered significantly different from control condition when p < 0.05.
3. Results
3.1. Substantial suppression of frataxin protein expression is required to extend lifespan in C. elegans
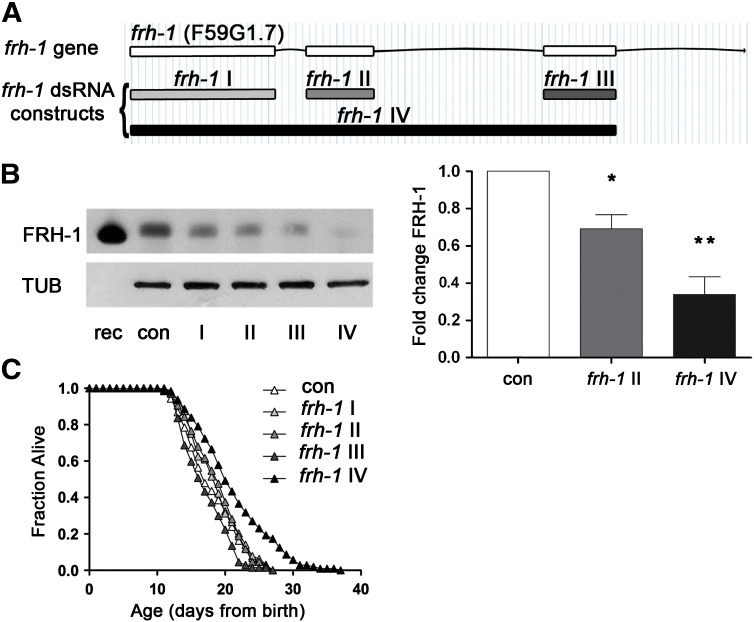
Residual levels of frataxin expression are crucial to maintain cell survival in FRDA disease, but how this is achieved is largely unknown. Moreover, contrasting evidence has been reported following frataxin suppression in modulating C. elegans lifespan (Vazquez-Manrique et al., 2006; Ventura et al., 2005; Zarse et al., 2007). To reconcile discrepancies in the field, and to shed light on mechanistic aspects of frh-1-suppression control of longevity, we generated four dsRNA constructs targeting different portions of the frh-1 gene and a specific antibody against C. elegans FRH-1. The four different dsRNA displayed different interference potency as revealed by western blotting of total C. elegans extracts (Figs. 1A–B; S1A–B). The anti-FRH-1 antibody revealed one or two bands (depending on protein extraction method) at the expected molecular weight of 15 kDa in wild-type lysate, likely corresponding, as in mammalian cells, to different forms of the protein (Fig. S1A-B). Lifespan extension was only achieved with the frh-1 IV dsRNA construct — referred to as frh-1 hereafter unless otherwise specified — which reduced FRH-1 expression to more than 50% of its wild-type level, without completely abolishing its expression (Fig. 1B–C; Table 1). Our results unambiguously demonstrated that a critical amount of frataxin protein must be suppressed to induce significant life-extending responses in C. elegans thus providing us as with a great tool to untangle protective pathways activated to compensate for frataxin suppression at the organismal level.
Fig. 1.
Substantial frataxin suppression is required to extend C. elegans lifespan. A) frh-1 gene and dsRNA interference constructs. One construct against the entire frataxin genomic CDS (frh-1IV), and three additional constructs targeting the three single exons, respectively named frh-1I, frh-1 II and frh-1III, were generated in this work. B) Frataxin protein (FRH-1) expression was quantified in 1-day-old adult wild-type (WT) animals fed bacteria expressing either empty-vector (con) or one of the four different frh-1 dsRNA. In the left panel FRH-1 expression was revealed through western blot with a specific antibody anti Ce-FRH-1 and normalized to tubulin (TUB). In the first lane 10 ng of FRH-1 recombinant protein (rec) were loaded. FRH-1 appears at the expected molecular weight ~ 15 kDa. In the right panel, bars and errors indicate mean FRH-1 expression ± standard error of the mean (SEM) after densitometric analysis from two independent experiments. *p < 0.05, **p < 0.001 t-test. C) Survival analysis of wild-type animals fed bacteria expressing either empty-vector (con) or the different frh-1 dsRNA.
Fig. S1.
Fig. 2.
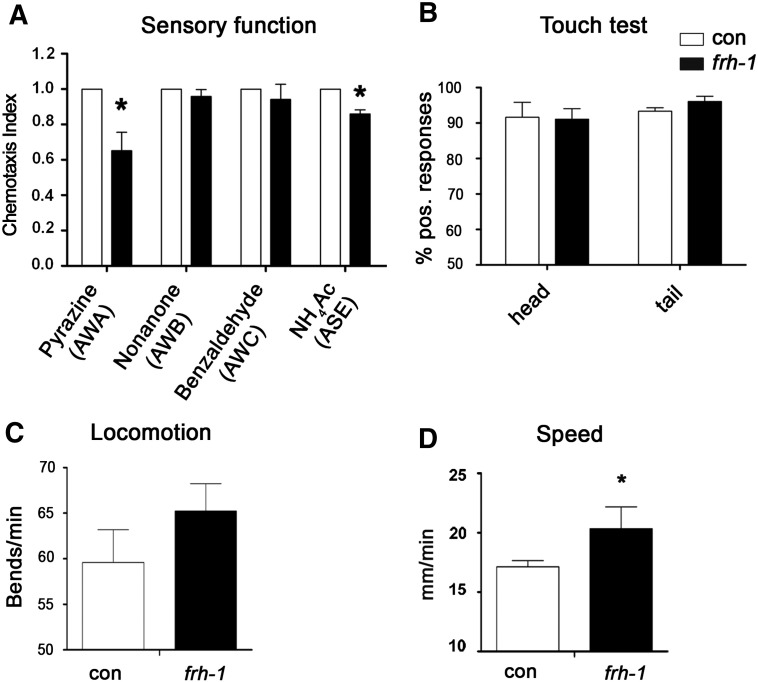
Characterization of neuronal features. A) Sensory function (chemotaxis index), B) proprioception (touch test) and C–D) locomotion activity (body bends and speed), in wild-type (1 day-old adults) animals fed empty-vector (con) or frh-1IV dsRNA (frh-1) expressing bacteria. Bars and errors indicate mean values + SEM from three independent replicates. *p < 0.05 vs control, t-test.
Table 1.
Lifespan analyses frh-1 dsRNA constructs — summary statistics.
| Genotype | RNAia | Mean Lifespan | ± SEM | p vs conb | Sample size (censored)c | N of replicates |
|---|---|---|---|---|---|---|
| Wild-type (N2) | con | 17.6 | 0.4 | 140(22) | 2 | |
| frh-1 I | 18.3 | 0.5 | 0.45750 | 130(10) | 2 | |
| frh-1 II | 17.2 | 0.3 | 0.37669 | 140(12) | 2 | |
| frh-1 III | 16.7 | 0.4 | 0.61293 | 130(6) | 2 | |
| frh-1 IV | 20.9 | 0.5 | 0.00001 | 140(36) | 2 |
Unless otherwise specified, all lifespan analyses in the paper were performed on 3rd generation worms cultured continuously on frh-1 RNAi.
p-values were calculated using the log-rank test between pooled populations.
Pooled worms from N replicate experiments (censored individuals were included in the log-rank analysis).
Residual levels of frataxin expression are crucial to maintain cell survival in FRDA disease, but how this is achieved is largely unknown. Moreover, contrasting evidence has been reported following frataxin suppression in modulating C. elegans lifespan (Vazquez-Manrique et al., 2006; Ventura et al., 2005; Zarse et al., 2007). To reconcile discrepancies in the field, and to shed light on mechanistic aspects of frh-1-suppression control of longevity, we generated four dsRNA constructs targeting different portions of the frh-1 gene and a specific antibody against C. elegans FRH-1. The four different dsRNA displayed different interference potency as revealed by western blotting of total C. elegans extracts (Figs. 1A–B; S1A–B). The anti-FRH-1 antibody revealed one or two bands (depending on protein extraction method) at the expected molecular weight of 15 kDa in wild-type lysate, likely corresponding, as in mammalian cells, to different forms of the protein (Fig. S1A-B). Lifespan extension was only achieved with the frh-1 IV dsRNA construct — referred to as frh-1 hereafter unless otherwise specified — which reduced FRH-1 expression to more than 50% of its wild-type level, without completely abolishing its expression (Fig. 1B–C; Table 1). Our results unambiguously demonstrated that a critical amount of frataxin protein must be suppressed to induce significant life-extending responses in C. elegans thus providing us as with a great tool to untangle protective pathways activated to compensate for frataxin suppression at the organismal level.
3.2. Reduced expression of frataxin affects sensory neurons and mitochondria functionality in C. elegans
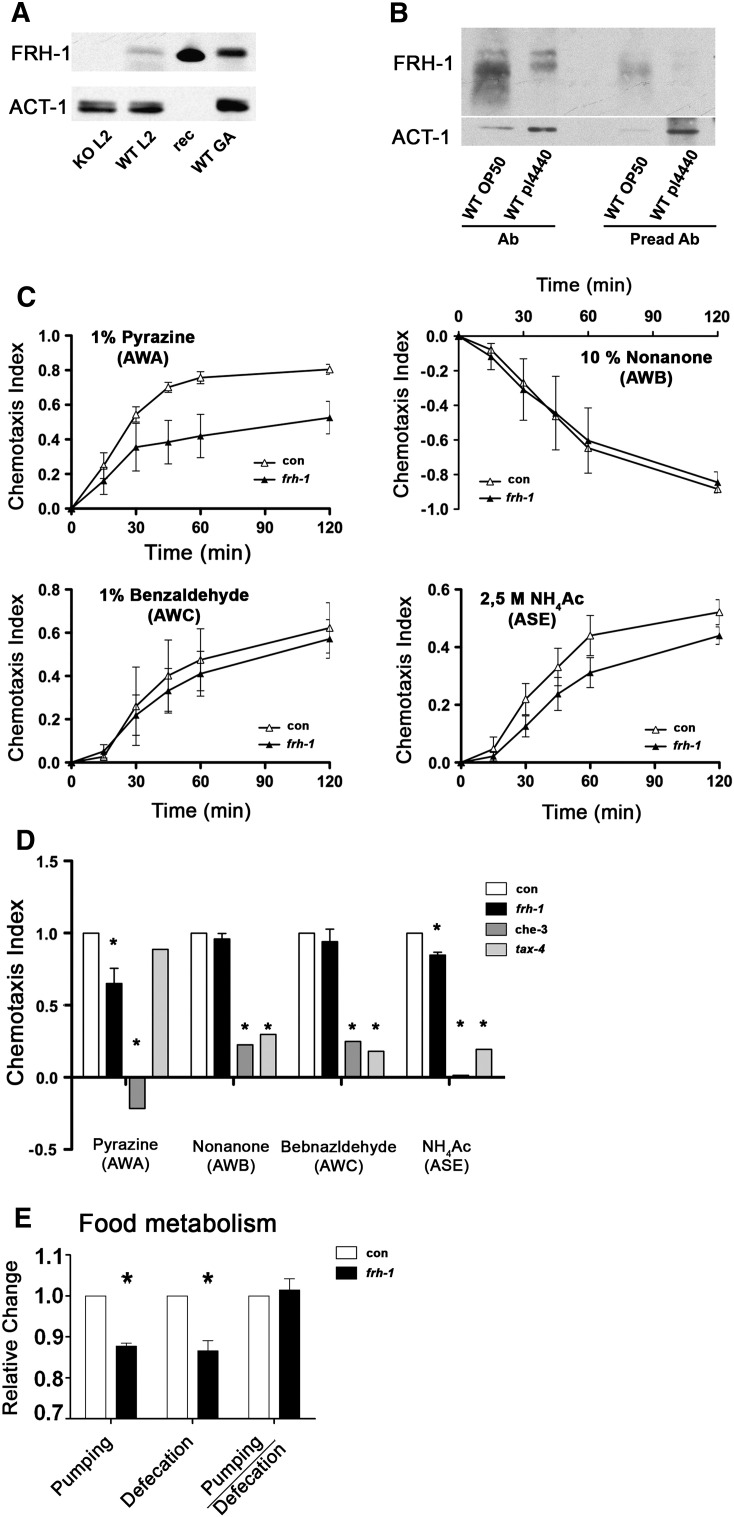
To shed light on mechanistic aspects of frataxin control of longevity, which can also be relevant for the human disease, we characterized in more details mitochondria and neuronal functions in response to frh-1 RNAi. As seen in mammals, reduced expression of frataxin protein in C. elegans specifically affected sensory neurons functionality: the reduced sensitivity of frh-1(RNAi) animals to specific chemicals, namely their attraction to pyrazine and to sodium acetate, reveals alteration of specific sensory neurons, AWA and ASE respectively (Figs. 2A; S1C–D). Interestingly the AWA pair of neurons participate in the regulation of C. elegans lifespan (Apfeld and Kenyon, 1999). These sensory defects are not ascribed to altered moving ability since frh-1(RNAi) worms have normal response to touch and locomotion activity (Fig. 2B–C), and they actually move slightly faster than control animals (Fig. 2D).
To shed light on mechanistic aspects of frataxin control of longevity, which can also be relevant for the human disease, we characterized in more detail mitochondria and neuronal functions in response to frh-1 RNAi. As seen in mammals, reduced expression of frataxin protein in C. elegans specifically affected sensory neurons functionality: the reduced sensitivity of frh-1(RNAi) animals to specific chemicals, namely their attraction to pyrazine and to sodium acetate, reveals alteration of specific sensory neurons, AWA and ASE respectively (Figs. 2A; S1C–D). Interestingly the AWA pair of neurons participate in the regulation of C. elegans lifespan (Apfeld and Kenyon, 1999). These sensory defects are not ascribed to altered moving ability since frh-1(RNAi) worms have normal response to touch and locomotion activity (Fig. 2B–C), and they actually move slightly faster than control animals (Fig. 2D).
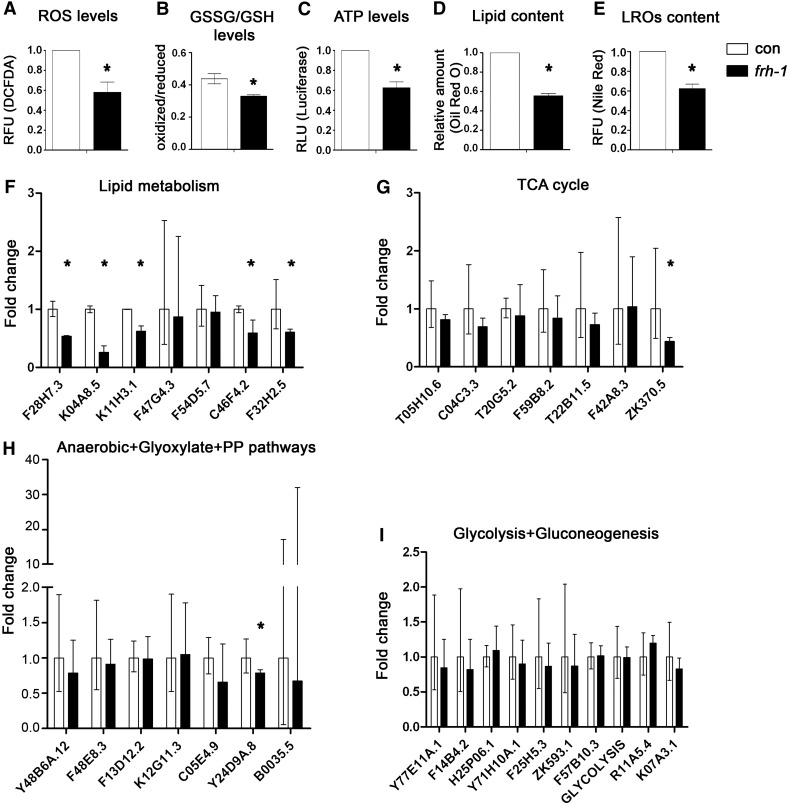
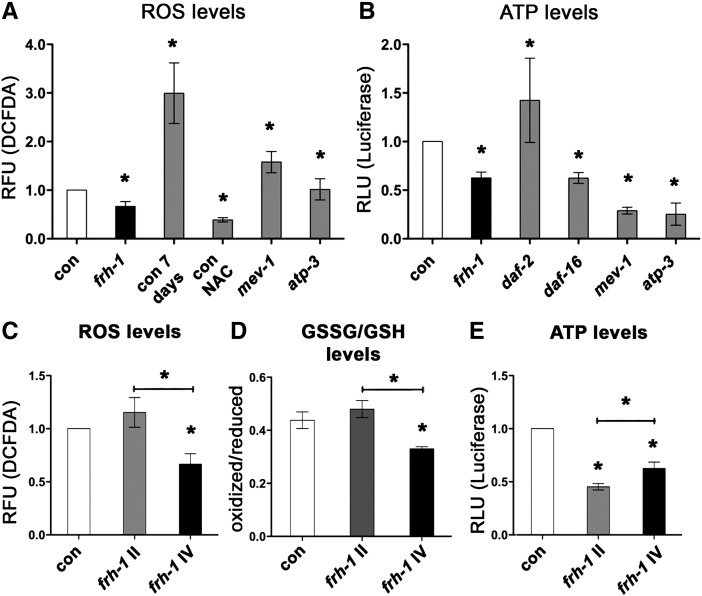
FRDA cells are more sensitive to oxidative stress (Condo et al., 2006) while long-lived frh-1(RNAi) animals, similar to other Mit mutants, are differentially sensitive to different pro-oxidants (Lee et al., 2003; Ventura et al., 2005). Here we found that frh-1 RNAi significantly decreased the levels of reactive oxygen species (ROS), of the oxidized pool of glutathione, and of ATP compared to control animals (Figs. 3A–C; S2A–B). Our data indicate that frataxin suppression in C. elegans recapitulates some of the features observed in the human disease (i.e. altered sensory neuron functionality and ATP production), while other features (i.e. reduced ROS and oxidized glutathione) may result from the activation of specific compensatory pathways.
Fig. 3.
Characterization of mitochondria and metabolic features. Levels of A) reactive oxygen species (ROS), B) oxidized/reduced glutathione (GSSG/GSH), C) ATP, D) lipid content and E) lysosome-related organelles (LROs) in 1-day-old adult WT animals fed bacteria expressing either empty-vector (con) or life-lengthening frh-1 dsRNA (frh-1). RFU, Relative Fluorescence Unit. RLU, Relative Luminescence Unit. F–I) Transcript expression of key metabolic enzymes in animals treated as in A. Bars and errors indicate mean fold changes and the 95% confidence interval of the mean, respectively, of cumulative data from three independent experiments. *p < 0.05 of each single gene vs control, t-test.
Fig. S2.
FRDA cells are more sensitive to oxidative stress (Condo et al., 2006) while long-lived frh-1(RNAi) animals, similar to other Mit mutants, are differentially sensitive to different pro-oxidants (Lee et al., 2003; Ventura et al., 2005). Here we found that frh-1 RNAi significantly decreased the levels of reactive oxygen species (ROS), of the oxidized pool of glutathione, and of ATP compared to control animals (Figs. 3A–C; S2A–B). Our data indicate that frataxin suppression in C. elegans recapitulates some of the features observed in the human disease (i.e. altered sensory neuron functionality and ATP production), while other features (i.e. reduced ROS and oxidized glutathione) may result from the activation of specific compensatory pathways.
3.3. Reduced frataxin expression specifically remodels lipid metabolism in C. elegans
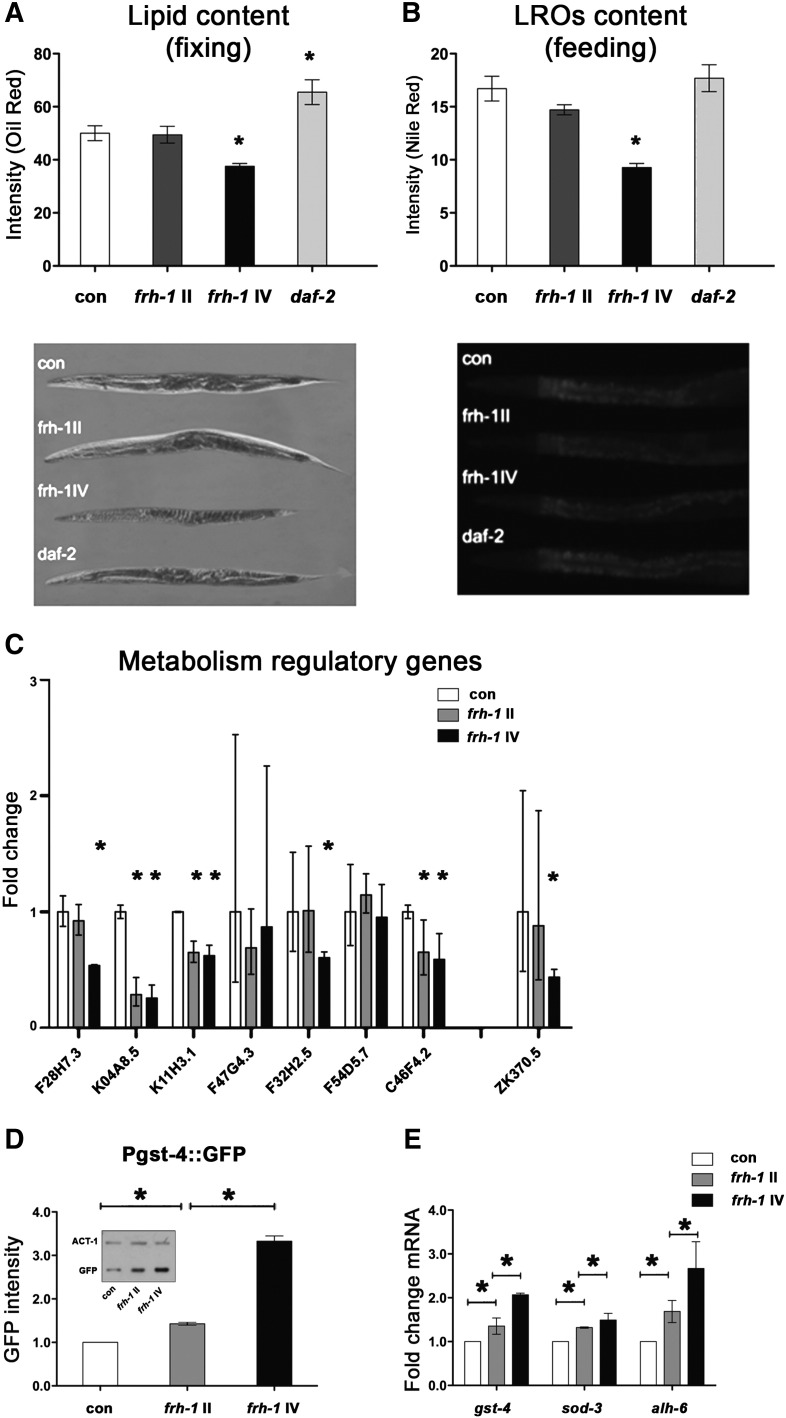
Long-lived frh-1(RNAi) worms, similar to other Mit mutants, are smaller and paler than wild-type animals. These animals, as previously observed but with a different frh-1 dsRNA construct (Vazquez-Manrique et al., 2006), have reduced pumping and defecation rate (Fig. S1E), and we found that they display a reduced content of lipid and of intestine-specific lysosome-related organelles (LROs) compared to control animals (Figs. 3D–E; S3A–B). The expression of genes involved in lipid metabolism was also selectively reduced upon frh-1 RNAi: specifically, the expression of enzymes regulating both lipid catabolism (gpdh-2/K11H3.1, lipl-4/K04A8.5 and another predicted lipase/F28H7.3) and lipid anabolism (acs-17/C46F4.2 and fasn-1/F32H2.5) were significantly reduced (Fig. 3F; Table SI). The transcript levels of key enzymes regulating other major metabolic pathways (glycolysis, gluconeogenesis, TCA cycle, anaerobic, glyoxylate and pentose phosphate pathways) were not significantly affected (Fig. 3G–I; Table SII), except for pdhk-2/ZK370.5 (Fig. 3G) and Y24D9A.8 (Fig. 3H).
Fig. S3.
Long-lived frh-1(RNAi) worms, similar to other Mit mutants, are smaller and paler than wild-type animals. These animals, as previously observed but with a different frh-1 dsRNA construct (Vazquez-Manrique et al., 2006), have reduced pumping and defecation rate (Fig. S1E), and we found that they display a reduced content of lipid and of intestine-specific lysosome-related organelles (LROs) compared to control animals (Figs. 3D–E; S3A–B). The expression of genes involved in lipid metabolism was also selectively reduced upon frh-1 RNAi: specifically, the expression of enzymes regulating both lipid catabolism (gpdh-2/K11H3.1, lipl-4/K04A8.5 and another predicted lipase/F28H7.3) and lipid anabolism (acs-17/C46F4.2 and fasn-1/F32H2.5) were significantly reduced (Fig. 3F; Table SI). The transcript levels of key enzymes regulating other major metabolic pathways (glycolysis, gluconeogenesis, TCA cycle, anaerobic, glyoxylate and pentose phosphate pathways) were not significantly affected (Fig. 3G–I; Table SII), except for pdhk-2/ZK370.5 (Fig. 3G) and Y24D9A.8 (Fig. 3H).
Interestingly, the frh-1 II dsRNA construct that did not extend animal lifespan, only marginally affected animal metabolic parameters (Figs S2C–D; S3A–C; Table SII) and the expression of antioxidant genes (Fig. S3D–E) previously shown to be induced upon life-lengthening frh-1 RNAi (Torgovnick et al., 2010). Taken together, these observations indicate that a critical threshold (50%) of FRH-1 suppression must be surpassed to concurrently extend lifespan and significantly alter mitochondria and lipid metabolism in C. elegans.
Fig. 4.
Frataxin suppression acts independently of the AMP kinases, in parallel to CR. A–F) Survival analysis of strains carrying mutant alleles of different AMP regulatory kinases fed bacteria expressing either empty-vector (con) or frh-1 dsRNA (frh-1). G–H) Lysosome-related organelles staining and food intake (normalized pumping rate) in WT and eat-2(ad465) mutants fed bacteria expressing either empty-vector (con) or frh-1 dsRNA (frh-1). Bars and errors indicate mean values ± SEM from different replica. *p < 0.05 vs control, t-test.
3.4. Reduced frataxin expression extends lifespan independently of AMP-kinases
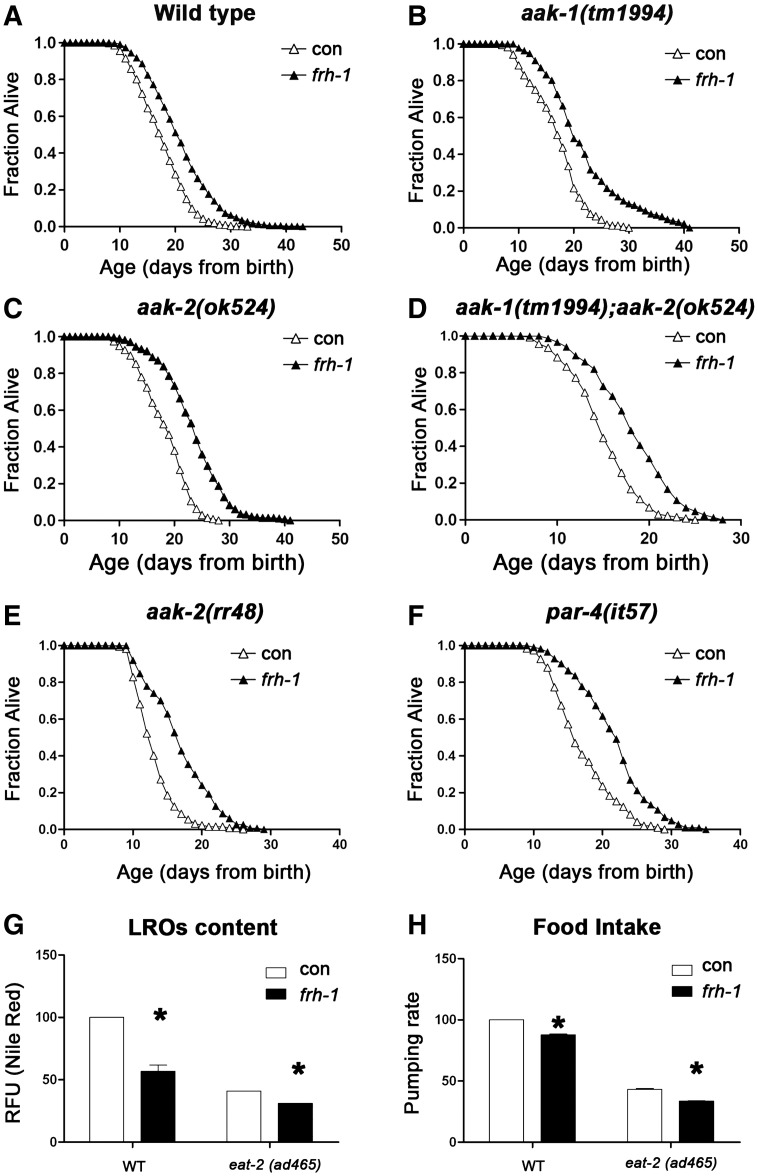
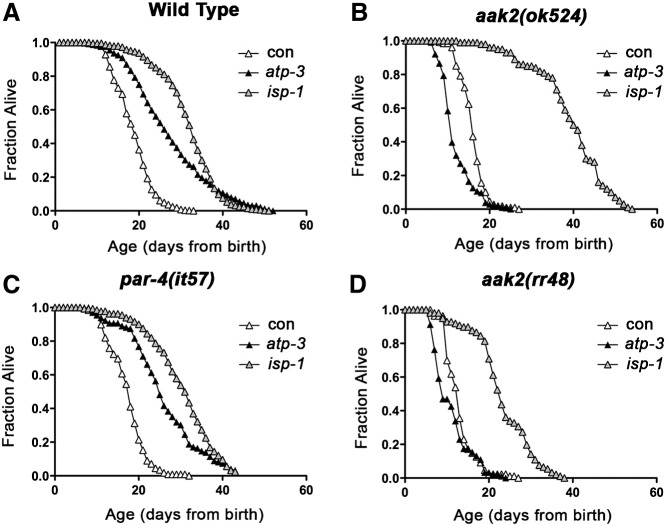
AMP-activated protein kinases (AMPKs) respond to variation in cellular ATP/AMP content and cooperate with p53 to modulate energy metabolism (Jones et al., 2005; Maddocks and Vousden, 2011; Mandal et al., 2005; Thoreen and Sabatini, 2005). AMPk and p53/cep-1 are in part required for the lifespan extension of C. elegans in response to mitochondrial disruption (Curtis et al., 2006; Ventura et al., 2009). We hypothesized that AMPKs are activated in response to frh-1 RNAi in turn modulating animal metabolism and longevity. Contrary to our expectations, none of the kinases involved in energy metabolism that we tested (aak-1, aak-2, par-4, sad-1, kin-29, or mml-1) affected lifespan in frh-1(RNAi) animals (Fig. 4A–F; Table 2) or in the majority of the RNAi-mediated Mit mutants analyzed (Fig. S4A–D; Table SIII) which also displayed reduced lipid accumulation (not shown). This is nevertheless consistent with a non-linear relationship between ATP levels and extension of lifespan in different RNAi-mediated Mit mutants (Fig. S2E and Dillin et al., 2002).
Table 2.
Lifespan analyses frh-1 RNAi on different kinase mutants — summary statistics.
| Genotype | RNAi | Mean Lifespan | + SEM | p vs con⁎ | p vs N2⁎ | Sample size (censored)a | N of replicates |
|---|---|---|---|---|---|---|---|
| Wild-type (N2) | con | 17.2 | 0.2 | 460 (40) | 7 | ||
| frh-1 | 20.6 | 0.3 | 4.9E − 17 | 468(100) | 7 | ||
| aak-2(rr48) | con | 12.6 | 0.3 | 1.06E − 26 | 198 (61) | 3 | |
| frh-1 | 16.3 | 0.4 | 1.5E − 12 | 0.17044 | 197 (43) | 3 | |
| aak-2(ok524) | con | 17.9 | 0.3 | 0.20326 | 319 (53) | 5 | |
| frh-1 | 23.1 | 0.3 | 1.5E − 27 | 1.39E − 34 | 330 (54) | 5 | |
| aak-1(tm1994) | con | 16.5 | 0.4 | 0.33077 | 190 (28) | 3 | |
| frh-1 | 21.5 | 0.6 | 3.6E − 08 | 1.06E − 10 | 191 (61) | 3 | |
| aak-1(tm1994); aak-2(ok524) | con | 14.6 | 0.3 | 3.85E − 01 | 191 (57) | 3 | |
| frh-1 | 17.9 | 0.3 | 6.9E − 10 | 0.38536 | 188 (38) | 3 | |
| par-4(it57) | con | 16.6 | 0.5 | 0.40960 | 140 (40) | 2 | |
| frh-1 | 21.2 | 0.5 | 1.6E − 08 | 2.44E − 10 | 140 (35) | 2 | |
| Wild-type (N2) | con | 16.4 | 0.3 | 200(10) | 3 | ||
| frh-1 | 19.8 | 0.4 | 1.2E − 08 | 1.17E − 08 | 200(32) | 3 | |
| sad-1(ky289) | con | 15.8 | 0.6 | 0.78698 | 120(46) | 2 | |
| frh-1 | 20.8 | 1.0 | 0.00003 | 0.00001 | 121(40) | 2 | |
| kin-29(gk270) | con | 12.1 | 0.2 | 1.29E − 18 | 139(22) | 2 | |
| frh-1 | 16.1 | 0.4 | 5.1E − 12 | 0.72663 | 141(30) | 2 | |
| mml-1(ok849) | con | 15.2 | 0.4 | 0.23012 | 200(40) | 3 | |
| frh-1 | 19.6 | 0.4 | 1.5E − 09 | 7.96E − 08 | 199(30) | 3 | |
p-values were calculated using the log-rank test between pooled populations.
Pooled worms from N replicate experiments (censored individuals were included in the log rank analysis).
Fig. S4.
AMP-activated protein kinases (AMPKs) respond to variation in cellular ATP/AMP content and cooperate with p53 to modulate energy metabolism (Jones et al., 2005; Maddocks and Vousden, 2011; Mandal et al., 2005; Thoreen and Sabatini, 2005). AMPk and p53/cep-1 are in part required for the lifespan extension of C. elegans in response to mitochondrial disruption (Curtis et al., 2006; Ventura et al., 2009). We hypothesized that AMPKs are activated in response to frh-1 RNAi in turn modulating animal metabolism and longevity. Contrary to our expectations, none of the kinases involved in energy metabolism that we tested (aak-1, aak-2, par-4, sad-1, kin-29, or mml-1) affected lifespan in frh-1(RNAi) animals (Fig. 4A–F; Table 2) or in the majority of the RNAi-mediated Mit mutants analyzed (Fig. S4A–D; Table SIII) which also displayed reduced lipid accumulation (not shown). This is nevertheless consistent with a non-linear relationship between ATP levels and extension of lifespan in different RNAi-mediated Mit mutants (Fig. S2E and Dillin et al., 2002).
Moreover, consistent with notion that frh-1 RNAi acts in parallel to caloric-restriction (CR), it further extends lifespan (Ventura and Rea, 2007) and decreased LROs content and pumping rate in the eat-2 mutant, a genetically defined CR model (Fig. 4G–H). Of note, reduced intestinal LROs content in frh-1(RNAi) worms and in eat-2 mutants was very similar; on the other hand the pumping rate was more severely affected in eat-2 mutants indicating that reduced LROs accumulation in frh-1(RNAi) animals is ascribed to mechanisms other than reduced food intake. Collectively, these findings indicate that frh-1 RNAi extends lifespan in parallel to CR-related pathways, independently of AMPKs, major regulators of energy metabolism.
3.5. Autophagy is induced by reduced expression of frataxin in C. elegans and in mammalian cells
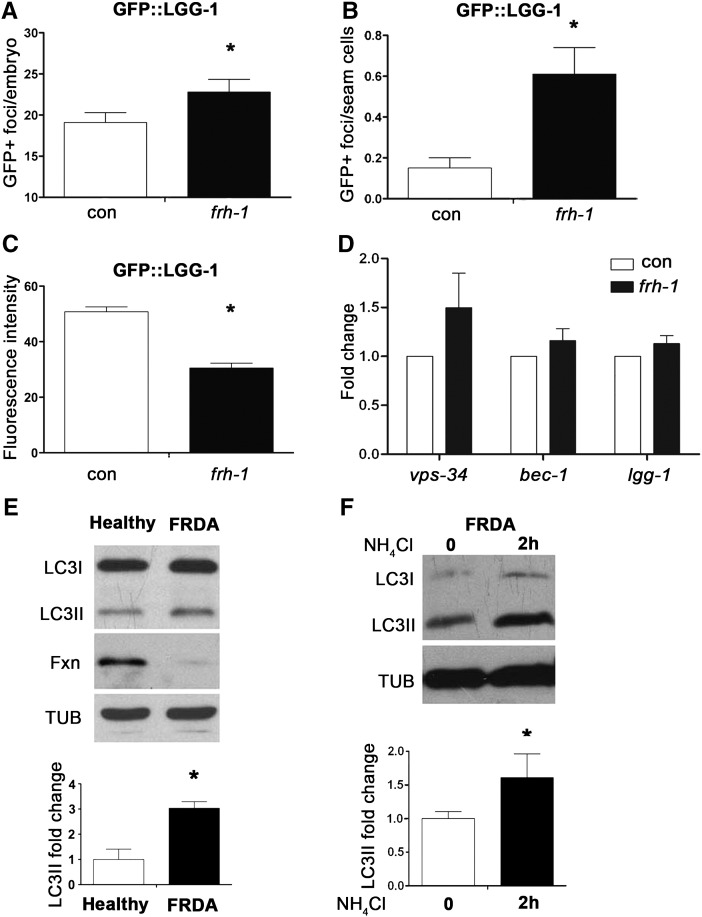
We next sought to investigate whether autophagy is induced in response to frataxin suppression as an adaptive process to mitochondrial damage (Lemasters, 2005). This could modulate lipid metabolism and lifespan (Lapierre et al., 2011), and suggest a critical pathogenetic target to prevent or postpone the established disease in humans. The gene lgg-1 encodes the C. elegans ortholog of Atg8/LC3, an ubiquitin-like, microtubule-associated protein required for autophagic vesicle growth. The change of Atg8/LC3/LGG-1 expression's pattern from diffuse to punctate represents a well-established marker for autophagosomes accumulation (Melendez et al., 2008). Taking advantage of a translational reporter strain expressing GFP::LGG-1 (Melendez et al., 2003) we found that frh-1 RNAi increases the number of GFP::LGG-1 positive foci in embryos (Figs. 5A; S5A) and in hypodermal seam cells in larvae (Figs. 5B; S5B). Moreover, the overall amount of GFP::LGG-1 expression in embryos of frh-1(RNAi) animals was decreased (Fig. 5C), a likely indication of an intact autophagic flux (Mizushima et al., 2010). The early steps of autophagosome formation involve, among others, Atg1/ULK1, Atg6/Beclin and the class III phosphoinositide 3-kinase (PI3K) Vps34. The transcriptional expression of vps-34, bec-1 and lgg-1 did not significantly change (Fig. 4D; Table SIV), suggesting frh-1 RNAi induces autophagy at the post-transcriptional level.
Fig. 5.
Frataxin suppression induces autophagy. A–B) Number of autophagosomal GFP+ foci in embryos (A) and seam cells of L3 larvae (B) of C. elegans GFP::LGG-1 transgenic animals fed bacteria expressing empty-vector (con) or frh-1 dsRNA (frh-1). C) Total amount of GFP::LGG-1 protein expression in embryos of animals treated as in A. D) Transcriptional changes of autophagy modulator genes in WT animals treated as in A. E) Autophagosome accumulation assessed by increased formation of LC3II in Friedreich's Ataxia patient derived lymphoblast (FRDA) compared to control lymphoblast derived from a healthy brother (Healthy). During the autophagic process the LC3/LGG-1 protein undergoes a posttranslational modification that accelerates its migration on acrylamide gel (from LC3I to LC3II). Fxn, human frataxin. F) Autophagosome accumulation in FRDA lymphoblasts left untreated or treated for two hours with the lysosome inhibitor NH4Cl. In each panel, bars and errors indicate mean values + SEM after densitometric analysis from three independent replicates. *p < 0.05 vs control, t-test.
Fig. S5.
We next sought to investigate whether autophagy is induced in response to frataxin suppression as an adaptive process to mitochondrial damage (Lemasters, 2005). This could modulate lipid metabolism and lifespan (Lapierre et al., 2011), and suggest a critical pathogenetic target to prevent or postpone the established disease in humans. The gene lgg-1 encodes the C. elegans ortholog of Atg8/LC3, an ubiquitin-like, microtubule-associated protein required for autophagic vesicle growth. The change of Atg8/LC3/LGG-1 expression's pattern from diffuse to punctate represents a well-established marker for autophagosomes accumulation (Melendez et al., 2008). Taking advantage of a translational reporter strain expressing GFP::LGG-1 (Melendez et al., 2003) we found that frh-1 RNAi increases the number of GFP::LGG-1 positive foci in embryos (Figs. 5A; S5A) and in hypodermal seam cells in larvae (Figs. 5B; S5B). Moreover, the overall amount of GFP::LGG-1 expression in embryos of frh-1(RNAi) animals was decreased (Fig. 5C), a likely indication of an intact autophagic flux (Mizushima et al., 2010). The early steps of autophagosome formation involve, among others, Atg1/ULK1, Atg6/Beclin and the class III phosphoinositide 3-kinase (PI3K) Vps34. The transcriptional expression of vps-34, bec-1 and lgg-1 did not significantly change (Fig. 4D; Table SIV), suggesting frh-1 RNAi induces autophagy at the post-transcriptional level.
To corroborate our findings in C. elegans, we assessed whether autophagy is affected in mammalian cells with reduced frataxin expression. Steady-state autophagy measurements indicated that the basal amount of autophagosomes accumulation is increased in lymphoblasts derived from FRDA patients compared to their healthy matched cells (Fig. 5E). This can be ascribed either to an increase in autophagosomes formation or to a block of lysosomal degradation. To discriminate between these two possibilities we treated FRDA cells with ammonium chloride (NH4Cl), which inhibits lysosome acidification, and found that the amount of autophagosomes is further increased (Fig. 5F), suggesting that the autophagic flux is not impaired. Collectively these findings suggest that induction of an intact autophagic flux is an evolutionarily conserved response to frataxin suppression, a notion with critical implication for the human disease.
3.6. Autophagy is required to extend longevity and reduce lipid accumulation in frh-1(RNAi) animals
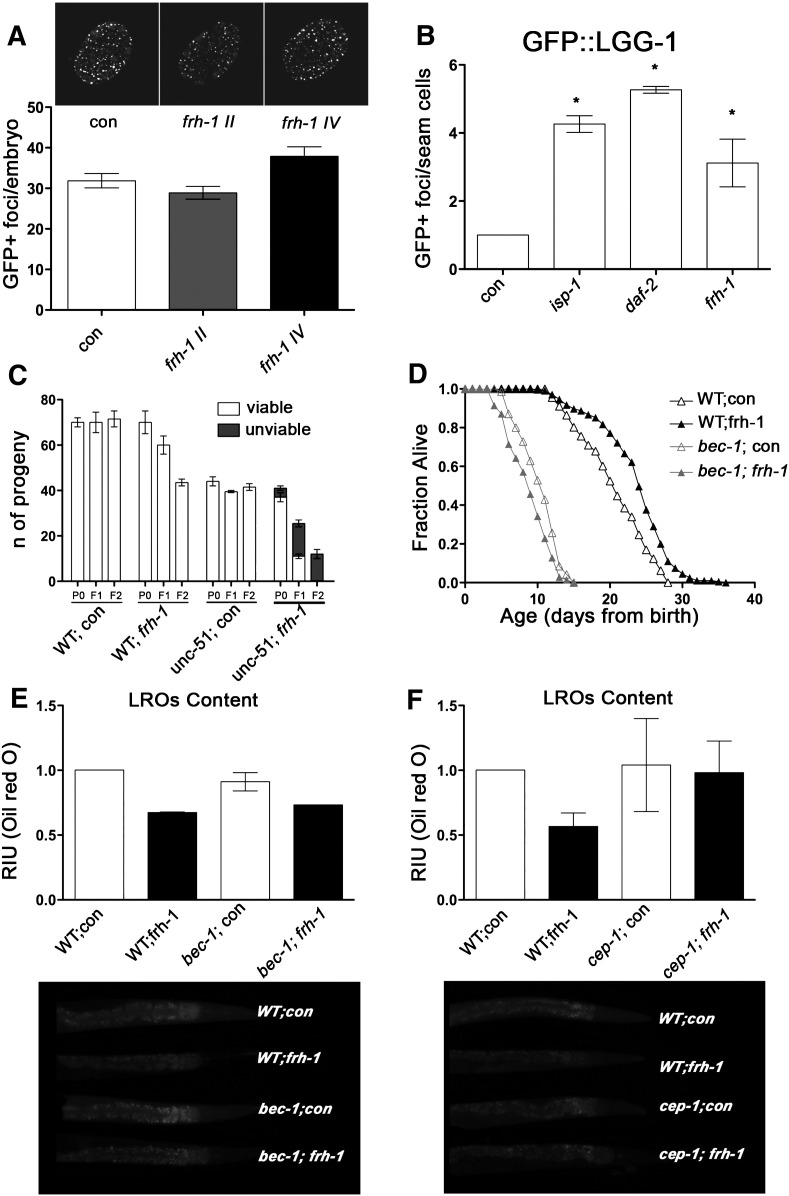
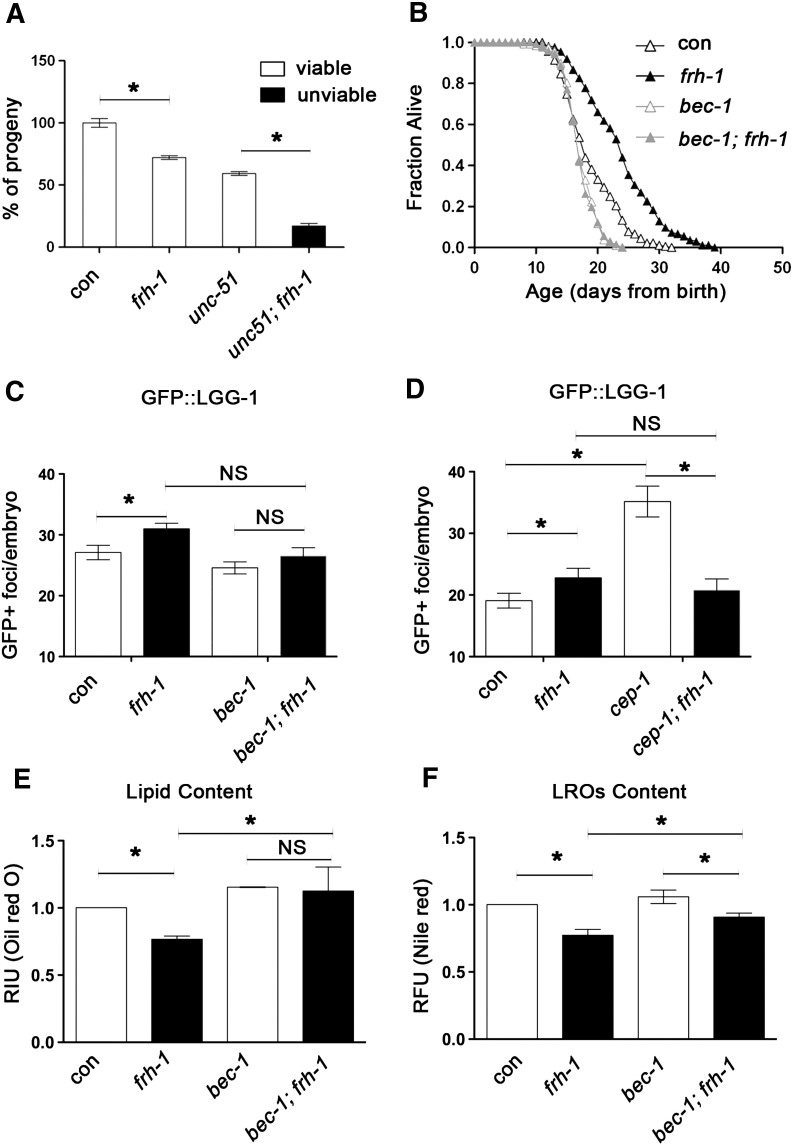
Autophagic genes are required to extend lifespan in different C. elegans genetic backgrounds but their role in controlling longevity in response to mitochondrial disruption has not been clearly elucidated (Hansen et al., 2008; Toth et al., 2008). We found that two core autophagy-regulating genes, unc-51 (the C. elegans ortholog of ULK1) and bec-1 are respectively required for survival and longevity of frh-1(RNAi) animals. Specifically, unc-51 mutants and frh-1(RNAi) animals had a reduced but viable number of progeny (Fig. 6A). On the other hand, unc-51 mutants fed frh-1 RNAi laid fewer and 100% non viable embryos (a mix of unhatched eggs and arrested larvae) (Figs. 6A; S5C). RNAi or genetic suppression of bec-1 shortened animal lifespan, and completely abolished the lifespan extension induced by frh-1 RNAi (Figs. 6B; S5D; Table 3). The lack of frh-1 RNAi pro-longevity effect was also observed in the bec-1 heterozygotes (unpublished observation). Of note, we found that frh-1 RNAi was not able to induce autophagy in bec-1(RNAi) animals (Fig. 6C). Interestingly, in support of a protective role for autophagy in response to frataxin suppression, the frh-1II dsRNA construct that did not extend lifespan was unable to trigger autophagy (Fig. S5A).
Fig. 6.
Autophagy is required to extend lifespan and reduce lipid accumulation in frh-1(RNAi) animals. A) Fertility and viability of WT and unc-51 mutants fed bacteria expressing either empty-vector (con) or frh-1 dsRNAi (frh-1). B) Survival curves of WT animals fed bacteria expressing empty-vector (con), an equal mix of empty-vector and frh-1 dsRNA (frh-1), an equal mix of empty-vector and bec-1 dsRNA (bec-1), or an equal mix of both dsRNA (bec-1;frh-1). C) Number of autophagosomal GFP+ foci in embryos of GFP::LGG-1 animals fed as in B. D) Number of autophagosomal GFP+ foci in embryos of GFP::LGG-1 and of GFP::LGG-1;cep-1(lg12501) strains fed bacteria expressing either empty-vector (con) or frh-1 dsRNA (frh-1). E–F) Content of lipid and of lysosomes-related organelles (LROs) were respectively measured by fixing animals and staining them with Oil-Red-O (E, RIU = relative intensity unit), and by feeding animals with the vital dye Nile Red (F, RFU = relative fluorescence unit) in WT animals treated as in B. Bars and errors indicate mean values + SEM from at least three independent replicates. *p < 0.05 vs control, t-test.
Table 3.
Beclin control of frh-1(RNAi) animals lifespan — summary statistics.
| Genotype | RNAi | Mean lifespan | + SEM | p vs con⁎ | p vs N2⁎ | Sample size (censored)a | N of replicates |
|---|---|---|---|---|---|---|---|
| Wild-type (N2) | con | 18.4 | 0.3 | 212(31) | 3 | ||
| frh-1b | 23.1 | 0.5 | 2.4E − 13 | 212(45) | 3 | ||
| bec-1b | 16.8 | 0.3 | 0.00065 | 140(36) | 2 | ||
| bec-1;frh-1 | 16.7 | 0.3 | 0.82070$ | 4.1E − 24$$ | 140(11) | 2 | |
| bec-1(ok691) | Con | 9.9 | 0.3 | 1.31E − 54 | 70(0) | 1 | |
| frh-1 | 8.4 | 0.4 | 0.02004 | 2.88E − 65 | 70(0) | 1 |
p-values were calculated using the log-rank test between pooled populations.
p-value versus bec-1.
p-value versus frh-1.
Pooled worms from N replicate experiments (censored individuals were included in the log rank analysis).
Lifespan analyses were performed on worms fed an equal mix of bacteria expressing empty-vector and frh-1 dsRNA or bec-1 dsRNA.
Autophagic genes are required to extend lifespan in different C. elegans genetic backgrounds but their role in controlling longevity in response to mitochondrial disruption has not been clearly elucidated (Hansen et al., 2008; Toth et al., 2008). We found that two core autophagy-regulating genes, unc-51 (the C. elegans ortholog of ULK1) and bec-1 are respectively required for survival and longevity of frh-1(RNAi) animals. Specifically, unc-51 mutants and frh-1(RNAi) animals had a reduced but viable number of progeny (Fig. 6A). On the other hand, unc-51 mutants fed frh-1 RNAi laid fewer and 100% non viable embryos (a mix of unhatched eggs and arrested larvae) (Figs. 6A; S5C). RNAi or genetic suppression of bec-1 shortened animal lifespan, and completely abolished the lifespan extension induced by frh-1 RNAi (Figs. 6B; S5D; Table 3). The lack of frh-1 RNAi pro-longevity effect was also observed in the bec-1 heterozygotes (unpublished observation). Of note, we found that frh-1 RNAi was not able to induce autophagy in bec-1(RNAi) animals (Fig. 6C). Interestingly, in support of a protective role for autophagy in response to frataxin suppression, the frh-1II dsRNA construct that did not extend lifespan was unable to trigger autophagy (Fig. S5A).
P53/cep-1 regulates autophagy and is required for the extended longevity of different Mit mutants including frh-1(RNAi) animals (Ventura et al., 2009). Consistent with work on a different cep-1 mutant allele (Tasdemir et al., 2008), we found that the accumulation of autophagosomes in cep-1(lg12501) mutants was significantly increased compared to wild-type (Fig. 6D). Notably, frh-1 RNAi did not increase the number of autophagosomes in the cep-1 mutant but rather reduced it (Fig. 6D), suggesting a reciprocal functional interplay between the two genes in regulating autophagosomes accumulation.
Autophagy regulates lipid metabolism in different biological contexts (Lapierre et al., 2011; Singh et al., 2009). We found that bec-1 RNAi significantly rescued the reduced content of lipids and of LROs in frh-1(RNAi) animals (Fig. 6E–F). Lipid and LROs content were similarly rescued in bec-1 heterozygotes and in the in cep-1 mutants, which like bec-1 RNAi, also reverted frh-1(RNAi) animals small size (Fig S5E and F title legend should be corrected to Lipid content. LROs content should be the title of panels G-H). Our data indicate that autophagy and cep-1 cooperate during animal development to exert beneficial effects in response to reduced frataxin levels.
4. Discussion
In this paper we unambiguously show that a substantial level of frataxin protein expression (below 50% of its wild-type level) must be reduced to extend C. elegans lifespan and remodel different aspects of animal metabolism. Importantly, we propose that this is achieved by inducing an adaptive response regulated by cep-1, namely autophagy, a fundamental cellular recycling program.
Different experimental conditions coupled with dose-dependent effects may therefore help to reconcile the opposite lifespan outcomes reported by other groups (Vazquez-Manrique et al., 2006; Zarse et al., 2007) in response to frataxin silencing in C. elegans. On the one hand, these studies might have not reached the threshold of frataxin suppression necessary to extend lifespan, and on the other hand the short lifespan could be ascribed to experimental conditions which lead to an overestimation of dead animals upon frh-1 RNAi compared to control. Namely, while Vazquez-Manrique and coworkers did not censor bagged animals in the survival analysis, the mean lifespan of control animals in the other study was 20 days at 20 °C on HT115 bacteria (as food source), which is slightly longer than average for a wild-type strain under normal laboratory conditions. Here we tested different constructs and generated a specific anti FRH-1 antibody, to specifically correlate level of fratxin protein expression, lifespan and other animal metabolic parameters and clearly showed that none of tested constructs actually shortened lifespan compared to control animals (which have a mean lifespan of almost 18 days, Table 1). Of note, also frh-1 knock-out animals, although arrested as L2/L3, live longer than wild-type (Ventura and Rea, 2007).
Neurons and myocytes are the cells mostly affected in FRDA patients. C. elegans neurons are relatively resistant to the effect of feeding RNAi, the technique routinely used for gene knock-down. Although this might lead to underestimation of specific neuronal effects, it notably allows one to studying the function of genes whose deficiency in the nervous system would be lethal. Nonetheless, frh-1(RNAi) animals displayed a mild but selective deficit in specific sensory neurons. An interesting possibility is that lifespan extension in response to frh-1 deficiency is achieved via disruption of specific subtypes of neurons, such as the AWA pair of neurons, which is known to extend lifespan in C. elegans (Apfeld and Kenyon, 1999). frh-1(RNAi) animals also have decreased pumping, a feature that might reflect altered functionality of the pharynx, interestingly the organ that more closely resembles mammalian heart's organogenesis (Haun et al., 1998). Here, we find that stimulation of autophagy is an evolutionarily conserved adaptive response to severe frataxin suppression. Autophagy induction in non-renewable cells (such as neurons and myocytes) might therefore represent an attempt to preserve cell viability upon frataxin deficiency in the human pathology. Importantly, although accumulation of autophagosomal foci was described in neurons of conditional frataxin KO mice (Simon et al., 2004), this could be ascribed to increase autophagy flux or block in autophagosomes degradation. Our results are indicative of an intact, and therefore increased autophagic flux, an observation with significant biological implications for Friedreich's ataxia disease pathogenesis and new potential therapeutic approaches.
Consistent with silencing of other MRC regulatory proteins (Dillin et al., 2002; Rea et al., 2007), we found that ATP or ROS levels in frh-1(RNAi) worms do not linearly correlate with lifespan. The induction of protective stress responses, such as autophagy, only upon severe frataxin suppression could account for these findings and explain the inability of the non-extending frh-1II dsRNA construct to decrease ROS and to maintain ATP levels. A threshold effect could also explain why contrary to the original observation (Simon et al., 2004), Palomo and coworkers did not observe any significant autophagy induction in neuronal-like cells treated with frataxin shRNA (Palomo et al., 2011).
Our findings indicate that frataxin suppression does not generally slow down animal metabolism, but rather plays a specific role in controlling lipid remodeling through the autophagic pathway. Interestingly, lipid metabolism is finely regulated within the autophagic-lysosomal compartments (Kiselyov et al., 2007; Singh et al., 2009) and although the two dyes utilized to quantify lipid and LROs content stain different compartments they can colocalize under altered metabolic conditions (such as peroxisomal β-oxidation alterations Zhang et al., 2010). The amount of lipid accumulation inversely correlates with lifespan extension but it remains to be established whether these are two independent consequences of autophagy induction or whether instead a direct causal relationship between lipid metabolism and mitochondrial control of longevity exists. We found that the expression of the C. elegans lipase LIPL-4, which is overexpressed in the glp-1 germline less mutant and cooperates with the autophagic pathway to extend its lifespan (Lapierre et al., 2011; Wang et al., 2008), is dramatically reduced by frh-1 RNAi. Overexpression of lipl-4 in the germline deficient mutant is daf-16 dependent while frh-1 RNAi extends lifespan in a daf-16 independent manner (Ventura et al., 2009); moreover the expression of different lipid-metabolizing enzymes, including lipl-4, is also reduced by the frh-1II dsRNA construct that does not extend longevity. Taken together these observations suggest that autophagy and lipid metabolism may differently coordinate longevity in a context-specific manner and that at least lipl-4 does not play a role in longevity specification in frh-1(RNAi) animals. More advanced analysis of animal metabolic composition (Butler et al., 2010; Mishur and Rea, 2011) will help to elucidate whether or not lipid metabolism plays a direct role in mitochondrial control of longevity.
Lifespan extension and autophagy in response to frh-1 RNAi required the C. elegans p53 orthologs, cep-1. Reciprocally, frataxin deficiency reduces the amount of autophagosomes in cep-1 mutants, leading to the interesting possibility that mitochondrial functionality is required to modulate autophagy in response to specific insults. Specifically, reduced frataxin expression (and thus mitochondrial functionality) could facilitate elimination of autophagosomes in conditions of impaired autophagic flux, as possibly revealed in the cep-1 KO mutant by the elevated total amount of GFP::LGG-1 in their embryos (our unpublished observation). In support of a functional interplay between frataxin and p53 we recently showed that frataxin participates in the hypoxia-induced response in mammalian tumors together with p53 (Guccini et al., 2011), suggesting that a similar p53-dependent control of autophagy could be orchestrated in frataxin-deficient mammalian cells.
5. Conclusions
In summary, our findings suggest that stimulation of an intact autophagic flux is an evolutionarily conserved mechanism activated in response to severe frataxin deficiency, which is required in C. elegans to extend lifespan and reduce lipid content in a p53-regulated manner. Since frataxin deficiency affects the expression of different MRC subunits (Rotig et al., 1997) and induces same phenotypic features observed in other RNAi-mediated Mit mutants, a similar autophagic control of lipid content and longevity is envisioned upon suppression of other mitochondrial proteins, as in part suggested by another study (Toth et al., 2008). Importantly, our results imply a role for autophagy as a protective compensatory response aimed at coping with dysfunctional mitochondria in frataxin-deficient cells, and provide a platform for testing novel therapeutic strategies for FRDA and possibly other human mitochondrial-associated diseases.
The following are the supplementary data related to this article.
Supplementary material
Acknowledgments
We thank all members of the Laboratory of Signal Transduction for constructive discussions and technical advices, and Joyce Chu for technical help. Some nematode strains were provided by the Caenorhabditis Genetics Center, funded by the National Institute of Health (NIH) National Center for Research Resources. We also thank Drs. Chris Link, Tom Johnson, Richard Roy, and Anton Gartner, for providing additional C. elegans strains. Financial support to carry out this study was provided by the National Ataxia Foundation and the Italian Association for Cancer Research (MFAG 11509) to N.V. and by Telethon grant GGP060059 and Ataxia UK to R.T. M.H. is funded by NIH/NIA (Grant numbers: R01 AG038664 and R01 AG039756).
References
- Apfeld J., Kenyon C. Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature. 1999;402:804–809. doi: 10.1038/45544. [DOI] [PubMed] [Google Scholar]
- Butler J.A., Ventura N., Johnson T.E., Rea S.L. Long-lived mitochondrial (Mit) mutants of Caenorhabditis elegans utilize a novel metabolism. FASEB J. 2010;24(12):4977–4988. doi: 10.1096/fj.10-162941. (Dec) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campuzano V., Montermini L., Molto M.D., Pianese L., Cossee M., Cavalcanti F., Monros E., Rodius F., Duclos F., Monticelli A., Zara F., Canizares J., Koutnikova H., Bidichandani S.I., Gellera C., Brice A., Trouillas P., De Michele G., Filla A., De Frutos R., Palau F., Patel P.I., Di Donato S., Mandel J.L., Cocozza S., Koenig M., Pandolfo M. Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- Condo I., Ventura N., Malisan F., Tomassini B., Testi R. A pool of extramitochondrial frataxin that promotes cell survival. J. Biol. Chem. 2006;281:16750–16756. doi: 10.1074/jbc.M511960200. [DOI] [PubMed] [Google Scholar]
- Curtis R., O'Connor G., DiStefano P.S. Aging networks in Caenorhabditis elegans: AMP-activated protein kinase (aak-2) links multiple aging and metabolism pathways. Aging Cell. 2006;5:119–126. doi: 10.1111/j.1474-9726.2006.00205.x. [DOI] [PubMed] [Google Scholar]
- Dillin A., Hsu A.L., Arantes-Oliveira N., Lehrer-Graiwer J., Hsin H., Fraser A.G., Kamath R.S., Ahringer J., Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- Guccini I., Serio D., Condò I., Rufini A., Tomassini B., Mangiola A., Maira G., Anile C., Fina D., Pallone F., Mongiardi M.P., Levi A., Ventura N., Testi R., Malisan F. Frataxin participates to the hypoxia-induced response in tumors. Cell Death Dis. 2011;24(2):e123. doi: 10.1038/cddis.2011.5. (Feb) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M., Chandra A., Mitic L.L., Onken B., Driscoll M., Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugen A.C., Di Prospero N.A., Parker J.S., Fannin R.D., Chou J., Meyer J.N., Halweg C., Collins J.B., Durr A., Fischbeck K., Van Houten B. Altered gene expression and DNA damage in peripheral blood cells from Friedreich's ataxia patients: cellular model of pathology. PLoS Genet. 2010;6:e1000812. doi: 10.1371/journal.pgen.1000812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haun C., Alexander J., Stainier D.Y., Okkema P.G. Rescue of Caenorhabditis elegans pharyngeal development by a vertebrate heart specification gene. Proc. Natl. Acad. Sci. U. S. A. 1998;95:5072–5075. doi: 10.1073/pnas.95.9.5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M.L., Becker E.M., Whitnall M., Rahmanto Y.S., Ponka P., Richardson D.R. Elucidation of the mechanism of mitochondrial iron loading in Friedreich's ataxia by analysis of a mouse mutant. Proc. Natl. Acad. Sci. U. S. A. 2009;106:16381–16386. doi: 10.1073/pnas.0906784106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R.G., Plas D.R., Kubek S., Buzzai M., Mu J., Xu Y., Birnbaum M.J., Thompson C.B. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol. Cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- Kiselyov K., Jennigs J.J., Jr., Rbaibi Y., Chu C.T. Autophagy, mitochondria and cell death in lysosomal storage diseases. Autophagy. 2007;3:259–262. doi: 10.4161/auto.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., Codogno P., Cuervo A.M., Deretic V., Elazar Z., Fueyo-Margareto J., Gewirtz D.A., Kroemer G., Levine B., Mizushima N., Rubinsztein D.C., Thumm M., Tooze S.A. A comprehensive glossary of autophagy-related molecules and processes. Autophagy. 2010;6 doi: 10.4161/auto.6.4.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G., Marino G., Levine B. Autophagy and the integrated stress response. Mol. Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre L.R., Gelino S., Melendez A., Hansen M. Autophagy and lipid metabolism coordinately modulate life span in germline-less C. elegans. Curr. Biol. 2011;21:1507–1514. doi: 10.1016/j.cub.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.S., Lee R.Y., Fraser A.G., Kamath R.S., Ahringer J., Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat. Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- Lemasters J.J. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005;8:3–5. doi: 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- Levine B., Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks O.D., Vousden K.H. Metabolic regulation by p53. J. Mol. Med. 2011;89:237–245. doi: 10.1007/s00109-011-0735-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal S., Guptan P., Owusu-Ansah E., Banerjee U. Mitochondrial regulation of cell cycle progression during development as revealed by the tenured mutation in Drosophila. Dev. Cell. 2005;9:843–854. doi: 10.1016/j.devcel.2005.11.006. [DOI] [PubMed] [Google Scholar]
- McDaniel D.O., Keats B., Vedanarayanan V.V., Subramony S.H. Sequence variation in GAA repeat expansions may cause differential phenotype display in Friedreich's ataxia. Mov. Disord. 2001;16:1153–1158. doi: 10.1002/mds.1210. [DOI] [PubMed] [Google Scholar]
- Melendez A., Talloczy Z., Seaman M., Eskelinen E.L., Hall D.H., Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- Melendez A., Hall D.H., Hansen M. Monitoring the role of autophagy in C. elegans aging. Methods Enzymol. 2008;451:493–520. doi: 10.1016/S0076-6879(08)03229-1. [DOI] [PubMed] [Google Scholar]
- Mishur R.J., Rea S.L. Applications of mass spectrometry to metabolomics and metabonomics: detection of biomarkers of aging and of age-related diseases. Mass Spectrom. Rev. 2011;31(1):70–95. doi: 10.1002/mas.20338. (Jan-Feb) [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T., Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z., Lee S.S. RNAi screens to identify components of gene networks that modulate aging in Caenorhabditis elegans. Brief. Funct. Genomics. 2010;9:53–64. doi: 10.1093/bfgp/elp051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomo G.M., Cerrato T., Gargini R., Diaz-Nido J. Silencing of frataxin gene expression triggers p53-dependent apoptosis in human neuron-like cells. Hum. Mol. Genet. 2011;20:2807–2822. doi: 10.1093/hmg/ddr187. [DOI] [PubMed] [Google Scholar]
- Puccio H., Simon D., Cossee M., Criqui-Filipe P., Tiziano F., Melki J., Hindelang C., Matyas R., Rustin P., Koenig M. Mouse models for Friedreich ataxia exhibit cardiomyopathy, sensory nerve defect and Fe–S enzyme deficiency followed by intramitochondrial iron deposits. Nat. Genet. 2001;27:181–186. doi: 10.1038/84818. [DOI] [PubMed] [Google Scholar]
- Rea S.L., Ventura N., Johnson T.E. Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in Caenorhabditis elegans. PLoS Biol. 2007;5:e259. doi: 10.1371/journal.pbio.0050259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotig A., de Lonlay P., Chretien D., Foury F., Koenig M., Sidi D., Munnich A., Rustin P. Aconitase and mitochondrial iron–sulphur protein deficiency in Friedreich ataxia. Nat. Genet. 1997;17:215–217. doi: 10.1038/ng1097-215. [DOI] [PubMed] [Google Scholar]
- Simon D., Seznec H., Gansmuller A., Carelle N., Weber P., Metzger D., Rustin P., Koenig M., Puccio H. Friedreich ataxia mouse models with progressive cerebellar and sensory ataxia reveal autophagic neurodegeneration in dorsal root ganglia. J. Neurosci. 2004;24:1987–1995. doi: 10.1523/JNEUROSCI.4549-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A.M., Czaja M.J. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle T. Maintenance of C. elegans. WormBook. 2006:1–11. doi: 10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasdemir E., Maiuri M.C., Galluzzi L., Vitale I., Djavaheri-Mergny M., D'Amelio M., Criollo A., Morselli E., Zhu C., Harper F., Nannmark U., Samara C., Pinton P., Vicencio J.M., Carnuccio R., Moll U.M., Madeo F., Paterlini-Brechot P., Rizzuto R., Szabadkai G., Pierron G., Blomgren K., Tavernarakis N., Codogno P., Cecconi F., Kroemer G. Regulation of autophagy by cytoplasmic p53. Nat. Cell Biol. 2008;10:676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen C.C., Sabatini D.M. AMPK and p53 help cells through lean times. Cell Metab. 2005;1:287–288. doi: 10.1016/j.cmet.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Torgovnick A., Schiavi A., Testi R., Ventura N. A role for p53 in mitochondrial stress response control of longevity in C. elegans. Exp. Gerontol. 2010;45(7-8):550–557. doi: 10.1016/j.exger.2010.02.007. (Aug) [DOI] [PubMed] [Google Scholar]
- Toth M.L., Sigmond T., Borsos E., Barna J., Erdelyi P., Takacs-Vellai K., Orosz L., Kovacs A.L., Csikos G., Sass M., Vellai T. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy. 2008;4:330–338. doi: 10.4161/auto.5618. [DOI] [PubMed] [Google Scholar]
- Vazquez-Manrique R.P., Gonzalez-Cabo P., Ros S., Aziz H., Baylis H.A., Palau F. Reduction of Caenorhabditis elegans frataxin increases sensitivity to oxidative stress, reduces lifespan, and causes lethality in a mitochondrial complex II mutant. FASEB J. 2006;20:172–174. doi: 10.1096/fj.05-4212fje. [DOI] [PubMed] [Google Scholar]
- Ventura N., Rea S.L. Caenorhabditis elegans mitochondrial mutants as an investigative tool to study human neurodegenerative diseases associated with mitochondrial dysfunction. Biotechnol. J. 2007;2:584–595. doi: 10.1002/biot.200600248. [DOI] [PubMed] [Google Scholar]
- Ventura N., Rea S., Henderson S.T., Condo I., Johnson T.E., Testi R. Reduced expression of frataxin extends the lifespan of Caenorhabditis elegans. Aging Cell. 2005;4:109–112. doi: 10.1111/j.1474-9726.2005.00149.x. [DOI] [PubMed] [Google Scholar]
- Ventura N., Rea S.L., Handerson S.T., Condo I., Testi R., Johnson T.E. C. elegans as a model for Friedreich ataxia. FASEB J. 2006;20:1029–1030. doi: 10.1096/fj.06-0505ufm. [DOI] [PubMed] [Google Scholar]
- Ventura N., Rea S.L., Schiavi A., Torgovnick A., Testi R., Johnson T.E. p53/CEP-1 increases or decreases lifespan, depending on level of mitochondrial bioenergetic stress. Aging Cell. 2009;8:380–393. doi: 10.1111/j.1474-9726.2009.00482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace D.C. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.C., O'Rourke E.J., Ruvkun G. Fat metabolism links germline stem cells and longevity in C. elegans. Science. 2008;322:957–960. doi: 10.1126/science.1162011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarse K., Schulz T.J., Birringer M., Ristow M. Impaired respiration is positively correlated with decreased life span in Caenorhabditis elegans models of Friedreich Ataxia. FASEB J. 2007;21:1271–1275. doi: 10.1096/fj.06-6994com. [DOI] [PubMed] [Google Scholar]
- Zhang S.O., Trimble R., Guo F., Mak H.Y. Lipid droplets as ubiquitous fat storage organelles in C. elegans. BMC Cell Biol. 2010;11:96. doi: 10.1186/1471-2121-11-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material