Abstract
Yin Yang 1 (YY1), a ubiquitously expressed transcription factor, plays a critical role in regulating cell development, differentiation, cellular proliferation and tumorigenesis. Previous studies identified many YY1-regulated target genes in both human and mouse. Emerging global mapping by Chromatin ImmnoPrecipitation (ChIP)-based high-throughput experiments indicate that YY1 binds to a vast number of loci genome-wide. However, the information is widely scattered in many disparate poorly cross-indexed literatures; a large portion was only published recently by the ENCODE consortium with limited annotation. A centralized database, which annotates and organizes YY1-binding loci and target motifs in a systematic way with easy access, will be valuable resources for the research community. We therefore implemented a web-based YY1 Target loci Database (YY1TargetDB). This database contains YY1-binding loci (binding peaks) from ChIP-seq and ChIP-on-chip experiments, computationally predicated YY1 and cofactor motifs within each locus. It also collects the experimentally verified YY1-binding motifs from individual researchers. The current version of YY1TargetDB contains 92 314 binding loci identified by ChIP-based experiments; 157 200 YY1-binding motifs in which 42 are experimentally verified and 157 158 are computationally predicted; and 130 759 binding motifs for 47 cofactors.
Database URL: http://www.myogenesisdb.org/YY1TargetDB
Introduction
Yin Yang 1 (YY1), also known as NF-E1, UCRBP, CF1 and δ, is a multifunctional zinc-finger transcription factor. It can act either as a transcriptional activator or a repressor depending on the cofactors that it interacts with, thus named Yin Yang 1 (1). YY1 is highly conserved among different species and ubiquitously expressed in various tissues and cells. Since its initial discovery, YY1 has been demonstrated to play vital roles in numerous biological processes and systems such as development, differentiation, cellular proliferation, invasion, apoptosis, tumorigenesis as well as regulation on viral gene expression (1).
The multifunctional property of YY1 mainly stems from its strong binding to a DNA sequence CGCCATNTT and its ability to physically interact with a large number of cellular factors. It was estimated that >7% of vertebrate and 24% of viral promoters contain the above YY1 consensus sites in their regulatory regions (2). YY1-interacting proteins range from co-repressor, co-activator to general transcription factors such as TATA-binding protein (TBP), TBP-associated factors and Transcription factor II B (TFIIB) (1). Various interacting modes determine its regulatory mechanisms at different target promoters. For example, we have demonstrated that YY1 recruits Ezh2 (Enhancer of Zeste Homolog 2, a histone 3 lysine 27 methyltransferase) containing Polycomb silencing complex to repress multiple genes expression in skeletal muscle cells (3–5). YY1 interaction with Smad3 leads to the repression of miR-29 promoter during myoblasts transdifferentiation into myofibroblasts (6). YY1 can also interact with CREB-binding protein (CBP) and E1A binding protein p300 to activate promoters (7). These findings highlight the fascinating complexity of YY1-involved gene regulation. Despite these advances, further identification of YY1 targets and its interacting partners are needed to gain a comprehensive view of its mechanisms of action.
Recent efforts on genome-wide mapping of YY1 binding using high-throughput technologies, including Chromatin ImmnoPrecipitation (ChIP)-seq and ChIP-on-chip, have provided us vast amount of information on YY1 global binding (8–12). However, it is not easy to obtain these data, as they are often hiding throughout different places. To integrate the available YY1-binding loci and their associated annotation information, we have created a web-based comprehensive database YY1 Target loci Database (YY1TargetDB, http://www.myogenesisdb.org/YY1TargetDB). This database mainly contains YY1-binding loci identified from 17 ChIP-based (i.e. ChIP-seq and ChIP-on-chip) high-throughput datasets. Computationally predicted YY1 and cofactor binding motifs within these loci were also collected; in addition, 42 YY1-binding motifs experimentally verified by individual laboratories were included. YY1TargetDB was implemented as a browsable database and integrated with a locally installed UCSC genome browser to facilitate the data exploration and visualization at the whole-genome level.
Data acquisition and database implementation
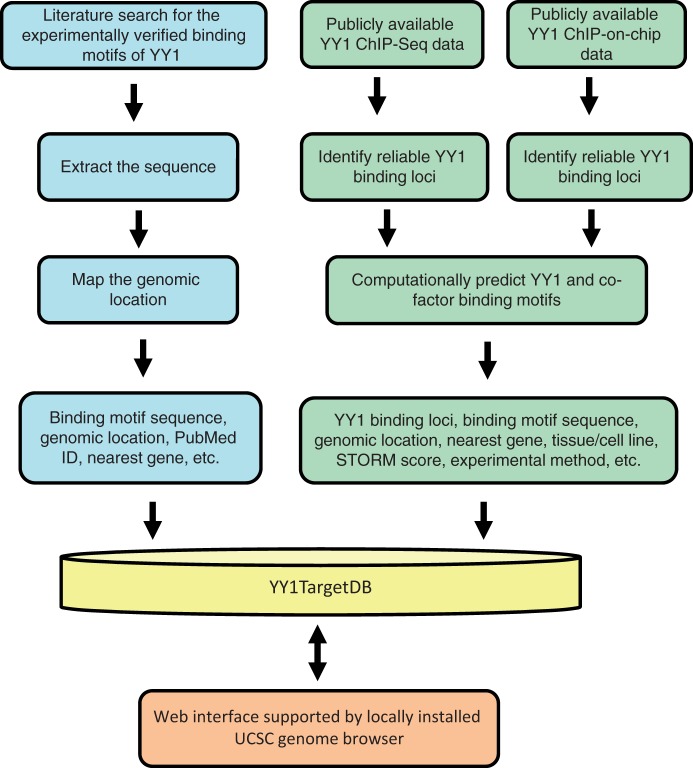
The current version of YY1TagetDB has collected YY1 binding and associated gene regulatory information for two species, human and mouse, in 15 different cell lines. The database is composed of three types of data: (i) experimentally verified YY1-binding motifs, which were collected manually from the literature and examined by individual laboratories by EMSA or ChIP-PCR assay in addition to reporter assay or expression analysis; these motifs are associated with target genes that are physically bound and directly regulated by YY1; (ii) YY1-binding loci identified from ChIP-based high-throughput methods, including ChIP-seq and ChIP-on-chip; binding motifs were computationally predicated from these loci and the associated putative target genes were identified to infer any cofactors that may cooperate with YY1 function. A work flow comprising two major data-analysis pipelines illustrates the data-processing steps (Figure 1).
Figure 1.
Schematic overview for YY1TargetDB data acquisition and database implementation. The experimentally verified YY1-binding motifs were collected from literatrues, mapped to the reference genomes and stored in the database. High-throughput (ChIP-seq or ChIP-on-chip) experimental datasets were downloaded from NCBI GEO server and processed by our analysis and annotation pipeline. The identifed YY1-binding loci, computationally predicted YY1 and its cofactors binding motifs were deposited into YY1TargetDB. The database in integrated with locally installed UCSC genome browser to visualize YY1-binding loci and assoicated annotation information.
Identification and mapping of experimentally verified YY1-binding targets
Collecting experimentally verified YY1-binding motifs from published articles and identifying relevant information by mining data were conducted mainly by manual inspection. A total of ∼70 YY1-binding motifs were reported in the literature with ∼60 from human and mouse. The motif sequence and coordinates given in the article were used when possible. In many cases where the exact genomic coordinates were not given, YY1-binding motif and flanking sequences (usually >50 bp) were used to retrieve the coordinates from the target gene promoter sequences available at UCSC. In a few cases, the claimed YY1-binding motifs from articles could not be located probably owing to the incorrect information reported. For simplicity, the database only contains the annotation for one genome assembly per species: hg19 was used for human and mm9 for mouse.
As a result, we have collected 42 experimentally verified YY1-binding motifs, corresponding to 21 target genes from human and 10 from mouse (Table 1 and Supplementary Tables S1–S2). The motif sequences, genomic coordinates, associated target genes, cell line information and PubMed Identifier (IDs) were all stored in the database. The effort will be continued in reviewing the literature and collecting these data periodically to ensure inclusion of recently published data.
Table 1.
Summary of experimentally verified YY1-binding motifs
| Species | Binding motifs | Target genes | Cell lines |
|---|---|---|---|
| Human | 29 | 21 | 24 |
| Mouse | 13 | 10 | 7 |
Identification and annotation of YY1-binding loci from ChIP-seq/ChIP-on-chip data
High-throughput methods have provided a promising way to study transcription factors and DNA interactions at the genome-wide level. We took advantage of the publicly available ChIP-seq and ChIP-on-chip data. A total of 16 ChIP-seq datasets corresponding to 14 cell lines were obtained, among which 5 were from individual researchers and the other 11 were recently published from the ENCODE consortium (13, 14) (Supplementary Table S3). To identify highly reliable YY1-binding loci from these datasets, we processed raw reads by aligning to hg19/mm9 reference genomes with SOAP2 (15) before using MACS (16) with stringent criteria (Supplementary Table S4) for peak identification. In a few cases where raw reads were unavailable for downloading and processing, a ‘liftover’ tool from UCSC was used to simply convert the genomic coordinates of the originally identified loci to Hg19 or Mm9 reference genomes. YY1-binding motifs within each of the above identified locus were further predicted using STORM program (17), and binding motifs for potential cofactors were identified with coMOTIF program (18) (Table 2). To further validate and prioritize the identified YY1 cofactors, we also used different programs in our pipeline for YY1-binding motif prediction and cofactor identification. For example, we used Tree-based Position weight matrix Discriminative approach (TPD) (19) instead of STORM for YY1-binding motif prediction. We also used W-ChIPMotifs (20) for the de novo identification of YY1 cofactors within the top-ranked YY1-binding loci. Only one ChIP-on-chip dataset was available and the YY1-binding loci were identified using a method described in (12). We should point out that binding loci (peaks) refer to the larger regions produced by ChIP-based experiments, while binding motifs refer to the predicted YY1-binding sequence within each locus, so binding loci have a one-to-many relationships with binding motifs, in other words, one locus may contain multiple binding motifs.
Table 2.
Summary of identified YY1-binding loci, predicted YY1 and cofactor binding motifs from ChIP based datasets
| No | Data sources | Species | Methods | Cell lines | Number of YY1-binding loci | Number of predicted YY1 motifs | % of loci with YY1 motifs | Number of Cofactors | Number of predicted cofactor motifs |
|---|---|---|---|---|---|---|---|---|---|
| 1 | ENCODE/HAIB | Human | ChIP-seq | A549 | 5855 | 8796 | 48.8% | 9 | 10 022 |
| 2 | ENCODE/HAIB | Human | ChIP-seq | GM12878 | 11 964 | 11 026 | 36.1% | 11 | 9813 |
| 3 | ENCODE/Stanford/Yale/USC/Harvard | Human | ChIP-seq | GM12878 | 103 | 527 | 91.3% | 6 | 261 |
| 4 | ENCODE/HAIB | Human | ChIP-seq | GM12891 | 3831 | 8150 | 64.4% | 7 | 5474 |
| 5 | ENCODE/HAIB | Human | ChIP-seq | GM12892 | 3967 | 9115 | 69.3% | 7 | 6808 |
| 6 | ENCODE/HAIB | Human | ChIP-seq | H1-hESC | 5416 | 13 143 | 70.4% | 13 | 9453 |
| 7 | ENCODE/HAIB | Human | ChIP-seq | HCT-116 | 9054 | 10 181 | 38.6% | 7 | 6819 |
| 8 | ENCODE/HAIB | Human | ChIP-seq | HepG2 | 4761 | 6498 | 47.4% | 10 | 8320 |
| 9 | ENCODE/HAIB | Human | ChIP-seq | K562 | 17 916 | 29 497 | 51.7% | 9 | 16 296 |
| 10 | ENCODE/HAIB | Human | ChIP-seq | SK-N-SH_RA | 4383 | 9043 | 64.4% | 8 | 5949 |
| 11 | ENCODE/Stanford/Yale/USC/Harvard | Human | ChIP-seq | NT2-D1 | 4505 | 9577 | 63.7% | 3 | 4217 |
| 12 | Cuddapah et al. Genomic profiling of HMGN1 reveals an association with chromatin at regulatory regions. Mol. Cell Biol. 2010 | Human | ChIP-seq | CD4+ T cells | 4280 | 5940 | 42.5% | 9 | 4441 |
| 13 | Hernandez/Herr Lab, University of Lausanne, Switzerland, 2012, unpublished. | Human | ChIP-seq | HeLa-S cells | 269 | 1164 | 88.8% | 8 | 558 |
| 14 | Li et al. YY1 regulates melanocyte development and function by cooperating with MITF. PLoS Genet. 2012. | Human | ChIP-seq | MALME-3M, Melanocyte | 7713 | 9337 | 54.9% | 14 | 22 701 |
| 15 | Gebhard et al. General transcription factor binding at CpG islands in normal cells correlates with resistance to De novo DNA methylation in cancer cells. Cancer Res. 2010. | Human | ChIP-on-chip | Mynocytes (CD14+) | 4472 | 7119 | 47.5% | 7 | 8801 |
| 16 | Mendenhall et al. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet. 2010. | Mouse | ChIP-seq | Murine ES cells | 1093 | 4994 | 83.3% | 10 | 2580 |
| 17 | Vella et al. Yin Yang 1 extends the Myc-related transcription factors network in embryonic stem cells. Nucleic Acids Res. 2012. | Mouse | ChIP-seq | Murine ES cells | 2732 | 13 051 | 83.5% | 11 | 8246 |
As a result, we have collected 92 314 YY1-binding loci from the above high-throughput data. In all, 157 158 consensus YY1-binding motifs were found in these loci, suggesting direct physical interactions with YY1. These sites were associated with 13 247 genes (human, 11 682; mouse, 1 565). Among these, 5 232 genes (human, 4118; mouse, 1 114) contain at least one YY1 motif in their proximal promoter region (−5K to +2K of the transcription start site). It is highly likely that these genes are direct regulatory targets of YY1. Nevertheless, expression data are needed to confirm their expression is indeed subjected to YY1 control. Many loci contain both YY1 and other transcription factor binding motifs nearby, in agreement with YY1’s co-operative nature with many cofactors.
Database organization
YY1TagetDB was designed based on an entity relationship model (21) (Supplementary Figure S1). It stores all the information in 5 tables ‘experiments’, ‘chip_loci’, ‘computed_bs’, ‘verified_bs’ and ‘genes’. The table ‘experiments’ collects important experimental details, e.g. experimental method, cell line, protocol, treatment and antibody. The ‘chip_loci’ table stores identified YY1-binding loci from ChIP-based experiments and related annotations such as the number of predicted motifs within each locus, the nearest genes and so forth. The table ‘computed_bs’ stores computationally predicted YY1 and cofactor binding motifs. The experimentally verified binding motifs were stored in the ‘verified_bs’ table. The table ‘genes’ collects the gene annotation originally from NCBI RefSeq gene (22) and the interaction types (i.e. direct, indirect, unknown) between YY1 and the associated genes. A ‘direct’ interaction type is defined if the promoter region (−5K ∼ +2K bp from transcription start site) of a given gene contains YY1-binding locus and at least one experimentally verified or computationally predicted YY1 motifs within the locus. An ‘indirect’ interaction type is established if the promoter region contains YY1-binding locus but not any motifs. In this case, the binding is probably mediated by another transcription factor indirectly. However, we cannot exclude the possibility that a novel binding motif mediates direct YY1 binding to this locus. If a gene associates with neither a binding locus nor motif, the interaction type is defined as ‘unknown’.
Web interface and data visualization
YY1TargetDB can be accessed at http://www.myogenesisdb.org/YY1TargetDB. It was implemented as a web-based relational database with user-friendly web interface for searching and browsing. MySQL was used as backend database, and a locally installed UCSC genome browser (23) was integrated with Common Gateway Interface (CGI) scripts (Python) for data visualization. All of the data are available for download as a MySQL dump file, along with the database schema, through the ‘Download’ link on the website. The search results can also be downloaded through the download function provided in the search result web page in text format.
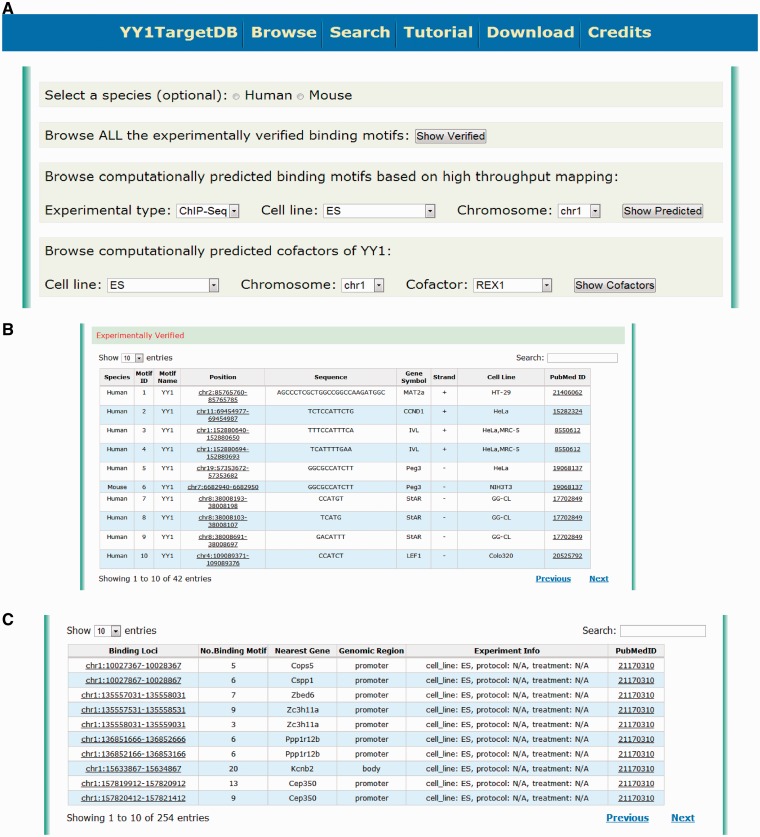
To query the database, users may retrieve YY1-binding information in several ways. First, they may browse the database through the ‘browse’ webpage. This feature enables users to explore our database conveniently by selecting some of the basic filters without knowing too much prior information. On this webpage (Figure 2A), users can first select the species (i.e. human, mouse or both without selecting). They may then browse either ‘experimentally verified binding motifs’ or ‘computationally predicted binding motifs based on high-throughput mapping’. The latter is further specified by three filters: experimental type (ChIP-seq or ChIP-chip), cell line and chromosome to locate the information more specifically. When browsing through ‘experimentally verified binding motifs’, the database table will provide detailed information about the genomic location of the verified YY1-binding motifs: sequence, target gene, cell line and publication source (PubMed ID). A hyperlink has been implemented for each binding motif leading to the visualization of the motif (Figure 2B). When browsing through ‘computationally predicted binding motifs’, the identified YY1-binding loci with the number of predicted motifs within this region and other related annotations will be presented. Users can further explore the details and the data visualization by clicking the hyperlink (Figure 2C). User can also browse the data by selecting the specific YY1 cofactors through the options provided by the ‘computationally predicted cofactors of YY1’ section (Figure 2A).
Figure 2.
Screen shot depicting the browsing interface. (A) Web interface for browsing the experimentally verified or computationally predicted YY1-binding motif. (B) Tabulated presentation for experimentally verified YY1-binding motifs. A hyperlink was implemented for further data visualization. (C) Tabulated presentation for identified YY1-binding loci. A hyperlink was implemented for further visualization of computationally predicted YY1-binding motifs under each locus.
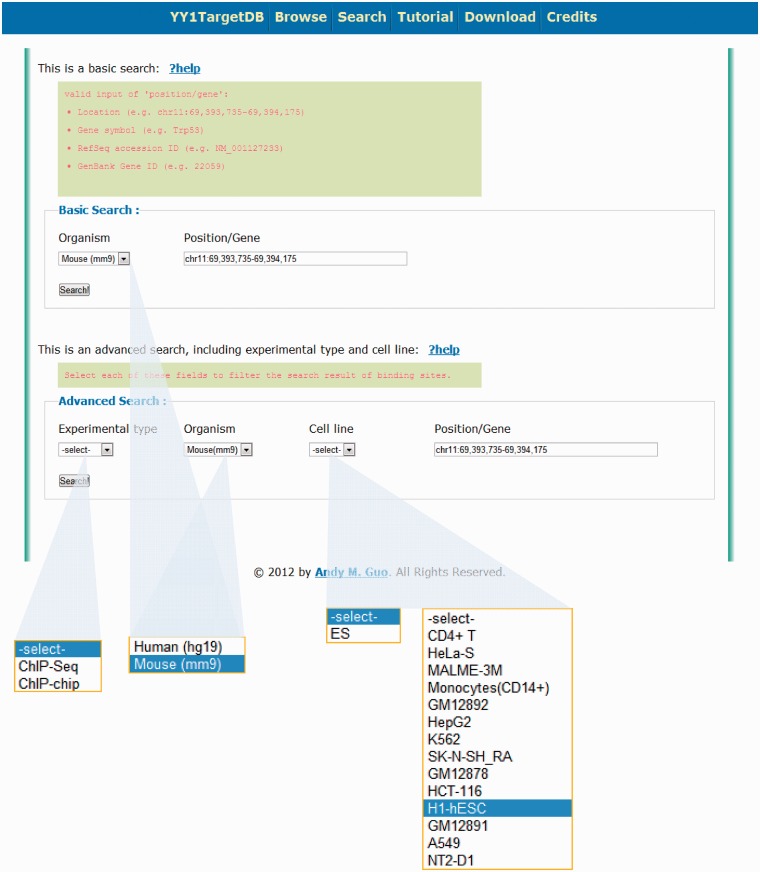
In addition to browsing the database, users may retrieve information through two types of searching. (i) Basic Search. This search option enables users to query the database for different species (i.e. human or mouse) together with one of the four keywords: (a) a specific genomic region; (b) gene symbol; (c) RefSeq accession number or (d) NCBI Gene ID (Figure 3). Similar to doing search on the UCSC genome browser, a genomic location in the format of chr1:1000–20 000 can be entered as a search keyword to retrieve all the data within this region. For example, a search for the region of chr11:69 393 735–69 394 175 in mouse genome will lead to a display of Trp53 gene, which contains one experimentally verified YY1-binding motif and two computationally predicted motifs within four YY1-binding loci in its promoter region. (ii) Advanced Search. This search option enables users to query the database with the same keyword as basic search but more specific criteria such as experimental type (i.e. ChIP-seq or ChIP-on-chip), species (i.e. human or mouse) and cell line (Figure 3).
Figure 3.
Search options for YY1 TargetDB. Users can perform basic search or advance search.
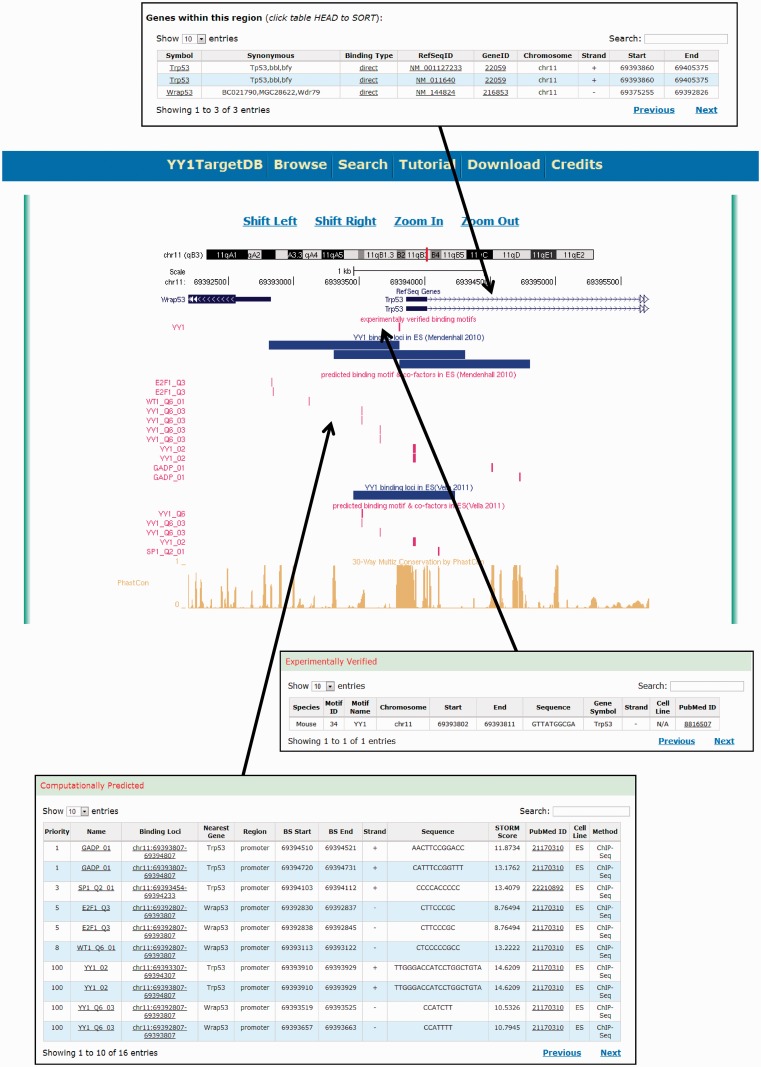
To visualize and present the searching results, we provide a graphic visualization supported by a locally installed customized UCSC genome browser and tabulated detailed information below (Figure 4). Each data source is assigned a separate track in the visualization. On the top, a RefSeq gene track displays the RefSeq gene transcripts falling into the searched region. The experimentally verified motifs and YY1-binding loci from ChIP mapping were also visualized as tracks, followed by the computationally predicted binding motif tracks for YY1 and cofactors as well as a cross-species conservation track. The data visualization also allows browsing the genome by shifting the regions left or right and zooming in and out.
Figure 4.
Screen shot depicting the data visualization using integrated UCSC genome browser and the tabulated information.
Below the graphic visualization, three tables display the details of each track. YY1-binding motifs in the searched genomic regions are presented as two tables: (i) ‘experimentally verified’ (Figure 4). It contains the following information: species, motif ID, motif name, chromosome number, start, end, motif sequence, nearest gene symbol, cell line, strand and PubMed ID. (ii) The computationally predicted binding motifs are presented in the second table named ‘computationally predicted’ (Figure 4). This table contains the priority of the YY1 cofactor, the cofactor, binding loci with cofactor’s binding motif, nearest gene, genomic region (promoter, body or intergenic), start, end, strand, sequence, STORM score, PubMed ID, cell line and methods (i.e. ChIP-seq or ChIP-on-chip). The priorities of the YY1 cofactors in this table were calculated based on the occurrences of the cofactors in all the cell lines included in the database. The highest priority, 100, was set for YY1 itself. The third table, ‘Genes within this region’, contains annotation information of the RefSeq gene transcripts found in the searchable region, e.g. gene symbol, synonymous, binding type, RefSeq ID, Gene ID, chromosome, strand, transcript start and end positions.
Discussion
In this study, we have developed the first and only available comprehensive data management system for the collection, identification and analysis of YY1-binding loci/motifs in human and mouse genomes. This database aims to facilitate the hypothesis generation through high-throughput data by the individual researchers. The current version collects 157 200 YY1-binding motifs with 42 verified experimentally and 157 158 predicted from high-throughput derived data.
In our study, we also identified 47 YY1 cofactors, which may work together with YY1 and regulate their target genes. To gain more confidence and prioritize our identified YY1 cofactors, we applied TPD, a more recent and sophisticated TF binding site tool, to predict the existence of YY1-binding motifs in the YY1-binding loci and then used coMOTIF program to identify the potential YY1 cofactors among the top 500 YY1-binding loci based on TPD prediction. We found that 72% (34/47) of our previously identified YY1 co-factors can also be identified through TPD followed by coMOTIF. Similarly, we have also used W-ChIPMotifs to de novo identify YY1 cofactors. The results indicated that ∼30% W-ChIPMotifs identified YY1 cofactors overlaps with previously identified YY1 cofactors with coMOTIF program. In this study, by using multiple programs for the identification of YY1 cofactors or ranking these cofactors based on their occurrences in different cell lines and predicting programs provides a scoring system that can be used to systematically prioritize the cofactors for the future downstream validation.
The first of several distinguishing features of YY1TargetDB is that it collects an unprecedented large number of genome-wide datasets. This not only includes 6 published YY1 ChIP-seq and ChIP-on-chip data from various biological systems but also 11 recently published datasets from ENCODE (13). The vast amount of high-throughput data generated by ENCODE has provided tremendously valuable resources but we realized that the numbers of YY1-binding loci provided in original ENCODE datasets are large. To reduce false positive peaks, we re-processed the raw data using stringent parameters to identify 92 314 high-confidence binding loci for 15 different cell lines, and 157 158 binding motifs were further predicted from these regions. The second feature is that we not only predicted YY1-binding motifs from the binding loci but also identified many cofactors that could function together with YY1. This is in agreement with the known interactive nature of YY1. Among the identified cofactors, some have been previously demonstrated such as Sp1 and E2F (24, 25). Many are unknown, suggesting novel cis-regulatory modules. This information will no doubt provide valuable basis for a biologist to generate new hypothesis in his/her research. The last feature is that we have integrated UCSC genome browser into our database. This guarantees access to important genome browser features and simultaneous availability of other genome browser tracks (annotated RefSeq gene, conservation, regulation and other tracks). In addition, the UCSC genome browser interface is familiar to biologists world-wide and requires minimal training to use.
Supplementary Data
Supplementary data are available at Database online.
Funding
This work was supported by the General Research Funds from the Research Grants Council of Hong Kong, China [CUHK476309, CUHK476310 to H.W., CUHK473211 to H.S.], and the Chinese University of Hong Kong direct grant [2041474 to H.S., 2041492, 2041662 to H.W] Funding for open access charge: CUHK473211.
Conflict of interest. None declared.
Supplementary Material
References
- 1.Gordon S, Akopyan G, Garban H, et al. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene. 2006;25:1125–1142. doi: 10.1038/sj.onc.1209080. [DOI] [PubMed] [Google Scholar]
- 2.Shi Y, Lee JS, Galvin KM. Everything you have ever wanted to know about Yin Yang 1. Biochim. Biophys. Acta. 1997;1332:F49–F66. doi: 10.1016/s0304-419x(96)00044-3. [DOI] [PubMed] [Google Scholar]
- 3.Wang H, Garzon R, Sun H, et al. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 2008;14:369–381. doi: 10.1016/j.ccr.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang H, Hertlein E, Bakkar N, et al. NF-kappaB regulation of YY1 inhibits skeletal myogenesis through transcriptional silencing of myofibrillar genes. Mol. Cell. Biol. 2007;27:4374–4387. doi: 10.1128/MCB.02020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu L, Zhou L, Chen EZ, et al. A Novel YY1-miR-1 regulatory circuit in skeletal myogenesis revealed by genome-wide prediction of YY1-miRNA network. PLoS One. 2012;7:e27596. doi: 10.1371/journal.pone.0027596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou L, Wang L, Lu L, et al. Inhibition of miR-29 by TGF-beta-Smad3 signaling through dual mechanisms promotes transdifferentiation of mouse myoblasts into myofibroblasts. PLoS One. 2012;7:e33766. doi: 10.1371/journal.pone.0033766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee HY, Chaudhary J, Walsh GL, et al. Suppression of c-Fos gene transcription with malignant transformation of human bronchial epithelial cells. Oncogene. 1998;16:3039–3046. doi: 10.1038/sj.onc.1201843. [DOI] [PubMed] [Google Scholar]
- 8.Cuddapah S, Schones DE, Cui K, et al. Genomic profiling of HMGN1 reveals an association with chromatin at regulatory regions. Mol. Cell. Biol. 2010;31:700–709. doi: 10.1128/MCB.00740-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J, Song JS, Bell RJ, et al. YY1 regulates melanocyte development and function by cooperating with MITF. PLoS Genet. 2012;8:e1002688. doi: 10.1371/journal.pgen.1002688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mendenhall EM, Koche RP, Truong T, et al. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet. 2010;6:e1001244. doi: 10.1371/journal.pgen.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vella P, Barozzi I, Cuomo A, et al. Yin Yang 1 extends the Myc-related transcription factors network in embryonic stem cells. Nucleic Acids Res. 2012;40:3403–3418. doi: 10.1093/nar/gkr1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gebhard C, Benner C, Ehrich M, et al. General transcription factor binding at CpG islands in normal cells correlates with resistance to de novo DNA methylation in cancer cells. Cancer Res. 2010;70:1398–1407. doi: 10.1158/0008-5472.CAN-09-3406. [DOI] [PubMed] [Google Scholar]
- 13.Rosenbloom KR, Dreszer TR, Long JC, et al. ENCODE whole-genome data in the UCSC genome browser: update 2012. Nucleic Acids Res. 2012;40:D912–D917. doi: 10.1093/nar/gkr1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunham I, Kundaje A, Aldred SF, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li R, Yu C, Li Y, et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 16.Feng J, Liu T, Zhang Y. Using MACS to identify peaks from ChIP-Seq data. Curr Protoc Bioinformatics. 2011 doi: 10.1002/0471250953.bi0214s34. Chapter 2, Unit 2 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schones DE, Smith AD, Zhang MQ. Statistical significance of cis-regulatory modules. BMC Bioinformatics. 2007;8:19. doi: 10.1186/1471-2105-8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu M, Weinberg CR, Umbach DM, et al. coMOTIF: a mixture framework for identifying transcription factor and a coregulator motif in ChIP-seq data. Bioinformatics. 2011;27:2625–2632. doi: 10.1093/bioinformatics/btr397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bi Y, Kim H, Gupta R, et al. Tree-based position weight matrix approach to model transcription factor binding site profiles. PLoS One. 2011;6:e24210. doi: 10.1371/journal.pone.0024210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin VX, Apostolos J, Nagisetty NS, et al. W-ChIPMotifs: a web application tool for de novo motif discovery from ChIP-based high-throughput data. Bioinformatics. 2009;25:3191–3193. doi: 10.1093/bioinformatics/btp570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kennedy BA, Gao W, Huang TH, et al. HRTBLDb: an informative data resource for hormone receptors target binding loci. Nucleic Acids Res. 2009;38:D676–D681. doi: 10.1093/nar/gkp734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pruitt KD, Tatusova T, Brown GR, et al. NCBI reference sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2011;40:D130–D135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujita PA, Rhead B, Zweig AS, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2010;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye J, Zhang X, Dong Z. Characterization of the human granulocyte-macrophage colony-stimulating factor gene promoter: an AP1 complex and an Sp1-related complex transactivate the promoter activity that is suppressed by a YY1 complex. Mol. Cell. Biol. 1996;16:157–167. doi: 10.1128/mcb.16.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schlisio S, Halperin T, Vidal M, et al. Interaction of YY1 with E2Fs, mediated by RYBP, provides a mechanism for specificity of E2F function. EMBO J. 2002;21:5775–5786. doi: 10.1093/emboj/cdf577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.