Abstract
Control of gene transcription is a major regulatory determinant for function of the endothelin pathway. Epigenetic mechanisms act on tissue-specific gene expression during development and in response to physiological stimuli. Most of the limited evidence available on epigenetic regulation of the endothelin pathway focuses on the EDN1 and EDNRB genes. Examination of whole genome databases suggests that both genes are influenced by histone modifications and DNA methylation. This interpretation is supported by studies directed at detecting epigenetic action on the two genes. The clearest illustration of epigenetic factors altering endothelin signalling is DNA methylation-associated EDNRB silencing during tumourigenesis. This review summarizes our current understanding of epigenetic regulation of the endothelin pathway genes.
LINKED ARTICLES
This article is part of a themed section on Endothelin. To view the other articles in this section visit http://dx.doi.org/10.1111/bph.2013.168.issue-1
Keywords: endothelin, endothelin receptor, epigenetics, histone modifications, DNA methylation
Introduction
The endothelin (ET) signalling pathway governs many physiological functions ranging from constriction of the vasculature to cell proliferation in development to salt retention by the kidney (reviewed by Barton and Yanagisawa, 2008; Kohan et al., 2011). The pathway is mediated by three ET proteins (ET-1, ET-2 and ET-3) and their interactions with two receptors [ET receptor type A (ETA) and ET receptor type B (ETB) receptor]. Each of the ET peptides is encoded by separate genes (EDN1, EDN2 and EDN3). The biologically active polypeptides differ from ET-1 by only two and six amino acids in ET-2 and ET-3 respectively. The EDNRA and EDNRB genes each encode a G protein-coupled receptor ETA receptor and ETB receptor. The two receptors share similar membrane topologies and 55% primary amino acid sequence identity (Arai et al., 1993). However, the two receptors display distinct ligand affinities. ETA receptor possesses a higher affinity for ET-1 and ET-2, whereas ETB receptor apparently does not discriminate between the three proteins (Kohan et al., 2011). Importantly, the two receptors activate different second messenger pathways and in specific instances may act in opposition to one another (Nelson et al., 2003).
Defects in the ET pathway contribute to a variety of pathogenic states. For example, mutations in the coding sequences of EDNRB and EDN3 are associated with Waardenburg syndrome, a genetic disease characterized by a lack of nerve cells in all or part of the colon (Puffenberger et al., 1994; Carrasquillo et al., 2002; Pingault et al., 2010). In various forms of cancer, somatic mutations can increase plasma levels of ET polypeptides or alter the ETA receptor to ETB receptor ratio (Bagnato et al., 2011). Increases in the ETA receptor : ETB receptor promote cell growth, angiogenesis, metastatic spread and inhibits apoptosis (Nelson et al., 2003; Bagnato et al., 2011). EDNRB is an established tumour suppressor gene and decreased expression of ETB receptor is seen in numerous tumour cells. Although chromosomal rearrangement accounts for a few examples of cancer-related changes in ET signalling (Huang et al., 2004), epigenetic regulation is emerging as an important factor affecting expression of the ET proteins and receptors in tumours.
The term ‘epigenetics’ refers to inheritable changes in gene expression that do not alter the DNA sequence (Margueron and Reinberg, 2010). The human genome contains more than 3 million base pairs organized into approximately 21 000 protein coding genes (Clamp et al., 2007; Pertea and Salzberg, 2010). Histone proteins constitute the nucleosome core particles that are largely responsible for packaging DNA into the nucleus. Epigenetic mechanisms that affect genes include insertion of histone variants, post-translational modifications of histones, expression of non-coding RNAs (ncRNAs) and methylation of DNA. These epigenetic effectors alter both the availability of genes for transcription and the rates of transcription (Figure 1). The present review will focus on our current understanding of the two epigenetic mechanisms known to regulate the genes of the ET pathway: DNA methylation and histone modification. Only two ET pathway genes, EDN1 and EDNRB, have been studied in any detail.
Figure 1.
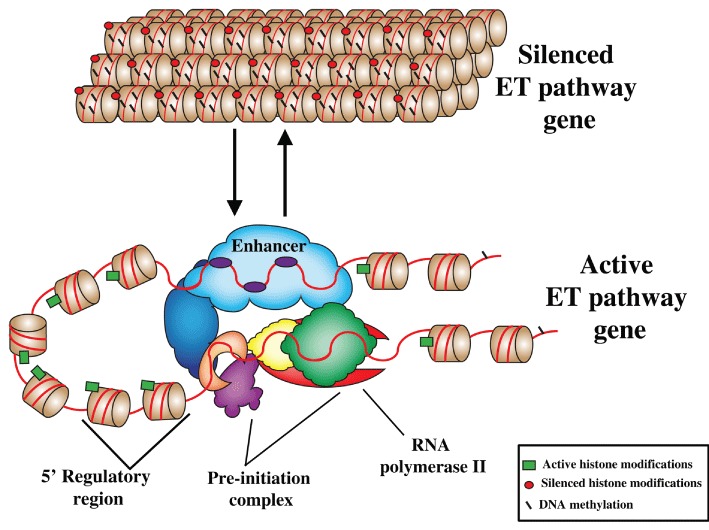
Activation of an ET pathway gene. Nucleosomes are shown as brown barrels and DNA as red lines. Histone modifications that repress transcription are represented by red circles and DNA methylation by black lines. In open chromatin, mediator (blue) interacts with the transcription factors (purple, yellow and green) bound at enhancers and within the 5′ regulatory region to organize the transcription pre-initiation complex containing RNA polymerase II (red). Acetylation of histones, indicative of actively transcribed genes, is shown by the green rectangles.
Regulation of EDN1
The structure of EDN1 is simple by human gene standards. The gene is located at 6p24.1, covers approximately 6 kbp and includes five exons (Figure 1). Transcription is initiated from a single site and maturation of the primary transcript results in a 2.8 kb mRNA. Aside from the expected evolutionary conservation within exon sequences, there are three other conserved regions: a region −1.6 to −1.2 kbp 5′ to the transcription start site, −0.5 to +0.2 kbp in the 5′ promoter proximal segment and an area +0.7 to +1.4 kbp within the first intron.
From a regulatory perspective, the EDN1 gene is not so simple. Transcription is thought to be the primary level of regulation for the EDN1 gene. It is known to be regulated by more than 20 different stimuli, each acting in selected cells and tissues (Stow et al., 2011). In response to those stimuli, at least 10 different transcription factors bind to the promoter proximal region. In general the response elements for these transcription factors display a very high level of nucleotide sequence conservation. For example, in pulmonary cells under hypoxic conditions hypoxia-inducible factor-1 (HIF-1) and activator protein-1 (AP-1) bind to specific regulatory elements to induce transcription of EDN1 (Yamashita et al., 2001). TGFβ supports the binding of the Smad family proteins and HIF-1 in endothelial cells (Rodríguez-Pascual et al., 2003). Furthermore, as many as five additional highly conserved sequences display appropriate conservation and length to suggest the actions of other as yet unidentified regulatory factors.
The histone code suggests that certain histone modifications are associated with open chromatin and active transcription, whereas others are indicative of closed chromatin and gene silencing. The advent of whole genome analysis provides a resource for considering expression of EDN1 and the other ET pathway genes. One of the new genome-wide approaches, chromatin immunoprecipitation sequencing (ChIP-seq), can be used to assay histone modifications across the entire genome. In the ChIP-seq protocol, histones are chemically cross-linked to DNA in situ and precipitated using a highly specific antibody for particular histone modifications (Johnson et al., 2007). The DNA is recovered from the immunoprecipitates and subjected to high-throughput DNA sequencing analysis. Numerous histone modifications have been assayed in this way and some of these data are accessible using the UCSC Genome Browser (http://www.genome.ucsc.edu). At the present time, the cell types studied in this manner are limited due to the extensive investment, expertise and effort required to perform a ChIP-seq experiment. However, whole genome analysis is continuing at a quickening pace, so one can expect data sets from additional cell types and tissue samples over the next decade.
Fortunately, genes of the ET pathway are expressed in a number of cell types that have already been studied. One cell line known to express ET-1 at high levels, the Human Umbilical Vein Endothelial Cell (HUVEC) line, has been examined using ChIP-seq. For example, histone modifications consistent with silenced genes, such as monomethylation of histone 3 lysine 9 (H3K9me1) and H3K27me3, are noticeably absent from the EDN1 locus in HUVECs (Fujita et al., 2011; Raney et al., 2011). Histone modifications, such as H3K4me1/2/3 and H3K36me3, are associated with active promoters, enhancers and genes undergoing active transcription. A massive ChIP-seq signal covering much of the EDN1 locus is observed for H3K4me3 (Figure 2). Interestingly, a gap is located in the immediate area of the transcription start site. This does not necessarily indicate a lack of histone modification, but more likely chromatin remodeling has excluded nucleosomes to make way for the assembly of the transcription pre-initiation complex. Acetylation of numerous lysines located in all the histone core proteins is commonly observed in areas of open chromatin and actively transcribed genes. The histone 3 acetyllysine 27 (H3K27Ac) ChIP-seq signal is widespread across the entire EDN1 locus. A very strong H3K4me1 signal, a histone modification that is typically abundant in enhancer elements (Ong and Corces, 2011), is seen −1.6 to −1.2 kbp upstream of the promoter and in an area of strong sequence conservation in HUVECs. Recently, Strait et al. (2010) showed nuclear factor of activated T-cells (NFAT) binds to two elements located −1563 bp and −1263 bp 5′ to the transcription start site in rat primary renal inner medullary collecting duct cell. The inner medullary collecting duct is the tissue of highest ET-1 expression in mammals (Kohan et al., 2011). Combining the whole genome evidence with the NFAT data suggest a strong enhancer active in cell types where ET-1 is expressed at very high levels.
Figure 2.
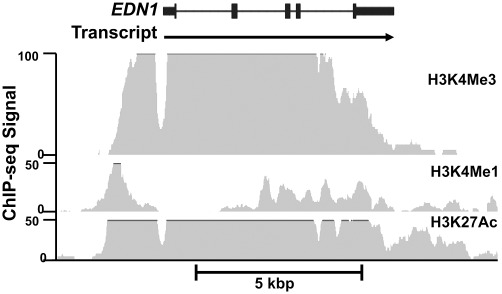
Selected histone modifications of the EDN1 locus in HUVEC. Histone markers associated with transcriptional activity of mammalian genes were determined in genome-wide studies of cultured HUVECs by the Broad Institute/Massachusetts General Hospital ENCODE consortium (Raney et al., 2011). The data were accessed and plotted using the UCSC Genome Browser (http://www.genome.ucsc.edu). Panels: EDN1 structure: the thin bar indicates intron sequence, the moderate bar shows exon sequence and the wide bar shows the preproendothelin open reading frame. Exon 2 contains the segment for mature ET-1. Transcript: the arrow shows the complete primary transcript. H3K4me3, H3K4me1 and H3K27ac; ChIP-seq signal strength for histone modifications plotted as a function of the human EDN1 gene sequence.
DNA methylation of a promoter is associated with transcriptionally silenced genes (Poetsch and Plass, 2011). Genome-wide studies have also provided information on the DNA methylation of the ET pathway genes. Maunakea et al. (2010) conducted an extensive survey of DNA methylation in human brain tissue. Evidence from that study suggests that EDN1 is transcriptionally active in the brain. Efficient digestion of the EDN1 gene with methylation-sensitive restriction endonucleases demonstrated hypomethylation at numerous sites within a 1.5 kb region surrounding the transcription start site. As expected, the DNA flanking the promoter region was not susceptible to methylation-sensitive restriction endonucleases and could be immunoprecipitated using an antibody to methylated DNA.
Epigenetic regulation of the EDN1 gene is cell and tissue specific. Dermal fibroblasts and chondrocytes that do not express ET-1 display methylation at multiple sites in the EDN1 promoter and first intron by bisulfite DNA sequence analysis (Vallender and Lahn, 2006). One of the areas of hypermethylation is within a putative Sp1 binding site and might be expected to affect binding of this common transcription factor. By comparison, a murine inner medullary collecting duct (mIMCD-3) cell line expresses ET-1 at very high levels and had substantial methylation at only two of the sites assayed in the same study. Another indicator of tissue specificity of epigenetic regulation can be found in histone modifications of the EDN1 gene. As discussed earlier, although the HUVEC line has histone modifications strongly indicating transcriptional activity (Figure 2), these histone modifications were not observed in a chronic myelogenous leukaemia cell line (K562). The K562 cells had a substantial level of H3K27me3 and little H3K27Ac suggesting repression of transcriptional activity. In support of this conclusion very little RNA polymerase II was found at the EDN1 locus in K562 cells.
Currently, direct evidence for changes in epigenetic regulation in response to physiological stimuli is very limited. Work performed in our laboratory showed that EDN1 is responsive to the mineralocorticoid hormone aldosterone (Stow et al., 2009). The mineralocorticoid and glucocorticoid receptors moved to the nucleus in an aldosterone concentration dependent manner. ChIP assays and DNA affinity purification assays showed the presence of a hormone response element functional within IMCD-3 cells. A substantial increase in H3K4me2 ChIP signal was seen upon exposure of cells to aldosterone, reflecting increased transcription of EDN1.
DNA methylation of EDNRB
The epigenetic mechanism studied in greatest detail among the ET pathway genes is DNA methylation of the 5′ regulatory region of the EDNRB gene. The human gene located at 13q22.3 is more complex than EDN1, in that it covers roughly 24 kbp. The primary transcript undergoes alternative splicing to produce three different receptor isoforms (Pruitt et al., 2009). Expression of the gene is dependent upon at least four different transcription start sites located at two distinct sites within the locus (Figure 3). The far upstream site is one of the rare examples of a functional mammalian promoter derived from an ancient insertion of a transposon (Medstrand et al., 2001; Landry and Mager, 2003). The regulation of this promoter can be expected to be unique, but has not been investigated. The other three transcription start sites are located within a 1200 bp segment of the 5′ regulatory region of EDNRB. An active promoter H3K4me3 signal can be seen in genome-wide ChIP-seq analyses of brain tissue (Maunakea et al., 2010). A highly CG-rich region, referred to as CpG island, is located −792 to +451 relative to the classically defined translation start site (Knight et al., 2009). The CpG island covers a portion of the promoter and extends into the first exon. Nelson et al. (1997) performed the seminal work on DNA methylation of the EDNRB regulatory region was performed in prostate cancer tissue about 15 years ago. This report spurred a series of publications describing EDNRB promoter methylation in normal tissue, tumours and tumour-derived cell lines.
Figure 3.
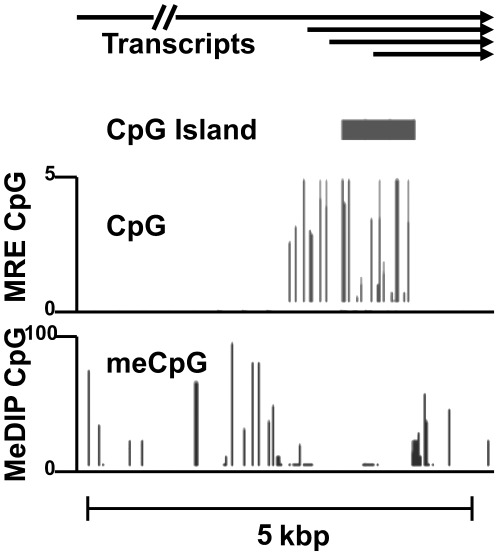
DNA methylation at the EDNRB promoter. Genome-wide CpG methylation data were collected from human brain tissue by Maunakea et al. (2010) and accessed using the UCSC Genome Browser (http://www.genome.ucsc.edu). Panels: Transcript: the arrows indicate the position of transcription initiation sites and direction of transcription for EDNRB. CpG island: the bar shows the extent of the CG rich element defined as a sequence of greater than 300 bp with a greater than 50% GC content and a 0.6 ratio of CG in comparison with the expected value CpG: unmethylated CpG sites (Gardiner-Garden and Frommer, 1987). MRE CpG: detected by digestion of genomic DNA with methylation-sensitive restriction endonucleases coupled to high throughput DNA sequence analysis. MeDIP CpG: position of methylated CpG sites indicated by high-throughput sequencing of DNA methylation-specific immunoprecipitates.
An extensive series of genome-wide microarray analyses found that eleven of 60 different tumour cell lines displayed a decrease in EDNRB expression (Ross et al., 2000). Individual cell lines derived from several melanomas, breast tumours and a single renal carcinoma all showed sharp drops in EDNRB mRNA. Studies directed at EDNRB also demonstrated reduced expression in cell lines from cancers of the nasopharynx, bladder and colon (Pao et al., 2001; Lo et al., 2002; Zhou et al., 2007). Knowledge of the reduced expression led to a search for the molecular mechanism. Early studies exhaustively mapped methylated CpG sites within the regulatory region at single base resolution by bisulfite sequencing (Pao et al., 2001; Lo et al., 2002). Comparison of data sets from several groups provided independent confirmation of heavy methylation at 16 CpG sites spread over 1 kbp. To illustrate the point, the extent of DNA methylation has been plotted for nine such sites (Figure 4) and CpG methylation was found to be at or close to 100% in many of the cancer cell lines. In contrast, normal control cell lines had little or no methylation. When the cancer cell lines were treated with a demethylating agent, 5′-aza-2′-deoxycytidine, the promoter region became hypomethylated and EDNRB expression was derepressed. The independently performed demethylation experiments directly linked transcription of EDNRB to the extent of DNA methylation at the promoter.
Figure 4.
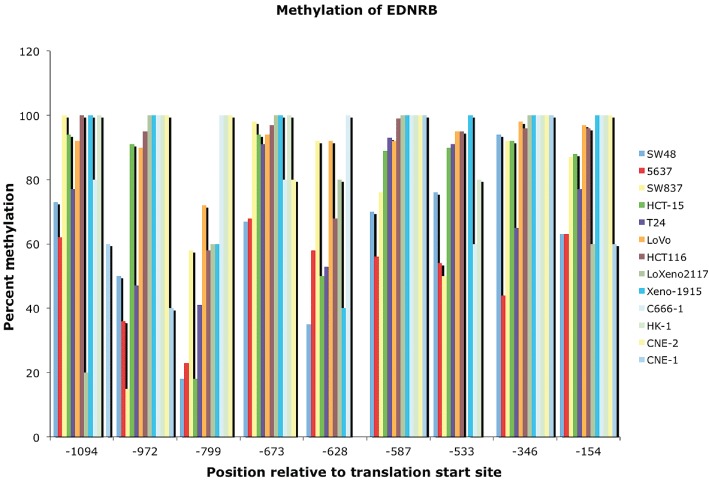
Methylated CpG site in tumour cell lines. Extent of methylation at nine specific sites in tumour and normal cells was assayed by bisulfite sequencing analysis (Pao et al., 2001; Lo et al., 2002). CpG sites are relative to the classical translational start site. The key for cell lines is on the right.
Identification of individual CpG sites methylated in tumour cells provided information needed for studying larger patient populations. For example, Knight et al. (2009) examined 24 CpG sites from 64 individuals with non-small cell lung cancer using pyrosequencing analysis. Greater than 50% of the patient tumour samples showed hypermethylation of the EDNRB promoter. An even higher percentage (>70%) of tissue samples from oral squamous cell carcinoma patients presented with increased DNA methylation in an experiment using the methylation-specific PCR (MS-PCR) approach (Kaur et al., 2010). Hypermethylation was also detected using MS-PCR in liver and prostate cancers (Jerónimo et al., 2003; Hsu et al., 2006; Rogers et al., 2006; Xie et al., 2007). As a result, DNA methylation of the EDNRB promoter is a candidate biomarker for prognosis and recurrence for several specific tumour types (Yates et al., 2007; Bastian et al., 2008; Ellinger et al., 2008; Demokan et al., 2010; Pattani et al., 2010; Vasiljevićet al., 2011). Recently, Viet et al. (2011) also demonstrated hypermethylation of the EDNRB promoter in oral squamous cell carcinoma using a state of the art microarray analysis. DNA samples were applied to a microarray and then analysed for methylation by a specialized mass spectrometry approach called MassARRAY (Sequenom, San Diego, CA, USA). This study presents a novel example of using overexpression of EDNRB to overcome a physiological phenotype that resulted from epigenetic silencing of the chromosomal gene. The EDNRB gene was reintroduced into a tumourigenic head and neck cancer cell line. Mice were inoculated with EDNRB expressing cells or negative control cells. Interestingly, the EDNRB positive animals showed an apparent decrease in tumour-related pain as compared with the control group.
Over the past decade it has become clear that DNA methylation of the EDNRB regulatory region is probably the predominant mechanism governing down-regulation of EDNRB. However, it should be noted that it is not the only mechanism for affecting EDNRB expression in cancer. A genetic mutation in EDNRB has been correlated to a familial predisposition for melanoma (Soufir et al., 2007).
Other ET pathway genes
If the information about epigenetic regulation of EDN1 is limited, even less is known about EDN2 (human 1p34.2) and EDN3 (human 20q13.32). EDN2 is expressed in the HUVEC line (O'Reilly et al., 1993), and the epigenetic marks detected by ChIP-seq indicate an active promoter. Human ET-2 has been shown to be involved in the immune response in intestinal mucosal cells, corpus luteum formation and ovulation (Takizawa et al., 2005; Klipper et al., 2010; Choi et al., 2011). Altered ET-2 expression has been observed in inflammatory bowel disease, breast cancer and hypertrophic cardiomyopathy (McCartney et al., 2002; Nagai et al., 2007). These tissues would be candidates for study of epigenetic markers governing expression of EDN2. Human EDN3 is known to be expressed in many tissues such as brain, lung and prostate. In breast cancer tissue samples expression of EDN3 is substantially reduced (Wiesmann et al., 2009). The down-regulation correlated to increased DNA methylation of the EDN3 gene
Like EDNRB, the human EDNRA gene (4q31.22) is complex. There are at least two transcription start sites and alternative splicing of the transcript gives rise to different receptor isoforms. The upstream promoter is equipped with a CpG island, but to date it has not been investigated beyond what is available in the whole genome databases. For example, the CpG island is unmethylated in brain tissue (Maunakea et al., 2010). A potential target for studying EDNRA epigenetic regulation is mammary tissue. The histone modifications seen in human mammary epithelial cells (HMEC) are indicative of high level expression (Fujita et al., 2011; Raney et al., 2011).
An illustration of the potential importance of epigenetic regulation can be seen with the human ET converting enzyme-1 gene, ECE1 (1p36.12). The gene has three authentic promoters spread across >50 kbp in the human genome (Schweizer et al., 1997; Valdenaire et al., 1999; Funke-Kaiser et al., 2000). Each gives rise to an mRNA yielding an active enzyme differing only in the extreme amino terminal primary sequence. Examination of H3K4me3 suggests that one, two or all three promoters can be active depending on cell type. Each of these promoters can be expected to be subject to differential epigenetic regulation providing a means for exquisitely sensitive regulation of ECE1 expression.
The future
Clearly epigenetic analysis of the ET pathway genes is in its infancy. The one area of substantial progress is the demonstration that epigenetic regulation acts as a central component in EDNRB expression in tumourigenesis. The marked change in DNA methylation of the CpG island and the dynamic nature of epigenetic mechanisms suggests that there are likely to be differences in histone modifications and histone variants at the EDNRB gene in pathological states.
First generation therapies targeting epigenetic regulation are currently in use for the treatment of specific tumours and more are in various phases of clinical trials. The general strategy is to reverse epigenetic markers in cancer cells by inhibiting histone deacetylases (HDACs) and DNA methyltransferases (DNMTs). Two DNMT inhibitors, 5-azacytidine and 2′-deoxy-5-azacytidine, have been approved for the treatment of myeloid leukaemia. Several HDAC inhibitors have also been approved for the treatment for a variety of cancers. For example, Istodax is a broad-range HDAC inhibitor approved by the FDA for the treatment of head and neck cancer among others (Mund and Lyko, 2010). Another pan-HDAC inhibitor, vorinostat, is approved for use in cutaneous T-cell lymphoma. Moreover, it is also being studied in phase I and II clinical trials for combination therapy to treat a variety of cancers including, but not limited to acute myelogenous leukaemia, colorectal cancer, non-squamous cell lung carcinoma and breast cancer (Wagner et al., 2010). It would be interesting to monitor the level of EDNRB expression and action of the ET pathway in response to such drug regimens.
Epigenetics dictates expression of the ET pathway genes in a tissue-specific manner. One of the current challenges is to define the epigenetic markers in normal tissues where the ET pathway is functional. In pathological states one can expect to see altered epigenetic modification of the genes. For example, one might reasonably expect to see differences in epigenetic markers between normal pulmonary vascular tissue and samples taken from individuals with pulmonary arterial hypertension. Another fallow area is the consideration of histone variants and non-coding RNAs (ncRNAs) in relation to the ET pathway genes. Histone variant H2A.Z is often located at the boundaries of DNA segments devoid of histones (Draker and Cheung, 2009). It would be informative to look for this histone variant at the gap in the H3K4me1 signal at the EDN1 locus in HUVECs. Finally, DNA methylation, ncRNAs, histone modification and variants are not isolated events, but act in concert with one another to control gene activity. Consideration of the interplay between the epigenetic mechanisms will be necessary to achieve a complete understanding of regulation of the ET pathway in normal and pathological states.
Acknowledgments
This work was supported by the US Public Health Service Grant RO1 DK 82680 to CSW and BDC. MEJ was supported by a pre-doctoral appointment on T32 DK 076541.
Glossary
- ChIP
chromatin immunoprecipitation
- ChIP-seq
chromatin immunoprecipitation sequencing
- CpG
DNA sequence CG
- DNMT
DNA methyltransferase
- ET
endothelin
- ETA receptor
endothelin receptor type A
- ETB receptor
endothelin receptor type B
- H3K27Ac
histone 3 acetyllysine 27
- H3K9me1
histone 3 monomethyllysine 9
- HDAC
histone deacetylase
- HMEC
human mammary epithelial cell
- HUVEC
human umbilical vein endothelial cell
- K562
chronic myelogenous leukaemia
- MS-PCR
methylation-specific PCR
- ncRNA
non-coding RNA
- NFAT
nuclear factor of activated T-cells
Conflict of interest
The authors have no conflict of interest.
References
- Arai H, Nakao K, Takaya K, Hosoda K, Ogawa Y, Nakanishi S, et al. The human endothelin-B receptor gene. Structural organization and chromosomal assignment. J Biol Chem. 1993;268:3463–3470. [PubMed] [Google Scholar]
- Bagnato A, Loizidou M, Pflug B, Curwen J, Growcott J. Role of the endothelin axis and its antagonists in the treatment of cancer. Br J Pharmacol. 2011;163:220–233. doi: 10.1111/j.1476-5381.2011.01217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton M, Yanagisawa M. Endothelin: 20 years from discovery to therapy. Can J Physiol Pharmacol. 2008;86:485–498. doi: 10.1139/Y08-059. [DOI] [PubMed] [Google Scholar]
- Bastian PJ, Palapattu GS, Yegnasubramanian S, Rogers CG, Lin X, Mangold LA, et al. CpG island hypermethylation profile in the serum of men with clinically localized and hormone refractory metastatic prostate cancer. J Urol. 2008;179:529–534. doi: 10.1016/j.juro.2007.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasquillo MM, McCallion AS, Puffenberger EG, Kashuk CS, Nouri N, Chakravarti A. Genome-wide association study and mouse model identify interaction between RET and EDNRB pathways in Hirschsprung disease. Nat Genet. 2002;32:237–244. doi: 10.1038/ng998. [DOI] [PubMed] [Google Scholar]
- Choi D-H, Kim EK, Kim K-H, Lee K-A, Kang D-W, Kim HY, et al. Expression pattern of endothelin system components and localization of smooth muscle cells in the human pre-ovulatory follicle. Hum Reprod. 2011;26:1171–1180. doi: 10.1093/humrep/der066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clamp M, Fry B, Kamal M, Xie X, Cuff J, Lin MF, et al. Distinguishing protein-coding and noncoding genes in the human genome. Proc Natl Acad Sci U S A. 2007;104:19428–19433. doi: 10.1073/pnas.0709013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demokan S, Chang X, Chuang A, Mydlarz WK, Kaur J, Huang P, et al. KIF1A and EDNRB are differentially methylated in primary HNSCC and salivary rinses. Int J Cancer. 2010;127:2351–2359. doi: 10.1002/ijc.25248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draker R, Cheung P. Transcriptional and epigenetic functions of histone variant H2AZ. Biochem Cell Biol. 2009;87:19–25. doi: 10.1139/O08-117. [DOI] [PubMed] [Google Scholar]
- Ellinger J, Bastian PJ, Jurgan T, Biermann K, Kahl P, Heukamp LC, et al. CpG island hypermethylation at multiple gene sites in diagnosis and prognosis of prostate cancer. Urology. 2008;71:161–167. doi: 10.1016/j.urology.2007.09.056. [DOI] [PubMed] [Google Scholar]
- Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funke-Kaiser H, Bolbrinker J, Theis S, Lemmer J, Richter CM, Paul M, et al. Characterization of the c-specific promoter of the gene encoding human endothelin-converting enzyme-1 (ECE-1) FEBS Lett. 2000;466:310–316. doi: 10.1016/s0014-5793(00)01086-3. [DOI] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Hsu L-S, Lee H-C, Chau G-Y, Yin P-H, Chi C-W, Lui WY. Aberrant methylation of EDNRB and p16 genes in hepatocellular carcinoma (HCC) in Taiwan. Oncol Rep. 2006;15:507–511. [PubMed] [Google Scholar]
- Huang Z, Desper R, Schäffer AA, Yin Z, Li X, Yao K. Construction of tree models for pathogenesis of nasopharyngeal carcinoma. Genes Chromosomes Cancer. 2004;40:307–315. doi: 10.1002/gcc.20036. [DOI] [PubMed] [Google Scholar]
- Jerónimo C, Henrique R, Campos PF, Oliveira J, Caballero OL, Lopes C, et al. Endothelin B receptor gene hypermethylation in prostate adenocarcinoma. J Clin Pathol. 2003;56:52–55. doi: 10.1136/jcp.56.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Kaur J, Demokan S, Tripathi SC, Macha MA, Begum S, Califano JA, et al. Promoter hypermethylation in Indian primary oral squamous cell carcinoma. Int J Cancer. 2010;127:2367–2373. doi: 10.1002/ijc.25377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klipper E, Levit A, Mastich Y, Berisha B, Schams D, Meidan R. Induction of endothelin-2 expression by luteinizing hormone and hypoxia: possible role in bovine corpus luteum formation. Endocrinology. 2010;151:1914–1922. doi: 10.1210/en.2009-0767. [DOI] [PubMed] [Google Scholar]
- Knight LJ, Burrage J, Bujac SR, Haggerty C, Graham A, Gibson NJ, et al. Epigenetic silencing of the endothelin-B receptor gene in non-small cell lung cancer. Int J Oncol. 2009;34:465–471. [PubMed] [Google Scholar]
- Kohan DE, Rossi NF, Inscho EW, Pollock DM. Regulation of blood pressure and salt homeostasis by endothelin. Physiol Rev. 2011;91:1–77. doi: 10.1152/physrev.00060.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry JR, Mager DL. Functional analysis of the endogenous retroviral promoter of the human endothelin B receptor gene. J Virol. 2003;77:7459–7466. doi: 10.1128/JVI.77.13.7459-7466.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo KW, Tsang YS, Kwong J, To KF, Teo PML, Huang DP. Promoter hypermethylation of the EDNRB gene in nasopharyngeal carcinoma. Int J Cancer. 2002;98:651–655. doi: 10.1002/ijc.10271. [DOI] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet. 2010;11:285–296. doi: 10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D'Souza C, Fouse SD, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney SA, Ballinger AB, Vojnovic I, Farthing MJG, Warner TD. Endothelin in human inflammatory bowel disease: comparison to rat trinitrobenzenesulphonic acid-induced colitis. Life Sci. 2002;71:1893–1904. doi: 10.1016/s0024-3205(02)01923-9. [DOI] [PubMed] [Google Scholar]
- Medstrand P, Landry JR, Mager DL. Long terminal repeats are used as alternative promoters for the endothelin B receptor and apolipoprotein C-I genes in humans. J Biol Chem. 2001;276:1896–1903. doi: 10.1074/jbc.M006557200. [DOI] [PubMed] [Google Scholar]
- Mund C, Lyko F. Epigenetic cancer therapy: proof of concept and remaining challenges. Bioessays. 2010;32:949–957. doi: 10.1002/bies.201000061. [DOI] [PubMed] [Google Scholar]
- Nagai T, Ogimoto A, Okayama H, Ohtsuka T, Shigematsu Y, Hamada M, et al. A985G polymorphism of the endothelin-2 gene and atrial fibrillation in patients with hypertrophic cardiomyopathy. Circ J. 2007;71:1932–1936. doi: 10.1253/circj.71.1932. [DOI] [PubMed] [Google Scholar]
- Nelson J, Lee WH, Nguyen SH, Jarrard DF, Brooks JD, Magnuson SR, et al. Methylation of the 5′ CpG island of the endothelin B receptor gene is common in human prostate cancer. Cancer Res. 1997;57:35–37. [PubMed] [Google Scholar]
- Nelson J, Bagnato A, Battistini B, Nisen P. The endothelin axis: emerging role in cancer. Nat Rev Cancer. 2003;3:110–116. doi: 10.1038/nrc990. [DOI] [PubMed] [Google Scholar]
- Ong CT, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet. 2011;12:283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly G, Charnock-Jones DS, Cameron IT, Smith SK, Davenport AP. Endothelin-2 mRNA splice variants detected by RT-PCR in cultured human vascular smooth muscle and endothelial cells. J Cardiovasc Pharmacol. 1993;22(Suppl. 8):S18–S21. doi: 10.1097/00005344-199322008-00007. [DOI] [PubMed] [Google Scholar]
- Pao MM, Tsutsumi M, Liang G, Uzvolgyi E, Gonzales FA, Jones PA. The endothelin receptor B (EDNRB) promoter displays heterogeneous site specific methylation patterns in normal and tumor cells. Hum Mol Genet. 2001;10:903–910. doi: 10.1093/hmg/10.9.903. [DOI] [PubMed] [Google Scholar]
- Pattani KM, Zhang Z, Demokan S, Glazer C, Loyo M, Goodman S, et al. Endothelin receptor type B gene promoter hypermethylation in salivary rinses is independently associated with risk of oral cavity cancer and premalignancy. Cancer Prev Res (Phila) 2010;3:1093–1103. doi: 10.1158/1940-6207.CAPR-10-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Salzberg SL. Between a chicken and a grape: estimating the number of human genes. Genome Biol. 2010;11:206–212. doi: 10.1186/gb-2010-11-5-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pingault V, Ente D, Dastot-Le Moal F, Goossens M, Marlin S, Bondurand N. Review and update of mutations causing Waardenburg syndrome. Hum Mutat. 2010;31:391–406. doi: 10.1002/humu.21211. [DOI] [PubMed] [Google Scholar]
- Poetsch AR, Plass C. Transcriptional regulation by DNA methylation. Cancer Treat Rev. 2011;37(Suppl. 1):S8–S12. doi: 10.1016/j.ctrv.2011.04.010. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status policy and new initiatives. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puffenberger EG, Hosoda K, Washington SS, Nakao K, deWit D, Yanagisawa M, et al. A missense mutation of the endothelin-B receptor gene in multigenic Hirschsprung's disease. Cell. 1994;79:1257–1266. doi: 10.1016/0092-8674(94)90016-7. [DOI] [PubMed] [Google Scholar]
- Raney BJ, Cline MS, Rosenbloom KR, Dreszer TR, Learned K, Barber GP, et al. ENCODE whole-genome data in the UCSC genome browser (2011 update) Nucleic Acids Res. 2011;39:D871–D875. doi: 10.1093/nar/gkq1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Pascual F, Redondo-Horcajo M, Lamas S. Functional cooperation between Smad proteins and activator protein-1 regulates transforming growth factor-beta-mediated induction of endothelin-1 expression. Circ Res. 2003;92:1288–1295. doi: 10.1161/01.RES.0000078491.79697.7F. [DOI] [PubMed] [Google Scholar]
- Rogers CG, Gonzalgo ML, Yan G, Bastian PJ, Chan DY, Nelson WG, et al. High concordance of gene methylation in post-digital rectal examination and post-biopsy urine samples for prostate cancer detection. J Urol. 2006;176:2280–2284. doi: 10.1016/j.juro.2006.07.047. [DOI] [PubMed] [Google Scholar]
- Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- Schweizer A, Valdenaire O, Nelböck P, Deuschle U, Dumas Milne Edwards JB, Stumpf JG, et al. Human endothelin-converting enzyme (ECE-1): three isoforms with distinct subcellular localizations. Biochem J. 1997;328:871–877. doi: 10.1042/bj3280871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufir N, Ollivaud L, Bertrand G, Lacapère JJ, Descamps V, Vitoux D, et al. A French CDK4-positive melanoma family with a co-inherited EDNRB mutation. J Dermatol Sci. 2007;46:61–64. doi: 10.1016/j.jdermsci.2006.11.016. [DOI] [PubMed] [Google Scholar]
- Stow LR, Gumz ML, Lynch IJ, Greenlee MM, Rudin A, Cain BD, et al. Aldosterone modulates steroid receptor binding to the endothelin-1 gene (edn1) J Biol Chem. 2009;284:30087–30096. doi: 10.1074/jbc.M109.030718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stow LR, Jacobs ME, Wingo CS, Cain BD. Endothelin-1 gene regulation. FASEB J. 2011;25:16–28. doi: 10.1096/fj.10-161612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strait KA, Stricklett PK, Kohan RM, Kohan DE. Identification of two nuclear factor of activated T-cells (NFAT)-response elements in the 5′-upstream regulatory region of the ET-1 promoter. J Biol Chem. 2010;285:28520–28528. doi: 10.1074/jbc.M110.153189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa S, Uchide T, Adur J, Kozakai T, Kotake-Nara E, Quan J, et al. Differential expression of endothelin-2 along the mouse intestinal tract. J Mol Endocrinol. 2005;35:201–209. doi: 10.1677/jme.1.01787. [DOI] [PubMed] [Google Scholar]
- Valdenaire O, Lepailleur-Enouf D, Egidy G, Thouard A, Barret A, Vranckx R, et al. A fourth isoform of endothelin-converting enzyme (ECE-1) is generated from an additional promoter molecular cloning and characterization. Eur J Biochem. 1999;264:341–349. doi: 10.1046/j.1432-1327.1999.00602.x. [DOI] [PubMed] [Google Scholar]
- Vallender TW, Lahn BT. Localized methylation in the key regulator gene endothelin-1 is associated with cell type-specific transcriptional silencing. FEBS Lett. 2006;580:4560–4566. doi: 10.1016/j.febslet.2006.07.017. [DOI] [PubMed] [Google Scholar]
- Vasiljević N, Wu K, Brentnall AR, Kim DC, Thorat MA, Kudahetti SC, et al. Absolute quantitation of DNA methylation of 28 candidate genes in prostate cancer using pyrosequencing. Dis Markers. 2011;30:151–161. doi: 10.3233/DMA-2011-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viet CT, Ye Y, Dang D, Lam DK, Achdjian S, Zhang J, et al. Re-expression of the methylated EDNRB gene in oral squamous cell carcinoma attenuates cancer-induced pain. Pain. 2011;152:2323–2332. doi: 10.1016/j.pain.2011.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner JM, Hackanson B, Lübbert M, Jung M. Histone deacetylase (HDAC) inhibitors in recent clinical trials for cancer therapy. Clin Epigenetics. 2010;1:117–136. doi: 10.1007/s13148-010-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesmann F, Veeck J, Galm O, Hartmann A, Esteller M, Knüchel R, et al. Frequent loss of endothelin-3 (EDN3) expression due to epigenetic inactivation in human breast cancer. Breast Cancer Res. 2009;11:R34. doi: 10.1186/bcr2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie CH, Naito A, Mizumachi T, Evans TT, Douglas MG, Cooney CA, et al. Mitochondrial regulation of cancer associated nuclear DNA methylation. Biochem Biophys Res Commun. 2007;364:656–661. doi: 10.1016/j.bbrc.2007.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita K, Discher DJ, Hu J, Bishopric NH, Webster KA. Molecular regulation of the endothelin-1 gene by hypoxia. Contributions of hypoxia-inducible factor-1 activator protein-1 GATA-2 AND p300/CBP. J Biol Chem. 2001;276:12645–12653. doi: 10.1074/jbc.M011344200. [DOI] [PubMed] [Google Scholar]
- Yates DR, Rehman I, Abbod MF, Meuth M, Cross SS, Linkens DA, et al. Promoter hypermethylation identifies progression risk in bladder cancer. Clin Cancer Res. 2007;13:2046–2053. doi: 10.1158/1078-0432.CCR-06-2476. [DOI] [PubMed] [Google Scholar]
- Zhou L, Feng X, Shan W, Zhou W, Liu W, Wang L, et al. Epigenetic and genetic alterations of the EDNRB gene in nasopharyngeal carcinoma. Oncology. 2007;72:357–363. doi: 10.1159/000113146. [DOI] [PubMed] [Google Scholar]


