Abstract
Iron is vital for almost all organisms because of its ability to donate and accept electrons with relative ease. It serves as a cofactor for many proteins and enzymes necessary for oxygen and energy metabolism, as well as for several other essential processes. Mammalian cells utilize multiple mechanisms to acquire iron. Disruption of iron homeostasis is associated with various human diseases: iron deficiency resulting from defects in acquisition or distribution of the metal causes anemia; whereas iron surfeit resulting from excessive iron absorption or defective utilization causes abnormal tissue iron deposition, leading to oxidative damage. Mammals utilize distinct mechanisms to regulate iron homeostasis at the systemic and cellular levels. These involve the hormone hepcidin and iron regulatory proteins, which collectively ensure iron balance. This review outlines recent advances in iron regulatory pathways, as well as in mechanisms underlying intracellular iron trafficking, an important but less-studied area of mammalian iron homeostasis.
Importance of iron in mammalian physiology
Iron is a d-block transition metal and like many such metals it can assume several oxidation states. The most common species are the divalent ferrous [(Fe2+)] and the trivalent ferric [(Fe3+)] iron. The redox potential of iron can be greatly modulated by the nature of attached ligands. This has significant physiological ramifications since other oxidation states, such as ferryl [(Fe4+)], can be transiently generated as key intermediates during metal-mediated oxidative transformations. Other transition elements such as copper and manganese are likewise capable of participating in biological redox reactions (1). Nevertheless, during evolution, organisms likely selected iron for following reasons: 1) Iron is the second most abundant metal on earth’s crust, falling closely behind aluminum; 2) Iron can exist in multiple oxidation states, which is essential for electron transfer, and for binding to biological ligands; 3) Iron’s redox potential ranges from +1000 mV to −550 mV, depending on the ligand environment, whereas the range for other transition elements is narrower; 4) By exploiting the oxidation states, electron spin state and redox potential, biological systems can adjust the chemical reactivity of iron to suit physiological needs (1, 2).
The indispensability of iron for living organisms is exemplified by the fact that it serves as a cofactor for several hemoproteins and non-heme iron-containing proteins, including many enzymes. Hemoproteins are involved in numerous biological functions such as oxygen binding and transport (hemoglobins), oxygen metabolism (catalases, peroxidases), cellular respiration and electron transport (cytochromes). Proteins containing non-heme iron are important for fundamental cellular processes such as DNA synthesis, cell proliferation and differentiation (ribonucleotide reductase), gene regulation, drug metabolism, and steroid synthesis. Early in evolution, before oxygen emerged as an abundant constituent of the atmosphere, anaerobic cells acquired soluble ferrous iron with relative ease (3). Later on, accumulation of oxygen oxidized ferrous to ferric iron, which is virtually insoluble at physiological pH. Moreover, the redox cycling of ferrous and ferric iron in the presence of H2O2 and O2·−, which are physiologically produced during aerobic respiration and enzymatic reactions, yields hydroxyl radicals (Fenton chemistry). These in turn readily attack and damage cellular macromolecules. Thus, despite its abundance, the acquisition and transport of iron poses a challenge for cells and organisms due to its low solubility and high toxicity. To overcome these problems, unicellular organisms such as bacteria synthesize “siderophores”, which are low molecular weight, iron-specific chelating agents that capture extracellular iron and transfer it into the cell (4, 5). In contrast, mammalian cells acquire iron from extracellular carrier proteins (6, 7).
Systemic Iron metabolism
Distribution of iron in the body
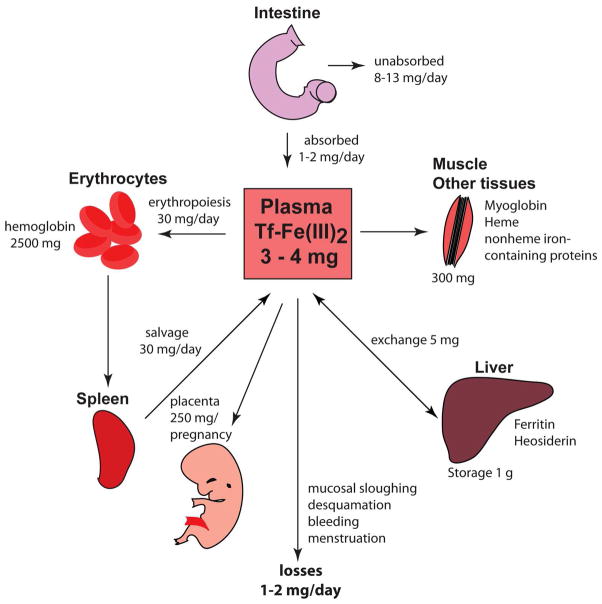
The human body contains ~3–5 g of iron (6). Most of it is present as heme in hemoglobin of erythroid cells (>2 g) or myoglobin of muscles (~300 mg). Macrophages in the spleen, liver and bone marrow maintain a transient fraction of iron (~600 mg), while excess of the metal is stored in the liver parenchyma within ferritin (~1000 mg). All other cellular iron-containing proteins and enzymes are estimated to bind a total of ~8 mg of iron (ref. 7; Fig. 1).
Fig. 1.
Iron absorption, distribution, and recycling in the body and quantitative exchange of iron between body iron sources. Body iron levels are maintained by daily absorption of ~1–2 mg of dietary iron to account for obligatory losses of a similar amount of iron through sloughing of mucosal and skin cells, hemorrhage, and other losses. Approximately 4 mg of iron is found in circulation bound to Tf, which accounts for 0.1% of the total body iron. Majority of the body iron is found in the erythroid compartment of bone marrow and in mature erythrocytes contained within the heme moiety of the hemoglobin. Splenic reticuloendothelial macrophages, which recycle iron from senescent red blood cells, provide iron for the new red blood cell synthesis. Tf delivers iron to developing erythroid precursors, as well as to other sites of iron utilization. Liver hepatocytes store iron in ferritin shells. During pregnancy, 250 mg of iron is transported across the placenta to the fetus. The distribution of iron in the body is altered in iron deficiency and iron overload (see text).
Iron is delivered to erythroblasts and to most tissues via circulating transferrin (Tf), which carries ~3 mg of the metal at steady state. Considering that Tf-bound iron turns over >10 times a day, it represents the most dynamic body iron pool. Plasma iron is predominantly replenished by reticuloendothelial macrophages, and to a small extent (~1–2 mg/day) by absorption from the diet, mediated by duodenal enterocytes. The macrophages acquire iron primarily via erythrophagocytosis, and the enterocytes by internalization of heme or inorganic iron from the intestinal lumen (8).
The absorption of inorganic iron involves reduction of Fe3+ to Fe2+ by ascorbate and/or membrane associated ferrireductases such as duodenal cytochrome B (DcytB; ref. 9), coupled to transport of Fe2+ across the apical membrane by divalent metal transporter 1 (DMT1/SLC11A2, solute carrier family 11 member 2, also known as natural resistance-associated macrophage protein 2 (NRAMP2) or divalent cation transporter (DCT1; refs. 6 & 7). The mechanism of heme internalization remains poorly defined and may involve either direct transport of heme or receptor-mediated endocytosis (10). Macrophages and enterocytes catabolize heme in a reaction catalyzed by heme oxygenases-1 and -2 (HO-1 and HO-2), which liberate inorganic iron (11). Both cell types export Fe2+ to plasma via the transmembrane transporter, ferroportin/SLC40A1 (6, 7), in a process coupled by the re-oxidation of Fe2+ to Fe3+. This is mediated by the circulating ferroxidase ceruloplasmin or its homologue, hephaestin, which is expressed on the basolateral membrane of duodenal enterocytes, and physically interacts with ferroportin (6, 7).
Exported iron is scavenged by plasma Tf, which maintains it in a redox inert state and delivers it to tissues. The loading of apo-Tf with iron may be facilitated by gastrins. These peptide hormones stimulate the secretion of gastric acid and have also been proposed to act as transient Fe3+ chaperones (12). Tf contains two ferric binding sites and is only partially (30%) saturated with iron under physiological conditions. The concentration of differic Tf in plasma is ~5 μmol/L, corresponding to approximately one tenth of total circulating Tf (13). The high abundance of unsaturated apo-Tf allows an efficient buffering of increased plasma iron levels and prevents the build-up of non-transferrin bound iron (NTBI), which is taken up by tissue parenchymal cells and promotes oxidative injury (14). NTBI is generated in iron overload states, such as in hereditary hemochromatosis, where transferrin gradually becomes fully saturated with iron and loses its buffering capacity.
Regulation of body iron metabolism
Hormonal regulation by hepcidin
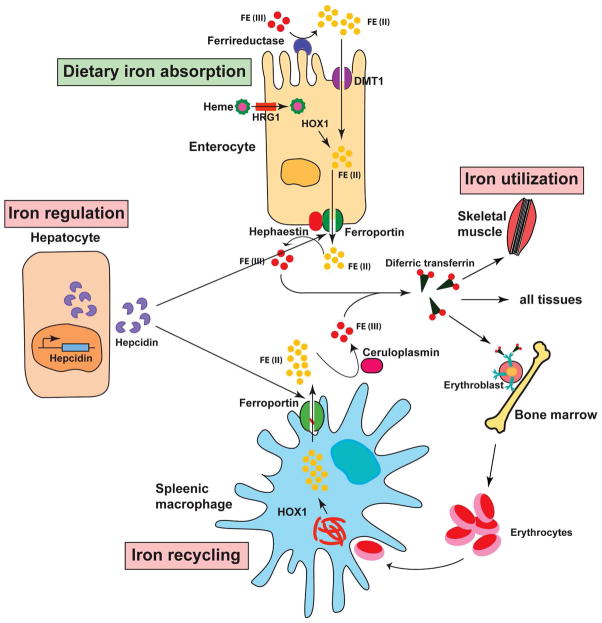
The trafficking of iron into plasma is largely controlled by the iron regulatory hormone hepcidin, which was first purified from plasma (15) and urine (16) on the basis of its antimicrobial activity. This liver-derived peptide binds to ferroportin on the plasma membrane of enterocytes, macrophages, hepatocytes and other cells, promoting its Jak-dependent phosphorylation and internalization that leads to its lysosomal degradation (17, 18). Ferroportin is the only exporter of inorganic iron in mammalian cells. Therefore inactivation of ferroportin causes intracellular iron retention (19, 20). Hepcidin has also been shown to promote proteasomal degradation of DMT1 (19). Pathological overexpression of hepcidin under prolonged inflammatory conditions is associated with the anemia of chronic disease, which is characterized by diversion of iron to storage sites, hypoferremia and reduced iron availability for erythropoiesis (ref. 20; also see Fig. 2). Even more severe iron redistribution occurs in patients with hereditary iron-refractory iron deficiency anemia (IRIDA), a disease caused by genetic disruption of the TMPRSS6 gene encoding the transmembrane serine protease matriptase-2 that negatively regulates hepcidin expression (21).
Fig. 2.
Regulation of systemic iron metabolism. Organs and cell types involved in systemic iron balance are shown. Duodenal enterocytes absorb dietary iron via DMT1 located on the apical surface upon reduction of Fe3+ to Fe2+ by DcytB. Spleenic reticuloendothelial macrophages recycle iron from senescent red blood cells. Both cell types release iron via ferroportin with the aid of hephaestin, which oxidizes Fe2+ to Fe3+. Iron is also oxidized by ceruloplasmin in the circulation. Plasma Tf captures and circulates iron in the body. Hepatic hormone, hepcidin regulates iron efflux from these cells by regulating the stability of ferroportin. Synthesis and secretion of hepcidin by hepatocytes is influenced by iron levels in the body as well as conditions that affect iron metabolism indirectly such as inflammation, ER stress, erythropoiesis, and hypoxia (see text for additional details).
Conversely, hepcidin deficiency, or in rare cases ferroportin resistance to hepcidin, are hallmarks of hemochromatosis, which is associated with uncontrolled dietary iron absorption and progressive tissue iron overload (22, 23). Patients with uncommon nonsense mutations in the HAMP (ref. 24) or high iron gene 2 (HFE2; ref. 25) genes, encoding hepcidin or its upstream activator hemojuvelin (HJV), respectively, develop early onset juvenile hemochromatosis (JH). The most frequent type of hemochromatosis is associated with mutations in the HFE gene (26), which leads to milder hepcidin insufficiency (27). Other types of hemochromatosis are caused by inactivation of the TFR2 gene encoding transferrin receptor 2 (TfR2; ref. 28), or by specific mutations in the SLC40A1 gene encoding ferroportin that prevent the binding of hepcidin (29, 30). The former is associated with inappropriately low hepcidin expression and the latter with hepcidin resistance (29, 30).
Hepcidin is generated in hepatocytes and other cell types, as a precursor propeptide of 84 amino acids (31). Following cleavage by the prohormone convertase furin (32), a biologically active mature 25-mer is secreted to plasma where it is thought to interact with α2-macroglobulin (33). The expression of hepcidin is regulated at the transcriptional level. C/EBPα (CCAAT enhancer-binding protein α) binds to a CCAAT motif within the HAMP promoter and maintains basal transcriptional activity (34). Alk3, a type I bone morphogenetic protein (BMP) receptor, also appears to be critical for basal hepcidin expression (35). Hepcidin transcription is induced by iron, inflammation or ER stress, and is inhibited by iron deficiency, increased erythropoietic drive or hypoxia (reviewed in ref. 22).
The responsiveness of hepcidin expression to iron was established during its initial discovery as an iron-induced peptide (22). Subsequently, oral iron administration was found to increase hepcidin mRNA in humans and mice (22). Nevertheless, hemochromatosis patients exhibit impaired hepcidin responses to iron ingestion (36, 37), associated with low hepcidin expression, disproportional to their iron load (27, 38). Likewise, mouse models of hemochromatosis demonstrate hepcidin insufficiency (39–41). There is evidence that hepatic and circulating iron induce hepcidin expression by distinct pathways (42, 43).
Regulation of hepcidin by hepatic iron
An increase in hepatic iron levels activates hepcidin transcription via the BMP signaling pathway (44). The binding of BMP6 and other BMP ligands to type I (Alk2 and Alk3) and type II BMP receptors promotes the phosphorylation of SMAD1, SMAD5 and SMAD8 proteins (35). The phosphorylated SMAD1/5/8 complex recruits SMAD4 and translocates to the nucleus, where it stimulates hepcidin transcription upon binding to BMP responsive elements (BMP-RE1 and BMP-RE2) at proximal and distal sites of the HAMP promoter (45). The liver-specific disruption of Alk2, Alk3 (35) or SMAD4 (46) in mice inhibits iron-induced hepcidin expression and leads to iron overload (22). Induction of inhibitory SMAD7, another target of BMP signaling, antagonizes the interaction of phosphorylated SMAD1/5/8 with SMAD4 and reduces hepcidin transcription in a feedback regulatory loop (47).
The expression of BMP6 is induced by hepatic iron and its mRNA levels reflect hepatic iron load (42). Liver-specific Bmp6−/− mice develop iron overload and fail to upregulate hepcidin (48, 49). Other endogenous BMPs do not appear capable of rescuing this phenotype, despite the capacity of BMP2, 5, 7 and 9 to activate hepcidin expression in cell culture experiments (44, 46, 50). Mice injected with recombinant BMP6 manifest increased hepcidin expression and hypoferremia (49). Conversely, a neutralizing BMP6 antibody (49) or the BMP inhibitor dorsomorphin (51) can reduce hepcidin expression and trigger an increase in serum iron levels. These data highlight a function of BMP6 as the principal upstream regulator of hepcidin.
Hjv is a BMP co-receptor that plays an essential role in iron-induced activation of hepcidin by forming complexes with type I and II BMP receptors, which enhances downstream signaling (44). It is primarily expressed in hepatocytes and skeletal muscle cells and associates with the plasma membrane via a C-terminal glycosylphosphatidylinositol (GPI) anchor. Soluble forms of Hjv have been detected in plasma (52), while recombinant soluble Hjv competes BMP signaling to hepcidin in vitro and in vivo (50, 52). Nevertheless, plasma Hjv seems unlikely to be of physiological importance for hepcidin regulation. Thus, mice with liver-specific disruption of Hjv develop quantitatively similar defects in hepcidin expression and iron overload as full Hjv−/− counterparts, while muscle-specific Hjv−/− animals exhibit normal iron homeostasis (53, 54; also see Table 1). These findings underline the role of hepatic Hjv as a major regulator of hepcidin.
Table 1.
Regulators of systemic iron metabolism: Animal models and human disorders.
| Metabolic Process | Protein | Gene | Function | Mouse model | Phenotype | Human disease | Refs |
|---|---|---|---|---|---|---|---|
| Iron absorption/recycling | Hepcidin (HAMP) | Hamp | Regulates intestinal iron absorption by controlling Ferroportin | Hamp−/− | Iron overload, no hepcidin | Juvenile hemochromatosis (HH type 2 B) | 24 |
| Transferrin receptor 2 (TfR2) | TfR2 | Sensor for diferric Tf; regulator of hepcidin | TfR2−/− | Increased iron absorption; Iron overload, decreased hepcidin | Hemochromatos is (HH type 3) | 61 | |
| SMAD4 | Smad4 | Regulates hepcidin expression | Smad4−/− | Iron overload; decreased hepcidin | Colorectal and pancreatic cancer predisposition | 44, 50 | |
| Bone morphogenic protein 6 (BMP6) | Bmp6 | Regulates hepcidin expression | Bmp6−/− | Iron overload; decreased hepcidin | Rare cases of hereditary hemochromatosis | 44, 50 | |
| Mariptase-2 (TMPRSS6) | Tmprs6 | Regulates hemojuvelin |
Tmprss6 msk/msk Tmprss6−/− |
Alopecia, microcytic anemia, infertility | Microcytic anemia; low serum iron; increased hepcidin | 21, 56, 57 | |
| Ferroportin (SLC40A1; MTP1) | Slc40a1 | Regulates iron efflux from enterocytes | Slc40a1−/− | Embryonic lethality; conditional deficiency – iron deficiency | Hemochromatos is, macrophage iron overload (HH type 4) | 214 | |
| Hephaestin (HEPH) | Heph | Ferroxidase, functions in concert with Fp |
Sla Heph−/− |
Microcytic anemia, iron deficiency | Unknown | 213 | |
| Ceruloplasmin (CP) | Cp | Plasma ferroxidase; closely related to Heph | Cp−/− | Iron overload, anemia, impaired motor coordination | Aceruloplasminemia | 213 | |
| High iron gene (HFE) | HFE | Regulator of HAMP; interacts with TfR1 and TfR2 | HFE−/− | Iron overload, decreased hepcidin | HLA-linked hemochromatosis (HH type 1) | 7.26, 61 | |
| Hemojuvelin (HJV) | Hjv | Bone morphogenic protein coreceptor | Hjv−/− | Iron overload, decreased hepcidin | Juvenile hemochromatosis (HH type 2 A) | 25, 53, 54 | |
| Divalent metal transporter 1 |
DMT1 Nramp1 Slc11a2 |
Transmembrane iron transporter |
mk/mk DMT−/− |
Hypochromic, microcytic anemia; impaired iron absorption | Iron overload in liver; anemia | 105, 108, 109 |
Biochemical data suggest that matriptase-2 negatively regulates hepcidin by proteolytic inactivation of Hjv (55). Nevertheless, Tmprss6−/− mice, a model of IRIDA with high hepcidin expression (56, 57; Table 1), were reported to have lower hepatic Hjv content compared to wild type animals (58). Matriptase-2 expression is induced by iron and BMP6 (59) consistent with this protein functioning as a negative feedback regulator of hepcidin.
Regulation of hepcidin by plasma iron
An increase in plasma iron levels activates hepcidin expression by an incompletely characterized pathway that likely involves hepatic HFE and TfR2. Evidence that these proteins are crucial for systemic iron homeostasis is provided by the iron overload that develops as a result of hepcidin suppression in mice with liver-specific disruption of either Hfe (60) or Tfr2 (61). The role of iron-loaded Tf in this pathway is illustrated by experiments where injection of holo-Tf into mice caused a specific increase in hepcidin mRNA within hours (42). Moreover, acute iron administration to mice increased Tf saturation and activated hepcidin expression without altering hepatic BMP6 levels (43).
Hemochromatosis patients carrying HFE or TFR2 mutations exhibit relatively higher basal hepcidin expression than healthy subjects; yet this increase is only modest by comparison to their iron burden, and does not prevent iron overload (37, 62). This suggests an at least partial preservation of hepcidin regulation by hepatic iron. Oral iron administration triggers a parallel increase in Tf saturation and hepcidin in healthy subjects, but a blunted hepcidin response in hemochromatosis patients, uncoupled from the increase in Tf saturation (37, 63). Likewise, Hfe−/− and Tfr2Y245X/Y245X mice mount an impaired hepcidin response to acute dietary iron challenge (42).
TfR1, which interacts with HFE (64), is also involved in hepcidin regulation by holo-Tf. This became evident in studies of mice bearing mutations, which either favor or prevent HFE/Tfr1 interactions. Animals in which HFE/Tfr1 interactions were constitutive developed iron overload and failed to appropriately increase hepcidin expression, similar to Hfe−/− mice (65). By contrast, animals in which HFE/Tfr1 interactions were abolished developed iron deficiency due to hepcidin overexpression, similar to hepatocyte-specific Hfe transgenic mice (65). These findings suggest that the binding of TfR1 compromises HFE signaling to hepcidin, while the dissociation of TfR1 induces it. Earlier biochemical studies showed that TfR1 utilizes the same binding site for its interaction with both HFE and holo-Tf, while TfR2 interacts with HFE and holo-Tf in a non-competitive manner (66–68). Moreover, TfR1 and TfR2 compete for binding to HFE (69). It should also be noted that holo-Tf stabilizes TfR2 (70, 71).
It is therefore thought that HFE and TfR2 form a signaling complex that leads to hepcidin activation. When plasma iron levels are low, signaling to hepcidin is blocked by TfR1, which sequesters HFE and prevents its interaction with TfR2. Conversely, when plasma iron levels (and Tf saturation) increase, holo-Tf displaces HFE from TfR1, allowing formation of the HFE/TfR2 signaling complex. A prediction of this model is that inactivation of either HFE or TfR2 would yield a quantitatively similar iron overload phenotype. Nevertheless, the lack of HFE usually causes a milder form of hemochromatosis than the absence of TfR2 in human patients (72) and in mice (73). Furthermore, compound pathogenic mutations in both HFE and TfR2 have been associated with early onset juvenile hemochromatosis in humans (98), while mice lacking both Hfe and Tfr2 accumulate more iron than mice lacking individual Hfe or Tfr2 genes (43, 73). Taken together, these data suggest that Hfe and Tfr2 also exhibit independent functions.
The downstream events that occur following activation of the putative HFE/TfR2 signaling complex are far from being understood. Biochemical experiments have detected a link between HFE/TfR2 signaling and the ERK/MAP kinase pathway. Thus, the treatment of primary murine hepatocytes with holo-Tf activates this pathway and hepcidin expression, which could be blocked by the ERK-specific inhibitor U0-126 (74). Along similar lines, the simultaneous silencing of HFE and TfR2 was associated with reduced ERK1/2 phosphorylation (75), while low hepatic phospho-ERK1/2 levels were also observed in Hfe−/−, Tfr2−/− and double Hfe−/−/Tfr2−/− mice and in HFE hemochromatosis patients (73).
There is also evidence for involvement of the SMAD signaling pathway in the regulation of hepcidin by HFE/TfR2. Thus, holo-Tf triggered enhanced SMAD1/5/8 phosphorylation in primary murine hepatocytes (74). In addition, SMAD1/5/8 phosphorylation was significantly reduced in mice lacking HFE or TfR2 or both, and in HFE-related hemochromatosis patients (43, 73, 76). In experiments with wild type mice, only the SMAD but not the ERK/MAPK pathway was activated following both acute (plasma) and chronic (hepatic) iron loading (43). Further work is required to clarify the exact role of these pathways in iron-dependent signaling to hepcidin.
Regulation of hepcidin expression during inflammation and ER stress
Hepcidin transcription is potently induced by IL-6 via a mechanism involving STAT3 phosphorylation, translocation to the nucleus and binding to a STAT-specific site within the HAMP promoter (77–79). This pathway leads to iron accumulation in macrophages and hypoferremia, which are considered as innate immune responses during systemic inflammation. However, persistent upregulation of hepcidin by IL-6 greatly contributes to the development of anemia of chronic disease (20). Under inflammatory conditions or during infection, myeloid cells (macrophages and neutrophils) are also stimulated to generate hepcidin via IL-1β, TLR2 or TLR4 signaling (6, 31). ER stress is another cue that leads to transcriptional activation of hepcidin via the CREBH and CHOP pathways, possibly a part of a further protective innate immune strategy (6, 31).
Regulation of hepcidin expression during anemia and hypoxia
Increased erythropoietic drive during anemia and ineffective erythropoiesis is known to suppress hepcidin (6). While erythropoietin (EPO) can reduce hepcidin expression by attenuating the binding of C/EBPα to the HAMP promoter, experiments with mice provided evidence that the inhibitory effects of EPO on hepcidin are indirect and require erythropoietic activity (80, 81). GDF15 (growth differentiation factor 15) and TWSG1 (twisted gastrulation-1) are potential mediators of bone marrow signaling, as they suppress hepcidin in thalassemias and other iron-loading anemias (82, 83). However, a general function of these proteins as “erythroid regulators” of hepcidin has been excluded (84). Thalassemic patients express low hepcidin levels despite their high iron load, indicating a dominance of the erythroid signal over iron (85, 86). In line with these data, experiments in mice confirmed that erythropoietic drive could inhibit hepcidin induction in response to iron or inflammation (6).
Hypoxia is also known to suppress hepcidin (6). Genetic studies in mice with liver-specific disruption of the Vhl gene (von Hippel-Lindau), a negative regulator of HIFα (hypoxia inducible factor α) subunit expression, suggested a key role for HIFs in hepcidin regulation (87). However, a direct transcriptional activity of HIFs on the HAMP promoter is not supported by biochemical data (88). It is conceivable that the inhibitory effects of hypoxia on hepcidin expression are primarily triggered by increased erythropoietic drive. Consistent with this view, recent experiments suggest that hepatic HIF2α inhibits hepcidin expression via EPO-mediated increase in erythropoiesis (89). Additionally, biochemical studies demonstrated that hypoxia downregulates hepcidin expression by inhibiting SMAD4 signaling (90).
Hepcidin-independent regulation of body iron metabolism
Hepcidin is the master regulator of both dietary iron absorption and systemic iron trafficking and homeostasis. However, these processes are fine-tuned by additional control mechanisms. Thus, the expression of DMT1 and DcytB in duodenal enterocytes is transcriptionally activated during iron deficiency via HIF2α (91, 92). Intestinal H-ferritin is also essential for proper dietary iron absorption, as the targeted ablation of this protein in the mouse intestine is associated with iron overload, despite functional preservation of the hepcidin/ferroportin axis (93). In addition, Ferroportin and DMT1 are subject to transcriptional (94, 95) and post-transcriptional regulation by the IRE/IRP system (96, 97).
Cellular iron metabolism
Cells have evolved metabolic strategies to import and utilize iron safely. Regulation of iron uptake, storage, intracellular trafficking and utilization is critical for the maintenance of cellular iron homeostasis (Fig. 3).
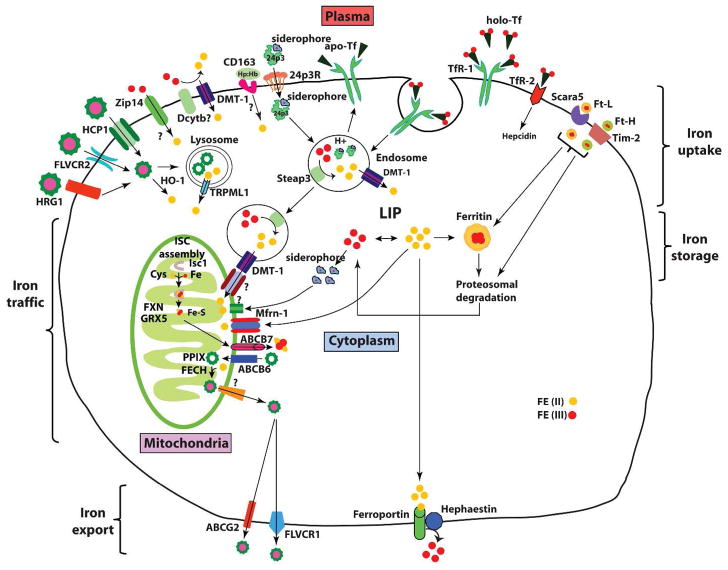
Fig. 3.
Cellular iron metabolism. Most cells in the body obtain iron from circulating differic Tf. Iron-loaded holo-Tf binds to TfR1 on the cell surface and the complex undergoes endocytosis via clathrin-coated pits. A proton pump acidifies the endosome resulting in the release of Fe3+, which is subsequently reduced to Fe2+ by Steap3 and transported across the endosomal membrane to the cytosol by DMT1. DMT1 also facilitates dietary iron absorption. Apo-Tf is recycled back to the cell surface and released from TfR1 to plasma to repeat another cycle. Newly acquired iron enters into cytosolic “labile iron pool” (LIP), which is redox-active. LIP is chelated by intracellular siderophore that facilitates intracellular iron trafficking to mitochondria via an unknown receptor for metabolic utilization (such as synthesis of heme and iron-sulfur clusters), and cellular iron that is not utilized is either stored in ferritin or exported via ferroportin. Cells also export iron contained in ferritin and heme.
Cellular uptake of iron
Cell membranes and tight junctions between cells impede free passage of iron. Cells therefore have transporters, chaperones, and chelators, which allow passage of iron across membranes and facilitate intracellular trafficking. With few exceptions, the majority of these transporters belong to the solute carrier (SLC) family of proteins. Most mammalian cells acquire iron from circulating Tf, upon binding to transferrin receptor 1 (TfR1) (6, 7). Tf, an 80-kDa glycoprotein, is synthesized and secreted mainly by the liver. Varying amounts are also produced in lymph nodes, thymus, spleen, salivary glands, bone marrow, and testis. Tf binds iron in plasma and extracellular fluids such as lymph and cerebrospinal fluid. It is equally distributed between plasma and extracellular fluids and has a half-life of 8 days. Tf is a homodimeric β-globulin, which binds two molecules of ferric iron with high affinity (kD = 10−23 M). Binding and release are accompanied by conformational change. Furthermore, the interaction between iron and Tf is pH dependent: at physiological plasma pH, Tf binds iron very strongly; whereas the interaction is essentially abolished at acidic pH (<5). Under physiological conditions, only a fraction of Tf (~30%) is saturated with iron. Catabolism of Tf takes place mainly in the liver by lysosomal degradation or glomerular filtration followed by reabsorption and degradation in renal tubules (99). Mice and humans bearing mutations in the Tf gene develop severe anemia (7). Interestingly, non-hematopoietic cells display massive iron overload (100). These studies highlight the importance of Tf-mediated iron delivery for erythropoiesis and imply the existence of alternative mechanisms for cellular iron uptake by non-hematopoietic cells.
TfR1 provides the physiological route for entry of Tf-bound iron into cells. It is expressed by most nucleated mammalian cells and its levels correlate with cellular iron requirements. TfR1 is a disulfide-linked transmembrane glycoprotein that forms a homodimer with a molecular mass of 180 kDa, where each subunit binds one Tf molecule (101). The efficiency of the interaction does not depend on temperature or energy but rather on the iron content of Tf, with diferric Tf having the greatest affinity, monoferric Tf intermediate and apo Tf the lowest. In contrast, endocytosis of Tf-TfR1 complexes is an energy-dependent process that involves endocytosis via clathrin-coated pits. Iron is released from Tf within endocytic vesicles due to acidification through the action of a v-ATPase proton pump. In a cooperative manner, Tf and TfR1 undergo conformational changes in the acidified endosome leading to iron release (102, 103). The released Fe3+ is then reduced to Fe2+ by STEAP 3 (Six transmembrane epithelial antigen of prostate-3), a member of the metalloreductase family (104). Fe2+ is transported across the endosomal membrane into the cytosol by DMT1/SLC11A2 in most cells or by its homologue Nramp 1 (Natural resistance-associated macrophage protein-1) in macrophages (105–107). In this regard, DMT1 is a dual-function protein, which regulates both systemic as well as cellular iron homeostasis. Dmt1 inactivation in mice and humans leads to iron deficiency anemia (105, 108, 109). Similarly, spontaneous mutations in Steap3 or its genetic ablation in mice cause iron deficiency in the erythroblasts but not systemically, suggesting the presence of alternative routes for iron uptake (110–112).
Newly assimilated cytosolic iron is transported to intracellular sites either for local use or for storage in ferritin (see below). The apoTf-TfR1 complex then returns to the cell surface where Tf dissociates from TfR1 (101). Targeted deletion of the mouse Tfr1 gene causes embryonic lethality at day E11.5 due to severe anemia, indicating that Tf-mediated iron delivery is critical for hematopoietic cells (113). Non-hematopoietic tissues develop normally up to day E11.5, again highlighting the existence of alternative mechanisms for iron delivery to cells in fetal life (113). Mutations in the TfR1 gene in humans have not been described. Nonetheless, an autoimmune-like disease with antibodies to TfR1 can lead to severe anemia (114).
Hepatocytes may take up Tf-bound iron via both TfR1 and TfR2. The latter shares 45% amino acid sequence similarity with TfR1 yet it differs significantly in terms of tissue distribution, affinity for Tf and iron regulation (115). TfR2 is also a type II transmembrane glycoprotein with cytoplasmic and ectodomains and the Tf-TfR2 complex is likewise internalized via clathrin-mediated endocytosis. However, while TfR1 is almost ubiquitously expressed, the expression of TfR2 is restricted to liver hepatocytes and to differentiated erythroblasts. In addition, TfR2 binds Tf with ~30-fold lower affinity than TfR1 (116), suggesting that it accounts for only a small fraction of iron uptake. Moreover, TfR2 is less stable than TfR1, is mainly regulated post translationally by protein degradation, and stabilized by differic Tf (117). The major function of TfR2 appears to be regulatory. TfR2 is involved in sensing iron-laden Tf and the control of hepcidin expression (118, 119). Thus, mutations in the TFR2 gene lead to non-HFE hemochromatosis due to hepcidin insufficiency in humans (29), a phenotype that is recapitulated in TfR2 mutant mice (120).
NTBI comprises all forms of plasma iron that are bound to ligands other than Tf (14, 121). It is undetectable under physiological conditions, but emerges in a variety of iron overload syndromes, when the iron-binding capacity of transferrin is saturated, or in atransferrinemia. Although NTBI is a key player in hemochromatotic tissue iron overload, the underlying mechanism remains unknown. Chelators that display lower affinity for iron than transferrin, such as lipocalin 24p3 are proposed to be NTBI ligands. This carrier protein sequesters iron-laden small hydrophobic molecules or siderophores (discussed in detail below). It has been proposed that 24p3-bound iron is important for epithelial morphogenesis, especially nephrogenesis (122, 123). Nonetheless, 24p3 null mice display no developmental abnormalities in kidney and apparently exhibit normal systemic and cellular iron metabolism (124–127). Experiments with double HFE and 24p3 knockout mice excluded a role of 24p3 in NTBI uptake during hepatic iron overload (127). Several biochemical and genetic studies identified other potential players of NTBI uptake, such as DMT1. Iron deficiency significantly enhances the expression of DMT1 and a gain-of-function mutation in DMT1 enhances NTBI uptake, suggesting the importance of DMT1 in the acquisition of elemental iron (128, 129). Additionally, in the microcytic (mk) mouse, mutant DMT1 behaves like a calcium channel to facilitate NTBI uptake (130), a notion that may have broader implications in understanding the functional relationship between iron transporters and other ion channels (discussed in detail below). Based on these studies, it was speculated that DMT1 is a major facilitator of NTBI uptake. However, the excessive build-up of iron in tissues of Dmt1 null mice points the involvement of additional players (108). Another candidate is the zinc transporter protein, Zip14/SLC39A14 (Zrt- and Irt-like protein 14), which facilitates NTBI-mediated iron delivery in cultured cells (131). Interestingly, HFE regulates Zip14-dependent NTBI uptake by controlling the half-life of Zip14 (132). Zip14/Slc39a14 null mice do not exhibit apparent abnormalities in iron metabolism (133), which may indicate redundancy of Zip14 in NTBI uptake. Nevertheless, it would be interesting to cross these animals with HFE−/− mice and examine the effects of Zip14 ablation in iron loading. Consistent with studies of mk mice suggesting that similar transmembrane pores conduct Fe2+ and Ca2+ across membranes, an involvement in NTBI uptake has been documented for the widely expressed L-type voltage gated calcium channels (LVGCCs), namely Cav1.1 to 1.4 (134, 135). Furthermore, calcium channel blockers have been reported to mitigate iron overload by enhancing the activity of DMT1 (136). However, these data have been disputed (137). Despite the evidence from in vitro studies supporting a role for LVGCCs in iron trafficking, mice lacking Cav1.2 to 1.3 do not exhibit alterations in iron metabolism (138). Combined, these studies suggest a significant redundancy in mechanisms for NTBI uptake and the lack of a single exclusive pathway.
Cells can also acquire iron complexed with proteins or small molecules. For instance, internalization of ferritin via ferritin-specific receptors such as T-cell immunoglobulin and mucin-domain containing protein-2 (TIM-2), or Scavenge receptor family class A, member 5 (Scara5) facilitates iron import (139, 140). However, Tim-2 null mice do not display any overt abnormalities in iron homeostasis (141). Silencing the H-ferritin receptor SCARA5, rather contributes to cancer development (142). Ingested heme also serves as an important source of iron. The importance of this route of iron uptake is exemplified by the enhanced heme uptake observed in iron deficiency. Cells are thought to internalize heme via a receptor-dependent process (directly) or indirectly with the aid of a heme carrier protein-1 (HCP-1; 143). HCP-1, also known as proton coupled folate transporter (PCFT), is a member of the solute carrier family (SLC46A1), which imports heme or folate into cultured cells or in isolated frog oocytes (143, 144). Imported heme is catabolized by heme oxygenases 1 and 2 in the endoplasmic reticulum (ER), and iron is released into the cytosol via DMT1 or the Transient receptor potential mucolipin 1 (TRPML1; refs 145, 146). Mice deficient for Hcp-1 do not exhibit iron deficiency but rather disturbances in folate metabolism (144). This phenotype is quite perplexing, given the fact that iron deficiency augments the expression of the HCP-1 (143), and may argue against a physiological role of this protein in dietary heme absorption. Heme can be transported from lysosomes to the cytosol by the heme response gene-1 (HRG-1/(SLC48A1; ref. 147). The in vivo functional relevance of HRG-1 in mammalian heme assimilation awaits the derivation of mice lacking this gene.
In summary, mammalian cells obtain iron via multiple routes tailored to meet their specific biochemical requirements.
Cellular iron utilization and intracellular iron trafficking pathways
One of the least understood aspects of cellular iron metabolism is the trafficking of iron within the cell. It is generally believed that iron taken up by the cell is routed to mitochondria via the cytoplasm. Facilitators of cytosolic iron trafficking to mitochondria have been unknown until recently.
Iron taken up by Tf-dependent or independent routes presumably enters into a labile intermediary pool or labile iron pool (LIP). The LIP is also referred to in the literature as “exchangeable”, “regulatory” or “chelatable” iron pool because its presence has been documented by using metal chelators (46). It is defined as a low-molecular weight pool of weakly chelated iron (148), including both Fe2+ and Fe3+, and represents a minor fraction of the total cellular iron (~3–5%). LIP links cellular iron uptake with iron utilization, storage, or export (148). The other source of iron for this pool comes from the degradation of non-heme and heme iron-containing proteins. Iron within the LIP is thought to be in steady state equilibrium, and is proposed to bind diverse low-molecular-weight chelates, such as organic anions (phosphates, citrates, carboxylates) and poly-functional ligands (polypeptides, siderophores; discussed in detail below). Organelles also contain LIP, which is estimated at ~ 6 to 16 μM (149). Mitochondria are the principal consumers of cellular iron. Even though mitochondrial iron is mostly bound to proteins or heme, chelatable iron has been detected within the organelle in cultured hepatocytes and cardiomyocytes (150). A rise in mitochondrial LIP is suspected under certain pathological conditions where heme export is impaired. A subpopulation of endosomes/lysosomes also contains a high amount of LIP, which is estimated at 16 μM. It may be derived from the degradation of iron carrier proteins (149).
The LIP is readily available for iron utilization and may also contribute to adverse side effects as a source of redox-active iron for the Fenton reaction (148, 151). In addition, the LIP operates as a mediator of apoptosis. Thus, iron scarcity in the cell leads to apoptosis, while similar effects are observed by targeting the LIP with chemical chelators such as deferoxamine (DFO) or biological chelators such as lipocalin 24p3 (see below) (152, 153). Moreover, apoptosis induced by these agents can be suppressed by exogenous supplementation with iron (152).
Mitochondria are central for the regulation of cellular iron metabolism and the majority of iron imported into the cell is utilized within this organelle for the synthesis of heme and iron sulfur clusters (ISCs; ref. 154). Very little is known about intracellular iron transport pathways that facilitate mitochondrial iron import. This is an important subject since iron in its free form must be shielded and escorted to the sites of utilization. Until recently, the nature and identity of molecules that chaperone iron and facilitate its intracellular trafficking was completely obscure. Nonetheless, early studies indicated the existence of low-molecular weight iron-binding compounds or siderophore-like molecules that were capable of binding iron in mammalian cells (155, 156). Interestingly, iron bound to siderophore-like molecules is a target for carrier proteins like 24p3.
Lipocalin 24p3, also known as lipocalin 2 (Lcn2) or siderocalin, is a member of the lipocalin family of proteins. This family includes over 20 small secreted proteins that have a highly conserved squat β-barrel enclosing a cavity, which consists of a continuous 8-stranded, anti-parallel β-sheet (157). The cavity within the β-barrel binds, transports and delivers small molecule ligands. By delivering this cargo via cell-surface receptors they are known to influence many cellular responses (123, 153). Lipocalin 24p3 is a unique iron binding protein, in that it lacks the ability to bind iron directly. Instead, iron binding is mediated by a cofactor. Bacteria-derived 24p3 forms a complex with enterobactin:iron in a 1:1:1 stoichiometry (158). E. coli enterobactin is the prototype of all bacterial siderophores (from the Greek, meaning “iron carriers” or “iron bearers”), low molecular weight, Fe3+-specific chelating agents that are hyperexcreted under low-iron conditions. These compounds scavenge iron from the environment and transport it into bacteria by specific receptor proteins. Enterobactin consists of three 2,3-DHBA (2,3-dihydroxybenzoic acid) and three L-serine molecules, cyclically linked to form a triserine-trilactone structure. Interestingly, 24p3 binds both 2,3-DHBA and enterobactin (158). Based on these observations it was proposed that 24p3 functions as a bacteriostat and operates in innate immunity (158). In line with this prediction, 24p3 null mice are hypersensitive to bacterial septicemia (124, 125). This mode of host defense is limited by the ability of 24p3 to sequester bacterial siderophores. In addition, certain bacteria biochemically modify their siderophores to evade capture by 24p3 (159).
Several studies have demonstrated the association of mammalian 24p3 with siderophore-like molecules. It has been proposed that iron delivery or sequestration by 24p3 is crucial for nephrogenesis (122), and mediates apoptosis (152). However, 24p3 deficient mice show normal kidney development and display no overt imbalances in iron metabolism but display multiple abnormalities in hematopoietic cells (126). A recent study identified 2,5-DHBA as a mammalian cofactor that facilitates iron loading onto 24p3. This molecule bears remarkable chemical similarity to 2,3-DHBA, which is generated by E. coli as a component of enterobactin (160). The biosynthesis of enterobactin is mediated by a two-step process: First, a series of reactions convert chorismate into 2,3-DHBA. Second, enterobactin peptide bonds are formed from three molecules each of 2,3-DHBA and L-serine, by non-ribosomal peptide synthetases (Fig. 1 and ref. 161). Six enzymes participate in the biosynthesis of enterobactin. Amongst them, EntA, a NAD+-dependent 2,3-dihydrobenzoate dehydrogenase, catalyzes the rate-limiting step. Elimination of EntA not only inhibits the synthesis of 2,3-DHBA but also enterobactin (161, 162). Interestingly, the mammalian siderophore (2,5-DHBA) is biosynthesized by an evolutionarily conserved pathway and BDH2 (3-hydroxy butyrate dehydrogenase) - a homologue of bacterial EntA - catalyzes the rate-limiting step. Elimination of bdh2 inhibits 2,5-DHBA biosynthesis (160). In contrast to 24p3, the majority of tissues synthesize 2,5-DHBA, suggesting that it has functions besides loading iron onto 24p3 (160). Siderophore-depleted mammalian cells, zebrafish, and yeast fail to synthesize heme, suggesting that 2,5-DHBA facilitates mitochondrial iron uptake (160). Mitochondria are evolutionary relics of aerobic bacteria, which supply host cell energy in the form of ATP generated in an oxygen-dependent manner. Iron is integral to all the protein subunits involved in the respiratory chain. Thus, the presence of a phylogenetically conserved siderophore-like molecule whose structure and biogenesis are evolutionarily linked to bacterial siderophores makes good sense. Although a siderophore-like molecule in mammals has been identified, the transporters or receptors that mediate siderophore-dependent mitochondrial iron uptake remain unknown. Additional cytosolic chaperones that traffic iron in the cell have been reported. For instance, evolutionarily conserved glutaredoxins Grx3 and Grx4 play an important role in iron sensing and intracellular iron trafficking, and their absence impairs mitochondrial iron import (163).
The mechanism of iron entry into mitochondria has been explored recently. Iron must traverse both outer and inner mitochondrial membranes to the site of heme synthesis, the matrix. Mitochondrial outer membrane facilitators of iron import were identified by computational screening, which yielded several members of the SLC family, namely SLC25a39 and SLC22A4 as well as a non-SLC transporter, TMEM14C (164). All these genes are consistently and specifically co-expressed with genes encoding enzymes of the heme synthesis pathway (164). Their targeted knockdown in the developing zebrafish embryo results in heme deficiency, thus validating their role in iron import into mitochondria (164). Interestingly, members of the SLC family also facilitate iron import across the mitochondrial inner membrane. Most notable are SLC25A37 or Mitoferrin-1 (Mfrn1) and SLC25A38 or Mitoferrin-2 (Mfrn2), which are important for mitochondrial iron import into erythroid and non-erythroid cells, respectively (165). Mitoferrin-1 levels in the developing erythron are stabilized by a member of the ATP-binding cassette transporter, ABCB10 (166). Although Mitoferrin-1 and Mitoferrin-2 are highly homologous, they exhibit no functional redundancy. Thus, overexpression of Mitoferrin-2 cannot rescue mitochondrial iron deficiency due to depletion of Mitoferrin-1 (167). Genetic ablation of Mitoferrin-1 confers embryonic lethality due to defective hemoglobinization (165). However, conditional deletion of Mitoferrin-1 in the hematopoietic system results in defective erythroblast formation, leading to anemia. In contrast, targeted deletion of Mitoferrin-1 in liver hepatocytes results in no discernible phenotypic or biochemical abnormalities under normal conditions (168). Despite these advances, how iron is transported across the outer mitochondrial membrane remains elusive. Siderophore-dependent iron transport supports mitochondrial heme biogenesis. The relationship between the SLC family of mitochondrial transporters and the mammalian siderophore is currently unknown.
An alternative to chaperone or siderophore-mediated iron delivery to the mitochondria has been proposed for the developing erythron. According to the “kiss-and-run” model, which is based on kinetic and imaging data, Tf-bound iron is delivered directly into the mitochondria upon contact of Tf-containing endosomes and mitochondria (169). The mediators that could facilitate this process are unclear.
Finally, the iron regulatory proteins (IRPs; discussed in detail below) whose primary function is to regulate cellular iron homeostasis, are also critical for ensuring adequate iron supplies for mitochondria, at least in liver hepatocytes (170). ISC limitations or even heme insufficiency are sensed as mitochondrial iron deficiency and these conditions activate IRPs in the cytosol. Activated IRPs increase cellular iron uptake and concomitantly reduce storage and export of iron (7, 170).
Mitochondrial handling of iron
As noted above, iron influences many aspects of mitochondrial metabolism. Imported iron is utilized for the biosynthesis of heme and ISCs, and excess iron is stored in the mitochondria-specific iron storage protein mitochondrial ferritin or mitoferritin (Ftmt; see below).
Heme synthesis begins and ends in mitochondria but intermediate steps occur in the cytoplasm (reviewed in ref. 171). The first step is the condensation of succinyl coenzyme A and glycine in the mitochondrial matrix to form 5-amino-levulinic acid (ALA), which is catalyzed by the erythroid specific ALA synthase 2 (ALAS2) or the housekeeping ALAS1 in non-erythroid cells. Mutations in ALAS2 result in anemia; heme deficiency and mitochondrial iron overload, underscoring the importance of this enzyme (172). ALA is then exported across mitochondrial membranes into the cytosol. The transporters or receptors that mediate ALA mitochondrial cross traffic are not known. Subsequently, ALA is converted to coproporphyrinogen in a series of reactions in the cytoplasm. The heme intermediate is then transported across the mitochondrial outer membrane for iron insertion by ABCB6, a member of the ATP-binding cassette (ABC) family (173). The final step, insertion of iron into protoporphyrin, is catalyzed by mitochondrial ferrochelatase (Fech). Mutations that render FECH inactive cause erythropoietic protoporphyria and liver damage (174). As discussed above, heme biosynthesis has been extensively studied, but the process by which heme is ferried across mitochondrial membranes into the cytosol for association with globins and apo cytochromes remains unknown. Likewise, no specific transporters or receptors that facilitate mitochondrial export of heme have been reported thus far.
Mitochondrial iron is also utilized for the synthesis of ISCs. These cofactors consist of iron cations and sulfide anions. ISCs are assembled on scaffold proteins and are then targeted to specific proteins via sulfur of cysteine residues. In mammals, the scaffold protein is “ISC assembly protein U” (IscU; ref. 175). Iron loading onto IscU is very likely mediated by mitochondrial frataxin (176), which thereby plays an important role in mitochondrial iron metabolism. Thus, mutations in the FRDA gene encoding frataxin result in Friedreich ataxia, a neurodegenerative disease characterized by profound mitochondrial abnormalities and iron toxicity (177). In addition, targeted disruption of this gene in mice causes embryonic lethality, while its conditional deletion in brain or muscle results in neuronal or cardiac symptoms (177). Although the majority of ISC biogenesis takes place inside mitochondria, ISCs may also be synthesized in the cytoplasm (178). The biological relevance of the mediators of cytosolic ISC biogenesis is not well understood. Nonetheless, the mitochondrial inner membrane protein, ABCB7, is important for the maturation of cytosolic ISCs since targeted deletion of Abcb7 results in altered cytosolic ISC levels and retards embryonic development (179, 180). ABCB7 may traffic ISCs (or ISC precursors) across mitochondrial membranes to the cytoplasm (179). Multiple mechanisms exist to ensure an adequate supply of iron to mitochondria. Perturbations of these processes affect iron homeostasis in both mitochondria and the cytoplasm (181). Human frataxin deficiency severely disrupts mitochondrial ISC biogenesis, promotes mitochondrial iron overload and causes increased sensitivity to oxidant stress (177). Mutations in human ABCB7 result in defective cytosolic ISC maturation and mitochondrial iron overload leading to X-linked sideroblastic anemia, which is also associated with developmental abnormalities in the brain (182). The specific mechanism by which ABCB7 deficiency contributes to anemia is unclear. Nonetheless, ABCB7 interacts with Fech, and ABCB7 plays a role in supplying iron to Fech for incorporation into protoporphyrin.
Cellular iron balance
While systemic iron metabolism is predominantly regulated at the transcriptional level, cellular iron balance is largely controlled by post-transcriptional mechanisms. The cytoplasmic iron regulatory proteins (IRPs) determine the fate of several mRNAs upon binding to their iron-responsive elements (IREs). These structural motifs are within their untranslated regions (UTRs; Fig. 4). The mRNAs encoding TfR1 and ferritin define prototypic examples of coordinate post-transcriptional regulation by IRE/IRP interactions (6, 7).
Fig. 4.
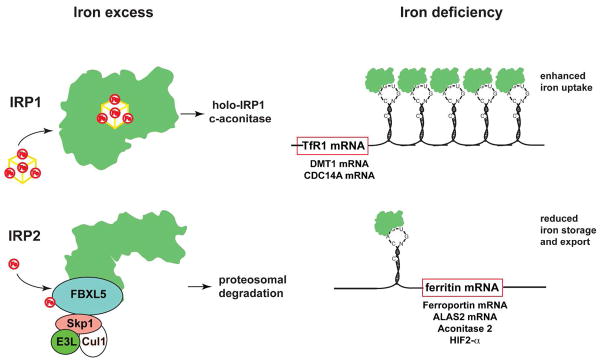
Cellular iron balance. A typical IRE motif consists of a hexanucleotide loop with the sequence 5′-CAGUGH-3′ (H could be A, C, or U) and a stem, interrupted by a bulge with an unpaired C residue. IREs post-transcriptional control expression of regulators of cellular iron metabolism in concert with IRPs. Translational-type IRE/IRP interactions in the 5′ UTR modulate the expression of the mRNAs encoding H- and L-ferritin, ALAS2, m-aconitase, ferroportin, and HIF-2α, which in turn control iron storage, erythroid iron utilization, energy homeostasis, iron efflux, and hypoxic responses, respectively. Conversely, IRE/IRP interactions in the 3′ UTR stabilize the mRNAs encoding TfR1, DMT1, and Cdc14A, which are involved in iron uptake, iron transport, and the cell cycle, respectively. Under physiological conditions, IRP1 is regulated by a reversible ISC switch. Iron deficiency, promotes ISC disassembly and a conformational rearrangement, resulting in conversion of IRP1 from c-aconitase to an IRE-binding protein. The ISC is regenerated in iron-replete cells. Hypoxia favors maintenance of the ISC, while H2O2 promotes its disassembly. When the ISC biogenesis pathway is not operational, iron leads to ubiquitination of apo-IRP1 by the FBXL5 E3 ligase complex (including Skp1, Cul1 and Rbx1), resulting in proteasomal degradation. IRP2 is stable in iron deficient and/or hypoxic cells; under these conditions FBXL5 undergoes ubiquitination and proteasomal degradation. An increase in iron and oxygen levels stabilizes FBXL5 by formation of an Fe-O-Fe center in its hemerythrin domain, triggering the assembly of an E3 ubiquitin ligase complex together with Skp1, Cul1 and Rbx1. This complex ubiquitinates IRP2, leading to its recognition by the proteasome and its degradation.
IREs
IREs are evolutionary conserved stem-loop structures of 25–30 nucleotides (183). The stem is divided into an upper and a lower segment, which are separated by an unpaired C residue or an asymmetric UGC/C bulge/loop. The loop consists of a conserved 5′-CAGUGH-3′ sequence (H denotes A, C or U), where the underlined C and G residues interact. TfR1 mRNA contains multiple IREs within its long 3′ UTR, while the mRNAs encoding H- and L-ferritin contain a single IRE in their 5′ UTRs.
In iron deficiency, IRPs bind with high affinity (Kd ≈ 10−12 M) to target IREs. This results in stabilization of TfR1 mRNA and steric inhibition of ferritin mRNA translation. Under these conditions, accumulation of TfR1 promotes cellular iron uptake from plasma Tf, while inhibition of ferritin biosynthesis prevents storage of iron, allowing its metabolic utilization. Conversely, in response to excess cellular iron IRPs are inactivated, which leads to TfR1 mRNA degradation and ferritin mRNA translation. This minimizes further internalization of iron via TfR1 and promotes the storage of excessive intracellular iron into ferritin. Interestingly, mutations in the L-ferritin IRE that prevent IRP-binding are causatively linked to the hereditary hyperferritinemia-cataract syndrome (HHCS), an autosomal dominant disorder characterized by overexpression of serum ferritin (without iron overload) and early-onset cataract (184). Mutations, in the IRE of H-ferritin have been associated with a rare form of autosomal dominant iron overload in a Japanese pedigree but the underlying pathogenic mechanism remains unclear (185).
Functional IRE motifs have also been identified in mRNAs encoding proteins of iron transport (DMT1, ferroportin) and erythroid heme synthesis (ALAS2), as well as others that appear less related to iron homeostasis (mitochondrial aconitase, Drosophila succinate dehydrogenase, HIF-2α, MRCKα, Cdc14A, β-amyloid precursor protein and α-synuclein; reviewed in ref. 6). It is notable that mRNAs encoding TfR2, Ftmt and the housekeeping ALAS1 as well as alternatively spliced transcripts of DMT1 and ferroportin, do not posses an IRE. The expression of IRE- or non-IRE isoforms of DMT1 and ferroportin mRNAs confers versatility and tissue specific regulation. For instance, the prevalence of the non-IRE isoform of ferroportin mRNA in duodenal enterocytes and erythroid precursor cells (186) allows its expression during iron deficiency (187) by evading the translational IRP block. It should also be noted that In erythroid cells, which require very high amounts of iron for hemoglobinization, the expression of TfR1 mRNA is induced by transcriptional mechanisms, and its stability is uncoupled from iron supply and IRP regulation (188, 189). Transcriptional regulation of TfR1 as well as ferritin mRNA expression has also been established in other settings (reviewed in 6, 7).
IRPs
IRP1 and IRP2 are homologous to mitochondrial aconitase and other members of the ISC isomerase family (6, 7), which possess a cubane ISC in their active site. In iron-replete cells, IRP1 assembles an ISC and functions as cytosolic aconitase (catalyzing the conversion of citrate to iso-citrate via a cis-aconitate intermediate). The ISC keeps IRP1 in a closed conformation, which precludes access of IREs. Iron deficiency promotes the loss of the ISC and a resulting structural rearrangement of the protein that acquires IRE-binding activity. Thus, IRP1 is a bifunctional protein that is regulated by an ISC switch.
Despite its extensive homology with IRP1, IRP2 does not assemble an ISC and is regulated by a distinct mechanism. It remains stable in iron-starved or hypoxic cells and undergoes ubiquitination and proteasomal degradation in response to iron and oxygen. The mechanism involves FBXL5, an E3 ubiquitin ligase that contains an N-terminal hemerythrin domain with a Fe-O-Fe bridge (190, 191). Iron deficiency and/or hypoxia lead to disassembly of Fe-O-Fe, which destabilizes FBXL5 and allows accumulation of IRP2. Interestingly, FBXL5 may also target apo-IRP1 for iron-dependent degradation; however, this occurs only as a backup mechanism under conditions where ISC assembly is defective (192, 193).
The ablation of both IRPs leads to early embryonic lethality at the blastocyst stage (194). Intestinal-specific disruption of both IRPs has been associated with growth defects, intestinal malabsorption, dehydration and weight loss (195). Likewise, disruption of IRPs in hepatocytes causes premature death due to liver damage, mitochondrial abnormalities and defects in heme and ISC biosynthetic pathways (170).
Single disruption of either IRP1 or IRP2 is not lethal, indicating a functional redundancy of these proteins. IRP1−/− mice do not develop any apparent pathology, but misregulate TfR1 and ferritin expression in the kidney and brown fat (196). IRP2−/− mice exhibit microcytosis and hypochromic anemia, associated with iron overload in the duodenum and the liver, and relative iron deficiency in the spleen (197, 198). The deregulation of tissue iron homeostasis in these animals can be explained by cell-autonomous functions of IRP2 (199). Some aged IRP2−/− mice develop brain iron overload and deposition in overexpressed ferritin (200). This leads to a progressive neurodegenerative disorder, possibly as a result of decreased iron availability (functional iron deficiency).
Mice lacking FBXL5 die early in embryogenesis, apparently from deleterious effects of iron accumulation and subsequent iron-induced damage to tissues. Their pathology resembles clinical features of neonatal hemochromatosis, which is fatal due to liver failure resulting from excessive iron deposition in this organ (201). Interestingly, ablation of IRP2 rescues the FBXL5 null phenotype, consistent with FBXL5 being a repressor of IRP2. Mice bearing liver-specific disruption of FBXL5 develop severe steatohepatitis, associated with hepatic iron overload and oxidative stress. Collectively, the above data highlight the significance of the IRE/IRP system in the control of mammalian iron homeostasis.
Cellular iron storage
Cellular iron in excess of immediate needs is stored as an iron oxide within the nanocavity of ferritin. Ferritin is an evolutionarily conserved, ubiquitous protein that can accommodate up to 4500 iron atoms. In mammals ferritin is a heteropolymer of 24 subunits of two types, heavy (H) and light (L), which assemble to make a hollow spherical shell (reviewed in ref. 202). L-ferritin is predominant in iron storing tissues, whereas H-ferritin is preferentially expressed in cells that take up and release iron rapidly. Different proportions of ferritin subunits give rise to the heterogeneity of the holo-protein in various tissue types. A glycosylated L-ferritin subunit circulates in serum and exhibits low saturation with iron. Its origin is debated. The levels of serum ferritin increase in response to systemic iron load or infection (6). Ferritin secretion may provide a mechanism to limit iron storage after a shift from high to low iron, and prior to activation of IRPs (202).
Channels in the ferritin shell may facilitate the entry and exit of iron. The ferroxidase activity of H-ferritin converts Fe2+ to Fe3+, which is necessary for iron deposition into the nanocage. L-ferritin induces iron nucleation and increases the turnover of the ferroxidase activity. How iron is delivered to ferritin remains elusive. Experimental evidence suggests the involvement of the cytosolic iron chaperone Poly(rC)-binding protein 1 (PCBP1; ref. 203). Iron release from ferritin is mediated by multiple mechanisms (reviewed in ref. 202). Physiologically, degradation of ferritin is coupled to supply of metabolic iron availability under iron-limiting conditions (204). Treatment with iron chelators or expression of ferroportin accelerates ferritin degradation (205, 206).
Changes in iron availability regulate ferritin gene expression mainly at the posttranscriptional level via the IRE/IRP system (discussed above). Additionally, ferritin expression is regulated transcriptionally, which determines the tissue distribution of H- and L- chains. Iron homeostasis is profoundly altered during inflammation and infection. Although multiple mechanisms collectively influence iron balance (such as induction of hepcidin by IL-6), alteration of ferritin expression by proinflammatory cytokines such as TNFα or IL-1α substantially contributes to reprogramming of iron homeostasis (6). Ferritin is transcriptionally activated under oxidative stress via an upstream antioxidant response element (ARE) in the promoter region of the ferritin genes (207). In contrast, extracellular H2O2 inhibits ferritin mRNA translation by activating the IRE/IRP regulatory system (refs. 6, 7).
By sequestering redox-active iron, ferritin plays an important antioxidant role and promotes cell survival. Thus, overexpression of ferritin decreases the LIP and the generation of ROS, and confers resistance to oxidative damage (208). Moreover, H-ferritin depletion triggers opposite effects (207). Genetic deletion of H-ferritin results in embryonic lethality, suggesting that its ferroxidase activity is critical (185). Conditional ablation of H-ferritin in liver hepatocytes results in iron-induced oxidative damage because these cells cannot sequester and detoxify iron (185). Mutations in L-ferritin are associated with an autosomal dominant neurological disorder (neuroferritinopathy), characterized by brain iron overload and entrapment of iron in mutant ferritin shells, which causes functional iron deficiency (185).
Mitochondrial ferritin (Ftmt) is encoded by a distinct nuclear gene (209) and possesses ferroxidase activity, by analogy to H-ferritin. In contrast to cytosolic ferritin, the expression of Ftmt is restricted to few tissues and is not iron-regulated (210). Ftmt serves as a molecular sink to prevent accumulation of unshielded iron in mitochondria, which protects the organelle against iron’s toxicity. Ftmt levels are increased in sideroblastic anemia (185). Forced overexpression or depletion of Ftmt results in profound alterations of mitochondrial as well as cytosolic iron levels (211).
Cellular iron export
Cellular iron export routinely occurs in specialized cells such as enterocytes and macrophages that are involved in iron absorption and recycling, respectively. The main purpose of iron export is to maintain adequate plasma iron levels and to meet systemic needs. Iron export involves coordination between many enzymes and proteins. Ferroportin, a member of the solute carrier family, facilitates iron export from enterocytes, macrophages, hepatocytes and from the extraembryonic visceral endoderm (ExVE). As discussed earlier, iron is reduced in endosomes prior to release into the cytoplasm. Thus, ferroportin exports Fe2+, which has to be oxidized to Fe3+ upon its release into plasma for binding to Tf (reviewed in ref. 212). This is mediated by the blue copper ferroxidases ceruloplasmin (soluble in serum or plasma membrane-associated in some cell types) and hephaestin (expressed on the plasma membrane of enterocytes and other cell types). These enzymes thus work in concert with ferroportin to coordinate iron export and oxidation. As copper is essential for ferroxidase activity, adequate levels of this metal are required for proper iron balance (reviewed in ref. 213).
Complete disruption of ferroportin in mice is associated with early embryonic lethality, while conditional ablation leads to iron accumulation in target cells, consistent with a function of this protein as the sole iron exporter (214). In humans, ferroportin autosomal dominant mutations lead to “ferroportin disease”. Mutations inhibiting iron efflux promote macrophage iron loading, low serum iron levels and transferrin saturation. On the other hand, mutations that impair the binding of hepcidin cause parenchymal iron overload, relatively high serum iron levels and transferrin saturation, as in classical hereditary hemochromatosis (23, 215)
Cells may also export iron bound to ferritin, especially in necroinflammation, or heme iron. The feline leukemia virus sub group C receptor 1 (FLVCR1) facilitates heme efflux from hematopoietic cells. Suppression or forced expression of FLVCR1 enhances or depletes cytoplasmic heme content, respectively (216, 217). Thus, it has been proposed that FLVCR1 mediates heme efflux from erythroblasts to ensure their survival and from macrophages to regulate hepatic and systemic iron homeostasis. Flvcr1−/− mice exhibit embryonic lethality and lack definitive erythropoiesis. In contrast, neonatal deletions of Flvcr1 cause severe macrocytic anemia with proerythroblast maturation arrest (216). Point mutations in human FLVCR1 are surprisingly not associated with erythroid disorders. Instead, affected patients exhibit neurological and vision abnormalities (218). Interestingly, the related protein FLVCR2, appears to promote heme import (219).
Adding to the list of heme exporters, an ATP binding cassette family member, ABCG2, has also been implicated in cellular heme export. Thus, genetic ablation of Abcg2 results in the accumulation of the heme synthesis intermediate, protoporphyrin IX (PIX) suggesting a function in heme or porphyrin export. Mice deficient for Abcg2 abnormally accumulate protoporphyrin, suggesting that Abcg2 aids in porphyrin export (220). Nevertheless, mutations in human ABCG2 are not associated with heme abnormalities, but rather with elevated levels of uric acid (221).
Conclusions
Iron is vital, yet poorly bioavailable and potentially toxic. Cells and multicellular organisms have evolved sophisticated mechanisms to cope with the challenges of iron acquisition and handling. Remarkable progress has been made in understanding regulation of mammalian iron metabolism. Mammals primarily control dietary iron absorption and systemic iron traffic via hepcidin, a hormonal peptide that responds to physiological stimuli. Cellular iron uptake, storage and utilization are coordinately regulated by the IRE/IRP system. Fine-tuning is achieved by further regulatory pathways, which operate at the systemic and cellular levels. While basic mechanisms for mammalian iron homeostasis are now fairly well understood, several issues that remain open include the molecular characterization of iron sensing by hepcidin and the IRE/IRP pathways, the elucidation of iron trafficking inside cells, and the clarification of the physiological significance of the mammalian siderophore/24p3 network.
Table 2.
Regulators of cellular iron metabolism: Animal models and human disorders.
| Metabolic Process | Protein | Gene | Function | Mouse model | Phenotype | Human disease | Refs |
|---|---|---|---|---|---|---|---|
| Cellular iron/heme uptake | Transferrin | Tf | Plasma, lymph, and CSF ferric iron carrier | hpx/hpx | Hypochromic, microcytic anemia | Atransferrinemia | 7, 100 |
| Transferrin receptor 1 | TfR1 | Facilitates Tf-dependent iron uptake | TfR1−/− | Embryonic lethality | Autoimmune-like with Abs to TfR1; anemia | 113 | |
| Transferrin receptor 2 | TfR2 | Regulates hepcidin; mediates Tf- bound and non-transferrin bound iron uptake | TfR2−/− | Iron overload | Hemochromatosis | 61 | |
| Six trans membrane epithelial antigen of the prostate | Steap3 | Erythroid endosomal ferrireductase |
nm:1054 Steap3−/− |
Microcytic, hypochromic anemia; Iron deficiency | Unknown | 110–112 | |
| Divalent metal transporter 1 |
DMT1 Nramp1 Slc11a2 |
Endosomal ferrous iron transporter; transmembrane non-transferrin iron transporter |
mk/mk DMT−/− |
Hypochromic, microcytic anemia; impaired iron absorption | Iron overload in liver; anemia | 105, 108, 109 | |
| Duodenal cytochrome B | DcytB | Intestinal ferrireductase | Dcytb−/− | Normal | Unknown | 9 | |
| Zrt- and Irt- like protein 14 | Zip14 | Non-transferrin iron transporter in liver | Slc39A14−/− | Impaired gluconeogenesis; no abnormalities in iron metabolism | Unknown | 133 | |
| L-type Ca2+ channels | Cav1.1 to 1.4 | Calcium/Iron transporters |
Cav1.2−/−
Cav1.3−/− |
No abnormalities in iron metabolism | Unknown | 138 | |
| T-cell Ig- domain and mucin domain protein-2 (TIM-2) | Tim-2 | Ferritin-H receptor | Tim2−/− | No overt abnormalities related to iron metabolism | Unknown | 141 | |
| Scavenger receptor class A member 5 | Scara5 | Ferritin-L receptor | Not available | Unknown | Predisposition to liver cancer | 142 | |
| Feline leukemia virus subgroup C receptor 2 | FLVCR2 | Heme importer | Not available | Unknown | Fowler syndrome | 219 | |
| Heme carrier protein 1 | HCP1 | Intestinal heme transporter | HCP−/− | No alterations in iron homeostasis | Folate malabsorption | 144 | |
| Transient receptor potential mucolipidosis-associated protein | TRPML1 | Iron transporter from endosomes and lysosomes | Not available | Unknown | Mucolipidosis | 146 | |
| Heme Responsive Gene 1 | HRG1 | Heme exporter into cytoplasm from lysosomes | Not available | Unknown | Unknown | 7 | |
| Heme oxygenase-1 | HO-1 | Heme iron reutilization | HO-1−/− | Iron overload | Iron overload, hemolytic anemia | 11 | |
| Heme oxygenase-2 | HO-2 | Heme iron reutilization | HO-2−/− | No disturbances in iron homeostasis | Unknown | 11 | |
| 24p3 | 24p3 | Facilitator of non-transferrin iron import | 24p3−/− | No observable alterations in iron homeostasis | Unknown | 127 | |
| Cellular iron storage | Ferritin-H (FT-H) | Fth | Iron storage protein subunit; ferroxidase | Fth−/− | Embryonic lethality | Iron overload | 185, 202 |
| Ferritin-L (FT-L) | Ftl | Iron storage protein subunit | Not available | Unknown | IRE mutation - Hyperferritenemi a-cataract syndrome; non- IRE mutation- Neuroferritinopathy | 185, 202 | |
| Intracellular iron trafficking | Ferrochelatase (FECH) | Fech | Insertion of iron into porphyrin |
Fechm1Pas Fech−/− |
Microcytic, hypochromic anemia; Embryonic lethality | Erythropoietic protoporphyria | 172, 174 |
| 5-amino- levulinic acid synthase (ALAS-2) | Alas-2 | Heme biogenesis | Alas2−/− | Embryonic lethality; iron overload | X-linked sideroblastic anemia | 171 | |
| ATP-binding cassette family member B 10 (ABCB10) | Abc-me or Abcb10 | Interacts with Fech and Mitoferrin-1; transport ligand is unknown | Abcb10−/− | Embryonic lethality; arrested erythroid differentiation | Unknown | 166 | |
| ATP-binding cassette family member B 7 (ABCB7) | Abcb7 | Interacts with Fech; Cytosolic ISC maturation | Abcb7−/− | Midgestational death; Defective hematopoiesis; hepatic iron overload, Ataxia | Hereditary X-linked sideroblastic anemia, Ataxia | 180, 182 | |
| ATP-binding cassette family member B 6 (ABCB6) | Abcb6 | Porphyrin import into mitochondria? | Unpublished | Porphyria? | Unknown | 173 | |
| Mitoferrin1 (MFRN1) | Mfrn1 | Mitochondrial iron import | Mfrn1−/− | Embryonic lethality; conditional deletion confers anemia and defective hematopoiesis | Erythropoietic protoporphyria | 165, 168 | |
| Glutaredoxin5 (GRX5) | Grx5 | Facilitator of ISC biogenesis | Not available | Unknown | Anemia; iron overload | 7 | |
| Frataxin (FXN) | Fxn | Mitochondrial iron chaperone? | Fxn−/− | Embryonic lethality; conditional deletion confers brain and heart deformities | Friedreich ataxia | 177 | |
| 3-OH butyrate dehydrogenase-2 (BDH2) | Bdh2 or Dhrs6 | Siderophore biosynthesis | Bdh2−/− | Anemia, iron overload, splenomegaly | Diabetes | Unpublished | |
| Cellular iron/heme export | Feline leukemia virus subgroup C receptor1 (FLVCR1) | FLVCR1 | Heme exporter | FLVCR1−/− | Embryonic lethal; systemic iron overload and erythroid abnormalities in conditional deletion | Diamond Blackfan anemia? Neurological abnormalities | 216, 218 |
| ATP-binding cassette family member C, G2 (ABCG2) | Abcg2 | Partially exports heme; PPIX exporter | Abcg2−/− | Elevated levels of PPIX; no iron overload | Gout | 220, 221 | |
| Ferroportin SLC40A1; MTP1) | Slc40a1 | Regulates iron efflux from cells |
Slc40a1−/−
pcm |
Embryonic lethality; conditional deletion – iron deficiency | Hemochromatosis, macrophage iron overload (HH type 4) | 19, 215 | |
| Cellular iron balance | Iron regulatory protein 1 (IRP 1) | IRP1 | Posttranscriptional regulation of target mRNAs via IREs; cytosolic aconitase | IRP1−/− | Unknown | 194–200 | |
| Iron regulatory protein 2 (IRP 2) | IRP2 | Posttranscription al regulation of target mRNAs via IREs | IRP2−/− | Unknown | 194–200 |
Acknowledgments
We thank John Protasiewicz for insightful comments on iron chemistry. This work is supported by K01CA113838, R01DK081395, and Case Western Reserve University start up funds to L.R.D. L.R.D. is also a recipient of career developmental awards from March of Dimes and American Society of Hematology. KP is supported by a grant from the Canadian Institutes for Health Research (CIHR; MOP-86514) and holds a Chercheur National career award from the Fonds de la Recherche en Santé du Quebéc (FRSQ).
References
- 1.Theil EC, Raymond KN. Bioinorganic chemistry. In: Bertini I, Gray HB, Lippard SJ, Valentine JS, editors. University Science Books; Mill Valley, CA: 1994. pp. 1–36. [Google Scholar]
- 2.Wigglesworth JM, Baum H. In: In Iron in Biochemistry and Medicine II. Jacobs A, Worwood M, editors. Academic press; New York, NY: 1980. pp. 29–86. [Google Scholar]
- 3.Pereira IAC, Teixeira M, Xavier AV. In: Bioinorganic chemistry: Trace element evolution from anaerobes to aerobes. Williams RJP, editor. Springer-Verlag; New York, NY: 2010. pp. 91–190. [Google Scholar]
- 4.Nielands JB. Siderophores: structure and functions of microbial iron transport compounds. J Biol Chem. 1995;27:26723–26726. doi: 10.1074/jbc.270.45.26723. [DOI] [PubMed] [Google Scholar]
- 5.Hider RC, Kong XL. Chemistry and biology of siderophores. Nat Products Rep. 2010;27:637–657. doi: 10.1039/b906679a. [DOI] [PubMed] [Google Scholar]
- 6.Wang J, Pantopoulos K. Regulation of cellular iron metabolism. Biochem J. 2011;434:365–381. doi: 10.1042/BJ20101825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hentze MW, Muckenthaler MU, Galy B, Camaschella C. Two to tango: regulation of Mammalian iron metabolism. Cell. 2010;142:24–38. doi: 10.1016/j.cell.2010.06.028. [DOI] [PubMed] [Google Scholar]
- 8.Knutson M, Wessling-Resnick M. Iron metabolism in the reticuloendothelial system. Crit Rev Biochem Mol Biol. 2003;38:61–88. doi: 10.1080/713609210. [DOI] [PubMed] [Google Scholar]
- 9.McKie AT. The role of DcytB in iron metabolism: an update. Biochem Soc Trans. 2008;36:1239–1241. doi: 10.1042/BST0361239. [DOI] [PubMed] [Google Scholar]
- 10.West AR, Oates PS. Mechanisms of heme iron absorption: current questions and controversies. World J Gastroenterol. 2008;14:4101–4110. doi: 10.3748/wjg.14.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryter SW, Alam J, Choi AM. Heme oxygenase-1/carbon monoxide: from basic science to therapeutic applications. Physiol Rev. 2006;86:583–650. doi: 10.1152/physrev.00011.2005. [DOI] [PubMed] [Google Scholar]
- 12.Kovac S, Anderson GJ, Baldwin GS. Gastrins, iron homeostasis and colorectal cancer. Biochim Biophys Acta. 2011;1813:889–895. doi: 10.1016/j.bbamcr.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gkouvatsos K, Papanikolaou G, Pantopoulos K. Regulation of iron transport and the role of transferrin. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbagen.2011.10.013. In press. [DOI] [PubMed] [Google Scholar]
- 14.Brissot P, Ropert M, Le Lan C, Loreal O. Non-transferrin bound iron: A key role in iron overload and iron toxicity. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbagen.2011.07.014. in press. [DOI] [PubMed] [Google Scholar]
- 15.Krause A, Neitz S, Magert HJ, Schulz A, Forssmann WG, Schulz-Knappe P, Adermann K. LEAP-1, a novel highly disulfide-bonded human peptide, exhibits antimicrobial activity. FEBS Lett. 2000;480:147–150. doi: 10.1016/s0014-5793(00)01920-7. [DOI] [PubMed] [Google Scholar]
- 16.Park CH, Valore EV, Waring AJ, Ganz T. Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J Biol Chem. 2001;276:7806–7810. doi: 10.1074/jbc.M008922200. [DOI] [PubMed] [Google Scholar]
- 17.Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, Ganz T, Kaplan J. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306:2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- 18.De Domenico I, Ward DM, Langelier C, Vaughn MB, Nemeth E, Sundquist WI, Ganz T, Musci G, Kaplan J. The molecular mechanism of hepcidin-mediated ferroportin down-regulation. Mol Biol Cell. 2007;18:2569–2578. doi: 10.1091/mbc.E07-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brasse-Lagnel C, Karim Z, Letteron P, Bekri S, Bado A, Beaumont C. Intestinal DMT1 cotransporter is down-regulated by hepcidin via proteasome internalization and degradation. Gastroenterol. 2011;140:1261–1271. doi: 10.1053/j.gastro.2010.12.037. [DOI] [PubMed] [Google Scholar]
- 20.Weiss G. Iron metabolism in the anemia of chronic disease. Biochim Biophys Acta. 2009;1790:682–693. doi: 10.1016/j.bbagen.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 21.Finberg KE, Heeney MM, Campagna DR, Aydinok Y, Pearson HA, Hartman KR, Mayo MM, Samuel SM, Strouse JJ, Markianos K, Andrews NC, Fleming MD. Mutations in TMPRSS6 cause iron-refractory iron deficiency anemia (IRIDA) Nat Genet. 2008;40:569–571. doi: 10.1038/ng.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ganz T, Nemeth E. Hepcidin and disorders of iron metabolism. Annu Rev Med. 2011;62:347–360. doi: 10.1146/annurev-med-050109-142444. [DOI] [PubMed] [Google Scholar]
- 23.Sebastiani G, Pantopoulos K. Disorders associated with systemic or local iron overload: from pathophysiology to clinical practice. Metallomics. 2011;3:971–986. doi: 10.1039/c1mt00082a. [DOI] [PubMed] [Google Scholar]
- 24.Roetto A, Papanikolaou G, Politou M, Alberti F, Girelli D, Christakis J, Loukopoulos D, Camaschella C. Mutant antimicrobial peptide hepcidin is associated with severe juvenile hemochromatosis. Nat Genet. 2003;33:21–22. doi: 10.1038/ng1053. [DOI] [PubMed] [Google Scholar]
- 25.Papanikolaou G, Samuels ME, Ludwig EH, MacDonald ML, Franchini PL, Dube MP, Andres L, MacFarlane J, Sakellaropoulos N, Politou M, Nemeth E, Thompson J, Risler JK, Zaborowska C, Babakaiff R, Radomski CC, Pape TD, Davidas O, Christakis J, Brissot P, Lockitch G, Ganz T, Hayden MR, Goldberg YP. Mutations in HFE2 cause iron overload in chromosome 1q-linked juvenile hemochromatosis. Nat Genet. 2004;36:77–82. doi: 10.1038/ng1274. [DOI] [PubMed] [Google Scholar]
- 26.Feder JN, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy DA, Basava A, Dormishian F, Domingo R, Jr, Ellis MC, Fullan A, Hinton LM, Jones NL, Kimmel BE, Kronmal GS, Lauer P, Lee VK, Loeb DB, Mapa FA, McClelland E, Meyer NC, Mintier GA, Moeller N, Moore T, Morikang E, Prass CE, Quintana L, Starnes SM, Schatzman RC, Brunke KJ, Drayna DT, Risch NJ, Bacon BR, Wolff RK. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- 27.Bridle KR, Frazer DM, Wilkins SJ, Dixon JL, Purdie DM, Crawford DH, Subramaniam VN, Powell LW, Anderson GJ, Ram GA. Disrupted hepcidin regulation in HFE-associated haemochromatosis and the liver as a regulator of body iron homoeostasis. Lancet. 2003;361:669–673. doi: 10.1016/S0140-6736(03)12602-5. [DOI] [PubMed] [Google Scholar]
- 28.Camaschella C, Roetto A, Cali A, De Gobbi M, Garozzo G, Carella M, Majorano N, Totaro A, Gasparini P. The gene TFR2 is mutated in a new type of haemochromatosis mapping to 7q22. Nat Genet. 2000;25:14–15. doi: 10.1038/75534. [DOI] [PubMed] [Google Scholar]
- 29.De Domenico I, Ward DM, Nemeth E, Vaughn MB, Musci G, Ganz T, Kaplan J. The molecular basis of ferroportin-linked hemochromatosis. Proc Natl Acad Sci USA. 2005;102:8955–8960. doi: 10.1073/pnas.0503804102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayr R, Griffiths WJ, Hermann M, McFarlane I, Halsall DJ, Finkenstedt A, Douds A, Davies SE, Janecke AR, Vogel W, Cox TM, Zoller H. Identification of mutations in SLC40A1 that affect ferroportin function and phenotype of human ferroportin iron overload. Gastroenterol. 2011;140:2056–2063. 2063. doi: 10.1053/j.gastro.2011.02.064. [DOI] [PubMed] [Google Scholar]
- 31.Ganz T. Hepcidin and iron regulation, 10 years later. Blood. 2011;117:4425–4433. doi: 10.1182/blood-2011-01-258467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valore EV, Ganz T. Posttranslational processing of hepcidin in human hepatocytes is mediated by the prohormone convertase furin. Blood Cells Mol Dis. 2008;40:132–138. doi: 10.1016/j.bcmd.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peslova G, Petrak J, Kuzelova K, Hardy I, Halada P, Kuchel PW, Soe-Lin S, Ponka P, Sutak R, Becker E, Huang ML, Suryo Rahmanto Y, Richardson DR, Vyoral D. Hepcidin, the hormone of iron metabolism, is bound specifically to alpha-2-macroglobulin in blood. Blood. 2009;113:6225–6236. doi: 10.1182/blood-2009-01-201590. [DOI] [PubMed] [Google Scholar]
- 34.Courselaud B, Pigeon C, Inoue Y, Inoue J, Gonzalez FJ, Leroyer P, Gilot D, Boudjema K, Guguen-Guillouzo C, Brissot P, Loreal O, Ilyin G. C/EBPalpha regulates hepatic transcription of hepcidin, an antimicrobial peptide and regulator of iron metabolism. Cross-talk between C/EBP pathway and iron metabolism. J Biol Chem. 2002;277:41163–41170. doi: 10.1074/jbc.M202653200. [DOI] [PubMed] [Google Scholar]
- 35.Steinbicker AU, Bartnikas TB, Lohmeyer LK, Leyton P, Mayeur C, Kao SM, Pappas AE, Peterson RT, Bloch DB, Yu PB, Fleming MD, Bloch KD. Perturbation of hepcidin expression by BMP type I receptor deletion induces iron overload in mice. Blood. 2011;118:4224–4230. doi: 10.1182/blood-2011-03-339952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piperno A, Girelli D, Nemeth E, Trombini P, Bozzini C, Poggiali E, Phung Y, Ganz T, Camaschella C. Blunted hepcidin response to oral iron challenge in HFE-related hemochromatosis. Blood. 2007;110:4096–4100. doi: 10.1182/blood-2007-06-096503. [DOI] [PubMed] [Google Scholar]
- 37.Girelli D, Trombini P, Busti F, Campostrini N, Sandri M, Pelucchi S, Westerman M, Ganz T, Nemeth E, Piperno A, Camaschella C. A time course of hepcidin response to iron challenge in patients with HFE and TFR2 hemochromatosis. Haematologica. 2011;96:500–506. doi: 10.3324/haematol.2010.033449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gehrke SG, Kulaksiz H, Herrmann T, Riedel HD, Bents K, Veltkamp C, Stremmel W. Expression of hepcidin in hereditary hemochromatosis: evidence for a regulation in response to serum transferrin saturation and non-transferrin-bound iron. Blood. 2003;102:371–376. doi: 10.1182/blood-2002-11-3610. [DOI] [PubMed] [Google Scholar]
- 39.Wallace DF, Summerville L, Lusby PE, Subramaniam VN. First phenotypic description of transferrin receptor 2 knockout mouse, and the role of hepcidin. Gut. 2005;54:980–986. doi: 10.1136/gut.2004.062018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niederkofler V, Salie R, Arber S. Hemojuvelin is essential for dietary iron sensing, and its mutation leads to severe iron overload. J Clin Invest. 2005;115:2180–2186. doi: 10.1172/JCI25683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang FW, Pinkus JL, Pinkus GS, Fleming MD, Andrews NC. A mouse model of juvenile hemochromatosis. J Clin Invest. 2005;115:2187–2191. doi: 10.1172/JCI25049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramos E, Kautz L, Rodriguez R, Hansen M, Gabayan V, Ginzburg Y, Roth MP, Nemeth E, Ganz T. Evidence for distinct pathways of hepcidin regulation by acute and chronic iron loading in mice. Hepatol. 2011;53:1333–1341. doi: 10.1002/hep.24178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corradini E, Rozier M, Meynard D, Odhiambo A, Lin HY, Feng Q, Migas MC, Britton RS, Babitt JL, Fleming RE. Iron regulation of hepcidin despite attenuated Smad1,5,8 signaling in mice without transferrin receptor 2 or Hfe. Gastroenterol. 2011;141:1907–1914. doi: 10.1053/j.gastro.2011.06.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Babitt JL, Huang FW, Wrighting DM, Xia Y, Sidis Y, Samad TA, Campagna JA, Chung RT, Schneyer AL, Woolf CJ, Andrews NC, Lin HY. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat Genet. 2006;38:531–539. doi: 10.1038/ng1777. [DOI] [PubMed] [Google Scholar]
- 45.Casanovas G, Mleczko-Sanecka K, Altamura S, Hentze MW, Muckenthaler MU. Bone morphogenetic protein (BMP)-responsive elements located in the proximal and distal hepcidin promoter are critical for its response to HJV/BMP/SMAD. J Mol Med. 2009;87:471–480. doi: 10.1007/s00109-009-0447-2. [DOI] [PubMed] [Google Scholar]
- 46.Wang RH, Li C, Xu X, Zheng Y, Xiao C, Zerfas P, Cooperman S, Eckhaus M, Rouault T, Mishra L, Deng CX. A role of SMAD4 in iron metabolism through the positive regulation of hepcidin expression. Cell Metab. 2005;2:399–409. doi: 10.1016/j.cmet.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 47.Mleczko-Sanecka K, Casanovas G, Ragab A, Breitkopf K, Muller A, Boutros M, Dooley S, Hentze MW, Muckenthaler MU. SMAD7 controls iron metabolism as a potent inhibitor of hepcidin expression. Blood. 2010;115:2657–2665. doi: 10.1182/blood-2009-09-238105. [DOI] [PubMed] [Google Scholar]
- 48.Meynard D, Kautz L, Darnaud V, Canonne-Hergaux F, Coppin H, Roth MP. Lack of the bone morphogenetic protein BMP6 induces massive iron overload. Nat Genet. 2009;41:478–481. doi: 10.1038/ng.320. [DOI] [PubMed] [Google Scholar]
- 49.Andriopoulos B, Jr, Corradini E, Xia Y, Faasse SA, Chen S, Grgurevic L, Knutson MD, Pietrangelo A, Vukicevic S, Lin HY, Babitt JL. BMP6 is a key endogenous regulator of hepcidin expression and iron metabolism. Nat Genet. 2009;41:482–487. doi: 10.1038/ng.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Babitt JL, Huang FW, Xia Y, Sidis Y, Andrews NC, Lin HY. Modulation of bone morphogenetic protein signaling in vivo regulates systemic iron balance. J Clin Invest. 2007;117:1933–1939. doi: 10.1172/JCI31342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu PB, Hong CC, Sachidanandan C, Babitt JL, Deng DY, Hoyng SA, Lin HY, Bloch KD, Peterson RT. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat Chem Biol. 2008;4:33–41. doi: 10.1038/nchembio.2007.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin L, Goldberg YP, Ganz T. Competitive regulation of hepcidin mRNA by soluble and cell-associated hemojuvelin. Blood. 2005;106:2884–2889. doi: 10.1182/blood-2005-05-1845. [DOI] [PubMed] [Google Scholar]
- 53.Chen W, Huang FW, de Renshaw TB, Andrews NC. Skeletal muscle hemojuvelin is dispensable for systemic iron homeostasis. Blood. 2011;117:6319–6325. doi: 10.1182/blood-2010-12-327957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gkouvatsos K, Wagner J, Papanikolaou G, Sebastiani G, Pantopoulos K. Conditional disruption of mouse Hfe2 gene: Maintenance of systemic iron homeostasis requires hepatic but not skeletal muscle hemojuvelin. Hepatol. 2011;54:1800–1807. doi: 10.1002/hep.24547. [DOI] [PubMed] [Google Scholar]
- 55.Silvestri L, Pagani A, Nai A, De Domenico I, Kaplan J, Camaschella C. The serine protease matriptase-2 (TMPRSS6) inhibits hepcidin activation by cleaving membrane hemojuvelin. Cell Metab. 2008;8:502–511. doi: 10.1016/j.cmet.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Du X, She E, Gelbart T, Truksa J, Lee P, Xia Y, Khovananth K, Mudd S, Mann N, Moresco EM, Beutler E, Beutler B. The serine protease TMPRSS6 is required to sense iron deficiency. Science. 2008;320:1088–1092. doi: 10.1126/science.1157121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Folgueras AR, Martin de Lara F, Pendas AM, Garabaya C, Rodriguez F, Astudillo A, Bernal T, Cabanillas R, Lopez-Otin C, Velasco G. The membrane-bound serine protease matriptase-2 (Tmprss6) is an essential regulator of iron homeostasis. Blood. 2008;112:2539–2545. doi: 10.1182/blood-2008-04-149773. [DOI] [PubMed] [Google Scholar]
- 58.Krijt J, Fujikura Y, Ramsay AJ, Velasco G, Necas E. Liver hemojuvelin protein levels in mice deficient in matriptase-2 (Tmprss6) Blood Cells Mol Dis. 2011;47:133–137. doi: 10.1016/j.bcmd.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 59.Meynard D, Vaja V, Sun CC, Corradini E, Chen S, Lopez-Otin C, Grgurevic L, Hong CC, Stirnberg M, Gutschow M, Vukicevic S, Babitt JL, Lin HY. Regulation of TMPRSS6 by BMP6 and iron in human cells and mice. Blood. 2011;118:747–756. doi: 10.1182/blood-2011-04-348698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vujic Spasic M, Kiss J, Herrmann T, Galy B, Martinache S, Stolte J, Grone HJ, Stremmel W, Hentze MW, Muckenthaler MU. Hfe acts in hepatocytes to prevent hemochromatosis. Cell Metab. 2008;7:173–178. doi: 10.1016/j.cmet.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 61.Wallace DF, Summerville L, Subramaniam VN. Targeted disruption of the hepatic transferrin receptor 2 gene in mice leads to iron overload. Gastroenterol. 2007;132:301–310. doi: 10.1053/j.gastro.2006.11.028. [DOI] [PubMed] [Google Scholar]
- 62.van Dijk BA, Laarakkers CM, Klaver SM, Jacobs EM, van Tits LJ, Janssen MC, Swinkels DW. Serum hepcidin levels are innately low in HFE-related haemochromatosis but differ between C282Y-homozygotes with elevated and normal ferritin levels. Br J Haematol. 2008;142:979–985. doi: 10.1111/j.1365-2141.2008.07273.x. [DOI] [PubMed] [Google Scholar]
- 63.Zimmermann MB, Troesch B, Biebinger R, Egli I, Zeder C, Hurrell RF. Plasma hepcidin is a modest predictor of dietary iron bioavailability in humans, whereas oral iron loading, measured by stable-isotope appearance curves, increases plasma hepcidin. Am J Clin Nutr. 2009;90:1280–1287. doi: 10.3945/ajcn.2009.28129. [DOI] [PubMed] [Google Scholar]
- 64.Lebrón JA, Bennet MJ, Vaughn DE, Chirino AJ, Snow PM, Mintier GA, Feder JN, Bjorkman PJ. Crystal structure of the hemochromatosis protein HFE and characterization of its interaction with transferrin receptor. Cell. 1998;93:111–123. doi: 10.1016/s0092-8674(00)81151-4. [DOI] [PubMed] [Google Scholar]
- 65.Schmidt PJ, Toran PT, Giannetti AM, Bjorkman PJ, Andrews NC. The transferrin receptor modulates hfe-dependent regulation of hepcidin expression. Cell Metab. 2008;7:205–214. doi: 10.1016/j.cmet.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.West AP, Jr, Giannetti AM, Herr AB, Bennett MJ, Nangiana JS, Pierce JR, Weiner LP, Snow PM, Bjorkman PJ. Mutational analysis of the transferrin receptor reveals overlapping HFE and transferrin binding sites. J Mol Biol. 2001;313:385–397. doi: 10.1006/jmbi.2001.5048. [DOI] [PubMed] [Google Scholar]
- 67.Chen J, Chloupkova M, Gao J, Chapman-Arvedson TL, Enns CA. HFE modulates transferrin receptor 2 levels in hepatoma cells via interactions that differ from transferrin receptor 1-HFE interactions. J Biol Chem. 2007;282:36862–36870. doi: 10.1074/jbc.M706720200. [DOI] [PubMed] [Google Scholar]
- 68.Giannetti AM, Bjorkman PJ. HFE and transferrin directly compete for transferrin receptor in solution and at the cell surface. J Biol Chem. 2004;279:25866–25875. doi: 10.1074/jbc.M401467200. [DOI] [PubMed] [Google Scholar]
- 69.Goswami T, Andrews NC. Hereditary hemochromatosis protein, HFE, interaction with transferrin receptor 2 suggests a molecular mechanism for mammalian iron sensing. J Biol Chem. 2006;281:28494–28498. doi: 10.1074/jbc.C600197200. [DOI] [PubMed] [Google Scholar]
- 70.Johnson MB, Enns CA. Diferric transferrin regulates transferrin receptor 2 protein stability. Blood. 2004;104:4287–4293. doi: 10.1182/blood-2004-06-2477. [DOI] [PubMed] [Google Scholar]
- 71.Robb A, Wessling-Resnick M. Regulation of transferrin receptor 2 protein levels by transferrin. Blood. 2004;104:4294–4299. doi: 10.1182/blood-2004-06-2481. [DOI] [PubMed] [Google Scholar]
- 72.Pietrangelo A. Hereditary hemochromatosis: pathogenesis, diagnosis, and treatment. Gastroenterol. 2010;139:393–408. 408. doi: 10.1053/j.gastro.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 73.Wallace DF, Summerville L, Crampton EM, Frazer DM, Anderson GJ, Subramaniam VN. Combined deletion of Hfe and transferrin receptor 2 in mice leads to marked dysregulation of hepcidin and iron overload. Hepatol. 2009;50:1992–2000. doi: 10.1002/hep.23198. [DOI] [PubMed] [Google Scholar]
- 74.Ramey G, Deschemin JC, Vaulont S. Cross-talk between the mitogen activated protein kinase and bone morphogenetic protein/hemojuvelin pathways is required for the induction of hepcidin by holotransferrin in primary mouse hepatocytes. Haematologica. 2009;94:765–772. doi: 10.3324/haematol.2008.003541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Poli M, Luscieti S, Gandini V, Maccarinelli F, Finazzi D, Silvestri L, Roetto A, Arosio P. Transferrin receptor 2 and HFE regulate furin expression via mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/Erk) signaling. Implications for transferrin-dependent hepcidin regulation. Haematologica. 2010;95:1832–1840. doi: 10.3324/haematol.2010.027003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ryan JD, Ryan E, Fabre A, Lawless MW, Crowe J. Defective bone morphogenic protein signaling underlies hepcidin deficiency in HFE hereditary hemochromatosis. Hepatol. 2010;52:1266–1273. doi: 10.1002/hep.23814. [DOI] [PubMed] [Google Scholar]
- 77.Wrighting DM, Andrews NC. Interleukin-6 induces hepcidin expression through STAT3. Blood. 2006;108:3204–3209. doi: 10.1182/blood-2006-06-027631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Verga Falzacappa MV, Vujic Spasic M, Kessler R, Stolte J, Hentze MW, Muckenthaler MU. STAT3 mediates hepatic hepcidin expression and its inflammatory stimulation. Blood. 2007;109:353–358. doi: 10.1182/blood-2006-07-033969. [DOI] [PubMed] [Google Scholar]
- 79.Pietrangelo A, Dierssen U, Valli L, Garuti C, Rump A, Corradini E, Ernst M, Klein C, Trautwein C. STAT3 is required for IL-6-gp130-dependent activation of hepcidin in vivo. Gastroenterol. 2007;132:294–300. doi: 10.1053/j.gastro.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 80.Pak M, Lopez MA, Gabayan V, Ganz T, Rivera S. Suppression of hepcidin during anemia requires erythropoietic activity. Blood. 2006;108:3730–3735. doi: 10.1182/blood-2006-06-028787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vokurka M, Krijt J, Sulc K, Necas E. Hepcidin mRNA levels in mouse liver respond to inhibition of erythropoiesis. Physiol Res. 2006;55:667–674. doi: 10.33549/physiolres.930841. [DOI] [PubMed] [Google Scholar]
- 82.Tanno T, Bhanu NV, Oneal PA, Goh SH, Staker P, Lee YT, Moroney JW, Reed CH, Luban NL, Wang RH, Eling TE, Childs R, Ganz T, Leitman SF, Fucharoen S, Miller JL. High levels of GDF15 in thalassemia suppress expression of the iron regulatory protein hepcidin. Nat Med. 2007;13:1096–1101. doi: 10.1038/nm1629. [DOI] [PubMed] [Google Scholar]
- 83.Tanno T, Porayette P, Sripichai O, Noh SJ, Byrnes C, Bhupatiraju A, Lee YT, Goodnough JB, Harandi O, Ganz T, Paulson RF, Miller JL. Identification of TWSG1 as a second novel erythroid regulator of hepcidin expression in murine and human cells. Blood. 2009;114:181–186. doi: 10.1182/blood-2008-12-195503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Santini V, Girelli D, Sanna A, Martinelli N, Duca L, Campostrini N, Cortelezzi A, Corbella M, Bosi A, Reda G, Olivieri O, Cappellini MD. Hepcidin levels and their determinants in different types of myelodysplastic syndromes. PLoS ONE. 2011;6:e23109. doi: 10.1371/journal.pone.0023109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Papanikolaou G, Tzilianos M, Christakis JI, Bogdanos D, Tsimirika K, MacFarlane J, Goldberg YP, Sakellaropoulos N, Ganz T, Nemeth E. Hepcidin in iron overload disorders. Blood. 2005;105:4103–4105. doi: 10.1182/blood-2004-12-4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kattamis A, Papassotiriou I, Palaiologou D, Apostolakou F, Galani A, Ladis V, Sakellaropoulos N, Papanikolaou G. The effects of erythropoetic activity and iron burden on hepcidin expression in patients with thalassemia major. Haematologica. 2006;91:809–812. [PubMed] [Google Scholar]
- 87.Peyssonnaux C, Zinkernagel AS, Schuepbach RA, Rankin E, Vaulont S, Haase VH, Nizet V, Johnson RS. Regulation of iron homeostasis by the hypoxia-inducible transcription factors (HIFs) J Clin Invest. 2007;117:1926–1932. doi: 10.1172/JCI31370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Volke M, Gale DP, Maegdefrau U, Schley G, Klanke B, Bosserhoff AK, Maxwell PH, Eckardt KU, Warnecke C. Evidence for a lack of a direct transcriptional suppression of the iron regulatory peptide hepcidin by hypoxia-inducible factors. PLoS ONE. 2009;4:e7875. doi: 10.1371/journal.pone.0007875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mastrogiannaki M, Matak P, Mathieu JR, Delga S, Mayeux P, Vaulont S, Peyssonnnaux C. Hepatic HIF-2 down-regulates hepcidin expression in mice through epo-mediated increase in erythropoiesis. Haematologica. 2011;96 doi: 10.3324/haematol.2011.056119. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chaston TB, Patak P, Pourvali K, Srai SK, McKie AT, Sharp PA. Hypoxia inhibits hepcidin expression in HuH7 hepatoma cells via decreased SMAD4 signaling. Am J Physiol Cell Physiol. 2011;300:C888–895. doi: 10.1152/ajpcell.00121.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shah YM, Matsubara T, Ito S, Yim SH, Gonzalez F. Intestinal Hypoxia- Inducible Transcription Factors Are Essential for Iron Absorption following Iron Deficiency. Cell Metab. 2009;9:152–164. doi: 10.1016/j.cmet.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mastrogiannaki M, Matak P, Keith B, Simon MC, Vaulont S, Peyssonnaux C. HIF-2alpha, but not HIF-1alpha, promotes iron absorption in mice. J Clin Invest. 2009;119:1159–1166. doi: 10.1172/JCI38499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vanoaica L, Darshan D, Richman L, Schumann K, Kuhn LC. Intestinal ferritin h is required for an accurate control of iron absorption. Cell Metab. 2010;12:273–282. doi: 10.1016/j.cmet.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 94.Ludwiczek S, Aigner E, Theurl I, Weiss G. Cytokine-mediated regulation of iron transport in human monocytic cells. Blood. 2003;101:4148–4154. doi: 10.1182/blood-2002-08-2459. [DOI] [PubMed] [Google Scholar]
- 95.Paradkar PN, Roth JA. Expression of the 1B isoforms of divalent metal transporter (DMT1) is regulated by interaction of NF-Y with a CCAAT-box element near the transcription start site. J Cell Physiol. 2007;211:183–188. doi: 10.1002/jcp.20932. [DOI] [PubMed] [Google Scholar]
- 96.Gunshin H, Allerson CR, Polycarpou-Schwarz M, Rofts A, Rogers JT, Kishi F, Hentze MW, Rouault TA, Andrews N, Hediger MA. Iron-dependent regulation of the divalent metal ion transporter. FEBS Lett. 2001;509:309–316. doi: 10.1016/s0014-5793(01)03189-1. [DOI] [PubMed] [Google Scholar]
- 97.Lymboussaki A, Pignatti E, Montosi G, Garuti C, Haile DJ, Pietrangelo A. The role of the iron responsive element in the control of ferroportin1/IREG1/MTP1 gene expression. J Hepatol. 2003;39:710–715. doi: 10.1016/s0168-8278(03)00408-2. [DOI] [PubMed] [Google Scholar]
- 98.Pietrangelo A, Caleffi A, Henrion J, Ferrara F, Corradini E, Kulaksiz H, Stremmel W, Andreone P, Garuti C. Juvenile hemochromatosis associated with pathogenic mutations of adult hemochromatosis genes. Gastroenterology. 2005;128:470–479. doi: 10.1053/j.gastro.2004.11.057. [DOI] [PubMed] [Google Scholar]
- 99.Aisen P. Transferrin, the transferrin receptor, and the uptake of iron by cells. Met Ions Biol Syst. 1998;35:585–631. [PubMed] [Google Scholar]
- 100.Andrews NC. Iron homeostasis: insights from genetics and animal models. Nat Rev Genetics. 2000;1:208–217. doi: 10.1038/35042073. [DOI] [PubMed] [Google Scholar]
- 101.Aisen P. Transferrin receptor 1. Int J Biochem & Cell Biol. 2004;36:2137–2143. doi: 10.1016/j.biocel.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 102.Dautry-Varsat A, Ciechanover A, Lodish HF. pH and the recycling of transferrin during receptor-mediated endocytosis. Proc Natl Acad Sci USA. 1983;80:2258–2262. doi: 10.1073/pnas.80.8.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bali PK, Zak O, Aisen P. A new role for the transferrin receptor in the release of iron from transferrin. Biochem. 1991;30:324–328. doi: 10.1021/bi00216a003. [DOI] [PubMed] [Google Scholar]
- 104.Ohgami RS, Campagna DR, McDonald A, Fleming MD. The Steap proteins are metalloreductases. Blood. 2006;108:1388–1394. doi: 10.1182/blood-2006-02-003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fleming MD, Tremor CC, 3rd, Su MA, Foernzler D, Beier DR, Dietrich WF, Andrews NC. Microcytic anaemia mice have a mutation in Nramp2, a candidate iron transporter gene. Nat Genet. 1997;16:383–386. doi: 10.1038/ng0897-383. [DOI] [PubMed] [Google Scholar]
- 106.Fleming MD, Romano MA, Su MA, Garrick ML, Garrick MD, Andrews NC. Nramp2 is mutated in anemia Belgrade (b) rat: Evidence for a role for Nramp2 in endosomal iron transport. Proc Natl Acad Sci USA. 1998;95:1148–1153. doi: 10.1073/pnas.95.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gunshin H, Mackenzie B, Berger UV, Gunshin Y, Romero MF, Boron WF, Nussberger S, Gollan JL, Hediger MA. Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature. 1997;388:482–488. doi: 10.1038/41343. [DOI] [PubMed] [Google Scholar]
- 108.Gunshin H, Fujiwara Y, Custodio AO, Direnzo C, Robine S, Andrews NC. Slx11a2 is required for intestinal iron absorption and erythropoiesis but dispensable in placenta and liver. J Clin Invest. 2005;115:1258–1266. doi: 10.1172/JCI24356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mims MP, Guan Y, Pospisilova D, Priwitzerova M, Indra K, Ponka P, Divoky Y, Prchal JT. Identification of a human mutation of DMT1 in a patient with microcytic anemia and iron overload. Blood. 2005;105:1337–1342. doi: 10.1182/blood-2004-07-2966. [DOI] [PubMed] [Google Scholar]
- 110.Ohgami RS, Campagna DR, Greer EL, Antiochos B, McDonald A, Chen J, Sharp JJ, Fujiwara Y, Barker JE, Fleming MD. Identification of a ferrireductase required for efficient transferrin-dependent iron uptake in erythroid cells. Nat Genet. 2005;37:1264–1269. doi: 10.1038/ng1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ohgami RS, Campagna DR, Antiochos B, Wood EB, Sharp JJ, Barker JE, Fleming MD. Nm1054: a spontaneous, recessive, hypochromic, microcytic anemia mutation in the mouse. Blood. 2005;106:3625–3631. doi: 10.1182/blood-2005-01-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lambe T, Simpson RJ, Dawson S, Bouriez-Jones T, Crockford TL, Lepherd M, Latunde-Dada GO, Robinson H, Raja KB, Campagna DR, Villarreal G, Jr, Ellroy JC, Goodnow CC, Fleming MD, McKie AT, Cornall RJ. Identification of Steap3 endosomal targeting motif essential for normal iron metabolism. Blood. 2009;113:1805–1808. doi: 10.1182/blood-2007-11-120402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Levy JE, Jin O, Fujiwara Y, Kuo F, Andews NC. Transferrin receptor is essential for development of erythrocytes and the nervous system. Nat Genet. 1999;21:396–399. doi: 10.1038/7727. [DOI] [PubMed] [Google Scholar]
- 114.Larrick J, Hyman E. Acquired iron-deficiency anemia caused by an antibody against the transferrin receptor. N Engl J Med. 1984;311:214–218. doi: 10.1056/NEJM198407263110402. [DOI] [PubMed] [Google Scholar]
- 115.Kawabata H, Yang R, Hirama T, Vuong PT, Kawano S, Gombart AF, Koeffler HP. Molecular cloning of transferrin receptor 2. A new member of the transferrin receptor-like family. J Biol Chem. 1999;274:20826–20832. doi: 10.1074/jbc.274.30.20826. [DOI] [PubMed] [Google Scholar]
- 116.West AP, Jr, Bennett MJ, Sellers VM, Andrews NC, Enns CA, Bjorkman PJ. Comparisons of the interactions of transferrin receptor with transferrin and hereditary hemochromatosis protein HFE. J Biol Chem. 2000;275:38135–38138. doi: 10.1074/jbc.C000664200. [DOI] [PubMed] [Google Scholar]
- 117.Johnson MB, Enns CA. Differic transferrin regulates transferrin receptor 2 protein stability. Blood. 2004;104:4287–4293. doi: 10.1182/blood-2004-06-2477. [DOI] [PubMed] [Google Scholar]
- 118.Robb A, Wessling-Resnick M. Regulation of transferrin receptor 2 protein levels by transferrin. Blood. 2004;104:4294–4299. doi: 10.1182/blood-2004-06-2481. [DOI] [PubMed] [Google Scholar]
- 119.Kawabata H, Fleming RE, Gui D, Moon SY, Saitoh T, O’Kelly J, Umehara Y, Wano Y, Said JW, Koeffler HP. Expression of hepcidin is down regulated in TfR2 mutant mice manifesting a phenotype of hereditary hemochromatosis. Blood. 2005;105:376–381. doi: 10.1182/blood-2004-04-1416. [DOI] [PubMed] [Google Scholar]
- 120.Wallace DF, Summerville L, Subramaniam VN. Targeted disruption of the hepatic transferrin receptor 2 gene in mice leads to iron overload. Gastroenterol. 2007;132:301–310. doi: 10.1053/j.gastro.2006.11.028. [DOI] [PubMed] [Google Scholar]
- 121.Breuer W, Hershko C, Cabantchik ZI. The importance of non-transferrin bound iron in disorders of iron metabolism. Transfusion Sci. 2000;23:185–192. doi: 10.1016/s0955-3886(00)00087-4. [DOI] [PubMed] [Google Scholar]
- 122.Yang J, Goetz D, Li Y-Y, Wang W, Mori K, Setlik D, Du T, Erdjument-Bromage H, Tempst P, Strong R, Barasch J. An iron delivery pathway mediated by a lipocalin. Mol Cell. 2002;10:1045–1056. doi: 10.1016/s1097-2765(02)00710-4. [DOI] [PubMed] [Google Scholar]
- 123.Yang J, Mori K, Li JY, Barasch J. Iron, lipocalin, and kidney epithelia. Am J Physiol Renal Physiol. 2003;285:F9–F18. doi: 10.1152/ajprenal.00008.2003. [DOI] [PubMed] [Google Scholar]
- 124.Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, Akira S, Aderem A. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestering iron. Nature. 2004;432:917–921. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- 125.Berger T, Togawa A, Duncan GS, Elia AJ, You-Ten A, Wakeham A, Fong HE, Cheung CC, Mak TW. Lipocalin-2-deficient mice exhibit increased sensitivity to Escherichia coli infection but not to ischemia-reperfusion injury. Proc Natl Acad Sci USA. 2006;103:1834–1839. doi: 10.1073/pnas.0510847103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Liu Z, Yang A, Wang Z, Bunting K, Davuluri G, Green MR, Devireddy LR. Multiple apoptotic defects in hematopoietic cells from mice lacking lipocalin 24p3. J Biol Chem. 2011;286:20606–20614. doi: 10.1074/jbc.M110.216549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Huang H, Akira S, Santos MM. Is the iron donor lipocalin 2 implicated in the pathophysiology of hereditary hemochromatosis? Hepatol. 2009;49:1012–1016. doi: 10.1002/hep.22699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Collins JF, Franck CA, Kowdley KA, Ghishan FK. Identificaiton of differentially expressed genes in response to dietary iron deprivation in rat duodenum. Am J Phsiol: Gasterointest Liver Physiol. 2005;288:G964–G971. doi: 10.1152/ajpgi.00489.2004. [DOI] [PubMed] [Google Scholar]
- 129.Shindo M, Torimoto Y, Saito H, Motomura W, Ikuta K, Sato K, Fujimoto Y, Kohgo Y. Functional role of DMT1 in transferrin-independent iron uptake by human hepatocyte and hepatocellular carcinoma cell, HLF. Hepatol Res. 2006;35:152–162. doi: 10.1016/j.hepres.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 130.Xu H, Jin J, DeFelice JL, Andrews NC, Clapham DE. A spontaneous, recurrent mutation in divalent metal transporter-1 exposes a calcium entry pathway. PLoS Biol. 2004;2:0378–0385. doi: 10.1371/journal.pbio.0020050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Liuzzi JP, Aydemir F, Nam H, Knutson MD, Cousins RJ. Zip14 (Slc39a14) mediates non-transferrin-bound iron uptake into cells. Proc Natl Acad Sci USA. 2006;103:13612–13617. doi: 10.1073/pnas.0606424103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gao J, Zhao N, Knutson MD, Enns CA. The hereditary hemochromatosis protein, HFE, inhibits iron uptake via down-regulation of Zip14 in HepG2 cells. J Biol Chem. 2008;283:21462–21468. doi: 10.1074/jbc.M803150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hojyo S, Fukada T, Shimoda S, Ohasi W, Bin BH, Koseki H, Hirano T. The zinc transporter Slc39a14/Zip14 controls G-protein coupled receptor-mediated signaling required for systemic growth. PLoS ONE. 2011;6:e18059. doi: 10.1371/journal.pone.0018059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tsushima RG, Wickenden AD, Bouchard RA, Oudit GY, Liu PP, Backx PH. Modulation of iron uptake in heart by L-type Ca2+ channel modifiers: possible implications in iron overload. Cir Res. 1999;84:1302–1309. doi: 10.1161/01.res.84.11.1302. [DOI] [PubMed] [Google Scholar]
- 135.Oudit GY, Sun H, Triveri MG, Koch SE, Dawood F, Ackerley C, Yazdanpanah M, Wilson GJ, Schwartz A, Liu PP, Backx PH. L-type Ca2+ channels provide a major pathway for iron entry into cardiomyocytes in iron-overload cardiomyopathy. Nat Med. 2003;9:1187–1194. doi: 10.1038/nm920. [DOI] [PubMed] [Google Scholar]
- 136.Ludwiczek S, Theurl I, Muckenthaler MU, Jakab M, Nair SM, Theurl M, Kiss J, Paulmichl M, Hentze MW, Ritter M, Weiss G. Ca2+ channel blockers reverse iron overload by a new mechanism via divalent metal transporter-1. Nat Med. 2007;13:448–454. doi: 10.1038/nm1542. [DOI] [PubMed] [Google Scholar]
- 137.Mackenzie B, Shawki A, Ghio AJ, Stonehuerner JD, Zhao L, Ghadersohi S, Garrick LM, Garrick MD. Calcium channel blockers do not affect iron transport mediated by divalent metal-ion tansporter-1. Blood. 2010;115:4148–4149. doi: 10.1182/blood-2010-03-274738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Moosmang M, Lenhardt P, Halder N, Hoffmann F, Wegener J. Mouse models to study L-type calcium channel function. Pharmacol Therapeutics. 2005;106:347–355. doi: 10.1016/j.pharmthera.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 139.Chen TT, Li L, Chung DH, Allen CD, Torti SV, Torti F, Cyster JG, Che CY, Brodsky FM, Niemi EC, Nakamura MC, Seaman WE, Daws MR. Tim-2 is expressed on B cells and in liver and kidney and is a receptor for H-ferritin endocytosis. J Exp Med. 2005;202:955–965. doi: 10.1084/jem.20042433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Li JY, Paragas N, Ned RM, Qiu A, Viltard M, Leete T, Drexler IR, Chen X, Sanna-Cherchi S, Mohammed F, Williams D, Lin CS, Schmidt-Ott KM, Andrews NC, Barasch J. Scara5 is a ferritin receptor mediating non-transferrin iron delivery. Dev Cell. 2009;16:35–46. doi: 10.1016/j.devcel.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rennert PD, Ichimura T, Sizing ID, Bailly V, Li Z, Rennard R, McCoon P, Pablo L, Miklasz S, Tarilonte L, Bonventre JV. T cell, Ig domain, mucin domain-2 gene-deficient mice reveal a novel mechanism for the regulation of Th2 immune responses and airway inflammation. J Immunol. 2006;177:4311–4321. doi: 10.4049/jimmunol.177.7.4311. [DOI] [PubMed] [Google Scholar]
- 142.Huang J, Zheng DL, Qin FS, Cheng N, Chen H, Wan BB, Wang YP, Xiao HS, Han ZG. Genetic and epigenetic silencing of Scara5 may contribute to human hepatocellular carcinoma activating FAK signaling. J Clin Invest. 2010;120:223–241. doi: 10.1172/JCI38012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shayegi M, Latunde-Dada GO, Oakhill JS, Laftah AH, Takeuchi K, Halliday N, Khan Y, Warley A, McCann FE, Hider RC, Frazer DM, Anderson GJ, Vulpe CD, Simpson RJ, McKie AT. Identification of an intestinal heme transporter. Cell. 2005;122:789–801. doi: 10.1016/j.cell.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 144.Qiu AD, Jansen M, Sakaris A, Min SH, Chattopadhay S, Tsai E, Sandoval C, Zhao RB, Akabas MH, Goldman ID. Identification of an intestinal folate transporter and the molecular basis for hereditary folate malabsorption. Cell. 2006;127:917–928. doi: 10.1016/j.cell.2006.09.041. [DOI] [PubMed] [Google Scholar]
- 145.Mims MP, Prchal JT. Divalent metal transporter 1. Hematol. 2005;10:339–345. doi: 10.1080/10245330500093419. [DOI] [PubMed] [Google Scholar]
- 146.Dong XP, Cheng X, Mills E, Delling M, Wang F, Kurz T, Xu H. The type IV mucolipidosis-associated protein TRPML1 is an endolysosomal iron release channel. Nature. 2008;455:992–996. doi: 10.1038/nature07311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rajagopal A, Rao AU, Amigo J, Tian M, Upadhay SK, Hall C, Uhm S, Mathew MK, Fleming MD, Paw BH, Krause M, Hamza I. Haem homeostasis is regulated by regulated by the conserved and concerted functions of HRG-1 proteins. Nature. 2008;453:1127–1131. doi: 10.1038/nature06934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kruszewski M. Labile iron pool: the main determinant of cellular response to oxidative stress. Mut Res. 2003;531:81–92. doi: 10.1016/j.mrfmmm.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 149.Petrat F, de Groot H, Sustmann R, Rauen U. The chelatable iron pool in living cells: a methodically defined quantity. Biol Chem. 2002;383:489–502. doi: 10.1515/BC.2002.051. [DOI] [PubMed] [Google Scholar]
- 150.Glickstein H, Ben El R, Shvartsman M, Cabantchik ZI. Intracellular labile iron pool as direct targets of iron chelators: a fluorescence study of chelator action in living cells. Blood. 2005;106:3242–3250. doi: 10.1182/blood-2005-02-0460. [DOI] [PubMed] [Google Scholar]
- 151.Meneghini R. Iron homeostasis, oxidative stress, and DNA damage. Free Radical Biol & Med. 1997;23:783–792. doi: 10.1016/s0891-5849(97)00016-6. [DOI] [PubMed] [Google Scholar]
- 152.Devireddy LR, Gazin C, Zhu X, Green MR. A cell surface receptor for lipocalin 24p3 selectively mediates apoptosis and iron uptake. Cell. 2005;123:1293–1305. doi: 10.1016/j.cell.2005.10.027. [DOI] [PubMed] [Google Scholar]
- 153.Richardson DR. 24p3 and its receptor: Dawn of a new iron age? Cell. 2005;123:1175–1177. doi: 10.1016/j.cell.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 154.Richardson DR, Lane DJ, Becker EM, Huang ML, Whitnall M, Rahmanto YS, Sheftel AD, Ponka P. Mitochondrial iron trafficking and the integration of iron metabolism between the mitochondrion and cytosol. Proc Natl Acad Sci USA. 2010;107:10775–10782. doi: 10.1073/pnas.0912925107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Fernandez-Pol JA. Isolation and characterization of a siderophore-like growth factor from mutants of SV40-transformed cells adapted to picolinic acid. Cell. 1977;14:489–499. doi: 10.1016/0092-8674(78)90235-0. [DOI] [PubMed] [Google Scholar]
- 156.Jones R, Peterson C, Grady R, Cerami A. Low molecular weight iron-binding factor from mammalian tissue that potentiates bacterial growth. J Exp Med. 1980;151:418–428. doi: 10.1084/jem.151.2.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Flower DR. Beyond the superfamily: the lipocalin receptors. Biochim Biophys Acta. 2000;1482:327–336. doi: 10.1016/s0167-4838(00)00169-2. [DOI] [PubMed] [Google Scholar]
- 158.Goetz D, Holmes MA, Borregaard N, Bluhm ME, Raymond KN, Strong RK. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol Cell. 2002;10:1033–1043. doi: 10.1016/s1097-2765(02)00708-6. [DOI] [PubMed] [Google Scholar]
- 159.Fischbach MA, Lin HN, Liu DR, Walsh CT. How pathogenic bacteria evade mammalian sabotage in the battle for iron? Nat Chem Biol. 2006;2:132–138. doi: 10.1038/nchembio771. [DOI] [PubMed] [Google Scholar]
- 160.Devireddy LR, Hart DO, Goetz DH, Green MR. A mammalian siderophore synthesized by an enzyme with a bacterial homolog involved in enterobactin production. Cell. 2010;141:1006–1017. doi: 10.1016/j.cell.2010.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Crosa JH, Walsh CT. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol Mol Biol Reviews. 2002;66:223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Raymond KN, Dertz EA, Kim SS. Enterobactin: an archetype for microbial iron transport. Proc Natl Acad Sci USA. 2003;100:3584–3588. doi: 10.1073/pnas.0630018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Muhlenhoff U, Molik S, Godoy JR, Uzarska MA, Richter N, Seubert A, Zhang Y, Stubbe J, Pierrel F, Herrero E, Liilig CH, Lill R. Cytosolic monothiol glutaredoxins function in intracellular iron sensing and trafficking via their bound iron-sulfur cluster. Cell Metab. 2010;12:373–385. doi: 10.1016/j.cmet.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Nilsson R, Schultz IJ, Pierce EL, Soltis KA, Naranutarat A, Ward DM, Baughman JM, Paradkar PN, Kingsley PD, Culotta VC, Kaplan J, Palis J, Paw BH, Mootha VK. Discovery of genes essential for heme biosynthesis through large-scale gene expression analysis. Cell Metab. 2009;10:119–130. doi: 10.1016/j.cmet.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Shaw GC, Cope JJ, Li L, Corson K, Hersey C, Ackerman GE, Gwynn B, Lambert AJ, Wingert RA, Traver D, Trede NS, Barut BA, Zhou Y, Minet E, Donovan A, Brownlie A, Balzan R, Weiss MJ, Peters LL, Kaplan J, Zon LI, Paw BH. Mitoferrin is essential for erythroid iron assimilation. Nature. 2006;440:96–100. doi: 10.1038/nature04512. [DOI] [PubMed] [Google Scholar]
- 166.Chen W, Paradkar PN, Li L, Pierce EL, Langer NB, Takahasi-Makise N, Hyde BB, Shirihai OS, Ward DM, Kaplan J, Paw BH. Abcb10 physically interacts with Mitoferrin-1 (Slc25a37) to enhance its stability and function in the erythroid mitochondria. Proc Natl Acad Sci USA. 2009;106:16263–16268. doi: 10.1073/pnas.0904519106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Paradkar PN, Zumbrennen KB, Paw BH, Ward DM, Kaplan J. Regulation of mitochondrial iron import through differential turnover of mitoferrin 1 and mitoferrin 2. Mol Cell Biol. 2009;29:1007–1016. doi: 10.1128/MCB.01685-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Troadec MB, Warner D, Wallace J, Thomas K, Spangrude GJ, Phillips J, Khalimonchuk O, Paw BH, Ward DM, Kaplan J. Targeted deletion of the mouse Mitoferrin1 gene: from anemia to protoporphyria. Blood. 2011;117:5494–5502. doi: 10.1182/blood-2010-11-319483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhang AS, Sheftel AD, Ponka P. Intracellular kinetics of iron in reticulocytes: Evidence for endosome involvement in iron targeting to mitochondria. Blood. 2005;105:368–375. doi: 10.1182/blood-2004-06-2226. [DOI] [PubMed] [Google Scholar]
- 170.Galy B, Ferring-Appel D, Sauer SW, Kaden S, Lyoumi S, Puy H, Kolker S, Grone HJ, Hentze MW. Iron regulatory proteins secure mitochondrial iron sufficiency and function. Cell Metab. 2010;12:194–201. doi: 10.1016/j.cmet.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 171.Ajioka RS, Phillips JD, Kushner JP. Biosynthesis of heme in mammals. Biochim Biophys Acta. 2006;1763:723–726. doi: 10.1016/j.bbamcr.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 172.Sheftel AD, Richardson DR, Prchal J, Ponka P. Mitochondrial iron metabolism and sideroblastic anemia. Acta Hematol. 2009;122:120–133. doi: 10.1159/000243796. [DOI] [PubMed] [Google Scholar]
- 173.Krishnamurthy PC, Du G, Fukuda Y, Sun D, Sampath J, Mercer KE, Wang J, Sosa-Pineda B, Murti KG, Schuetz JD. Identification of a mammalian mitochondrial porphyrin transporter. Nature. 2006;443:586–589. doi: 10.1038/nature05125. [DOI] [PubMed] [Google Scholar]
- 174.Rufenacht U, Gregor A, Gouya L, Tarczynska-Nosal S, Schneider-Yin X, Deybach JC. New missense mutation in the human ferrochelatase gene in a family with erythropoietic protoporphyria: Functional studies and correlation of genotype and phenotype. Clin Chem. 2001;47:1112–1113. [PubMed] [Google Scholar]
- 175.Tong WH, Rouault TA. Functions of mitochondrial ISCU and cytosolic ISCU in mammalian iron-sulfur cluster biogenesis and iron homeostasis. Cell Metab. 2006;3:199–210. doi: 10.1016/j.cmet.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 176.Stemmler TL, Lesuisse E, Pain D, Dancis A. Frataxin and mitochondrial FeS cluster biogenesis. J Biol Chem. 2010;285:26737–26743. doi: 10.1074/jbc.R110.118679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Puccio H. Multicellular models of Friedreich ataxia. J Neurol. 2009;256:18–24. doi: 10.1007/s00415-009-1004-1. [DOI] [PubMed] [Google Scholar]
- 178.Lill R, Muhlenhoff U. Maturation of iron-sulfur proteins in eukaryotes: Mechanisms, connected process, and diseases. Annu Rev Biochem. 2008;77:669–700. doi: 10.1146/annurev.biochem.76.052705.162653. [DOI] [PubMed] [Google Scholar]
- 179.Kispal G, Csere P, Prohl C, Lill R. The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 1999;18:3981–3989. doi: 10.1093/emboj/18.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Pondarre C, Antiochos BB, Campagna DR, Clarke SL, Greer EL, Deck KM, McDonald A, Han AP, Medlock A, Kutok JL, Anderson SM, Eisenstein RS, Fleming MD. A mitochondrial ATP-binding cassette transporter Abcb7 is essential in mice and participates in cytosolic iron-sulfur cluster biogenesis. Hum Mol Genet. 2006;15:953–964. doi: 10.1093/hmg/ddl012. [DOI] [PubMed] [Google Scholar]
- 181.Napier I, Ponka P, Richardson DR. Iron trafficking in the mitochondrion: Novel pathways revealed by disease. Blood. 2005;105:1867–1874. doi: 10.1182/blood-2004-10-3856. [DOI] [PubMed] [Google Scholar]
- 182.Allikmets R, Raskind WH, Hutchinson A, Schueck ND, Dean M, Koeller DM. Mutation of a putative mitochondrial iron transporter gene (ABC7) in X-linked sideroblastic anemia and ataxia (XLSA/A) Hum Mol Genet. 1999;8:743–749. doi: 10.1093/hmg/8.5.743. [DOI] [PubMed] [Google Scholar]
- 183.Piccinelli P, Samuelsson T. Evolution of the iron-responsive element. RNA. 2007;13:952–966. doi: 10.1261/rna.464807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Roetto A, Bosio S, Gramaglia E, Barilaro MR, Zecchina G, Camaschella C. Pathogenesis of hyperferritinemia cataract syndrome. Blood Cells Mol Dis. 2002;29:532–535. doi: 10.1006/bcmd.2002.0590. [DOI] [PubMed] [Google Scholar]
- 185.Arosio P, Levi S. Cytosolic and mitochondrial ferritins in the regulation of cellular iron homeostasis and oxidative damage. Biochim Biophys Acta. 2010;1800:783–792. doi: 10.1016/j.bbagen.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 186.Zhang DL, Hughes RM, Ollivierre-Wilson H, Ghosh MC, Rouault TA. A ferroportin transcript that lacks an iron-responsive element enables duodenal and erythroid precursor cells to evade translational repression. Cell Metab. 2009;9:461–473. doi: 10.1016/j.cmet.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.McKie AT, Marciani P, Rolfs A, Brennan K, Wehr K, Barrow D, Miret S, Bomford A, Peters TJ, Farzaneh F, Hediger MA, Hentze MW, Simpson RJ. A novel duodenal iron-regulated transporter IREG1, implicated in the basolateral transfer of iron to the circulation. Mol Cell. 2000;5:299–309. doi: 10.1016/s1097-2765(00)80425-6. [DOI] [PubMed] [Google Scholar]
- 188.Schranzhofer M, Schifrer M, Cabrera JA, Kopp S, Chiba P, Beug H, Mullner EW. Remodeling the regulation of iron metabolism during erythroid differentiation to ensure efficient heme biosynthesis. Blood. 2006;107:4159–4167. doi: 10.1182/blood-2005-05-1809. [DOI] [PubMed] [Google Scholar]
- 189.Lok CN, Ponka P. Identification of an erythroid active element in the transferrin receptor gene. J Biol Chem. 2000;275:24185–24190. doi: 10.1074/jbc.M000944200. [DOI] [PubMed] [Google Scholar]
- 190.Vashisht AA, Zumbrennen KB, Huang X, Powers DN, Durazo A, Sun D, Bhaskaran N, Persson A, Uhlen M, Sangfelt O, Spruck C, Leibold EA, Wohlschlegel JA. Control of iron homeostasis by an iron-regulated ubiquitin ligase. Science. 2009;326:718–721. doi: 10.1126/science.1176333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Salahudeen AA, Thompson JW, Ruiz JC, Ma HW, Kinch LN, Li Q, Grishin NV, Bruick RK. An E3 ligase possessing an iron-responsive hemerythrin domain is a regulator of iron homeostasis. Science. 2009;326:722–726. doi: 10.1126/science.1176326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Clarke SL, Vasanthakumar A, Anderson SA, Pondarre C, Koh CM, Deck KM, Pitula JS, Epstein CJ, Fleming MD, Eisenstein RS. Iron-responsive degradation of iron-regulatory protein 1 does not require the Fe-S cluster. EMBO J. 2006;25:544–553. doi: 10.1038/sj.emboj.7600954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Wang J, Fillebeen C, Chen G, Biederbick A, Lill R, Pantopoulos K. Iron-dependent degradation of apo-IRP1 by the ubiquitin-proteasome pathway. Mol Cell Biol. 2007;27:2423–2430. doi: 10.1128/MCB.01111-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Smith SR, Ghosh MC, Ollivierre-Wilson H, Hang Tong W, Rouault TA. Complete loss of iron regulatory proteins 1 and 2 prevents viability of murine zygotes beyond the blastocyst stage of embryonic development. Blood Cells Mol Dis. 2006;36:283–287. doi: 10.1016/j.bcmd.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 195.Galy B, Ferring-Appel D, Kaden S, Grone HJ, Hentze MW. Iron regulatory proteins are essential for intestinal function and control key iron absorption molecules in the duodenum. Cell Metab. 2008;7:79–85. doi: 10.1016/j.cmet.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 196.Meyron-Holtz EG, Ghosh MC, Iwai K, LaVaute T, Brazzolotto X, Berger UV, Land W, Ollivierre-Wilson H, Grinberg A, Love P, Rouault TA. Genetic ablations of iron regulatory proteins 1 and 2 reveal why iron regulatory protein 2 dominates iron homeostasis. EMBO J. 2004;23:386–395. doi: 10.1038/sj.emboj.7600041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Cooperman SS, Meyron-Holtz EG, Ollivierre-Wilson H, Ghosh MC, McConnell JP, Rouault TA. Microcytic anemia, erythropoietic protoporphyria, and neurodegeneration in mice with targeted deletion of iron-regulatory protein 2. Blood. 2005;106:1084–1091. doi: 10.1182/blood-2004-12-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Galy B, Ferring D, Minana B, Bell O, Janser HG, Muckenthaler M, Schumann K, Hentze MW. Altered body iron distribution and microcytosis in mice deficient in iron regulatory protein 2 (IRP2) Blood. 2005;106:2580–2589. doi: 10.1182/blood-2005-04-1365. [DOI] [PubMed] [Google Scholar]
- 199.Ferring-Appel D, Hentze MW, Galy B. Cell-autonomous and systemic context-dependent functions of iron regulatory protein 2 in mammalian iron metabolism. Blood. 2009;113:679–687. doi: 10.1182/blood-2008-05-155093. [DOI] [PubMed] [Google Scholar]
- 200.LaVaute T, Smith SR, Cooperman S, Iwai K, Land W, Meyron-Holtz EG, Drake SK, Miller G, Abu-Asab M, Tsokos M, Switzer R3rd, Grinberg A, Love P, Tresser N, Rouault TA. Targeted deletion of the gene encoding iron regulatory protein-2 causes misregulation of iron metabolism and neurodegenerative disease in mice. Nat Genet. 2001;27:209–0214. doi: 10.1038/84859. [DOI] [PubMed] [Google Scholar]
- 201.Moroishi T, Nishiyama M, Takeda Y, Iwai K, Nakayama K. The FBXL5-IRP2 axis is integral to control of iron metabolism in vivo. Cell Metab. 2011;14:339–351. doi: 10.1016/j.cmet.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 202.Arosio P, Ingrassia R, Cavadini P. Ferritins: A family of molecules for iron storage, antioxidation and more. Biochim Biophys Acta. 2006;1790:589–599. doi: 10.1016/j.bbagen.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 203.Shi H, Bencze KZ, Stemmler TL, Philpott CC. A cytosolic iron chaperone that delivers iron to ferritin. Science. 2009;320:1207–1210. doi: 10.1126/science.1157643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zhang Y, Mikhael M, Xu D, Li Y, Soe-Lin B, Li W, Nie G, Zhao Y, Ponka P. Lysosomal proteolysis is the primary degradation pathway for cytosolic ferritin and cytosolic ferritin degradation is necessary for iron exit. Antioxid Redox Signal. 2010;13:999–1009. doi: 10.1089/ars.2010.3129. [DOI] [PubMed] [Google Scholar]
- 205.De Domenico I, Vaughn MB, Li L, Bagley D, Musci G, Ward DM, Kaplan J. Ferroportin-mediated mobilization of ferritin iron precedes ferritin degradation by the proteasome. EMBO J. 2006;25:5396–5404. doi: 10.1038/sj.emboj.7601409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.De Domenico I, Ward DM, Kaplan J. Specific iron chelators determine the route of ferritin degradation. Blood. 2009;114:4546–4551. doi: 10.1182/blood-2009-05-224188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Cozzi A, Corsi B, Levi S, Santambrogio P, Biasiotto G, Arosio P. Analysis of the biologic functions of H-and L-ferritins in HeLa cells by transfection with siRNAs and cDNAs: evidence of proliferative role of L-ferritin. Blood. 2004;103:2377–2383. doi: 10.1182/blood-2003-06-1842. [DOI] [PubMed] [Google Scholar]
- 208.Epsztein S, Glickstein H, Picard V, Slotki IN, Breuer W, Beaumont C, Cabantchik ZI. H-ferritin subunit overexpression in erythroid cells reduces the oxidative stress response and induces multidrug resistance properties. Blood. 1999;94:3593–3603. [PubMed] [Google Scholar]
- 209.Levi S, Corsi B, Bosisio M, Invernizzi R, Volz A, Sanford D, Arosio P, Drysdale J. A human mitochondrial ferritin encoded by an intronless gene. J Biol Chem. 2001;276:24437–24440. doi: 10.1074/jbc.C100141200. [DOI] [PubMed] [Google Scholar]
- 210.Santambrogio P, Biasiotto G, Sanvitto F, Olivieri S, Arosio P, Levi S. Mitochondrial ferritin expression in adult mouse tissues. J Histochem Cytochem. 2007;55:1129–1137. doi: 10.1369/jhc.7A7273.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Nie G, Chen G, Sheftel AD, Pantopoulos K, Ponka P. In vivo tumor growth is inhibited by cytosolic iron deprivation caused by the expression of mitochondrial ferritin. Blood. 2006;108:2428–2434. doi: 10.1182/blood-2006-04-018341. [DOI] [PubMed] [Google Scholar]
- 212.Ganz T. Cellular iron: ferroportin is the only way out. Cell Metab. 2005;1:155–157. doi: 10.1016/j.cmet.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 213.De Domenico I, Ward DM, Kaplan J. Regulation of iron acquisition and storage. Consequences for iron-linked disorders. Nat Rev Mol Cell Biol. 2008;9:72–81. doi: 10.1038/nrm2295. [DOI] [PubMed] [Google Scholar]
- 214.Donovan A, Lima CA, Pinkus JL, Pinkus GS, Zon LI, Robine S, Andrews NC. The iron exporter ferroportin/Slc40a1 is essential for iron homeostasis. Cell Metab. 2005;1:191–200. doi: 10.1016/j.cmet.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 215.Montosi G, Donovan A, Totaro A, Garuti C, Pignatti E, Cassanelli S, Trenor CC, Gasparini P, Andrews NC, Pietrangelo A. Autosomal dominant hemochromatosis is associated with a mutation in the Ferroportin (SLC11A3) gene. J Clin Invest. 2002;108:619–623. doi: 10.1172/JCI13468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Keel SB, Doty RT, Yang Z, Quigley JG, Chen J, Knoblaugh S, Kingsley PD, De Domenico I, Vaughn MB, Kaplan J, Palis J, Abkowitz JL. A heme export protein is required for red blood cell differentiation and iron homeostasis. Sicence. 2008;319:825–828. doi: 10.1126/science.1151133. [DOI] [PubMed] [Google Scholar]
- 217.Quigley JG, Yang Z, Worthington MT, Phillips JD, Sabo KM, Sabath DE, Berg CL, Sassa S, Wood BL, Abkowitz JL. Identification of a heme exporter that is essential for erythropoiesis. Cell. 2004;118:757–766. doi: 10.1016/j.cell.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 218.Rajadhyaksha AM, Elemento O, Puffenberger EG, Schierberl KC, Xiang JZ, Putorti ML, Berciano J, Poulin C, Brais B, Michaelides M, Weleber RG, Higgins JJ. Mutations in FLVCR1 cause posterior column ataxia and retinitis pigmentosa. Am J Hum Genet. 2002;87:643–654. doi: 10.1016/j.ajhg.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Duffy SP, Shing J, Saraon P, Berger LC, Eiden MV, Wilde A, Tailor CS. The fowler syndrome-associated protein FLVCR2 is an importer of heme. Mol Cell Biol. 2010;30:5318–5324. doi: 10.1128/MCB.00690-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Jonker JW, Buitelaar M, Wagenaar E, van der Valk MA, Scheffer GL, Scheper RJ, Plosch T, Kuipers F, Oude Elferink RPJ, Rosing H, Beijnen JH, Schinkel AH. The breast cancer resistance protein protects against a major chlorophyll-derived dietary phototoxin and protoporphyria. Proc Natl Acad Sci USA. 2002;99:15649–15654. doi: 10.1073/pnas.202607599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Woodward OM, Kottgen A, Coresh J, Boerwinkle E, Guggino WB, Kottgen M. Identification of a urate transporter, ABCG2, with a common functional polymorphism causing gout. Proc Natl Acad Sci USA. 2009;106:10338–10342. doi: 10.1073/pnas.0901249106. [DOI] [PMC free article] [PubMed] [Google Scholar]