Abstract
With the sequencing of an eukaryotic genome, it is possible to inventory the predicted proteome for proteins that carry one or more Src Homology 3 (SH3) domains. Due to the current ease of cloning and gene synthesis, these short domains can be readily overexpressed and manipulated for the purpose of characterizing their specificity and affinity for peptide ligands, as well as solving the three-dimensional structures of the domains. This information can be used to predict and confirm their cellular interacting partners, in the effort to understand the function of a eukaryotic protein by focusing on its SH3 domain. Finally, capitalizing on our mature understanding about protein–protein interacting modules, like the SH3 domain, it is possible to use directed evolution to enhance or change the specificity and affinity of an SH3 domain for the purpose of creating reagents to be used in biochemical purification or cell perturbation studies.
Keywords: Affinity reagents, Combinatorial peptide libraries, Directed evolution, Gene synthesis, Mapping protein–protein interactions, Peptide arrays, Phage-display, Proline-rich peptides, Protein–protein interaction module, Scaffold
1. Overview
Our understanding of Src Homology 3 (SH3) domains has come long way since the early 1988, when they were identified as a 60 amino acid segments shared among diverse signaling and cytoskeletal proteins of eukaryotes [1-3]. Since then, their three-dimensional structure has been determined [4,5], and it was recognized that they, along with other protein interaction modules [6], bind short proline-rich peptide segments within proteins [7,8]. Since these seminal discoveries, the scientific community has busily cataloged SH3 domains in the various genomes [9] of eukaryotes, and defined their potential cellular, interacting partners, either directly through biochemical characterization [10] or indirectly by defining their specificity with combinatorial peptide libraries [11-14].
The three-dimensional structure of SH3 domains is highly conserved, even though the amino acid similarity of any two SH3 domains is typically 25%. Like many other protein interaction modules, the N-terminus and C-terminus of this 60 amino acid long domain are next to each other, on the side opposite to its binding surface [4]. The domain is composed of two perpendicular, three-stranded anti-parallel beta sheets, with a shallow groove formed by the RT and the n-Src loops [15]. It has been proposed that the conformational and sequence variability created by these two surface loops accounts for the ligand specificity of SH3 domains [16].
With the availability of complete genome sequences, one can now today use a variety of techniques to characterize and compare the specificity of SH3 domains from any organism (Fig. 1). For example, one can predict the ‘SH3 domain interactome’ (i.e., the interaction of SH3 domains with proteins within a certain cell type) by screening large arrays of synthetic peptides, corresponding to proteins predicted from genomics sequences [17], affinity selection of phage-displayed SH3 domains [18] with individual peptide sequences, or through large-scale yeast two-hybrid screening [19]. Additionally, with the implementation of high-throughput molecular biology techniques and liquid handling robotics, it is straightforward to amplify their ~180 nucleotide long coding regions from mRNA or genomic DNA by the polymerase chain reaction (PCR); alternatively, due the decreased cost of oligonucleotides, one can order their coding regions synthesized. Thus, large sets of SH3 domains can be constructed, and probed with various labeled proteins or peptides [20-22]. In the future, it may be possible to use solid-phase peptide synthesis and chemical ligation to generate the SH3 domain domains chemically [23], both in large number and in an economical fashion, and define their specificity accordingly. In parallel to experimental effort, computational approaches are also being pursued for predicting the SH3 domain interactome of organisms [19,24,25].
Fig. 1.
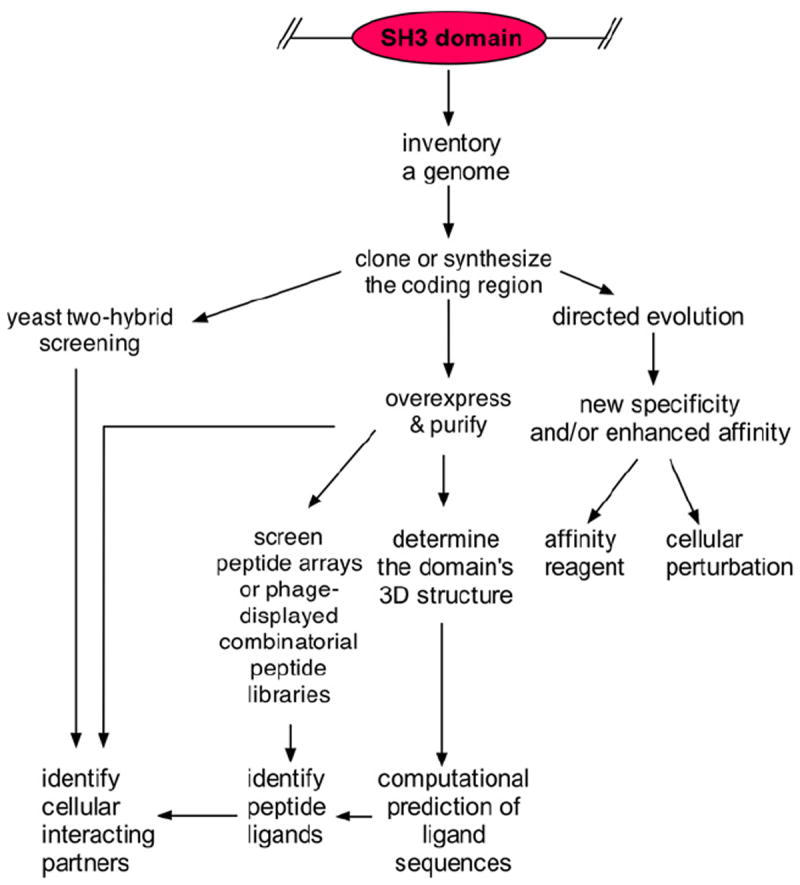
The current workflow for characterizing SH3 domains. Through bioinformatics, an inventory of SH3 domains can be created from a genome’s sequence, and coding regions for domains of interest can be recovered through cloning or by gene synthesis. The domains can then be overexpressed and purified from E. coli, and used to screen peptide arrays or phage-displayed combinatorial peptide libraries. Once peptide ligands for an SH3 domain have been identified, the cellular interacting partner can be predicted. Alternatively, the SH3 domain coding region can be used in yeast two-hybrid screening experiments or the domain can be used to pull-down interacting proteins from cell/tissue lysates. The three-dimensional (3D) structure can be deduced by X-ray diffraction or nuclear magnetic resonance, and computational techniques can be applied to predict their peptide ligands. It is also fruitful to used directed evolution methods to change the specificity and/or affinity of a particular SH3 domain for the purpose of generating a novel recombinant affinity reagent or tool for cellular perturbation studies.
Fortunately, most SH3 domains are well expressed in the cytosol of Escherichia coli, and remain stably folded after purification. The domains are thermal stable, with melting temperature in the range of 60–75 °C [26]. The ability to express the small, stable domain easily in E. coli has prompted many lines of investigation (Fig. 1). First, their affinity, which is typically low micromolar to mid-nanomolar for peptide ligands and cellular interacting proteins, respectively, can been improved at least 40-fold by directed evolution [27]. Second, by randomizing the residues in the RT and n-Src loops, it has been possible to generate new SH3 domains with novel specificities [28-30]. Such engineered SH3 domains offer interesting promise as a new class of recombinant affinity reagents [31,32] that can be used in proteomic and cellular perturbation studies. Third, SH3 domains can be combined with recombinant antibody fragments to generate reagents that bind avidly to cellular targets [33].
In conclusion, through advancements in molecular biology, genomic sequencing, and bioinformatics, it is now straight-forward for any laboratory to characterize from one to hundreds of SH3 domains at a time, and predict genome-wide protein–protein interactions in any organism. Furthermore, they can utilized in bio-technological applications for protein engineering and synthetic biology. SH3 domains have truly ‘come of age’!
References
- 1.Stahl ML, Ferenz CR, Kelleher KL, Kriz RW, Knopf JL. Sequence similarity of phospholipase C with the non-catalytic region of src. Nature. 1988;332:269–272. doi: 10.1038/332269a0. [DOI] [PubMed] [Google Scholar]
- 2.Mayer BJ, Hamaguchi M, Hanafusa H. A novel viral oncogene with structural similarity to phospholipase C. Nature. 1988;332:272–275. doi: 10.1038/332272a0. [DOI] [PubMed] [Google Scholar]
- 3.Lehto VP, Wasenius VM, Salven P, Saraste M. Transforming and membrane proteins. Nature. 1988;334:388. doi: 10.1038/334388a0. [DOI] [PubMed] [Google Scholar]
- 4.Yu H, Rosen MK, Shin TB, Seidel-Dugan C, Brugge JS, Schreiber SL. Solution structure of the SH3 domain of Src and identification of its ligand-binding site. Science. 1992;258:1665–1668. doi: 10.1126/science.1280858. [DOI] [PubMed] [Google Scholar]
- 5.Koyama S, Yu H, Dalgarno DC, Shin TB, Zydowsky LD, Schreiber SL. Structure of the PI3K SH3 domain and analysis of the SH3 family. Cell. 1993;72:945–952. doi: 10.1016/0092-8674(93)90582-b. [DOI] [PubMed] [Google Scholar]
- 6.Kay BK, Williamson MP, Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000;14:231–241. [PubMed] [Google Scholar]
- 7.Cicchetti P, Mayer BJ, Thiel G, Baltimore D. Identification of a protein that binds to the SH3 region of Abl and is similar to Bcr and GAP-rho. Science. 1992;257:803–806. doi: 10.1126/science.1379745. [DOI] [PubMed] [Google Scholar]
- 8.Ren R, Mayer BJ, Cicchetti P, Baltimore D. Identification of a tenamino acid proline-rich SH3 binding site. Science. 1993;259:1157–1161. doi: 10.1126/science.8438166. [DOI] [PubMed] [Google Scholar]
- 9.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. PNAS. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suen KL, Bustelo XR, Pawson T, Barbacid M. Molecular cloning of the mouse grb2 gene: differential interaction of the Grb2 adaptor protein with epidermal growth factor and nerve growth factor receptors. Mol Cell Biol. 1993;13:5500–5512. doi: 10.1128/mcb.13.9.5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheadle C, Ivashchenko Y, South V, Searfoss GH, French S, Howk R, Ricca GA, Jaye M. Identification of a Src SH3 domain binding motif by screening a random phage display library. JBC. 1994;269:24034–24039. [PubMed] [Google Scholar]
- 12.Rickles RJ, Botfield MC, Weng Z, Taylor JA, Green OM, Brugge JS, Zoller MJ. Identification of Src, Fyn, Lyn, PI3K and Abl SH3 domain ligands using phage display libraries. EMBO J. 1994;13:5598–5604. doi: 10.1002/j.1460-2075.1994.tb06897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sparks AB, Quilliam LA, Thorn JM, Der CJ, Kay BK. Identification and characterization of Src SH3 ligands from phage-displayed random peptide libraries. J Biol Chem. 1994;269:23853–23856. [PubMed] [Google Scholar]
- 14.Rickles RJ, Botfield MC, Zhou XM, Henry PA, Brugge JS, Zoller MJ. Phage display selection of ligand residues important for Src homology 3 domain binding specificity. Proc Natl Acad Sci U S A. 1995;92:10909–10913. doi: 10.1073/pnas.92.24.10909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Musacchio A, Wilmanns M, Saraste M. Structure and function of the SH3 domain. Prog Biophys Mol Biol. 1994;61:283–297. doi: 10.1016/0079-6107(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 16.Kaneko T, Sidhu SS, Li SS. Evolving specificity from variability for protein interaction domains. Trends Biochem Sci. 2011;36:183–190. doi: 10.1016/j.tibs.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Reiss DJ, Schwikowski B. Predicting protein–peptide interactions via a network-based motif sampler. Bioinformatics. 2004;20(Suppl 1):I274–I282. doi: 10.1093/bioinformatics/bth922. [DOI] [PubMed] [Google Scholar]
- 18.Karkkainen S, Hiipakka M, Wang JH, Kleino I, Vaha-Jaakkola M, Renkema GH, Liss M, Wagner R, Saksela K. Identification of preferred protein interactions by phage-display of the human Src homology-3 proteome. EMBO Rep. 2006;7:186–191. doi: 10.1038/sj.embor.7400596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tonikian R, Xin X, Toret CP, Gfeller D, Landgraf C, Panni S, Paoluzi S, Castagnoli L, Currell B, Seshagiri S, Yu H, Winsor B, Vidal M, Gerstein MB, Bader GD, Volkmer R, Cesareni G, Drubin DG, Kim PM, Sidhu SS, Boone C. Bayesian modeling of the yeast SH3 domain interactome predicts spatiotemporal dynamics of endocytosis proteins. PLoS Biol. 2009;7:e1000218. doi: 10.1371/journal.pbio.1000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang R, Fang P, Kay BK. Isolation of monobodies that bind specifically to the SH3 domain of the Fyn tyrosine protein kinase. N Biotechnol. 2011;29:526–533. doi: 10.1016/j.nbt.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Espejo A, Cote J, Bednarek A, Richard S, Bedford MT. A protein–domain microarray identifies novel protein–protein interactions. Biochem J. 2002;367:697–702. doi: 10.1042/BJ20020860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asbach B, Kolb M, Liss M, Wagner R, Schaferling M. Protein microarray assay for the screening of SH3 domain interactions. Anal Bioanal Chem. 2010;398:1937–1946. doi: 10.1007/s00216-010-4202-x. [DOI] [PubMed] [Google Scholar]
- 23.Mende F, Beisswenger M, Seitz O. Automated Fmoc-based solid-phase synthesis of peptide thioesters with self-purification effect and application in the construction of immobilized SH3 domains. J Am Chem Soc. 2010;132:11110–11118. doi: 10.1021/ja101732a. [DOI] [PubMed] [Google Scholar]
- 24.Hou T, Li N, Li Y, Wang W. Characterization of domain–peptide interaction interface. prediction of SH3 domain-mediated protein–protein interaction network in yeast by generic structure-based models. J Proteome Res. 2012;11:2982–2995. doi: 10.1021/pr3000688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yip KY, Utz L, Sitwell S, Hu X, Sidhu SS, Turk BE, Gerstein MB, Kim PM. Identification of specificity determining residues in peptide recognition domains using an information theoretic approach applied to large-scale binding maps. BMC Biol. 2011;9:53. doi: 10.1186/1741-7007-9-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wales TE, Engen JR. Partial unfolding of diverse SH3 domains on a wide timescale. J Mol Biol. 2006;357:1592–1604. doi: 10.1016/j.jmb.2006.01.075. [DOI] [PubMed] [Google Scholar]
- 27.Hiipakka M, Poikonen K, Saksela K. SH3 domains with high affinity and engineered ligand specificities targeted to HIV-1 Nef. J Mol Biol. 1999;293:1097–1106. doi: 10.1006/jmbi.1999.3225. [DOI] [PubMed] [Google Scholar]
- 28.Hiipakka M, Saksela K. Versatile retargeting of SH3 domain binding by modification of non-conserved loop residues. FEBS Lett. 2007;581:1735–1741. doi: 10.1016/j.febslet.2007.03.044. [DOI] [PubMed] [Google Scholar]
- 29.Wrenn SJ, Weisinger RM, Halpin DR, Harbury PB. Synthetic ligands discovered by in vitro selection. J Am Chem Soc. 2007;129:13137–13143. doi: 10.1021/ja073993a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grabulovski D, Kaspar M, Neri D. A novel, non-immunogenic Fyn SH3-derived binding protein with tumor vascular targeting properties. J Biol Chem. 2006;282:3196–3204. doi: 10.1074/jbc.M609211200. [DOI] [PubMed] [Google Scholar]
- 31.Boersma YL, Pluckthun A. DARPins and other repeat protein scaffolds: advances in engineering and applications. Curr Opin Biotechnol. 2011;22:849–857. doi: 10.1016/j.copbio.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 32.Gronwall C, Stahl S. Engineered affinity proteins–generation and applications. J Biotechnol. 2009;140:254–269. doi: 10.1016/j.jbiotec.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 33.Bouchet J, Herate C, Guenzel C, Verollet C, Jarviluoma A, Mazzolini J, Rafie S, Chames P, Baty D, Saksela K, Niedergang F, Maridonneau-Parini I, Benichou S. Single-domain antibody-SH3 fusions for efficient neutralization of HIV-1 Nef functions. J Virol. 2012;86:4856–4867. doi: 10.1128/JVI.06329-11. [DOI] [PMC free article] [PubMed] [Google Scholar]


