Abstract
Boundary Element Associated Factor-32 (BEAF-32) is an insulator protein predominantly found near gene promoters and thought to play a role in gene expression. We find that mutations in BEAF-32 are lethal, show loss of epithelial morphology in imaginal discs and cause neoplastic growth defects. To investigate the molecular mechanisms underlying this phenotype, we carried out a genome-wide analysis of BEAF-32 localization in wing imaginal disc cells. Mutation of BEAF-32 results in miss-regulation of 3850 genes by at least 1.5-fold, 794 of which are bound by this protein in wing imaginal cells. Up-regulated genes encode proteins involved in cell polarity, cell proliferation and cell differentiation. Among the down-regulated genes are those encoding components of the wingless pathway, which is required for cell differentiation. Miss-regulation of these genes explains the unregulated cell growth and neoplastic phenotypes observed in imaginal tissues of BEAF-32 mutants.
Keywords: Transcription, Chromatin, Epigenetics, Cancer
Introduction
Chromatin insulators are DNA–protein complexes that regulate enhancer–promoter interactions and/or delimit chromatin boundaries. Several different insulators have been studied in Drosophila, each of which is characterized by a different DNA binding protein that associates with one or more components common to all insulators. Insulator DNA binding proteins include Suppressor of Hair–wing [Su(Hw)], Drosophila CTCF (dCTCF), Boundary Element Associated Factor 32 (BEAF-32), Zeste-white 5 (Zw5), and GAGA factor (GAF). These DNA binding proteins interact with Centrosomal Protein 190 (CP190) and Modifier of mdg4 [(Mod(mdg4)], which contain BTB domains capable of multimerization and responsible for interactions among individual insulator sites (Capelson and Corces, 2005; Gurudatta and Corces, 2009). Contrary to Drosophila, vertebrate cells appear to mostly rely on the CTCF insulator, since CTCF has been shown to play a broad role in mediating interactions between distant genomic sites in higher organisms (Handoko et al., 2011). More recently, the TFIIIC protein, which is present in tRNA genes and is involved in RNA Polymerase III-mediated transcription, has also been shown to possess insulator activity in human cells (Raab et al., 2012). These observations raise the question of whether the different Drosophila insulators play similar or distinct roles in the regulation of gene expression.
To address this issue, we decided to examine the precise role of BEAF-32 insulators in controlling transcription in Drosophila cells. BEAF-32 was originally identified as a component of the scs’ insulator and found to localize to the boundaries between bands and interbands on polytene chromosomes (Zhao et al., 1995). The BEAF-32 gene encodes two isoforms, BEAF-32A and BEAF-32B, which differ from each other in an 80 amino acid region that contains different atypical C2H2 zinc fingers termed BED fingers (Aravind, 2000; Hart et al., 1997). The two isoforms can interact and form multimers, although the ratio between the two proteins appears to be different in different sites on polytene chromosomes. A map of the distribution of BEAF-32 genome-wide has been determined from ChIP-chip and ChIP-seq experiments (Bushey et al., 2009; Jiang et al., 2009; Negre et al., 2010). The results indicate the presence of BEAF-32B at several thousand sites (1800–4700) in the Drosophila genome whereas BEAF-32A is present at 33 sites by itself and overlaps with BEAF-32B at an additional 735 sites. Analysis of these data suggests that most BEAF-32 sites are present within 1 kb upstream of transcription start sites (Jiang et al., 2009; Negre et al., 2010).
The location of BEAF-32 with respect to genes is similar to that of CTCF and CP190 but different from that of Su(Hw), which tends to be located far away from gene promoters (Bushey et al., 2009; Jiang et al., 2009; Negre et al., 2010; Wood et al., 2011). This observation suggests that BEAF-32, and perhaps dCTCF, insulators, may affect gene expression by a different mechanism than Su(Hw). Alternatively, BEAF-32 may regulate the expression of a distinct subset of genes. BEAF-32 preferentially associates with highly transcribed genes and loss of BEAF-32 in Drosophila embryos carrying the BEAFAB-KO allele results in reduction of transcription of 19 out of 23 genes tested (Jiang et al., 2009). The role of BEAF-32 in transcription has also been studied at a specific subset of genomic loci that show a distinct arrangement of BEAF binding sites. These sites, called BEAF dual-core binding sites, contain 5–6 BEAF binding motifs flanking 200 bp of AT-rich nuclease-resistant spacers. BEAF dual-cores are preferentially located next to genes involved in cell cycle control and chromosome organization, and depletion of BEAF-32 using siRNA leads to an increase in tetraploid cells, suggesting chromosome segregation defects (Emberly et al., 2008).
Here we have analyzed the role of BEAF-32 in transcription by examining developmental defects in animals carrying a null mutation in the BEAF-32 gene. We find that mutations in BEAF-32 have a larval lethal phenotype characterized by an increase in the larval time of development and overgrowth of imaginal discs. To explain these phenotypes we carried out a transcriptome analysis of wild type and BEAF-32 mutant wing imaginal discs. In addition, we determined the genome-wide location of BEAF-32B in wing imaginal disc cells by ChIP-seq. We identify 3850 genes that are miss-regulated by at least 1.5-fold normal transcript levels. Of these, 794 genes have BEAF-32 adjacent to the promoter region, suggesting that they may be direct targets of this protein and the alteration of their expression may be a direct consequence of the mutation. Proteins encoded by these genes are enriched in components of various signaling pathways that control tissue growth and cell polarity. In particular, the bazooka gene is up-regulated, which may explain the loss of cell polarity in BEAF-32 mutants. In addition, Insulin receptor-1 and Unpaired 3 are up-regulated, leading to activation of the MAP kinase and JAK-STAT signaling pathways and increased cell proliferation. These results highlight the requirement for the BEAF-32 insulator in cell growth and development.
Results
Loss of BEAF-32 causes neoplastic growth
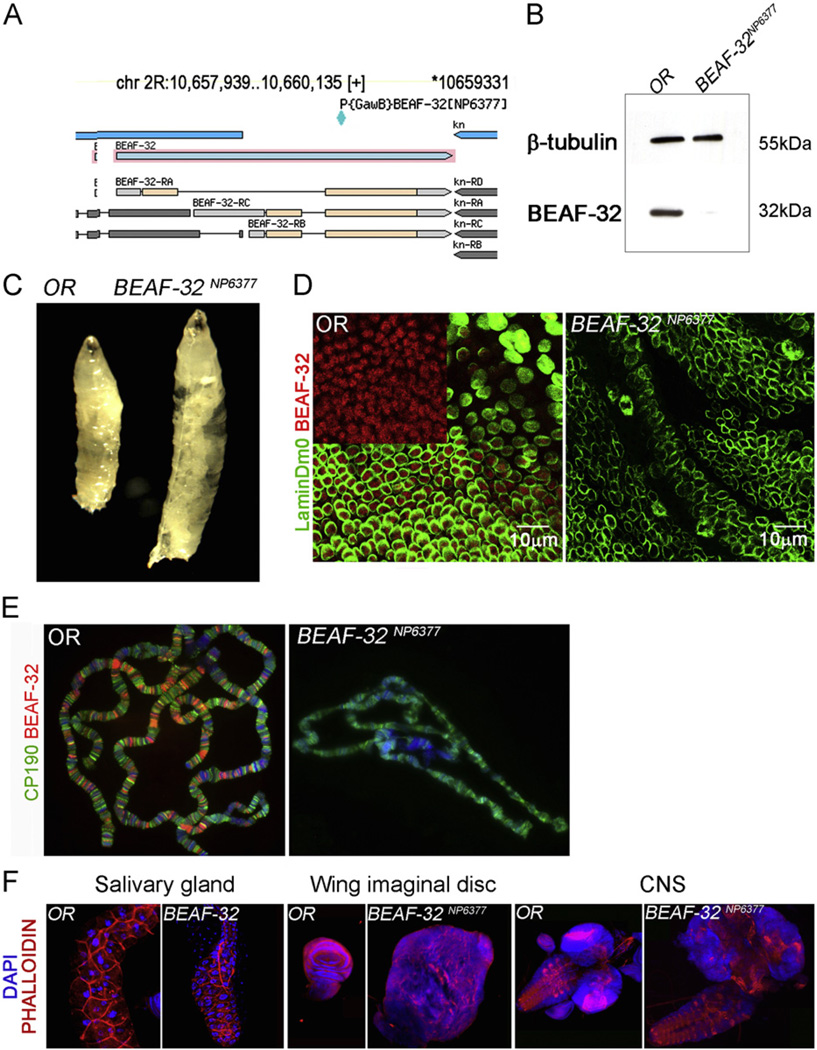
In order to understand the role of BEAF-32 in development we first characterized an existing P-element allele of BEAF-32 named BEAF-32NP6377. This mutation is caused by an insertion of the P{GawB} element at position 106,593,311 in chromosome 2R. Although this information is available in FlyBase, the actual location of the insertion in GBrowse is incorrect. The P-element is in fact inserted in a protein coding exon common to the BEAF-32A and BEAF-32B proteins (Fig. 1A). Western analysis of protein extracts from third instar larvae indicate complete lack of both BEAF-32 isoforms in BEAF-32NP6377 mutants (Fig. 1B). Animals homozygous for this mutation show larval lethality and abnormal growth. They also develop slower, requiring approximately 4 additional days at 25 1C to become third instar larva, compared to wild type animals. These larvae show a dramatic increase in size, lack fat body tissue, and fail to pupate (Fig. 1C). This phenotype is similar to that of mutations in the lethal giant larvae and scribs genes (Dow et al., 2003). Immunofluorescence microscopy of BEAF-32 distribution using an antibody that recognizes both protein isoforms shows that this protein is present at foci in the nucleus of wing imaginal disc cells, whereas BEAF-32 is missing in cells from mutant larvae (Fig. 1D). BEAF-32 is present at band/interband boundaries of polytene chromosomes colocalizing with CP190. Chromosomes from BEAF-32NP6377 mutant larvae show a complete absence of BEAF-32 while CP190 is still bound normally. The morphology of the polytene chromosomes is abnormal, appearing small and condensed, perhaps due to an effect on endoreplication (Fig. 1E). The imaginal discs and central nervous system of BEAF-32NP6377 larvae show tumor-like massive growth, making large structures that, in the case of the imaginal discs, are miss-shaped with respect to the normal tissues and unrecognizable as such (Fig. 1F).
Fig. 1.
Mutations in BEAF-32 result in neoplastic growth. (A) Diagram showing the BEAF-32 locus adapted from the Flybase genome browser. The location of the P{GAW} transposon insertion is indicated; this insertion disrupts an exon common to both BEAF-32A and BEAF-32B isoforms. (B) Western blot for BEAF-32 (−32 kDa) showing the levels of this protein in wild type and mutant imaginal disc tissue; tubulin (55 kDa) was used as loading control. (C) Wild type and BEAF-32NP6377 mutant larvae showing the dramatic overgrowth of the later at the third instar stage. (D) Immunofluorescence microscopy using antibodies to the BEAF-32 protein in imaginal tissue; BEAF-32 (red) is present in nuclear foci while the peripheral location of lamin Dm0 (green) marks the nuclear lamina. (E) Immunofluorescence microscopy of polytene chromosomes showing the location of CP190 (green) and BEAF-32 (red). (F) Effect of BEAF-32 mutations in developing tissues such as salivary glands, wing imaginal discs and central nervous system. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Loss of BEAF-32 in wing epithelium shows defective epithelial architecture, cell identity and patterning
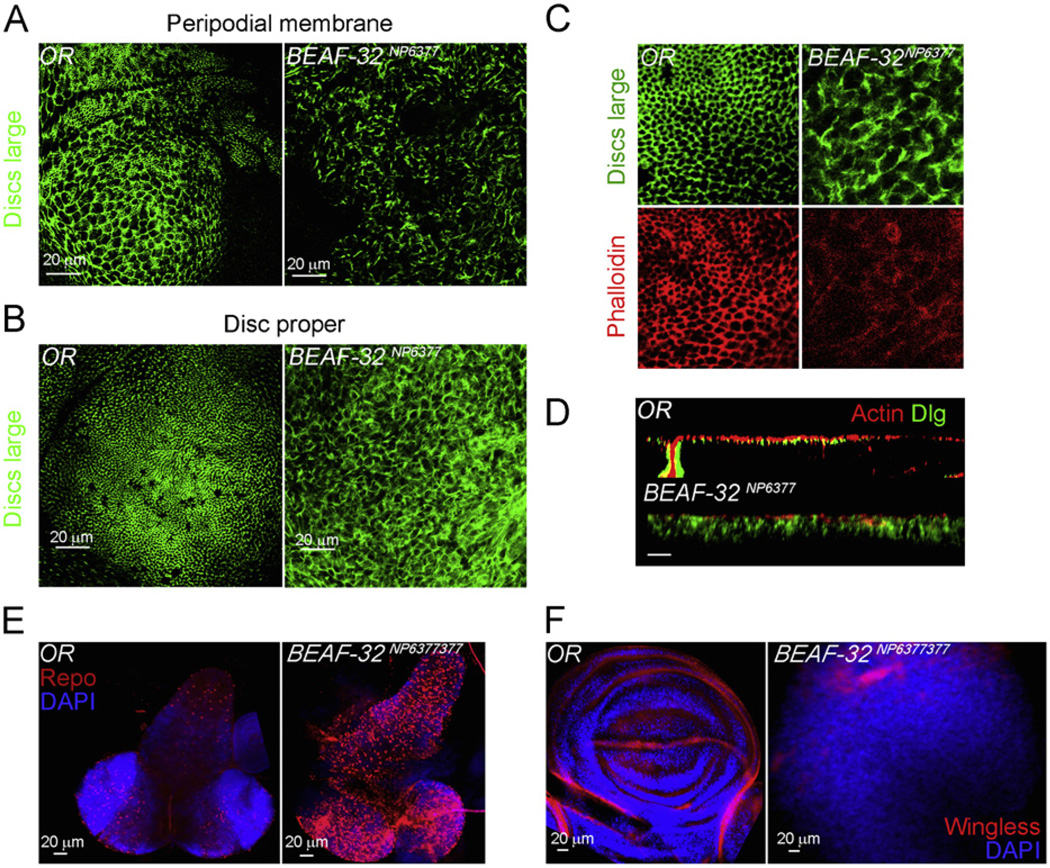
The defects in the BEAF-32NP6377 mutant larvae appear to be due to change in cell size or shape but architecture and morphology of the tissue also appear to be strongly compromised. Hence we investigated if the epithelial architecture is affected in BEAF-32NP6377. We visualized the epithelial structures either by labeling f-actin or examining the localization of the Discs Large (Dlg) protein in the wing epithelial tissue by immunofluorescence. The wing epithelium forms honeycomb structures demarcated by Dlg in both peripodial membrane and disc proper epithelium (Woods et al., 1996). The organized array of Dlg is absent in these two tissues in mutant larvae (Fig. 2A and B). In addition, proper adherent junctions marked by actin–phalloidin show the cell–cell contacts in the wild type epithelium whereas they are lacking in cells from BEAF-32NP6377 larvae (Fig. 2C). The loss of cell contacts could be a consequence of the loss of apico-basal polarity. This is exemplified in the X–Z confocal cross sections of wild type and mutant wing disc. In wild type tissue, disc cells are ordered with proper polarity, while in the mutant tissue, cells are scrambled, indicating loss of such organization (Fig. 2D). In addition, wing disc morphology is disrupted in the mutant with no detectable structures such as pouch and notum. For example, wingless, a patterning protein, is normally present in the dorso–ventral boundary of the wing disc; however in the mutant disc no dorso-ventral boundary or wingless expressing cells are observed, indicating loss of lineage identity (Fig. 2E). The larval central nervous system (CNS) also shows increased proliferation in comparison with the wild type tissue. The CNS is slightly affected, with the ventral nerve cord appearing larger but similar in morphology to wild type. The increased proliferation also affects CNS cell fate determination during differentiation. When stained with Repo to mark the glial cells, the mutant CNS shows a larger number of Repo positive cells than the wild type CNS. These changes in cell fate, morphology, and growth could be a consequence of miss-regulation of specific genes in cells lacking BEAF-32.
Fig. 2.
BEAF-32 mutants show loss of epithelial architecture and defects in patterning and cell differentiation. (A) Immunofluorescence microscopy using antibodies to the Discs Large (Dlg) protein (green) in the wing imaginal tissue of wild type and BEAF-32NP6377 mutant larvae. The panels show the projections of confocal sections from the peripodial membrane. (B) Same as in (A) but showing the disc proper cells in the wing pouch. (C) The adherens junctions marked by the Dlg protein (green) and actin marked by pholloidin (red). (D) Longitudinal cross section (X–Z) of wing epithelium in the pouch region of the wing imaginal disc; actin in red marks the cytoskeleton and Dlg in green marks adherent junctions. (E) The central nervous system from wild type and mutant larvae stained with the glial cell marker Repo (red) and DAPI (blue). Wing imaginal tissues stained for wingless (red) showing the dorso-ventral boundary in wild type discs and its absence in the mutant; DAPI stains DNA (blue). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
BEAF-32 is enriched around transcription start sites of genes highly expressed in wing imaginal tissue
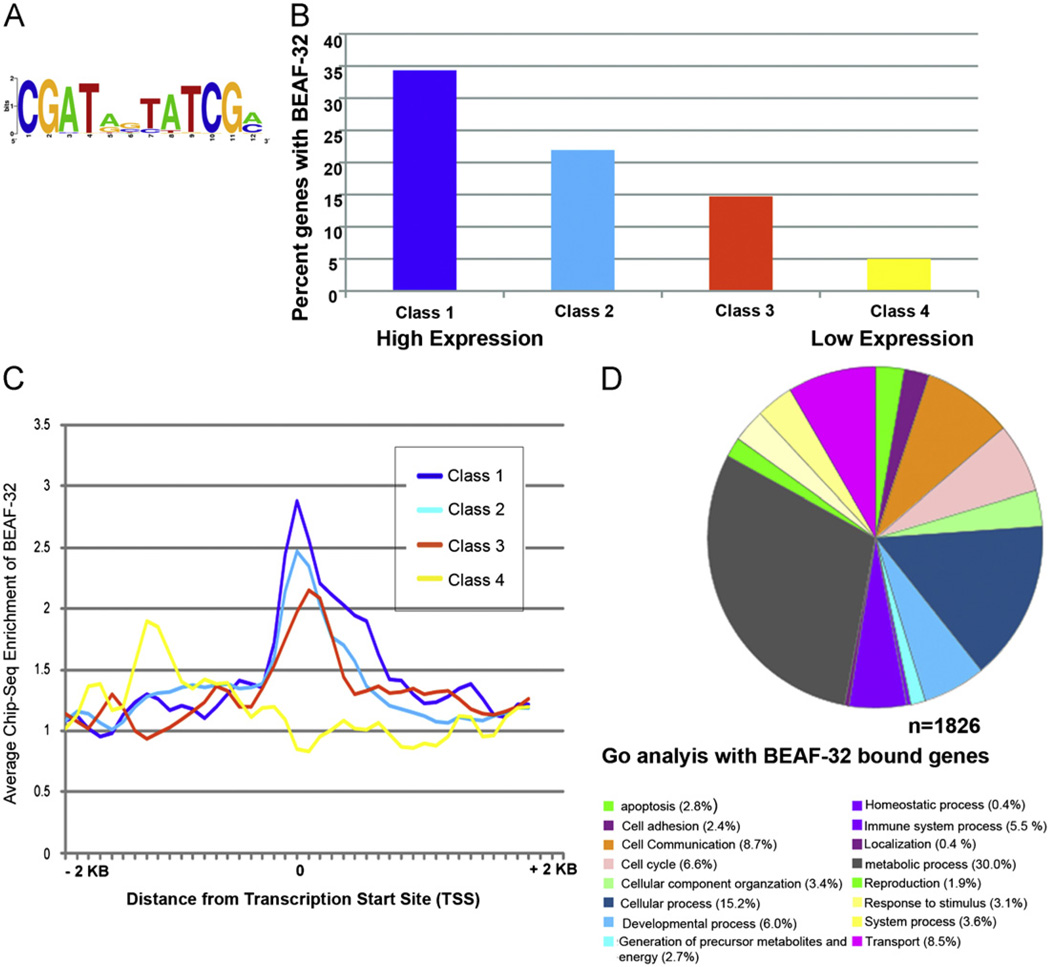
In order to examine the role of BEAF-32 in the expression of genes in the wing imaginal discs, we carried out ChIP-Seq experiments with wing imaginal tissue and BEAF-32 antibodies. We found that BEAF-32 is bound at 2633 sites in the genome of wing imaginal disc cells. Motif analysis of these wing-specific BEAF-32 binding sites indicates the presence of the core consensus CGATA (Fig. 3A). We then parsed out the BEAF-32 bound genes, defined as genes with BEAF-32 occupancy in the region between −200 bp upstream of the transcription start site (TSS) and the transcription termination site. Using these criteria, we identified 1826 genes as containing BEAF-32. We then carried out microarray analyses to determine the transcriptome of wing imaginal disc cells and grouped genes into four different classes based on their expression level in wing tissue. These four expression classes were obtained by dividing the normalized intensity levels in the microarray into four equal groups. Interestingly, there is a strong correlation between gene expression levels and the presence of BEAF-32, with a higher percentage of BEAF-32-containing genes belonging to the class of highest expressed genes (Fig. 3B). BEAF-32 is present around the TSSs of genes in the three classes with the highest transcription but this protein is not arranged in a discernible pattern with respect to the lowest expressed genes (Fig. 3C).
Fig. 3.
Genome wide distribution of BEAF-32 in wing imaginal tissue. (A) BEAF-32 recognition motif predicted using data from ChIP-seq experiments. (B) Histograms indicating the percent distribution of BEAF-32 bound genes and their gene expression levels in wing imaginal tissue. The percentages refer to the fraction of BEAF-bound genes within each class. (C) Distribution of BEAF-32 around transcription start sites in genes with different expression levels. (D) Gene ontology analysis of BEAF-32 bound genes enriched in different cellular processes.
Earlier studies have shown that BEAF-32 plays a role in the expression of genes encoding cell cycle regulators in Drosophila S2 cells (Emberly et al., 2008). Surprisingly, based on gene ontology (GO) term predictions, BEAF-32 bound genes in the wing disc are involved in cellular (29.6%) and metabolic processes (58.5%) with a small fraction involved in cell cycle (5.5%), cell adhesion (4.7%) and developmental processes (14.0%) (Fig. 3D). It is possible that the phenotypes observed in BEAF-32 mutants are due to miss-expression of genes in one or more of these categories.
Mutation of BEAF-32 results in changes in transcription of BEAF-32-bound genes
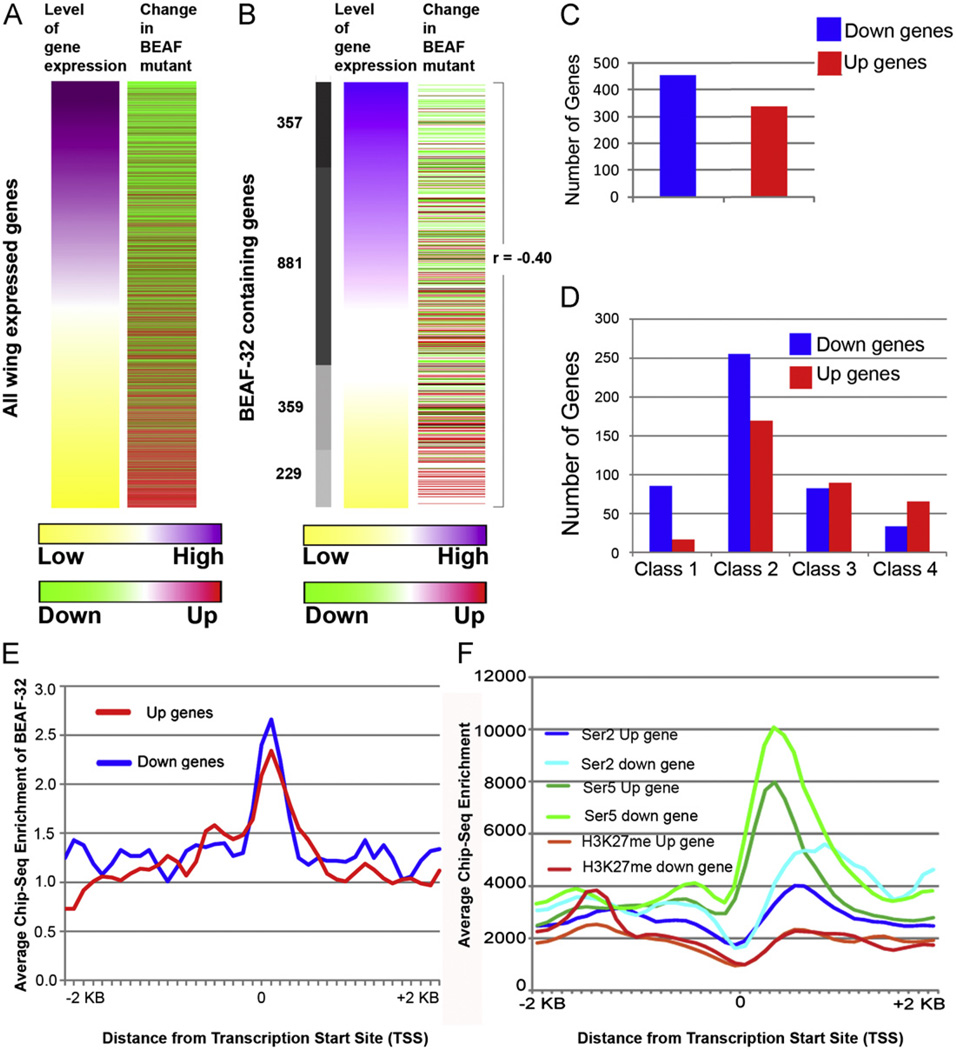
In order to examine the role of BEAF-32 in transcription, we carried out microarray analyses using RNA isolated from wing imaginal disc cells of larvae mutant for the BEAF-32NP6377 allele. Comparison of the transcriptome from wild type and mutant wing disc cells indicates that a total of 3850 genes are miss-regulated by more than 1.5-fold in BEAF-32 mutant cells. Of these, 1932 are up-regulated and 1918 are down-regulated (Table S1). Interestingly, most of the down-regulated genes are those that are normally highly transcribed whereas the up-regulated genes are normally expressed at low levels (Fig. 4A). This trend is maintained when we restrict the analysis to those genes that contain BEAF-32 binding sites and whose transcription is therefore more likely to be the primary target of this protein. When considering these genes, those with the highest expression levels largely show down regulation while the genes with low expression show up regulation (Pearson correlation r=−0.40) (Fig. 4B). There are a total of 287 up-regulated and 360 down-regulated BEAF-32-bound genes with at least a 1.5-fold change in the BEAF-32NP6377 mutant (Fig. 4C). Since both groups of genes contain BEAF-32, it is surprising that they respond differently to the absence of this protein in mutant cells. Most downregulated genes belong to Classes 1 and 2 as defined in Fig. 3B and Fig. 4D. To gain further insights into possible mechanisms responsible for this differential response, we examined the location of the BEAF-32 protein with respect to the affected genes. The distribution of BEAF-32 at these genes is significantly enriched near the TSS (Fig. 4E). We then examined the distribution of RNA polymerase II (RNAPII) and H3K27me3 with respect to the TSSs and coding regions of the genes. Genes down-regulated in BEAF-32 mutant cells are enriched in RNAPII phosphorylated in Ser5 and Ser2 (Fig. 4F). Both groups of genes have equal levels of H3K27me3, a modification characteristic of silenced chromatin (Fig. 4F). These results suggest that, in spite of its presence close to TSSs, the role of BEAF-32 in transcription is complex and differs from that of classical transcription factors. The effect of BEAF-32 on gene expression in the wing imaginal tissue must relate to its ability to mediate interactions between different regions in the genome as has been suggested for other insulator proteins.
Fig. 4.
Genome-wide changes in BEAF-32 dependent transcription. (A) Heatmaps representing genome wide gene expression changes in wing imaginal tissue upon loss of BEAF-32. Levels of gene expression are ranked from high (purple) to low (yellow) according to wild type expression. The second column indicates changes seen in mutant cells, either up-regulated (towards red) or down-regulated (towards green). (B) BEAF-32 bound genes sorted based on levels of gene expression and their respective changes in mutant tissue. The Pearson correlation values are indicated to signify the changes in BEAF-32 mutant tissue. (C) Number of BEAF-32 bound genes showing transcription changes by a factor of more than 1.5-fold in the BEAF-32NP6377 mutant. (D) Number of BEAF-32 bound genes that are either up-regulated or down-regulated at different expression levels. (E) Distribution of BEAF-32 around the TSS of up- and down-regulated genes. (F) Genomic landscape of CTD-phosphorylated RNA polymerase II and histone H3 trimethylated at Lysine 27 in wild type wing tissue around up- and down-regulated genes. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
BEAF-32-regulated genes are involved in pathways controlling neoplastic cell growth
In order to explain the phenotypes observed in BEAF-32 mutants, we examined the nature of the BEAF-32 bound genes whose transcription is altered by loss of BEAF-32. Earlier studies with cell lines in which BEAF-32 was knockdown suggested 5 out of 6 genes tested were affected by lack of BEAF-32, including cell cycle genes such as cdk7 and mei-s332 (Emberly et al., 2008). On the other hand, mutation of BEAF-32 in flies appears to have a dramatic effect on transcription of a large number of genes as described above. To analyze the role of these genes in various cellular processes, we used the Ingenuity Pathway Analysis program to predict pathways affected in the BEAF-32 mutant (Table 1) (Mayburd et al., 2006). Results from this analysis reveal an overrepresentation of genes implicated in cancer, developmental disorders, oxidative stress response, amino acid metabolism and DNA repair. Proteins encoded by these genes play roles in various pathways including IGF, Insulin receptor, CDK5, PTEN, FAK, ILK, Integrin, ERK-MAP kinase, and JNK signaling. Other affected pathways are involved in amino acid metabolism, protein ubiquitination and cell survival pathways such as DNA double-strand break repair by non-homologous end joining, NRF2-mediated oxidative stress response and hypoxia (Table 1). These pathways are significantly compromised in the mutant tissue. For example, in the case of insulin growth factor signaling, the IGF1 receptor and the p70-S6 kinase are up-regulated in mutant cells. The apical polarity determinant gene bazooka was also found up-regulated in the BEAF-32 mutant. The unpaired 3 ligand and Socs65E, a target of the JNK pathway, are also up-regulated. These pathways are known to affect increased proliferation and loss of cell morphology in epithelial tissues (Table 1). The down-regulated pathways include proteins involved in ubiquitin-mediated proteasome degradation, wingless signaling, amino acid degradation and hypoxia signaling (Table 2). These results suggest that miss-regulation of these key molecules that attenuate signaling pathways may result in the BEAF-32 mutant phenotype. The observed phenotypes could also be due to miss-expression of genes not bound by BEAF-32 as a consequence of secondary events. For example, transcript levels of lethal giant larvae, wingless2, and archipelago are decreased in the mutant, while the levels of Pi3Kinase, which is part of the FAK, integrin and IGF1 signaling pathways, is increased (Supplementary Tables 1 and 2).
Table 1.
Pathways involving BEAF-32-bound genes up-regulated by transcription changes in cells lacking BEAF-32 in wing imaginal tissue. The classification and analysis is based on gene id and nature of response seen in mutant tissue. The p value (in parenthesis) is calculated based on significant representation of genes with respect to the total number of genes involved in the pathway using the Ingenuity Pathway Analysis program (IPA).
| Name of the upregulated pathway | Pathway component, gene name |
|---|---|
| Glutathione metabolism (2.86E00) | Zw, Gclc, Tina-1 |
| CDK5 Signaling (2.6E00) | CG6191, mys, Mbs, SNF4Agamma |
| DNA double-strand break repair by non-homologous end joining (2.48E00) | Irbp, Lig4 |
| IGF-1 signaling (2.43E00) | Pxn, S6k, InR, SNF4Agamma |
| PTEN signaling (1.5E00) | mys, S6k, InR |
| Insulin receptor signaling (1.98E00) | S6k, Ptp61F, Mbs, SNF4Agamma |
| FAK signaling (1.73E00) | mys, Pxn, CalpA |
| ILK signaling (1.53E00) HIF1α signaling (1.55E00) Integrin signaling (1.44E00) ERK/MAPK Signaling (1.52E00) | mys, Pxn, Mmp1, SNF4Agamma, Flo, sima, Hph |
| NRF2-mediated oxidative stress response (1.58E00) | MRP, CG5001, MRP |
| Caveolar-mediated endocytosis signaling (1.92E00) | mys, Ptp61F, Flo |
| Amyloid processing (1.33E00) | SNF4Agamma, CalpA |
| Actin cytoskeleton signaling (1.33E00) | mys, Pxn, DIAPH2, Mbs |
| Cell polarity genes | Baz |
| JNK signaling | Upd3, Socs35 |
Table 2.
Pathways involving BEAF-32-bound genes down-regulated by transcription changes in cells lacking BEAF-32 in wing imaginal tissue. The classification and analysis is based on gene id and nature of response seen in mutant tissue. The p value (in parenthesis) is calculated based on significant representation of genes with respect to the total number of genes involved in the pathway using the Ingenuity Pathway Analysis program (IPA).
| Name of the down regulated pathway | Pathway component, gene name |
|---|---|
| Protein ubiquitination pathway (1.02E01) | Prosbeta3, Roc1a, Prosbeta5, CG7375, Rpn9, CG7375, Prosalpha7, P58IPK, CG7872, CG12096, CG12096, Rpn2, CG7656, Rpt3, Elongin-C, l(2)05070, Rpn11, Pros54, Rpt1, UbcD2 |
| Hypoxia signaling (3.15E00) | Pdi, CG7375, CG7656, UbcD2, tgo |
| Mitochondrial dysfunction (2.86E00) | ATPsyn-gamma, ND75, levy, ND23, CG9314, CG17856 |
| Oxidative phosphorylation (2.17E00) | ATPsyn-gamma, ND75, levy, ND23, CG3560 |
| NRF2-mediated oxidative stress response (2.51E00) | Roc1a, Mgstl, CG2852, Hop, Cat, UbcD4, P58IPK |
| Fatty acid metabolism (2.05E00) | Fdh, CG31075, CG3961, CG3961, CG3415, CG12262, CG4389 |
| Mismatch repair in eukaryotes (1.82E00) | mus209, tos |
| Aryl hydrocarbon receptor signaling (1.76E00) | cathD, Mgstl, CG31075, Cdk4, Tango9 |
| Nucleotide sugars metabolism (1.73E00) | UGP, CG12030 |
| Wnt/β-catenin signaling (1.45E00) | nmo, gish, CG7115, CkIIbeta-PE, dod |
| Valine, leucine and isoleucine degradation (1.69E00) tryptophan metabolism (1.4E00) β-alanine metabolism (1.39E00) arginine and proline metabolism (1.35E00) |
CG31075, CG3415, CG12262, CG4389 Cat, CG5220 CG12262 Pdi, slgA, SamDC |
Discussion
The BEAF-32 insulator protein is predominantly enriched near transcription start sites and associates with highly expressed genes (Bushey et al., 2009; Jiang et al., 2009; Negre et al., 2010). The role of this protein in transcription has only been examined previously at a specific subset of genes. Knockdown of BEAF-32 in S2 cells using RNAi results in reduced transcription of five out of six genes tested (Emberly et al., 2008; Roy and Hart, 2010; Roy et al., 2007b), whereas loss of BEAF-32 in Drosophila embryos carrying the BEAFAB-KO allele results in reduction of transcription of 19 out of 23 genes tested (Jiang et al., 2009). Analysis of flies carrying the BEAFAB-KO mutation suggests that BEAF-32B is required for viability of adult files. This allele is female sterile and the maternal BEAF-32 protein is sufficient to drive development to adult. Additionally, these flies show abnormal X polytene chromosome morphology but structural defects are not pronounced, suggesting BEAFAB-KO could be a hypomorphic allele. In support of this, these mutants appear to have residual BEAF-32 protein as detected by western blot analysis (Roy et al., 2007a). Here we examine the effect of BEAF-32 on the transcriptome of wing imaginal disc cells by analyzing a null allele of BEAF-32 and show that loss of this protein has a dramatic effect on transcription in wing imaginal disc cells, resulting in up-regulation and down-regulation of a large number of genes and suggesting a general role for this protein in gene expression.
The mechanisms by which BEAF-32 affects gene expression during wing development are not clear. Based on the results described here it appears that BEAF-32 does not act as a classical transcription factor, since lack of BEAF-32 results in both up- and down-regulation of bound genes without an obvious correlation to levels of RNAPII or silencing histone modifications. It is then likely that, in spite of its location close to TSSs, BEAF-32 plays a role more directly related to its hypothesized function as a chromatin insulator. In mammalian cells the insulator protein CTCF has been identified in complexes associated with active RNAPII (Chernukhin et al., 2007; Melnik et al., 2011) and has been shown to help target enhancers to the appropriate promoter (Phillips and Corces, 2009). It is possible that BEAF-32 plays a similar role in Drosophila, helping recruit genes involved in specific cellular processes to transcription factories. In the absence of BEAF-32, these genes may fail to be recruited to these factories or may be recruited to alternative subnuclear compartments by other insulator proteins, resulting in the observed down- or up-regulation of their expression.
Loss of function mutations in BEAF-32 result in developmental abnormalities characterized by neoplastic growth. This phenotype is similar to that of mutations in the lethal giant larvae and scribs genes (Woods et al., 1997). The loss of function BEAF-32NP6377 allele shows defects in development, cell identity and patterning. The wing imaginal discs lack features such as notum or pouch cells. The discs also lack patterning features such as anterior posterior axis (A-P) or markers such as wingless, normally expressed by organizer cells on the A-P axis. We have identified genes whose expression is affected by a factor of 1.5-fold or more in BEAF-32 mutants and examined the role of the encoded proteins in the context of these phenotypes. If we only consider those genes bound by BEAF-32, which are likely direct targets of regulation by this protein, the nature of the affected genes can explain the observed phenotypes. These include genes involved in cell polarity, various signaling pathways, cell survival, cell patterning, and amino acid metabolism. Mechanisms involved in the establishment of cell-polarity are responsible not only for the diversification of cell shape but also participate in the regulation of asymmetric cell divisions of stem cells that are crucial for their correct self-renewal and differentiation. Disruption of cell polarity is a hallmark of cancer, and its establishment requires localization of the crumbs, stardust, Par6, Bazooka and aPKCs proteins to the apical membrane to specify the apical domain (Margolis and Borg, 2005; Suzuki and Ohno, 2006). The up-regulation of Bazooka in BEAF-32 mutant imaginal tissue could compete with aPkc for binding to PAR6, resulting in loss of epithelial morphogenesis (Morais-de-Sá et al., 2010). The Drosophila cell polarity proteins Scrib, Dlg and Lethal giant larvae (Lgl) are powerful tumor suppressors and loss of these proteins cause neoplastic transformation (Bilder, 2004). These results suggest that mutation of BEAF-32 could affect cell polarity, epithelial morphology and Dlg localization. Perturbation of cell morphology is also known to activate the JNK pathway (Igaki et al., 2006; Zhu et al., 2010). Molecular profiling of BEAF-32 mutant tissue also shows miss-regulation of key mitogenic pathways, including Insulin growth factor, CDK5, Integrin, FAK, ERK/MAP kinase and JNK signaling (Feldmann et al., 2010; Harrison et al., 1998; Igaki et al., 2006; Pollak, 2008; Wary et al., 1998). Insulin growth factor signaling is crucial for malignant transformation. The regulation of the IGF receptor is a link between oncogene and tumor suppressor pathways that has substantial impact on metabolic and proliferative pathways (Werner, 2011). Key molecules in these pathways are Insulin receptor 1 and p70-S6 kinase. These signaling events can activate other pathways such as PI3 kinase, AKT, PTEN and MAP kinase in BEAF-32 mutant cells (Siddle, 2011). Activation of the JNK pathway in mutant cells appears to promote a proliferative role as suggested by the over-expression of unpaired 3 ligand and the target gene Socs35. The activated pathways causing increased cell proliferation could increase cell survival by counteracting hypoxia or oxidative stress through the activation of antioxidant response element (ARE) dependent genes (Yu et al., 2000) or by inducing DNA double strand break response genes (Bozulic et al., 2008).
Mutations in other insulator proteins, such as dCTCF and Mod(mdg4), result in homeotic transformations (Gerasimova et al., 2007). The only other insulator protein that shows a strong tumor like growth phenotype is l(3)mbt, which has recently been shown to be required for CTCF/CP190-mediated function of the Fab8 insulator (Bonasio et al., 2010; Richter et al., 2011). However, unlike l(3)mbt, which plays a role in hyperplastic cell growth by activating Hippo pathway target genes, BEAF-32 mutations result in neoplastic overgrowth. BEAF-32 proteins are restricted to Drosophila species and have not been identified in higher organisms (Aravind, 2000; Schoborg and Labrador, 2010). Nevertheless, a possible role of insulators in the expression of tumor suppressor genes in mammalian cells should be explored further.
Materials and methods
Fly strains and antibodies
The BEAF-32NP6377 fly line was obtained from the DGRC-NIG Kyoto collection. Mouse monoclonal antibodies to BEAF-32, Wingless (4D4), Armadillo (N), Discs Large (F9), and beta-tubulin (e7) were obtained from the Developmental Studies Hybridoma Bank (DSHB). Rabbit polyclonal BEAF-32B is described in (Bushey et al., 2009) and was used for the Chip-Seq experiments in the wing imaginal tissue. Fly stocks were maintained at 25 °C.
Protein lysates and western analysis
Larval brain and imaginal discs were dissected in PBS, lysed in 2X Laemmli’s lysis buffer (Biorad) containing mercapetoethanol and boiled at 100 1C for 5 min (Nowak et al., 2003). The debris was spundown at 15 K at 4 1C for 10 min. The supernatant was transferred into a new tube and stored at −20 °C. Total protein quantity was determined using Qubit (Invitrogen). Proteins were then electrophoresed on an acrylamide gel (10% Tris-glycine, Invitrogen) and transferred on to PVDF membranes. Blocking was done using non fat dry milk for 30 min at room temperature. Membranes were incubated with primary antibodies (1:5000 for beta-tubulin, 1:100 for BEAF-32) at 4 1C, washed twice with TBS (1X) containing Tween-20 (0.01%) for 10 min and incubated with secondary antibodies HRP-conjugated anti-mouse (1:3000) for 45 min at room temperature. Proteins were detected using the chemi-luminiscence reagents Thermo-Pico or Thermo-Dura (Thermo Scientific).
Gene expression analysis by microarrays
RNA was purified from wing imaginal disc tissue with the RNeasy kit (Qiagen) and cDNA synthesis was performed with the Megascript reverse transcription kit (Sigma). cDNA was digested with RNase A for 30 min at 37 °C and cleaned through a PCR purification column (Qiagen). Sample labeling, hybridization, and data extraction were performed by the FSU-Nimblegen Microarray facility (http://www.bio.fsu.edu/nimblegen_microarray.php) using Nimblegen 12×135 K arrays. Two biological replicates were performed for each sample. To determine genes that change in expression in wild type and mutant tissues we used Partek software at a statistical cutoff of 1% FDR generated with a fold change of 1.5.
Chip-Seq in wing imaginal tissue
Chromatin immunoprecipitation was performed as described (Bushey et al., 2009) with a few modifications. Two hundred pairs of wing imaginal discs were dissected in PBS, fixed in CCM3 media for 20 min, and the reaction was stopped by adding glycine to a final concentration of 0.125 mM. Chromatin was prepared by shearing the chromatin using sonication. Chromatin was immunoprecipitated using specific antibodies and the DNA was used for library preparation using the Illumina TruSEQ kit. ChIP libraries were sequenced at the HudsonAlpha Institute for Biotechnology, using an Illumina HiSeq system. Sequences were mapped to the dm3 genome with Bowtie 0.12.3 (Langmead et al., 2009) using default settings. Peaks were then called with MACS 1.4.0 alpha2 (Zhang et al., 2008) using equal numbers of unique reads for input and ChIP samples.
Gene ontology, pathway analysis and other bioinformatics tools
Ontology analysis of genes associated with BEAF-32 was carried out using the Panther Classification System (http://www.pantherdb.org). The list of genes and fold change values were used as input data for the Ingenuity Pathway Analysis (IPA) (http://www.ingenuity.com). Genome distribution analysis was performed using Galaxy (http://main.g2.bx.psu.edu). To determine the position of a peak relative to genomic elements, the middle of a peak was used as its location in the genome and genome annotation dm3 (D. melanogaster Apr. 2006 BDGP R5/dm3). Genomic coordinates were obtained using the flybase batch download tables (http://flybase.org/static_pages/downloads/ID.html). To determine BEAF-bound genes we included an additional 200 bp upstream to the transcription start site and the transcription termination site.
Supplementary Material
Acknowledgments
We would like to thank the Developmental Studies Hybridoma Bank for monoclonal antibodies, Dr Subhabrata Sanyal for sharing the imaging system and Dr. Ravi Dyvarshetty for help with the IPA analysis. We also thank The Genomic Services Lab at the HudsonAlpha Institute for Biotechnology for their help in performing Illumina sequencing of ChIP-Seq samples. This work was supported by U.S. Public Health Service Award GM35463 from the National Institutes of Health.
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.ydbio.2012.06.013.
Footnotes
Accession numbers
ChIP-seq and gene expression data are deposited in NCBI’s Gene Expression Omnibus (GEO) under accession number GSE36737.
References
- Aravind L. The BED finger, a novel DNA-binding domain in chromatin-boundary-element-binding proteins and transposases. Trends. Biochem. Sci. 2000;25:421–423. doi: 10.1016/s0968-0004(00)01620-0. [DOI] [PubMed] [Google Scholar]
- Bilder D. Epithelial polarity and proliferation control: links from the Drosophila neoplastic tumor suppressors. Genes Dev. 2004;18:1909–1925. doi: 10.1101/gad.1211604. [DOI] [PubMed] [Google Scholar]
- Bonasio R, Lecona E, Reinberg D. MBT domain proteins in development and disease. Semin. Cell Dev. Biol. 2010;21:221–230. doi: 10.1016/j.semcdb.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozulic L, Surucu B, Hynx D, Hemmings BA. PKBalpha/Akt1 acts downstream of DNA-PK in the DNA double-strand break response and promotes survival. Mol. Cell. 2008;30:203–213. doi: 10.1016/j.molcel.2008.02.024. [DOI] [PubMed] [Google Scholar]
- Bushey AM, Ramos E, Corces VG. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev. 2009;23:1338–1350. doi: 10.1101/gad.1798209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M, Corces VG. The ubiquitin ligase dTopors directs the nuclear organization of a chromatin insulator. Mol. Cell. 2005;20:105–116. doi: 10.1016/j.molcel.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Chernukhin I, Shamsuddin S, Kang SY, Bergstrom R, Kwon YW, Yu W, Whitehead J, Mukhopadhyay R, Docquier F, Farrar D, Morrison I, Vigneron M, Wu SY, Chiang CM, Loukinov D, Lobanenkov V, Ohlsson R, Klenova E. CTCF interacts with and recruits the largest subunit of RNA polymerase II to CTCF target sites genome-wide. Mol. Cell. Biol. 2007;27:1631–1648. doi: 10.1128/MCB.01993-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow LE, Brumby AM, Muratore R, Coombe ML, Sedelies KA, Trapani JA, Russell SM, Richardson HE, Humbert PO. hScrib is a functional homologue of the Drosophila tumour suppressor scribble. Oncogene. 2003;22:9225–9230. doi: 10.1038/sj.onc.1207154. [DOI] [PubMed] [Google Scholar]
- Emberly E, Blattes R, Schuettengruber B, Hennion M, Jiang N, Hart CM, Kas E, Cuvier O. BEAF regulates cell-cycle genes through the controlled deposition of H3K9 methylation marks into its conserved dual-core binding sites. PLoS Biol. 2008;6:2896–2910. doi: 10.1371/journal.pbio.0060327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann G, Mishra A, Hong SM, Bisht S, Strock CJ, Ball DW, Goggins M, Maitra A, Nelkin BD. Inhibiting the cyclin-dependent kinase CDK5 blocks pancreatic cancer formation and progression through the suppression of Ras–Ral signaling. Cancer Res. 2010;70:4460–4469. doi: 10.1158/0008-5472.CAN-09-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimova TI, Lei EP, Bushey AM, Corces VG. Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila. Mol. Cell. 2007;28:761–772. doi: 10.1016/j.molcel.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurudatta BV, Corces VG. Chromatin insulators: lessons from the fly. Brief Funct. Genomic. Proteomic. 2009;8:276–282. doi: 10.1093/bfgp/elp032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, Lee CW, Ye C, Ping JL, Mulawadi F, Wong E, Sheng J, Zhang Y, Poh T, Chan CS, Kunarso G, Shahab A, Bourque G, Cacheux-Rataboul V, Sung WK, Ruan Y, Wei CL. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat. Genet. 2011;43:630–638. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DA, McCoon PE, Binari R, Gilman M, Perrimon N. Drosophila unpaired encodes a secreted protein that activates the JAK signaling pathway. Genes Dev. 1998;12:3252–3263. doi: 10.1101/gad.12.20.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart CM, Zhao K, Laemmli UK. The scs’ boundary element: characterization of boundary element-associated factors. Mol. Cell. Biol. 1997;17:999–1009. doi: 10.1128/mcb.17.2.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igaki T, Pagliarini RA, Xu T. Loss of cell polarity drives tumor growth and invasion through JNK activation in Drosophila. Curr. Biol. 2006;16:1139–1146. doi: 10.1016/j.cub.2006.04.042. [DOI] [PubMed] [Google Scholar]
- Jiang N, Emberly E, Cuvier O, Hart CM. Genome-wide mapping of boundary element-associated factor (BEAF) binding sites in Drosophila melanogaster links BEAF to transcription. Mol. Cell. Biol. 2009;29:3556–3568. doi: 10.1128/MCB.01748-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis B, Borg JP. Apicobasal polarity complexes. J. Cell. Sci. 2005;118:5157–5159. doi: 10.1242/jcs.02597. [DOI] [PubMed] [Google Scholar]
- Mayburd AL, Martlinez A, Sackett D, Liu H, Shih J, Tauler J, Avis I, Mulshine JL. Ingenuity network-assisted transcription profiling: identification of a new pharmacologic mechanism for MK886. Clin. Cancer Res. 2006;12:1820–1827. doi: 10.1158/1078-0432.CCR-05-2149. [DOI] [PubMed] [Google Scholar]
- Melnik S, Deng B, Papantonis A, Baboo S, Carr IM, Cook PR. The proteomes of transcription factories containing RNA polymerases. I, II or III. Nat. Methods. 2011;8:963–968. doi: 10.1038/nmeth.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais-de-Sá E, St Mirouse V, Johnston D. aPKC phosphorylation of Bazooka defines the apical/lateral border in Drosophila epithelial cells. Cell. 2010;141:509–523. doi: 10.1016/j.cell.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre N, Brown CD, Shah PK, Kheradpour P, Morrison CA, Henikoff JG, Feng X, Ahmad K, Russell S, White RA, Stein L, Henikoff S, Kellis M, White KP. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 2010;6:e1000814. doi: 10.1371/journal.pgen.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak SJ, Pai CY, Corces VG. Protein phosphatase 2A activity affects histone H3 phosphorylation and transcription in Drosophila melanogaster. Mol. Cell. Biol. 2003;23:6129–6138. doi: 10.1128/MCB.23.17.6129-6138.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak M. Insulin and insulin-like growth factor signalling in neoplasia. Nat. Rev. Cancer. 2008;8:915–928. doi: 10.1038/nrc2536. [DOI] [PubMed] [Google Scholar]
- Raab JR, Chiu J, Zhu J, Katzman S, Kurukuti S, Wade PA, Haussler D, Kamakaka RT. Human tRNA genes function as chromatin insulators. EMBO J. 2012;31:330–350. doi: 10.1038/emboj.2011.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter C, Oktaba K, Steinmann J, Muller J, Knoblich JA. The tumour suppressor L(3)mbt inhibits neuroepithelial proliferation and acts on insulator elements. Nat. Cell. Biol. 2011;13:1029–1039. doi: 10.1038/ncb2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Gilbert MK, Hart CM. Characterization of BEAF mutations isolated by homologous recombination in Drosophila. Genetics. 2007;176:801–813. doi: 10.1534/genetics.106.068056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Hart CM. Targeted gene replacement by homologous recombination in Drosophila stimulates production of second-site mutations. Fly (Austin) 2010;4:12–17. doi: 10.4161/fly.4.1.10995. [DOI] [PubMed] [Google Scholar]
- Roy S, Tan YY, Hart CM. A genetic screen supports a broad role for the Drosophila insulator proteins BEAF-32A and BEAF-32B in maintaining patterns of gene expression. Mol. Genet. Genomics. 2007;277:273–286. doi: 10.1007/s00438-006-0187-8. [DOI] [PubMed] [Google Scholar]
- Schoborg TA, Labrador M. The phylogenetic distribution of non-CTCF insulator proteins is limited to insects and reveals that BEAF-32 is Drosophila lineage specific. J. Mol. Evol. 2010;70:74–84. doi: 10.1007/s00239-009-9310-x. [DOI] [PubMed] [Google Scholar]
- Siddle K. Signalling by insulin and IGF receptors: supporting acts and new players. J. Mol. Endocrinol. 2011;47:R1–R10. doi: 10.1530/JME-11-0022. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Ohno S. The PAR-aPKC system: lessons in polarity. J. Cell. Sci. 2006;119:979–987. doi: 10.1242/jcs.02898. [DOI] [PubMed] [Google Scholar]
- Wary KK, Mariotti A, Zurzolo C, Giancotti FG. A requirement for caveolin-1 and associated kinase Fyn in integrin signaling and anchorage-dependent cell growth. Cell. 1998;94:625–634. doi: 10.1016/s0092-8674(00)81604-9. [DOI] [PubMed] [Google Scholar]
- Werner H. Tumor suppressors govern insulin-like growth factor signaling pathways: implications in metabolism and cancer. Oncogene. 2011 doi: 10.1038/onc.2011.447. [DOI] [PubMed] [Google Scholar]
- Wood AM, Van Bortle K, Ramos E, Takenaka N, Rohrbaugh M, Jones BC, Jones KC, Corces VG. Regulation of chromatin organization and inducible gene expression by a Drosophila insulator. Mol. Cell. 2011;44:29–38. doi: 10.1016/j.molcel.2011.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods DF, Hough C, Peel D, Callaini G, Bryant PJ. Dlg protein is required for junction structure, cell polarity, and proliferation control in Drosophila epithelia. J. Cell. Biol. 1996;134:1469–1482. doi: 10.1083/jcb.134.6.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods DF, Wu JW, Bryant PJ. Localization of proteins to the apico-lateral junctions of Drosophila epithelia. Dev. Genet. 1997;20:111–118. doi: 10.1002/(SICI)1520-6408(1997)20:2<111::AID-DVG4>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Yu R, Chen C, Mo YY, Hebbar V, Owuor ED, Tan TH, Kong AN. Activation of mitogen-activated protein kinase pathways induces antioxidant response element-mediated gene expression via a Nrf2-dependent mechanism. J. Biol. Chem. 2000;275:39907–39913. doi: 10.1074/jbc.M004037200. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Hart CM, Laemmli UK. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell. 1995;81:879–889. doi: 10.1016/0092-8674(95)90008-x. [DOI] [PubMed] [Google Scholar]
- Zhu M, Xin T, Weng S, Gao Y, Zhang Y, Li Q, Li M. Activation of JNK signaling links lgl mutations to disruption of the cell polarity and epithelial organization in Drosophila imaginal discs. Cell. Res. 2010;20:242–245. doi: 10.1038/cr.2010.2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.