Abstract
Understanding how cells move, change shape, and alter cellular behaviors to form organs, a process termed morphogenesis, is one of the great challenges of developmental biology. Formation of the C. elegans vulva is a powerful, simple, and experimentally accessible model for elucidating how morphogenetic processes produce an organ. In the first step of vulval development, three epithelial precursor cells divide and differentiate to generate 22 cells of seven different vulval subtypes. The 22 vulval cells then rearrange from a linear array into a tube, with each of the seven cell types undergoing characteristic morphogenetic behaviours that construct the vulva. Vulval morphogenesis entails many of the same cellular activities that underlie organogenesis and tissue formation across species, including invagination, lumen formation, oriented cell divisions, cell-cell adhesion, cell migration, cell fusion, extracellular matrix remodelling and cell invasion. Studies of vulval development have led to pioneering discoveries in a number of these processes and are beginning to bridge the gap between the pathways that specify cells and their connections to morphogenetic behaviors. The simplicity of the vulva and the experimental tools available in C. elegans will continue to make vulval morphogenesis a powerful paradigm to further our understanding of the largely mysterious mechanisms that build tissues and organs.
Introduction
The architect Louis Sullivan famously wrote that “form ever follows function,” a principal that is as true in biology as it is in architecture. Organisms have evolved body plans that are uniquely suited to carrying out the tasks necessary for survival and reproduction. In many multicellular organisms, the functional whole is divided into tissues (e.g., muscles, nerves), organs (skin, lung), and systems (reproductive, digestive). The formation of such ordered structures, a process termed morphogenesis, remains one of the great mysteries of biology. The inherent difficulty of understanding morphogenesis is that it encompasses a broad range of interconnected cellular activities that occur in precise spatial locations at precise times 1. Despite the experimental challenges presented by morphogenesis, a greater understanding of how tissues and organs form holds the promise of improving human health by enhancing our ability to diagnose and treat developmental diseases, engineer tissues to replace diseased body parts 2, and arrest the development of tumors that use morphogenetic processes to mimic native tissues 3. A useful experimental system to study morphogenesis would feature three attributes: (1) Display a range of conserved cellular behaviors that underlie morphogenesis across species; (2) Allow for the examination of the entire morphogenetic process in vivo, in real-time, with high visual resolution; (3) Be experimentally tractable so that the system can be manipulated and examined. One model that fits these criteria is development of the C. elegans vulva, which offers a number of experimental advantages to the researcher while still featuring complex morphogenetic behaviors. This review provides an overview of the morphogenetic basis of vulval development, highlighting advances in our understanding of oriented cell division, cell movement, invagination, cell fusion, cell invasion, and the coordinated attachment of tissues. This array of cellular behaviors results in the formation of the simple, yet elegantly constructed C. elegans vulva.
OVERVIEW OF VULVAL DEVELOPMENT
The C. elegans vulva is a hermaphrodite-specific ectodermal organ that develops post-embryonically and functions to connect the internal reproductive system with the external environment. The vulva is required for mating, as males inject sperm through it, and for deposition of embryos after internal fertilization (Fig. 1A). Vulval development has emerged as a paradigm of morphogenesis because it offers a simple model of tissue rearrangement in which a tube forms from an epithelial sheet of only 22 cells. The vulva also serves as an ideal system for genetic screens 4, since mutations that affect mating and egg-laying do not prevent hermaphrodites from producing viable progeny. Plate-level genetic screens have been conducted to identify vulval development mutants, such as animals that are egg laying defective (Egl, retain developing embryos in the uterus), have a protruding vulva (Pvl), or have multiple vulvae (Muv). Higher magnification compound microscopic analysis can further determine if these plate level mutants lack a vulva (Vul), have specific defects in the connection of the vulva with the gonad (Cog), have a squashed vulva (Sqv), or have defects in specific cellular behaviors such as cell fusion, migration, or invasion. A further advantage to studying vulval morphogenesis is that a number of the signaling pathways that specify vulval cell fates have been characterized 5, 6. This makes the vulva an emerging model to understand how cell specification pathways connect to the morphogenetic processes that shape tissues. These efforts are bringing a more holistic level of understanding to the process of organogenesis.
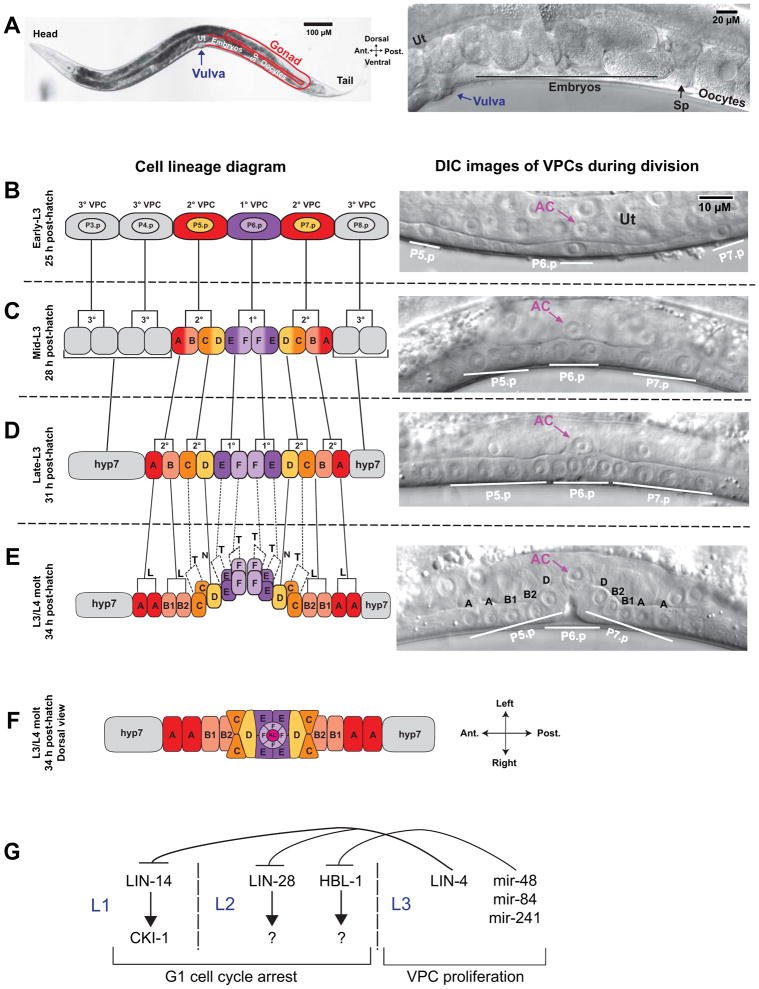
Figure 1.
Vulval cell divisions. (A) Adult worm at low (left) and high (right) magnification showing components of the reproductive system. Oocytes in the gonad (outlined in red) move in a distal to proximal direction and are fertilized in the spermatheca (Sp). Embryos pass into the uterus (Ut) and undergo the first rounds of cell division. Muscle contractions push embryos through the vulva to the external environment, where embryogenesis continues until hatching. The posterior gonad is shown; the anterior gonad has an identical arrangement. For additional images of the C. elegans reproductive system, see WormAtlas 14. (B–E) Cell divisions in the L3 larval stage. Left is schematic diagram; right is differential interference microscopy (DIC) images of live animals. Lateral view with anterior left and dorsal top. (B) The P6.p cell is induced to the 1° VPC fate, P5.p and P7.p are induced to the 2° VPC fate, and P3.p, P4.p, and P8.p take on the non-vulval 3° VPC fate. The anchor cell (AC) is located in the uterine epithelium (Ut) and positioned dorsal to P6.p (right DIC image). (C) All six VPCs divide in the mid-L3 larval stage, after which the 3° VPCs arrest cell divisions and initiate fusion with the hyp7 syncytium. The 1° VPCs differentiate to vulE and vulF cell types; the 2° VPCs differentiate to vulA, vulB, vulC, and vulD cell types. (D) The third VPC divisions take place in mid to late L3 and produces 12 cells in a longitudinal array. (E) The final VPC divisions occur during the L3/L4 molt. vulC, vulE, and vulF cell divisions occur in the transverse (T) orientation, which is indicated by dashed lines in the lineage diagram. vulA and vulB divide in the longitudinal orientation (L), and vulD does not divide (N). In the DIC image, only the longitudinally divided cells and vulD are in the plane of focus. (F) Dorsal view of the vulval primordium following terminal cell divisions, with the AC at the vulval apex. Modeled after 8. For additional schematics of vulval development and a time series of vulval images from the L3/L4 molt to adulthood, see WormAtlas 14. (G) Heterchronic genes regulate the timing of VPC divisions. LIN-14, acting through the cell cycle repressor cki-1, inhibits cell divisions in L1, and LIN-28 and HBL-1 inhibit L2 cell divisions, acting through unknown targets. In L3, the miRNA lin-4 represses lin-14 and lin-28 mRNA translation to promote cell division. Analogously, the miRNAs mir-48, mir-84, and mir-241 redundantly repress hbl-1 translation.
Vulval cell induction and differentiation
During larval development, six epidermal cells on the ventral surface of the hermaphrodite, P3.p–P8.p, are competent to form vulval tissue. Through a series of inductive interactions involving the LIN-3/epidermal growth factor (EGF) and LIN-12/Notch pathways, P6.p is induced to adopt the 1° vulval precursor cell (VPC) fate, and P5.p and P7.p are induced as 2° VPCs (Fig. 1B). In wild-type animals, P4.p, P4.p and P8.p are not induced to a vulval fate and contribute to the hypodermis, which generates the cuticle that covers the animal. The 1° VPC produces progeny cells of type vulE and vulF, and the 2° VPCs produce progeny cells of type vulA, vulB1, vulB2, vulC and vulD (Fig. 1C–E). Each of the different vulval cell types undergoes a distinct morphogenetic program 7, 8.
Cell division
Vulval induction determines the subsequent pattern of cell divisions, which occur during the L3 larval stage. The 1° VPC, P6.p, undergoes three rounds of cell division to produce eight cells, and the 2° VPCs, P5.p and P7.p, divide three times to produce seven cells each (Fig. 1C–E). After cell divisions, the vulval primordium has 22 cells displaying mirror-image symmetry, in the sequence AAB1B2CCDEEFFFFEEDCCB2B1AA (Fig. 1E–F). The first two rounds of cell division occur in the longitudinal (anterior-posterior) orientation. The terminal cell divisions of vulC, vulE, and vulF occur in the transverse (left-right) orientations, and the terminal cell divisions of vulA and vulB occur in the longitudinal direction. vulD does not undergo a third cell division (Fig. 1E–F).
Cell arrangements, invagination, toroid formation, and cell-cell fusion
After the second cell division, the 12 VPCs are arranged as a linear array (Fig. 2A). As the final cell divisions occur, the central 1° VPCs invaginate by detaching from the ventral cuticle and move dorsally (Fig. 2B). In a proximal to distal temporal wave (inner to outermost), cells on each lateral half extend apical membrane processes that form rings with homotypic cells from the other lateral half, and undergo short-range cell migrations toward the midline (Fig. 2C–D) 8. Distal cells migrate along the ventral surface of the inner proximal cell, which displaces cells dorsally and expands the invagination as cells compact around the vulval midline (Fig. 2E) 9. In addition to short-range migrations, expansion of the vulval lumen also requires VPC expression of glycosaminoglycans, which are thought to shape or maintain the lumen formed by invaginating cells (Fig. 2E) 10, 11. Through the L4 larval stage, ring formation and invagination continue, giving rise to a characteristic “Christmas tree” appearance formed by the dorsal-ventral stack of cells (Fig. 2F). These ringed structures are termed toroids, circular groups of cells with a central hole, analogous to a bagel (Fig. 2G). Cell fusions occur during toroid formation to generate multinucleate syncytia. First, sister cells on each half of the midline fuse (i.e., vulA with vulA; Fig. 2B), then cells of the same subtype from contralateral halves fuse (i.e., anterior vulA with posterior vulA; Fig. 2F). All homotypic cells fuse with the exception of vulB1 and vulB2 (Fig. 2E) 8.
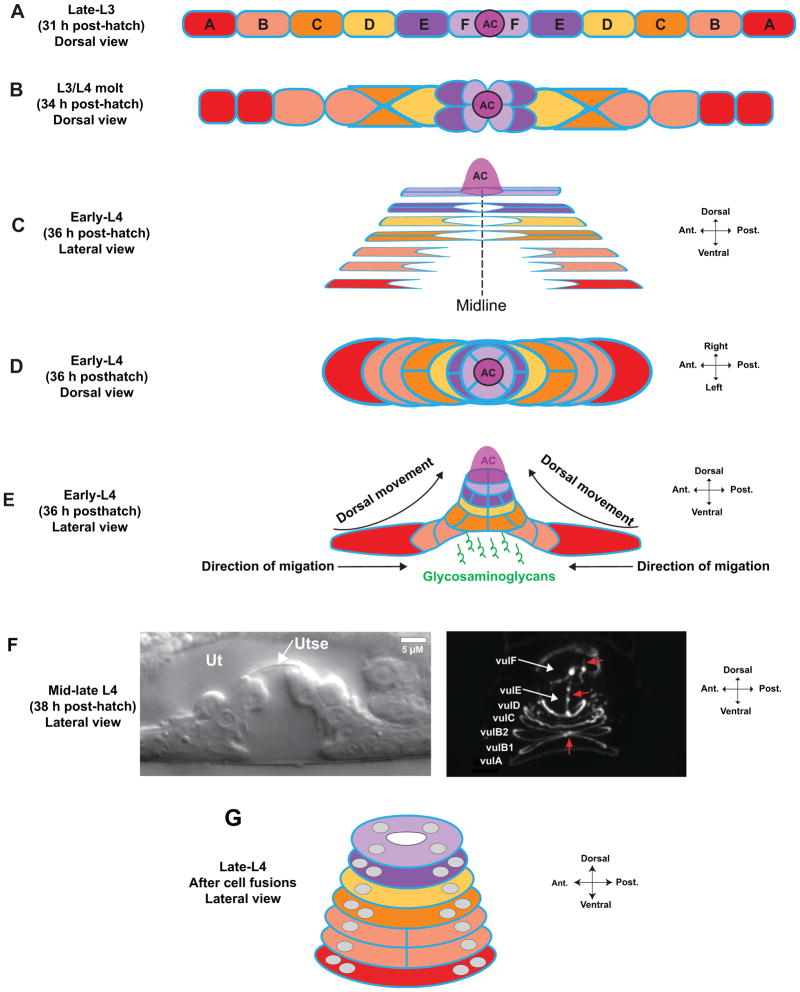
Figure 2.
Vulval invagination and toroid formation. (A) After the second cell divisions, the VPCs are arranged as a longitudinal array. The anchor cell (AC) is positioned at the junction of the vulF cells at the vulval midline. Apical domains are in blue here and in other panels. (B) Concurrent with the final cell divisions, the 1° VPCs invaginate by moving dorsally. vulF occupies the most dorsal position, with vulE positioned ventral to it. (C) VPCs extend apical membrane processes toward the midline and establish junctions with homotypic cells from the contralateral half, forming rings that encircle a tube in the center of the vulval primordium. In this schematic, vulC–vulF have formed rings. Sister vulA cells on each lateral half fuse shortly after the final cell division 8, shown as a single cell boundary. (D) Dorsal view of early-L4 VPCs, showing the lateral migration of vulval cells toward the midline. (E) Invagination is driven by distal VPCs migrating toward the midline along the ventral surface of inner cells, which forces cells to move dorsally 9. Glycosaminoglycans are expressed by the VPCs and are thought to maintain and shape the forming lumen 10, 11. (F) As the L4 stage continues, vulval cells continue to migrate and homotypic cells undergo fusions. A uterine lumen forms, and the uterine and vulval epithelia become separated by the thin membrane of the utse syncytium (left DIC image). The right image shows labeling of apical junctions with the apical membrane marker AJM-1::GFP 134. At this stage, the sister cells on each lateral half have fused, and contralateral vulA and vulD cells have fused at the midline. Red arrows indicate apical borders between vulB1, vulB2, vulC, vulE, and vulF cells. (G) Schematic of vulval toroids in late-L4 after fusion has completed. Grey circles represent nuclei. All homotypic cells use except vulB1 and vulB2. For additional images, see refs. 8, 9 and WormAtlas 14.
Attachments to other tissues
The vulva ultimately forms a connection and a continuous lumen with the uterus to allow for mating and egg-laying. The connection of these two tissues is initiated by a cell invasion event from the uterine anchor cell (AC), which breaches the basement membrane separating these tissues and contacts the central 1° VPCs (Fig. 6A) 12. The uterine-vulval attachment matures by further removal of the basement membrane, allowing direct attachment of vulF cells with the overlying uv1 cells of the ventral uterus (Fig. 3) 13. After invasion, the AC fuses with neighboring uterine cells, generating the multinucleated utse cell that forms a thin cellular membrane between the vulval and uterine lumens (Fig. 2F; Fig. 3). The vulva also forms attachments with hypodermal and muscle tissues. vulE cells extend processes that contact hypodermal seam cells on the lateral sides of the animal, which is thought to structurally stabilize the vulva. Eight sex muscle cells, four vm1 and four vm2, contact the vulval lips to control opening during mating and egg-laying (Fig. 3). The muscles in turn are innervated by motorneurons that are guided to synaptic locations by cues from the vulva. The final step in vulval formation is eversion, during which the vulva turns partially inside out and the lumen closes into a slit (Fig. 1A) 8, 14.
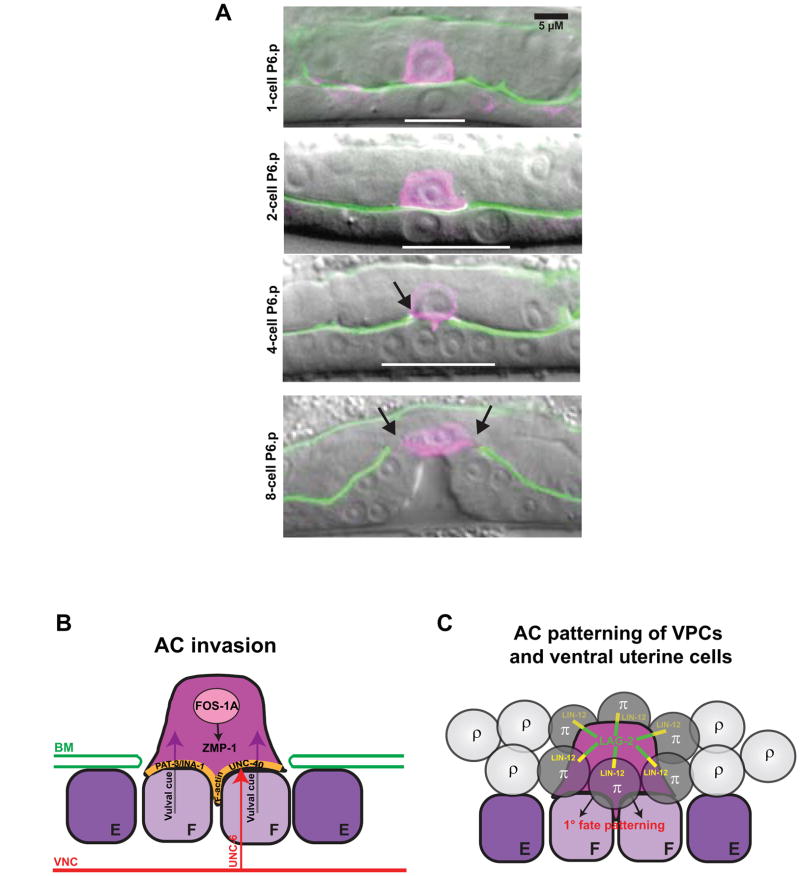
Figure 6.
The anchor cell (AC) connects the uterine and vulval tissues and patterns cell fates. (A) AC invasion into the vulval epithelium. At the one-cell P6.p stage (top panel), the AC is positioned dorsal to P6.p (indicated with white bar) and separated from it by basement membrane (green). Following the first VPC division (second panel), the AC initiates invasion across the basement membrane. By the four-cell P6.p stage (third panel), a breach is evident in the basement membrane (indicated with arrow), and the AC has contacted the 1° VPCs. The AC remains at the vulval apex following the final VPC division (bottom panel) and the gap in the basement membrane continues to expand beyond the border of the AC membrane (arrows). The AC expresses a reporter for the PH domain of phospholipase Cδ fused to mCherry under an AC-specific cdh-3 promoter 95. The basement membrane expresses GFP-tagged laminin β subunit under its own promoter 135. (B) Molecular pathways regulating AC invasion. The transcription factor FOS-1A is required for invasion, and regulates the expression of the matrix metalloprotease ZMP-1. The basal (ventral) AC membrane is enriched for cytoskeletal components F-actin and the INA-1/PAT-3 integrin receptor complex. The UNC-6/netrin ligand is released by the ventral nerve cord (VNC) and regulates invasion through its receptor UNC-40/DCC. A vulval cue from the 1° VPCs is also required for complete invasion. Modeled after 101. (C) The AC patterns uterine and vulval fates. The AC expresses LAG-2/Delta ligand, which signals to the LIN-12/Notch receptor on six surrounding ventral uterine cells to induce the π fate rather than the default ρ fate. The presence of the AC between the vulF cells is required for 1° fate patterning.
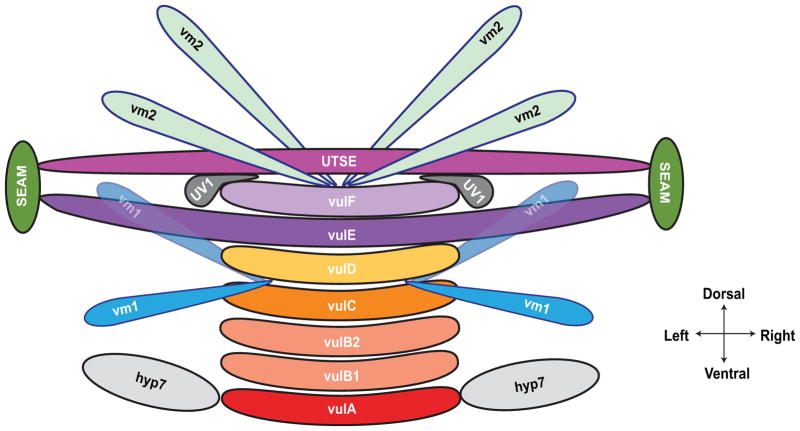
Figure 3.
Attachments of the vulva to other tissues. (A) The vulA cells on the ventral surface contact the hyp7 hypodermal syncytium; vulE cells extend laterally to contact the hypodermal seam cells; and vulF cells contact the uv1 cells and are overlaid by the utse (see Fig. 2F). Eight vulval muscles, four vm1 and four vm2, control vulval opening. vm1 cells contact the vulva between vulC and vulD toroids, and vm2 cells contact between vulF and the uterus. Modeled after 8, 102. For additional images of late vulval development, see WormAtlas 14.
VULVAL MORPHOGENESIS
Vulval cell induction and differentiation
Vulval induction has been extensively studied [for reviews, see 15–18], and will be summarized briefly here. During the L1 larval stage, 12 epidermal cells (P1.p–P12.p) are born as the posterior daughters of P ectoblast cell divisions and are positioned along the ventral midline. Six of these cells (P1.p–P2.p, P9.p–P12.p) fuse with the surrounding hypodermal syncytium, hyp7, soon after birth 19. The remaining unfused cells, P3.p–P8.p, are termed vulval precursor cells (VPCs), because all possess the potential to be specified as vulval cells 7. In wild-type animals, however, only P5.p–P7.p assume vulval identities due to an inductive LIN-3/EGF signal from the AC, which is positioned dorsal to P6.p (Fig. 1B). P6.p is thought to receive the highest levels of AC-secreted LIN-3 and through Ras activation assumes the 1° VPC fate. P5.p and P7.p receive moderate LIN-3 and assume the 2° fate. The distal P3.p, P4.p and P8.p cells receive low EGF and assume the non-vulval 3° fate, which is fusion with the hyp7 syncytium 20. Recent work has supported a model in which higher levels of Ras activation in the P6.p lineage results in the engagement of the LIN-45/Raf effector, which promotes the 1° fate, whereas lower Ras activation in the P5.p and P7.p lineages utilizes the RalGEF effector, which signals a pro-2° fate21. In addition to a gradient of LIN-3, vulval fates are also patterned by sequential induction in which the 1°-fated VPC is first specified, and as a result of this induction expresses transmembrane and secreted DSL proteins that signal via the LIN-12/Notch receptor on P5.p and P7.p to promote the 2° fate 22. In addition to graded and sequential induction, the 1° and 2° fates are also patterned through pathway quenching. In the 1°-fated VPC, activation of the EGF/Ras/Raf pathway antagonizes the LIN-12/Notch pathway in part by downregulating the LIN-12 receptor 21–23, while in the 2°-fated VPCs, activation of LIN-12/Notch upregulates numerous inhibitors of the EGF signaling pathway 22, 24, 25. Other mechanisms appear to further contribute to the precise patterning of the VPCs, including an early role for HOX genes in regulating VPC induction competency, and apparent roles for the Wnt pathway and the ROM-1 protease in VPC induction (reviewed in 16). The extensive levels of redundancy in this well studied system of VPC induction likely reflects more generally on how cell fate specification systems are wired to ensure robust patterning of tissues during development.
Cell division
During the L1 larval stage, P3.p–P8.p VPCs are born and immediately arrest in an extended G1 phase26. The VPCs remain in this quiescent state until the mid-L3 stage, when they resume division. Proper vulval formation requires the precise control of cell division timing, cell cycle number, spindle orientation, and polarity. Many classes of genes and genetic pathways have been found to regulate these distinct aspects of vulval cell divisions and are discussed below.
Heterochronic genes regulate vulval cell division timing
Genes that regulate developmental timing are referred to as heterochronic, as their loss or gain of activity leads to changes in the timing of specific developmental events in relation to development as a whole 27. Heterochronic genes that regulate the timing of VPC divisions include the transcription factors LIN-14 28, the RNA binding protein LIN-28 29, and the microRNA lin-4 (Fig. 1G) 30. LIN-14 functions to promote L1 larval stage cell fates, whereas LIN-28 specifies aspects of L2 fates 27, 29. Loss of lin-14 or lin-28 leads to an early G1 to S transition and precocious VPC divisions, presumably as a result of skipping the L1 or L2 stages, respectively. Although the vulva is precociously induced in lin-14 and lin-28 mutants, it displays normal lineage patterns and marker gene expression. The vulva fails to connect properly to the uterus, however, likely due to mistiming in the development of the two tissues 12, 26, 27. One downstream effector of LIN-14 is cki-1, a C. elegans p21/p27 cyclin-dependent kinase inhibitor, which maintains a G1 state and inhibits cell cycle progression31. cki-1 is first expressed in the late L1 stage and is absent in the L3 stage upon initiation of cell divisions. lin-14 is required for the early larval expression of cki-1, and loss of cki-1 results in precocious VPC divisions similar to lin-14 mutants 31. During VPC proliferation in L3, LIN-14 and LIN-28 protein levels are reduced through binding of lin-4 miRNA to the 3′ UTRs of lin-14 and lin-28 mRNAs, leading to their translational repression (Fig. 1G) 26, 29, 32. In lin-4 loss-of-function mutants, the first VPC divisions occur at the correct time in mid-L3, but subsequent cell divisions are missing or delayed 26, 33. lin-4 also has a role in specifying vulval fates, as P5.p and P7.p fail to adopt 2° VPC identities in lin-4 mutants. This is due to the continued presence of LIN-14 protein, which inhibits the activity of LIN-12 necessary for 2° fate specification 34.
Similar to lin-28, the transcription factor hbl-1/Hunchback also regulates timing of VPC divisions by promoting L2 cell fates (Fig. 1G). A mutant allele of hbl-1 or RNAi depletion of the gene causes precocious VPC divisions at or before the L2/L3 molt, leading to Pvl and Egl phenotypes 35. hbl-1 is negatively regulated in a redundant manner by let-7 family microRNAs mir-48, mir-84, and mir-241 36. Overexpression of mir-48 phenocopies the precocious VPC divisions of hbl-1 reduction of function, possibly by repressing hbl-1 mRNA translation during L2 (Li et al., 2005). The mechanisms by which inhibition of HBL-1, LIN-14, and LIN-28 lead to promotion of VPC divisions in L3 is not well understood and cannot be accounted for solely by cki-1 inhibition, as the phenotype of cki-1 RNAi reduction of function is a single extra VPC cell division in L2 31. Activation of the cell cycle in L3 thus appears to involve other targets that function downstream of heterochronic gene repression.
General cell cycle machinery and control of vulval cell divisions
The C. elegans cell-cycle machinery, built around the regulation of expression and activity of cyclin dependent kinases (Cdks) and cyclins, is highly conserved with other metazoans 37. Many general regulators of the cell cycle have been found to influence the number and timing of vulval cell divisions. As mentioned above, cki-1 is a key regulator of the G1 quiescent state of VPCs and its expression is positively regulated by the phosphatase cdc-14 38, 39. The C. elegans retinoblastoma (Rb) gene ortholog lin-35 functions redundantly with the putative SCF complex regulator lin-23 and the Cdh1/Hct1/FZR ortholog fzr-1 to limit VPC cell division in the G1 quiescent state in the L1/L2 larval stages, possibly by reducing cyclin levels 40, 41.
Regulators of the cell cycle during the proliferative L3 stage have also been identified. The CUL-4/DDB-1 E3 ubiquitin ligase complex, which downregulates CKI-1 protein expression in hypodermal seam cells, also likely downregulates CKI-1 in VPCs. ddb-1::GFP is expressed in all vulval cells, and a ddb-1 null mutant has a reduced number of VPC progeny, consistent with its loss leading to the continued expression of CKI-1 42. An additional general regulator of vulval cell divisions is cye-1/Cyclin E, which promotes the G1- to S-phase transition43. In cye-1 mutant animals the VPC cell cycle in the L3 stage is lengthened and the third cell division frequently does not occur. cul-1 is an ortholog of mammalian cullins, which target Cyclin E for degradation as part of the SCF E3 ubiquitin ligase complex 44. Consistent with a role in limiting cell divisions through degradation of CYE-1 protein, cul-1 mutants display vulval hyperplasia, producing an average of 82 vulval cells 45. Similar to lin-14 and lin-28 mutants, perturbations in cell cycle do not affect vulval cell differentiation, suggesting that cell cycle regulation and differentiation can be uncoupled 31, 41, 43, 45. One exception to this finding is the overproliferation defects associated with loss of lin-35/Rb, in which vulval fate patterning and morphogenesis are also aberrant 40. This suggests that lin-35/Rb might have additional cell cycle-independent roles in vulval differentiation 46, similar to the roles of Rb in vertebrate muscle and bone differentiation 47.
Vulval-specific regulators of cell division
How developmental pathways regulate precise cell cycle progression during tissue formation and organogenesis remains largely unknown 48. Although some transcriptional regulators of vulval cell divisions have been identified, the precise links between these genes and the cell cycle remains unclear. During VPC quiescence in L1 and L2, the transcription factors LIN-1/Ets and LIN-31/FoxB promote the expression of CKI-1 to prevent precocious VPC divisions 39. Upon exit from quiescence in the L3 stage, the Hox gene lin-39 promotes initiation of VPC division 49. Still other transcription factors regulate the number of cell divisions upon initiation of cell division. Loss-of-function mutants of cog-1, an Nkx6 homeodomain transcription factor, display defects in which the final cell divisions of vulB, vulC, and vulF sometimes do not occur, and the vulD cells undergo a third cell division 50. cog-1 also regulates vulval gene expression and the proper connection of uterine and vulval tissues, indicating that it is required for several aspects of vulval differentiation 5, 50. Mutations in the zinc-finger transcription factor bed-3 cause a cell division defect similar to cog-1, suggesting that the two transcription factors form part of a regulatory network required for differentiation of a subset of vulval cells 51. The LIM homeodomain transcription factor lin-11 also regulates multiple aspects of vulval identity, including cell division arrest of vulD 52. A future challenge will be to understand how these vulva-specific transcriptional regulators direct the general cell cycle machinery in specific cell lineages.
Oriented cell divisions
Oriented cell division is a common morphogenetic mechanism to shape organs 53. After the first two rounds of longitudinal cell divisions, six of the ten VPC granddaughters that divide a third time (vulC, vulE, vulF precursors) change their mitotic spindle orientation to divide in the transverse direction (Fig. 1E). Our current understanding of the mechanisms that regulate mitotic spindle reorientation during these cells divisions is rudimentary. Combined loss of the Rac-like GTPases ced-10 and mig-2 alters the terminal cell divisions of vulC and vulE, such that they occur longitudinally or obliquely rather than transversely. Interestingly, the vulF cell divisions are still transverse after mig-2 and ced-10 loss, suggesting that vulF may be regulated differently from vulC and vulE 54. Mutations in the guanine nucleotide exchange factor (GEF) unc-73/Trio produce a similarly penetrant defect to combined loss of ced-10 and mig-2, indicating that unc-73 is likely the GEF for both genes 54. Mutations in lin-40/MTA, a component of the NuRD transcriptional co-repression complex, also transform transverse to longitudinal divisions, suggesting that changes in gene expression may be required to properly orient the mitotic spindle 55. It will be important in the future to determine whether these Rac genes or lin-40 ultimately influence spindle orientation through conserved extrinsic (e.g., planar cell polarity, Fat/Dachsous) or intrinsic (e.g., LGN/Numa/ Gαi) pathways known to regulate spindle orientation 53.
An evolutionary comparison of 51 rhabditid species showed that the terminal cell divisions of distal VPCs (vulA and vulB) always occur longitudinally, and terminal cell divisions of proximal VPCs (vulE and vulF) always occur transversely. Terminal vulC divisions are variable: transverse in Caenorhabditis species but longitudinal in diplogastrids such as Pristionchus pacificus 56, 57. An analysis of ring formation in 12 nematode species by Kolotuev and Podbilewicz (2008) found that longitudinal divisions generate two rings (e.g., vulB1 and vulB2 in C. elegans), whereas one ring forms from transverse or no cell divisions (e.g., vulC, vulD, vulE, vulF). While VulA is an apparent exception to this rule as it forms one ring despite dividing longitudinally 58, the divided VulA cells fuse with each other in most species (including C. elegans) prior to toroid formation, thus precluding two rings from forming. Blocking vulA fusions in C. elegans results in the formation of an additional toroid ring. Further, an additional ring forms in lin-11 mutants in which vulC undergoes a longitudinal division 58. Thus, the number of toroid rings and the structure of the mature vulva appears to be dictated by the orientation of cell divisions and the number of dividing cells.
Establishing mirror-image symmetry
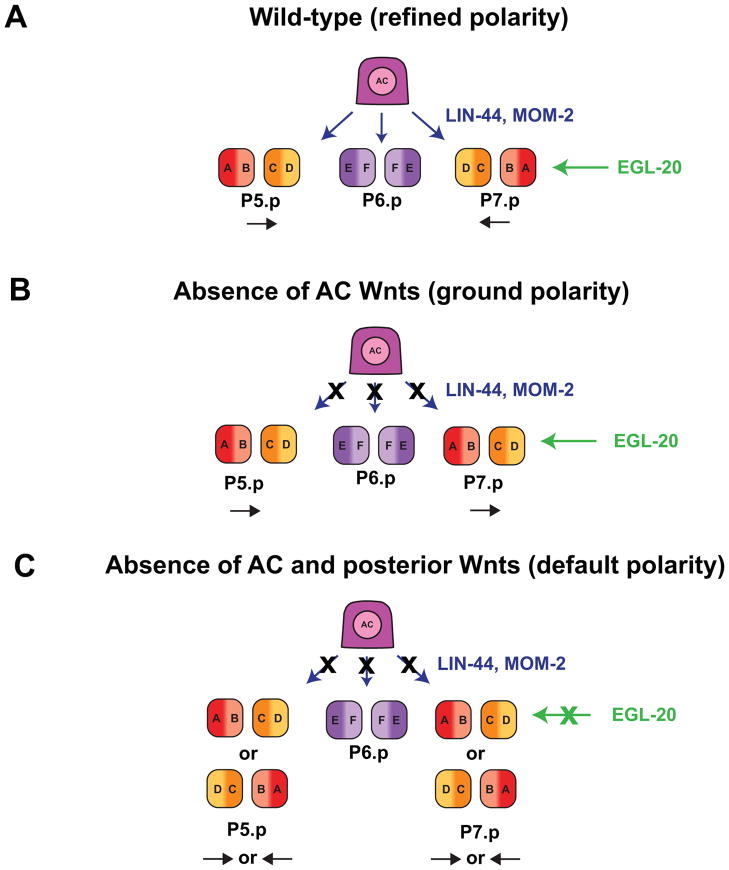
The final order of cell types in the vulva has mirror-image symmetry around the vulF cells (Fig. 1E). Although the molecular mechanism is unclear, the AC is required for patterning of E-F-F-E fates in the 1° VPC lineage (Fig. 6C). Ablation of the AC at the two- or four-cell 1° VPC stage causes mispatterning of vulE and vulF fates. A similar mispatterning defect is observed in dig-1 mutants in which the AC is displaced dorsally, demonstrating that proximity of the AC with the 1° VPCs is required for proper patterning 59. The Wnt pathway is required to polarize the 2° VPC cell fates (P5.p and P7.p) and establish the mirror-image symmetry of these lineages (A-B-C-D/D-C-B-A) (Fig. 1C). A Wnt ligand, EGL-20, is expressed in posterior cells and acts at a distance as a directional cue through a Ror tyrosine kinase receptor, CAM-1, and the planar cell polarity core pathway component VANG-1/Van Gogh, to generate a posterior-oriented polarity in VPC cell divisions 60, 61. A second group of Wnt ligands refines the EGL-20-induced posterior polarity to orient VPC cell divisions toward the center of the vulval primordium, thus reversing P7.p polarity. This second group of Wnt ligands, LIN-44 and MOM-2, are expressed in the AC and function redundantly via their putative VPC-expressed receptors, LIN-17/Frizzled and LIN-18/Ryk, respectively (Fig. 4A). A third Wnt, CWN-2, also appears to have a modest, redundant function with LIN-44 and MOM-2 62–64. The phenotype of combined mutants of lin-44 and mom-2 is that VPCs divide normally and appear to be properly specified, but do so with reversed P7.p polarity (Fig. 4B), producing one P5.p and P6.p-derived vulva and a second P7.p-derived pseudovulva 61–63. Reduction of all Wnts, including posterior localized EGL-20, leads to randomized P5.p and p7.p polarity, which is the apparent default state (Fig. 4C). The precise time of action and downstream effectors used by LIN-17/LIN-18 and CAM-1/VANG-1 to influence the orientation of P5.p and P7.p remain an area for future study.
Figure 4.
Wnt signaling determines the polarity of VPC cell divisions. (A) In the presence of posterior-expressed Wnt ligand egl-20 and AC-expressed Wnt ligands lin-44 and mom-2, P5.p and P7.p have mirror-image symmetry in their cell division patterns, which is termed “refined polarity” 61. (B) When lin-44 and mom-2 are absent, egl-20 directs a posterior-oriented “ground polarity”, such that both P5.p and P7.p divide in the same orientation. (C) In the absence of lin-44, mom-2, and egl-20, P5.p and P7.p divide with random polarity. This is considered the “default polarity” 61.
Cell rearrangements, invagination, toroid formation and cell-cell fusion
Cell rearrangements and invagination
Invagination initiates in the late-L3 stage after the anchor cell has invaded and just prior to the terminal VPC divisions. The vulF cells detach from the ventral cuticle and move dorsally to generate a small invagination at the midline (Fig. 2B). In a temporal order starting with vulE, vulval cells extend unidirectional apical membrane processes that make homotypic contacts with their contralateral partner cells (i.e., vulD with vulD) at the midline, which results in ring formation around a central lumen (Fig. 2C) 8. As they form rings, vulval cells migrate laterally and ventrally toward the midline relative to proximal cells, which pushes inner cells dorsally and expands the invagination (Fig. 2D–E). Ring formation has been proposed as the driving force for invagination, moving the vulva dorsally as cells migrate toward the midline 8, 9. The cellular behaviors of vulval invagination are notably similar to those of ventral enclosure in C. elegans embryogenesis, in which the coordinated movement and extension of cellular processes towards a contralateral partner at the ventral midline is key to forming an epithelial monolayer that covers the embryo 65. Initial studies suggest that a coordinated action of guidance signaling pathways and cytoskeletal components regulate both processes.
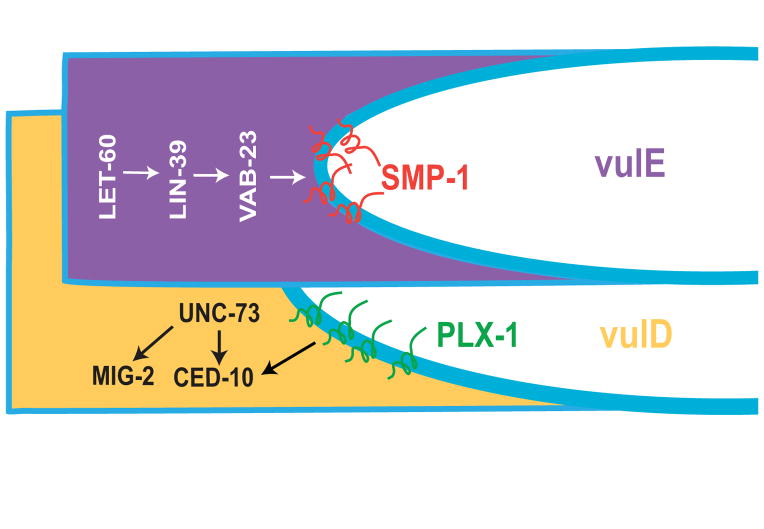
Two reports have shown roles in vulval morphogenesis for the guidance molecule SMP-1/Semaphorin and its receptor PLX-1/Plexin 66, 67. Following the cell divisions in L1 that produce the Px.p cells, the VPCs extend longitudinally until establishing contacts with neighboring cells. In smp-1 and plx-1 mutant animals, VPCs sometimes extend past their neighbors and make contact on their lateral sides rather than at their anterior-posterior ends. This causes mispositioning of VPCs and later defects in vulval morphogenesis 67. These results suggest an inhibitory effect of SMP-1 on the outgrowth of PLX-1 expressing VPCs, which is analogous to the role played by the semaphorin MAB-20 in inhibiting ectopic contacts between adjacent cells during ventral enclosure 68.
There are instances in Drosophila and mouse in which Semaphorin molecules attract Plexin-expressing cells 69, 70. Dalpe et al. (2005) examined the role of the SMP-1/PLX-1 pathway during later stages of vulval development and identified an attractive role during vulval cell migrations in the L4 stage 66. SMP-1 is initially expressed on the lumen-facing surface of proximal VPCs, and attracts distal cells expressing PLX-1 on their apical membranes (Fig. 5) 66. As distal cells migrate into the lumenal area, they switch on expression of SMP-1 to attract the PLX-1-expressing cell ventral to it. SMP-1 expression is downstream of LET-60/Ras, LIN-39/Hox, and the zinc finger transcription factor VAB-23 (Fig. 5) 71. In the absence of either smp-1 or plx-1, VPCs fail to migrate and sometimes detach from neighboring cells, causing Egl, Pvl, and Muv phenotypes 66. The two described roles for the SMP-1/PLX-1 signaling pathway during vulval development demonstrate that a ligand-receptor interaction can be both attractive and repulsive within the same cell lineage during distinct temporal events.
Figure 5.
Molecular pathways of vulval cell migration. Cells that are proximal to the midline (vulE in this example) express SMP-1 on the apical membrane, downstream of LET-60/Ras, LIN-39/Hox, and the zinc-finger transcription factor VAB-23. More distal cells (vulD) express PLX-1, which regulates cell migrations in part through the Rac GTPase CED-10 and its GEF UNC-73. A second pathway involving MIG-2 also regulates migration. Modeled after 66.
The ced-10 and mig-2 Rac-like GTPases and their GEF unc-73, which are required to orient VPC division planes (see above), also regulate VPC migration 54, 66. ced-10 and mig-2 function redundantly: animals with single gene mutations have negligible vulval cell migration defects whereas ced-10; mig-2 double mutants are 77% defective. Genetic evidence suggests that ced-10 regulates VPC migration exclusively in a pathway with plx-1; mig-2, however, has roles outside the plx-1 pathway, since a mig-2; plx-1 double mutant enhances the migration defect of a plx-1 null mutant. unc-73 mutants also enhance the plx-1 defect, consistent with it serving as the GEF for mig-2 (Fig. 5) 66. There is therefore at least one parallel pathway of vulval cell migration involving unc-73 and mig-2 that has yet to be elucidated.
Toroid formation
Cell-cell adhesion plays a critical role in morphogenesis by generating changes in cell shape and cell-cell association 72. Highly regulated adhesions occur during development of the vulva, as homotypic cells form toroid rings (Fig. 2B). The mechanisms that mediate these complex morphogenetic behaviors are poorly understood. Only a single gene has been identified that is specifically required for the midline adhesion of vulval cells: par-1, a membrane-associated EMK-family serine/threonine kinase 73. In par-1 RNAi-treated animals, vulval cell specification, invagination, and migration are normal, but cells fail to make contact with or maintain attachments to their homotypic partners. This results in a gap between rings from each half-vulva and a Pvl phenotype. Fusions between sister cells on each lateral half are normal, demonstrating that the ability to fuse is not compromised by reduction of par-1 73. It is currently unclear how par-1 functions to regulate adhesion of vulval cells. Additional insight will come from the identification of par-1 downstream targets.
The seven vulval cell types form only homotypic adhesions, despite the close proximity of differently fated cells. Even in cye-1 mutants with fewer than the correct number of vulval cells or in plx-1 and smp-1 mutants with defects in migration, ring formation between nonhomologous cells is not known to occur 43, 66, 67. This cell-type matching is also observed in ventral enclosure in C. elegans and in Drosophila dorsal closure, in which two epithelial sheets adhere at the midline, with only similarly patterned cells forming adhesions. Filopodia extension has been observed in leading edge cells during these processes and also in vulval ring formation 9, 66, 74, 75. It is thought that filopodia sample the environment until encountering a match, although the mechanisms of matching are not well understood 75.
Cell-cell fusions
Cell-cell fusions occur within five of the seven vulval toroids during the L4 stage (Fig. 2F–G). Sister vulA, vulC, vulE, and vulF cells first fuse on each lateral half, then homotypic cells from each half-vulva fuse at the midline, in the order vulD, vulA, vulC, vulF, and vulE 8. vulB1 and vulB2 toroids remain unfused. Fusion is a common event in C. elegans—300 of 1090 somatic cells undergo fusion, generating multinucleate syncytia in the hypodermis, uterus, and vulva 76. All C. elegans fusions are executed by two proteins, EFF-1 and AFF-1, which function as homotypic fusogens (i.e., mediate fusion between two cells expressing the same fusogen). EFF-1 and AFF-1 are type I transmembrane proteins, and appear to be the first identified membranes of a divergent, ancient family of fusogens 77. Most vulval cell fusions require eff-1, although fusions of vulA and vulD require aff-1 instead of or in addition to eff-1 78, 79. In eff-1 mutant animals, vulval specification, cell division, migration, and midline attachment are normal, but membranes fail to fuse, causing exploded or protruding vulva phenotypes 78. aff-1 mutants also appear normal for vulva formation but are Egl, possibly due to a failure to properly connect the uterus with the vulva (see below) 80.
Ectopic expression of eff-1 or aff-1 in cultured insect cells is sufficient to induce cell-cell fusions 80, 81. Given this intrinsic fusogenic cabability, expression of eff-1 and aff-1 is thought to be tightly controlled to prevent premature fusion. In P3.p–P8.p VPCs, the Hox gene lin-39 inhibits the expression of eff-1 prior to LIN-3 induction of VPC fate. After induction, lin-39 is downregulated in 3°-fated VPCs, which results in eff-1 expression and fusion to the neighboring hyp7 cell 49. In the 1° and 2°-fated VPCs, LET-60/Ras acting downstream of LIN-3 maintains the expression of lin-39, which through the expression of the zinc-finger protein vab-23 suppresses eff-1 expression 71. The transcription factor nhr-67/Tailless also represses eff-1 expression, and RNAi reduction of nhr-67 causes inappropriate fusions between vulE and vulF cells 82. An interesting question for future studies is how the repressive pathways are shut down to allow expression of eff-1 and aff-1 during fusions.
Role of sqv mutants in invagination and lumen formation
In a screen for mutants with defective vulval morphogenesis, Herman et al. (1999) identified eight genes whose mutants have a squashed vulva (Sqv) phenotype, as well as apparent defects is oogenesis and early embryogenesis 10. In the sqv mutants, VPCs detach from the cuticle and form an invagination, but it is narrower and shorter than in wild-type animals. Other aspects of VPC differentiation, including cell division, ring formation, and cell fusion, are normal in sqv mutants, indicating that the defect is limited to the invagination process 10. Characterization of the sqv genes revealed that all eight encode enzymes that function to produce chondroitin and heparan sulfate glycosaminoglycans (GAGs)—large, unbranched oligosaccharides that covalently modify proteins in the Golgi apparatus 10, 83–87. The modified proteins are secreted into the extracellular matrix or, if transmembrane, located at the plasma membrane with glycosylated regions on the extracellular face (Fig. 2D) 88. Chondroitin appears to be the specific modification necessary for vulval morphogenesis, as sqv-5 encodes a glycosyltransferase specific for the production of chondroitin 87, and a mutant allele of rib-2, a gene required for initiation and elongation of heparan sulfate, does not have a Sqv phenotype 89. The ways in which GAGs regulate vulval morphogenesis have not been ascertained, but a possible explanation is that the presence of glycosylated proteins in the extracellular space between the plasma membrane and cuticle generates hydrostatic forces that allows expansion of the forming lumen. GAGs are negatively charged molecules that attract cations and water, generating turgor pressure to resist external force 88. By analogy, chondroitin sulfate is secreted into the extracellular space during sea urchin gastrulation, and blocking secretion by drug treatment impaired the timing and extent of invagination 90. Other explanations have been posited for the role of GAGs in invagination, including regulation of cell-cell adhesiveness, extracellular matrix remodeling during migration, and apical constriction 88, 91.
The expression pattern of SQV proteins is consistent with a cell-autonomous role in vulval invagination. SQV-1, SQV-5, and SQV-7 proteins are expressed in cells throughout the vulva, and SQV-4 is expressed in vulC–vulF cells 84, 85, 87. Upstream regulators of sqv gene expression in the vulva have not been identified to date. Interestingly, a Sqv phenotype was observed in combined mutants of lin-35/Retinoblastoma and spr-1, an ortholog of the CoREST transcriptional repressor 89. The cause of the Sqv phenotype in lin-35; spr-1 is not clear, as mutant animals have wild-type levels of chondroitin in the vulval cells 89. Thus, there appears to exist at least one pathway that functions in parallel to the sqv genes during vulval invagination.
Attachments to other tissues
Organogenesis requires functional connections between different tissues in the body. The vulva connects with three different tissues: (1) the uterine epithelium, to generate a continuous lumen between the tissues; (2) hypodermal cells, to structurally stabilize the vulva by physically tethering it to the body wall; and (3) muscle cells, to move embryos through the vulva to the outside environment (Fig. 3).
The anchor cell and the uterine-vulval connection
The uterine epithelium initially develops independently of the vulval epithelium and is separated from it by gonadal and epidermal basement membranes. During the mid-L3 stage, the anchor cell (AC), a specialized ventral uterine cell, breaches the basement membrane separating the tissues and contacts the vulval cells (Fig. 6A) 12. The AC is a striking example of a single cell performing pleitropic functions during development: in the late-L2 larval stage, the AC induces VPC fates via LIN-3 expression 20; in L3, the AC expresses Wnt ligands to orient VPC planar polarity (Fig. 4) 61, expresses the LIN-12/Notch ligand LAG-2/Delta to induce the surrounding ventral uterine cells to adopt specific fates 92, patterns the 1° VPC lineage 59, and breaches the basement membrane (Fig. 6). The critical role for the AC in vulval morphogenesis was appreciated by Judith Kimble and David Hirsh, who named the cell for its ability to “anchor” the gonad to the vulva 93.
Cell invasion across basement membrane occurs during several developmental events and is also a key cell biological process in tumor metastasis and leukocyte trafficking 94. AC invasion integrates a stimulatory signal from the developing vulva with a cell autonomous invasion program (Fig. 6B). The 1° VPCs secrete a diffusible chemotactic cue that triggers invasion and guides the invading AC to centrally located 1° VPCs. The presence of a diffusible cue was demonstrated using genetic backgrounds in which the 1° VPCs are displaced from the AC, which results in the AC extending membrane processes toward the ectopic VPCs and through the basement membrane 12. Although the vulval cells are required for complete invasion, the AC still invades 20% of the time in Vul animals, demonstrating that AC invasion through basement membrane may be triggered by other cues or driven cell autonomously 12.
The AC initiates its invasion program at around the L2 molt by concentrating F-actin and other cytoskeletal components at the basal (ventral) surface in contact with the basement membrane 95. Among the genes involved in cytoskeletal polarization are unc-6/netrin, a guidance molecule expressed in the ventral nerve cord (VNC), its receptor unc-40/DCC, and pat-3/ina-1, whose protein products heterodimerize to form the integrin receptor complex (Fig. 6B). In the mid-L3 stage, the basal membrane of the AC extends actin-enriched membrane protrusions that penetrate the basement membrane at a location between the two vulF cells (Fig. 6A) 95, 96. AC invasion requires the transcription factor fos-1a, an ortholog of vertebrate Fos transcription factors that are expressed in invasive tumor cells 97. A downstream target of FOS-1A in the AC is zmp-1 (Fig. 6B) 98, a member of the matrix metalloprotease (MMP) family of genes that have been implicated in proteolytic breakdown of basement membrane and are expressed downstream of Fos proteins during tumor cell invasion 99. Analogous to the situation in vertebrates, in which single or double knockdown of MMPs results in only minor developmental defects 100, a null allele of zmp-1 has no invasion defect 98. The simplified genetic architecture of C. elegans, with only six MMPs, makes the AC model of invasion particularly useful for achieving greater clarity on the involvement of proteases during cell invasion. More broadly, studies on AC invasion are facilitating the identification of novel genes that regulate the process of cell invasion. A recent whole-genome RNAi screen identified 99 genes required to promote invasion, most of which were newly identified regulators of this process. Knockdown of human orthologs of two novel regulators, the NEMO-like kinase lit-1 and the CCT chaperonin complex component cct-5 reduced invasiveness in metastatic human carcinoma cells 101. Thus, AC invasion is an effective model to understand mechanisms that regulate invasion and to identify new therapeutic targets to block invasive behavior in cancer.
Following basement membrane invasion, the AC remains at the vulval apex, where its basal region contacts and moves between the central vulF cells to complete the invasion process 8, 102. Direct contact of the AC with the vulF cells appears to be required to pattern of E-F-F-E fates in the 1° VPC lineage (Fig. 6C) 59. Morphogenesis of the vulva also requires the complete invasion of the AC between the vulF cells 103. Animals mutant for fos-1 or unc-6 have defective AC invasion through the basement membrane and incomplete AC invasion into vulF, which results in a closed dorsal vulval lumen, suggesting that AC invasion between the vulF cells is required for dorsal lumen formation 103. Lumen formation in the vulF cells also depends on the NlpC/P60 family protein EGL-26, which may function as a palmitoyltransferase 104, 105. EGL-26 is expressed in the vulE lineage and localizes to the apical membrane where it might modify a lipid signaling molecule required for proper vulF morphology.
The AC also patterns the ventral uterine (VU) cells surrounding it, all of which are involved in establishing the uterine-vulval connection 13. During the L3 larval stage, the AC expresses LAG-2/Delta, which signals via LIN-12/Notch on six adjacent VU cells, inducing them to adopt the π (pi) cell fate rather the default ρ (rho) cell fate (Fig. 6C) 92. Specification of π fate is required for proper uterine-vulval connection, as ablation of all π cells causes a completely penetrant Egl phenotype, as do gene mutations that result in the failure of VU cells to adopt the π fate 92, 106–108. Following specification, the six π cells divide to generate twelve cells, eight of which fuse to form the uterine-seam (utse) syncytium, which stabilizes the uterus through attachment to the lateral epithelial cells (Fig. 3) 102. The remaining four π cells are specified as uv1 cells, which contact the underlying vulF cells through adherens junctions and stabilize the uterine-vulval connection (Fig. 3). LIN-3/EGF signaling from vulF cells via LET-23 receptor tyrosine kinase on uv1 cells is required for adoption of uv1 fate 109. egl-38, a homolog of Pax5 transcription factor, is also required for adoption of uv1 fate, functioning in both vulF and uv1 cells 109–111.
A question that until recently remained unsolved was how the uv1 cells and vulF cells make direct contact with each other, as basement membrane initially separates these cells. A recent study revealed that the gap in the basement membrane widens quickly beyond the cell boundaries of the AC during the mid and late L3 larval stages (Fig. 6A), thus allowing uv1 and vulF cells to make direct contact 112. To understand how the basement membrane is removed, animals expressing the tagged basement membrane components laminin::Dendra and type IV collagen::Dendra were examined. Dendra is a highly-stable, photoconvertible fluorescent protein that changes irreversibly from a green to a red fluorescent state following exposure to short-wavelength light 113. Optical highlighting of laminin::Dendra and type IV collagen::Dendra revealed that laminin and type IV collagen are removed through a novel mechanism of basement membrane sliding, and that this shift is dependent on the dividing and invaginating vulval cells. Further, an RNAi expression screen identified the integrin receptor complex INA-1/PAT-3 and VAB-19, a homolog of the tumour suppressor Kank, as regulators of basement membrane opening. Both concentrate within vulval cells at the basement membrane gap boundary and halt expansion of the shifting basement membrane 112. Basement membrane sliding followed by targeted adhesion represents a newly identified mechanism for creating precise basement membrane breaches that is likely used by cells to break down compartment boundaries not only in tissue morphogenesis, but also disease states such as cancer.
Following its invasion, the AC is positioned at the vulval apex (Fig. 6A), from which is must be removed to form a continuous lumen between uterine and vulval tissues. Removal of the AC is achieved through AC fusion with the uterine utse cell during the early L4 stage. After fusion, the AC nucleus and cytoplasm are cleared, leaving a thin plasma membrane between the uterine and vulval lumens that is broken by first embryo passage (Fig. 2F, Fig. 3) 13. aff-1 is expressed in the AC and utse and required for fusion. aff-1 expression is downstream of the FOS-1A transcription factor, which is expressed throughout the uterine tissue with strongest expression in the AC 80, 98. Also required for AC-utse fusion is the AC-expressed ATPase NSF-1 114 and the π-cell expressed LIM domain transcription factor LIN-11 115. LIN-11 expression levels and retention in the nucleus are regulated in π cells by the small ubiquitin-like modifier SMO-1 116.
Vulval cell attachments to hypodermal cells and muscles
In addition to attaching to uterine cells, the vulva also contacts hypodermal and muscle cells, which provides structural stability and controls the opening of the vulva during mating and egg laying, respectively (Fig. 3). The hypodermal connections occur through vulA cell contact with the hyp7 syncytium at the ventral surface of the vulva, and vulE contact with lateral seam cells near the dorsal surface. The transcription factor lin-29 is expressed in seam cells, and mosaic analysis suggests that loss of lin-29 in the subset of seam cells closest to the vulva causes vulval morphology defects, possibly through failure to attach to vulE cells 117. Little else is known about the vulE-seam cell connection, such as the molecular pathways that induce vulE membrane extension and the mechanisms of cell-cell attachment.
More is known about the mechanisms guiding muscle attachment to the vulva. The vulval musculature is derived from two sex myoblasts (SM), which are born in the posterior of the animal during the L1 stage and migrate anteriorly during L2 and L3 to flank the developing gonad near the vulva region. Following migration, the SM cells each divide three times in the L3 stage to generate 16 total cells. Eight of these, the um cells, induce contractions that move eggs through the uterus. The other eight cells, four vm1 and four vm2, extend processes in a diagonal configuration that contact the vulval lips and control opening during mating and egg-laying. The four vm2 cells connect between the uterus and vulF; the four vm1 cells connect between vulC and vulD toroids (Fig. 3) 8, 13.
The muscles that control vulval opening are innervated by six ventral cord (VC) motorneurons and two hermaphrodite-specific neuons (HSN). vm2 muscles are directly contacted by neurons, whereas vm1 muscles are electrically linked to vm2 118. The six VC neurons, VC1–6, run anterior-posterior along the ventral nerve cord (VNC) and extend branches dorsally at the vulval opening. VC4 and VC5 have cell bodies closest to the vulva and extend the longest projections into the vulval epithelium (Fig. 7). The HSN cell bodies are dorsal and posterior to the vulva, with axons that extend ventrally to the VNC, run anterior toward the head, defasciculate dorsally into the vulval region, and branch to synpase onto vm2, VC4 and VC5 (Fig. 7) 118. Branching of VC and HSN neuronal processes and their correct synapse formation requires signals from the vulva. In Vul animals, VC neurons do not send dorsal processes, and HSN axon extension toward the VNC and branching at the vulva are defective. These defects are not observed in animals lacking the gonad, SM cells, or other neurons, demonstrating a singular role for the vulval cells in regulating neuronal outgrowth branching and synapse formation 119–121.
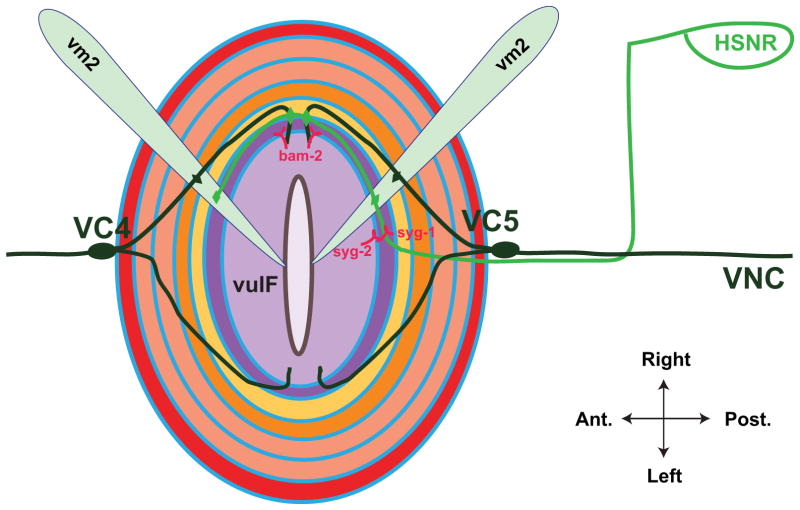
Figure 7.
Patterning of neuronal connections in the vulval region. Dorsal view of everted vulva. The two HSN neurons (shown is the right HSN in green) extend axons ventrally to the nerve cord, then anteriorly to the vulva. The interaction between HSN-expressed SYG-1 and vulF-expressed SYG-2 serves to guide the future sites of synaptic contact, which occur on vm2 muscle cells (shown are two of four vm2 cells, light blue) and VC neurons. VC4 and VC5 have cell bodies at the anterior and posterior of the vulval epithelium, respectively, and extend axons around the vulva and dorsally to synapse on vm2 cells. vulF-expressed BAM-2 is required to terminate axon branches. Modeled after 121 and 122.
Genetic screens have begun to provide insight into the mechanisms by which the vulval cells direct VC and HSN neurite outgrowth. VC4 and VC5 axonal processes extend around the vulva without crossing the midline. This pattern of outgrowth is dependent on BAM-2, a neurexin-like transmembrane protein that is expressed in vulF (Fig. 7). In bam-2 mutants, axon branches cross the midline, demonstrating that BAM-2 functions in vulF as a signal to terminate axon branching 122. HSN axon guidance and synapse formation are also established by signals from the 1° VPCs. One 1° VPC-expressed signal is UNC-6/netrin, which appears to combine with VNC-expressed UNC-6 to coordinate the complex trajectory of HSN axon growth. Specifically, 1° VPC-derived UNC-6 is the main determinant of HSN dorsal growth from the VNC 123. The 1° VPCs also direct HSN synapse formation onto muscles and neurons, which occurs near the dorsal region of the vulva. SYG-2, a transmembrane protein of the immunoglobulin (Ig) superfamily, is expressed in 1° VPCs and acts as a receptor for SYG-1, a second Ig transmembrane protein expressed on HSN axons. SYG-2 serves as a guidepost to localize SYG-1, which establishes sites of synaptic vesicle clustering (Fig. 7). In syg-1 and syg-2 mutants, HSN synaptic vesicle clusters are located in ectopic anterior locations 121, 124. These findings highlight the key role of the vulva in organizing both the outgrowth and neuronal contacts necessary for egg-laying.
Bridging the gap between cell fate specification and morphogenesis
How cell fate specification is linked to morphogenetic behaviors that shape tissues remains largely unknown 125. Five transcription factors have been identified as major regulators of a GRN that help to specify the fates of the seven vulval cell types. These are the LIM homeodomain protein LIN-11, the C2H2-type zinc finger protein LIN-29, the Nkx6 homeoprotein COG-1, the Pax2/5/8 ortholog EGL-38, and the conserved nuclear hormone receptor NHR-67 5, 6, 82. How this GRN is regulated by VPC induction and whether these transcription factors control the effectors of vulval morphogenesis is unclear. Recently, a transcription factor, the C4H2 zinc-finger protein VAB-23, was identified that bridges the gap between cell fate specification and the effectors of morphogenesis 71. During vulval induction, the Hox gene lin-39 is upregulated in the induced VPCs by EGF/Ras signaling 126. Pellegrino et al. (2011) found that LIN-39 and its conserved co-factor CEH-20 127 directly control VAB-23 expression in the vulval cells, with highest levels in the 1° lineage where EGF/Ras signaling is strongest. VAB-23 functions to repress eff-1 expression in vulE and vulF cells and promote smp-1 expression in vulF, vulE, and vulD cells 71. vab-23 mutants have ectopic eff-1 expression, inappropriate cell-cell fusions, a reduction in toroid number, and defects in migration. VAB-23 is also required for the correct differentiation of the vulF and vulE cell fates, as determined by reporter gene expression 71. These results demonstrate the connection between vulval induction (EGF/Ras), cell fate differentiation (LIN-39/VAB-23), and morphogenetic effectors (EFF-1/SMP-1), providing a paradigm for understanding how cells link specification with morphogenesis.
Conclusion
The vulva has served as a model of organogenesis for more than two decades, providing insight into developmental mechanisms of tissue patterning. Studies on the vulva have expanded our knowledge of important cellular processes, including oriented cell division, migration, invasion, adhesion, and fusion. The experimental advantages of the vulva—limited number of cells, in vivo visual accessibility, and ease of genetic and transgenic analysis—can now be combined with advanced microscopic techniques such as live cell imaging, convertible fluorophores, force sensors, and optogenetics to provide insight into cellular dynamics. The vulva is also amenable to the systems-level approaches available in C. elegans, including interactome, localizome, expression profiling, modEncode, and RNAi databases 128. Thus, future studies on vulval development will yield greater knowledge about fundamental mechanisms underlying morphogenesis. Among the interesting future subjects to explore are:
Molecular basis of lumen formation
Lumen formation allows movement of materials between different tissues and the outside environment. How lumens achieve their correct size and shape is poorly understood 129. Vulval lumen formation involves cell shape changes and migrations that generate an invagination, and proteoglycan secretion that is thought to create and maintain a lumen. Several questions remain unanswered about the mechanisms of lumen formation, including the cues that trigger the initial invagination, the upstream regulators of sqv gene expression, and the identity of the proteins modified by oligosaccharide chains.
Regulation of adhesion and fusion
VPCs extend membrane processes that meet their matching partners at the midline to form adhesions, but whether this recognition is based on cell-type specific receptor complexes or different mechanisms has not been determined. Fusions between homotypic cells (other than vulB) follow their adhesion at the midline. It is not well understood how the expression and subcellular localization of EFF-1 and AFF-1 fusogens are regulated to control the precise timing of fusions between matching cells 130.
Anchor cell invasion
AC invasion into the vulval epithelium offers a single-cell, in vivo model to study the cellular processes of invasion. Genetic screens and candidate gene approaches have led to the identification of more than 100 genes required for normal AC invasion 95, 96, 98, 101, 131, 132. Many of the genes are uncharacterized, and understanding how they function in AC invasion should provide insights into this conserved biological process. Among the areas of focus are the involvement of proteases, transcriptional regulation of the invasive program, the identity of exogenous stimulatory cues that target membrane processes to specific sites for invasion, and the mechanisms by which a cell shifts from cell-BM to cell-cell attachments.
Axon branching and synapse formation
During development of the egg-laying system, the vulva serves as an organizer of neuronal circuitry without receiving direct innervations itself. It terminates VC4 and VC5 axon branches by expression of the transmembrane protein BAM-2, but the VC-expressed binding partner of BAM-2 has not been identified. The SYG-1/SYG-2 interaction between 1° VPCs and the HSN neuron is an example of guidepost signaling, in which a cell acts as a scaffold for synapse formation. The downstream effectors of the SYG-1/SYG-2 signaling pathway that regulate guidepost signaling have yet to be determined.
Gene regulatory networks of vulval cell specification and morphogenesis
The gene regulatory networks (GRNs) that connect the adoption of cellular identity with gene expression and cell biological behaviors sit at the heart of understanding developmental mechanisms. Only a few developmental GRNs are understood at even a rudimentary level 133. The two-step patterning of vulval cell fates—induction by the gonad followed by differentiation into seven different cellular subtypes—is a model for how cell specification connects with gene expression and morphogenesis. Future work can build on the knowledge of vulval specification pathways and the genes involved in morphogenesis to make connections between gene expression and morphogenesis.
Acknowledgments
We thank members of the Sherwood Lab, Judith Kimble and Sunita Kramer for advice and suggestions. This work was supported by an American Cancer Society Postdoctoral Fellowship to AJS and Basil O’Connor Scholars Research Award, The Pew Scholars Program in the Biomedical Sciences, and NIH Grants GM079320 and GM100083 to D.R.S.
Contributor Information
Adam J Schindler, Duke University.
David R Sherwood, Email: david.sherwood@duke.edu, Duke University.
References
- 1.Gilbert SF, Singer SR, Tyler MS, Kozlowski RN. Developmental biology. 8. Sunderland, Mass: Sinauer Associates; 2006. [Google Scholar]
- 2.Langer R, Vacanti JP. Tissue engineering. Science. 1993;260:920–926. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 3.Egeblad M, Nakasone ES, Werb Z. Tumors as organs: complex tissues that interface with the entire organism. Dev Cell. 2010;18:884–901. doi: 10.1016/j.devcel.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jorgensen EM, Mango SE. The art and design of genetic screens: caenorhabditis elegans. Nat Rev Genet. 2002;3:356–369. doi: 10.1038/nrg794. [DOI] [PubMed] [Google Scholar]
- 5.Inoue T, Wang M, Ririe TO, Fernandes JS, Sternberg PW. Transcriptional network underlying Caenorhabditis elegans vulval development. Proc Natl Acad Sci U S A. 2005;102:4972–4977. doi: 10.1073/pnas.0408122102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ririe TO, Fernandes JS, Sternberg PW. The Caenorhabditis elegans vulva: a post-embryonic gene regulatory network controlling organogenesis. Proc Natl Acad Sci U S A. 2008;105:20095–20099. doi: 10.1073/pnas.0806377105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sternberg PW, Horvitz HR. Pattern formation during vulval development in C. elegans. Cell. 1986;44:761–772. doi: 10.1016/0092-8674(86)90842-1. [DOI] [PubMed] [Google Scholar]
- 8.Sharma-Kishore R, White JG, Southgate E, Podbilewicz B. Formation of the vulva in Caenorhabditis elegans: a paradigm for organogenesis. Development. 1999;126:691–699. doi: 10.1242/dev.126.4.691. [DOI] [PubMed] [Google Scholar]
- 9.Shemer G, Kishore R, Podbilewicz B. Ring formation drives invagination of the vulva in Caenorhabditis elegans: Ras, cell fusion, and cell migration determine structural fates. Dev Biol. 2000;221:233–248. doi: 10.1006/dbio.2000.9657. [DOI] [PubMed] [Google Scholar]
- 10.Herman T, Hartwieg E, Horvitz HR. sqv mutants of Caenorhabditis elegans are defective in vulval epithelial invagination. Proc Natl Acad Sci U S A. 1999;96:968–973. doi: 10.1073/pnas.96.3.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herman T, Horvitz HR. Three proteins involved in Caenorhabditis elegans vulval invagination are similar to components of a glycosylation pathway. Proc Natl Acad Sci U S A. 1999;96:974–979. doi: 10.1073/pnas.96.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sherwood DR, Sternberg PW. Anchor cell invasion into the vulval epithelium in C. elegans. Dev Cell. 2003;5:21–31. doi: 10.1016/s1534-5807(03)00168-0. [DOI] [PubMed] [Google Scholar]
- 13.Newman AP, Sternberg PW. Coordinated morphogenesis of epithelia during development of the Caenorhabditis elegans uterine-vulval connection. Proc Natl Acad Sci U S A. 1996;93:9329–9333. doi: 10.1073/pnas.93.18.9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lints R, Hall DH. Reproductive system, egglaying apparatus. WormAtlas. 2009 doi: 10.3908/wormatlas.1.24. [DOI] [Google Scholar]
- 15.Sundaram MV. The love-hate relationship between Ras and Notch. Genes Dev. 2005;19:1825–1839. doi: 10.1101/gad.1330605. [DOI] [PubMed] [Google Scholar]
- 16.Sternberg PW. Vulval development. WormBook. 2005:1–28. doi: 10.1895/wormbook.1.6.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fay DS, Yochem J. The SynMuv genes of Caenorhabditis elegans in vulval development and beyond. Dev Biol. 2007;306:1–9. doi: 10.1016/j.ydbio.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang M, Sternberg PW. Pattern formation during C. elegans vulval induction. Curr Top Dev Biol. 2001;51:189–220. doi: 10.1016/s0070-2153(01)51006-6. [DOI] [PubMed] [Google Scholar]
- 19.Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- 20.Katz WS, Hill RJ, Clandinin TR, Sternberg PW. Different levels of the C. elegans growth factor LIN-3 promote distinct vulval precursor fates. Cell. 1995;82:297–307. doi: 10.1016/0092-8674(95)90317-8. [DOI] [PubMed] [Google Scholar]
- 21.Zand TP, Reiner DJ, Der CJ. Ras effector switching promotes divergent cell fates in C. elegans vulval patterning. Dev Cell. 2011;20:84–96. doi: 10.1016/j.devcel.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoo AS, Bais C, Greenwald I. Crosstalk between the EGFR and LIN-12/Notch pathways in C. elegans vulval development. Science. 2004;303:663–666. doi: 10.1126/science.1091639. [DOI] [PubMed] [Google Scholar]
- 23.Levitan D, Greenwald I. LIN-12 protein expression and localization during vulval development in C.elegans. Development. 1998;125:3101–3109. doi: 10.1242/dev.125.16.3101. [DOI] [PubMed] [Google Scholar]
- 24.Berset T, Hoier EF, Battu G, Canevascini S, Hajnal A. Notch inhibition of RAS signaling through MAP kinase phosphatase LIP-1 during C. elegans vulval development. Science. 2001;291:1055–1058. doi: 10.1126/science.1055642. [DOI] [PubMed] [Google Scholar]
- 25.Berset TA, Hoier EF, Hajnal A. The C. elegans homolog of the mammalian tumor suppressor Dep-1/Scc1 inhibits EGFR signaling to regulate binary cell fate decisions. Genes Dev. 2005;19:1328–1340. doi: 10.1101/gad.333505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Euling S, Ambros V. Heterochronic genes control cell cycle progress and developmental competence of C. elegans vulva precursor cells. Cell. 1996;84:667–676. doi: 10.1016/s0092-8674(00)81045-4. [DOI] [PubMed] [Google Scholar]
- 27.Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- 28.Ruvkun G, Giusto J. The Caenorhabditis elegans heterochronic gene lin-14 encodes a nuclear protein that forms a temporal developmental switch. Nature. 1989;338:313–319. doi: 10.1038/338313a0. [DOI] [PubMed] [Google Scholar]
- 29.Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- 30.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 31.Hong Y, Roy R, Ambros V. Developmental regulation of a cyclin-dependent kinase inhibitor controls postembryonic cell cycle progression in Caenorhabditis elegans. Development. 1998;125:3585–3597. doi: 10.1242/dev.125.18.3585. [DOI] [PubMed] [Google Scholar]
- 32.Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 33.Chalfie M, Horvitz HR, Sulston JE. Mutations that lead to reiterations in the cell lineages of C. elegans. Cell. 1981;24:59–69. doi: 10.1016/0092-8674(81)90501-8. [DOI] [PubMed] [Google Scholar]
- 34.Li J, Greenwald I. LIN-14 inhibition of LIN-12 contributes to precision and timing of C. elegans vulval fate patterning. Curr Biol. 2010;20:1875–1879. doi: 10.1016/j.cub.2010.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, Miller EA, Rougvie AE. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell. 2003;4:625–637. doi: 10.1016/s1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- 36.Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, Horvitz HR, Ambros V. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell. 2005;9:403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kipreos ET. C. elegans cell cycles: invariance and stem cell divisions. Nat Rev Mol Cell Biol. 2005;6:766–776. doi: 10.1038/nrm1738. [DOI] [PubMed] [Google Scholar]
- 38.Saito RM, Perreault A, Peach B, Satterlee JS, van den Heuvel S. The CDC-14 phosphatase controls developmental cell-cycle arrest in C. elegans. Nat Cell Biol. 2004;6:777–783. doi: 10.1038/ncb1154. [DOI] [PubMed] [Google Scholar]
- 39.Clayton JE, van den Heuvel SJ, Saito RM. Transcriptional control of cell-cycle quiescence during C. elegans development. Dev Biol. 2008;313:603–613. doi: 10.1016/j.ydbio.2007.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fay DS, Keenan S, Han M. fzr-1 and lin-35/Rb function redundantly to control cell proliferation in C. elegans as revealed by a nonbiased synthetic screen. Genes Dev. 2002;16:503–517. doi: 10.1101/gad.952302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kipreos ET, Gohel SP, Hedgecock EM. The C. elegans F-box/WD-repeat protein LIN-23 functions to limit cell division during development. Development. 2000;127:5071–5082. doi: 10.1242/dev.127.23.5071. [DOI] [PubMed] [Google Scholar]
- 42.Kim Y, Kipreos ET. The Caenorhabditis elegans replication licensing factor CDT-1 is targeted for degradation by the CUL-4/DDB-1 complex. Mol Cell Biol. 2007;27:1394–1406. doi: 10.1128/MCB.00736-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fay DS, Han M. Mutations in cye-1, a Caenorhabditis elegans cyclin E homolog, reveal coordination between cell-cycle control and vulval development. Development. 2000;127:4049–4060. doi: 10.1242/dev.127.18.4049. [DOI] [PubMed] [Google Scholar]
- 44.Winston JT, Chu C, Harper JW. Culprits in the degradation of cyclin E apprehended. Genes Dev. 1999;13:2751–2757. doi: 10.1101/gad.13.21.2751. [DOI] [PubMed] [Google Scholar]
- 45.Kipreos ET, Lander LE, Wing JP, He WW, Hedgecock EM. cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell. 1996;85:829–839. doi: 10.1016/s0092-8674(00)81267-2. [DOI] [PubMed] [Google Scholar]
- 46.Boxem M, van den Heuvel S. C. elegans class B synthetic multivulva genes act in G(1) regulation. Curr Biol. 2002;12:906–911. doi: 10.1016/s0960-9822(02)00844-8. [DOI] [PubMed] [Google Scholar]
- 47.Buttitta LA, Edgar BA. Mechanisms controlling cell cycle exit upon terminal differentiation. Curr Opin Cell Biol. 2007;19:697–704. doi: 10.1016/j.ceb.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaldis P, Richardson HE. When cell cycle meets development. Development. 2012;139:225–230. doi: 10.1242/dev.073288. [DOI] [PubMed] [Google Scholar]
- 49.Shemer G, Podbilewicz B. LIN-39/Hox triggers cell division and represses EFF-1/fusogen-dependent vulval cell fusion. Genes Dev. 2002;16:3136–3141. doi: 10.1101/gad.251202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Palmer RE, Inoue T, Sherwood DR, Jiang LI, Sternberg PW. Caenorhabditis elegans cog-1 locus encodes GTX/Nkx6. 1 homeodomain proteins and regulates multiple aspects of reproductive system development. Dev Biol. 2002;252:202–213. doi: 10.1006/dbio.2002.0850. [DOI] [PubMed] [Google Scholar]
- 51.Inoue T, Sternberg PW. C. elegans BED domain transcription factor BED-3 controls lineage-specific cell proliferation during organogenesis. Dev Biol. 2010;338:226–236. doi: 10.1016/j.ydbio.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Freyd G, Kim SK, Horvitz HR. Novel cysteine-rich motif and homeodomain in the product of the Caenorhabditis elegans cell lineage gene lin-11. Nature. 1990;344:876–879. doi: 10.1038/344876a0. [DOI] [PubMed] [Google Scholar]
- 53.Gillies TE, Cabernard C. Cell division orientation in animals. Curr Biol. 2011;21:R599–609. doi: 10.1016/j.cub.2011.06.055. [DOI] [PubMed] [Google Scholar]
- 54.Kishore RS, Sundaram MV. ced-10 Rac and mig-2 function redundantly and act with unc-73 trio to control the orientation of vulval cell divisions and migrations in Caenorhabditis elegans. Dev Biol. 2002;241:339–348. doi: 10.1006/dbio.2001.0513. [DOI] [PubMed] [Google Scholar]
- 55.Chen Z, Han M. Role of C. elegans lin-40 MTA in vulval fate specification and morphogenesis. Development. 2001;128:4911–4921. doi: 10.1242/dev.128.23.4911. [DOI] [PubMed] [Google Scholar]
- 56.Kiontke K, Barriere A, Kolotuev I, Podbilewicz B, Sommer R, Fitch DH, Felix MA. Trends, stasis, and drift in the evolution of nematode vulva development. Curr Biol. 2007;17:1925–1937. doi: 10.1016/j.cub.2007.10.061. [DOI] [PubMed] [Google Scholar]
- 57.Kolotuev I, Podbilewicz B. Pristionchus pacificus vulva formation: polarized division, cell migration, cell fusion, and evolution of invagination. Dev Biol. 2004;266:322–333. doi: 10.1016/j.ydbio.2003.10.029. [DOI] [PubMed] [Google Scholar]
- 58.Kolotuev I, Podbilewicz B. Changing of the cell division axes drives vulva evolution in nematodes. Dev Biol. 2008;313:142–154. doi: 10.1016/j.ydbio.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 59.Wang M, Sternberg PW. Patterning of the C. elegans 1 degrees vulval lineage by RAS and Wnt pathways. Development. 2000;127:5047–5058. doi: 10.1242/dev.127.23.5047. [DOI] [PubMed] [Google Scholar]
- 60.Green JL, Inoue T, Sternberg PW. The C. elegans ROR receptor tyrosine kinase, CAM-1, non-autonomously inhibits the Wnt pathway. Development. 2007;134:4053–4062. doi: 10.1242/dev.005363. [DOI] [PubMed] [Google Scholar]
- 61.Green JL, Inoue T, Sternberg PW. Opposing Wnt pathways orient cell polarity during organogenesis. Cell. 2008;134:646–656. doi: 10.1016/j.cell.2008.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Inoue T, Oz HS, Wiland D, Gharib S, Deshpande R, Hill RJ, Katz WS, Sternberg PW. C. elegans LIN-18 is a Ryk ortholog and functions in parallel to LIN-17/Frizzled in Wnt signaling. Cell. 2004;118:795–806. doi: 10.1016/j.cell.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 63.Gleason JE, Szyleyko EA, Eisenmann DM. Multiple redundant Wnt signaling components function in two processes during C. elegans vulval development. Dev Biol. 2006;298:442–457. doi: 10.1016/j.ydbio.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 64.Deshpande R, Inoue T, Priess JR, Hill RJ. lin-17/Frizzled and lin-18 regulate POP-1/TCF-1 localization and cell type specification during C. elegans vulval development. Dev Biol. 2005;278:118–129. doi: 10.1016/j.ydbio.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 65.Chisholm AD, Hardin J. Epidermal morphogenesis. WormBook. 2005:1–22. doi: 10.1895/wormbook.1.35.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dalpe G, Brown L, Culotti JG. Vulva morphogenesis involves attraction of plexin 1-expressing primordial vulva cells to semaphorin 1a sequentially expressed at the vulva midline. Development. 2005;132:1387–1400. doi: 10.1242/dev.01694. [DOI] [PubMed] [Google Scholar]
- 67.Liu Z, Fujii T, Nukazuka A, Kurokawa R, Suzuki M, Fujisawa H, Takagi S. C. elegans PlexinA PLX-1 mediates a cell contact-dependent stop signal in vulval precursor cells. Dev Biol. 2005;282:138–151. doi: 10.1016/j.ydbio.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 68.Roy PJ, Zheng H, Warren CE, Culotti JG. mab-20 encodes Semaphorin-2a and is required to prevent ectopic cell contacts during epidermal morphogenesis in Caenorhabditis elegans. Development. 2000;127:755–767. doi: 10.1242/dev.127.4.755. [DOI] [PubMed] [Google Scholar]
- 69.Wu Z, Sweeney LB, Ayoob JC, Chak K, Andreone BJ, Ohyama T, Kerr R, Luo L, Zlatic M, Kolodkin AL. A combinatorial semaphorin code instructs the initial steps of sensory circuit assembly in the Drosophila CNS. Neuron. 2011;70:281–298. doi: 10.1016/j.neuron.2011.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chauvet S, Cohen S, Yoshida Y, Fekrane L, Livet J, Gayet O, Segu L, Buhot MC, Jessell TM, Henderson CE, et al. Gating of Sema3E/PlexinD1 signaling by neuropilin-1 switches axonal repulsion to attraction during brain development. Neuron. 2007;56:807–822. doi: 10.1016/j.neuron.2007.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pellegrino MW, Farooqui S, Frohli E, Rehrauer H, Kaeser-Pebernard S, Muller F, Gasser RB, Hajnal A. LIN-39 and the EGFR/RAS/MAPK pathway regulate C. elegans vulval morphogenesis via the VAB-23 zinc finger protein. Development. 2011;138:4649–4660. doi: 10.1242/dev.071951. [DOI] [PubMed] [Google Scholar]
- 72.Lynch AM, Hardin J. The assembly and maintenance of epithelial junctions in C. elegans. Front Biosci. 2009;14:1414–1432. doi: 10.2741/3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hurd DD, Kemphues KJ. PAR-1 is required for morphogenesis of the Caenorhabditis elegans vulva. Dev Biol. 2003;253:54–65. doi: 10.1006/dbio.2002.0866. [DOI] [PubMed] [Google Scholar]
- 74.Williams-Masson EM, Malik AN, Hardin J. An actin-mediated two-step mechanism is required for ventral enclosure of the C. elegans hypodermis. Development. 1997;124:2889–2901. doi: 10.1242/dev.124.15.2889. [DOI] [PubMed] [Google Scholar]
- 75.Millard TH, Martin P. Dynamic analysis of filopodial interactions during the zippering phase of Drosophila dorsal closure. Development. 2008;135:621–626. doi: 10.1242/dev.014001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Podbilewicz B. Cell fusion. WormBook. 2006:1–32. doi: 10.1895/wormbook.1.52.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Avinoam O, Fridman K, Valansi C, Abutbul I, Zeev-Ben-Mordehai T, Maurer UE, Sapir A, Danino D, Grunewald K, White JM, et al. Conserved eukaryotic fusogens can fuse viral envelopes to cells. Science. 2011;332:589–592. doi: 10.1126/science.1202333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mohler WA, Shemer G, del Campo JJ, Valansi C, Opoku-Serebuoh E, Scranton V, Assaf N, White JG, Podbilewicz B. The type I membrane protein EFF-1 is essential for developmental cell fusion. Dev Cell. 2002;2:355–362. doi: 10.1016/s1534-5807(02)00129-6. [DOI] [PubMed] [Google Scholar]
- 79.Sapir A, Avinoam O, Podbilewicz B, Chernomordik LV. Viral and developmental cell fusion mechanisms: conservation and divergence. Dev Cell. 2008;14:11–21. doi: 10.1016/j.devcel.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sapir A, Choi J, Leikina E, Avinoam O, Valansi C, Chernomordik LV, Newman AP, Podbilewicz B. AFF-1, a FOS-1-regulated fusogen, mediates fusion of the anchor cell in C. elegans. Dev Cell. 2007;12:683–698. doi: 10.1016/j.devcel.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Podbilewicz B, Leikina E, Sapir A, Valansi C, Suissa M, Shemer G, Chernomordik LV. The C. elegans developmental fusogen EFF-1 mediates homotypic fusion in heterologous cells and in vivo. Dev Cell. 2006;11:471–481. doi: 10.1016/j.devcel.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 82.Fernandes JS, Sternberg PW. The tailless ortholog nhr-67 regulates patterning of gene expression and morphogenesis in the C. elegans vulva. PLoS Genet. 2007;3:e69. doi: 10.1371/journal.pgen.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bulik DA, Wei G, Toyoda H, Kinoshita-Toyoda A, Waldrip WR, Esko JD, Robbins PW, Selleck SB. sqv-3, -7, and -8, a set of genes affecting morphogenesis in Caenorhabditis elegans, encode enzymes required for glycosaminoglycan biosynthesis. Proc Natl Acad Sci U S A. 2000;97:10838–10843. doi: 10.1073/pnas.97.20.10838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hwang HY, Horvitz HR. The Caenorhabditis elegans vulval morphogenesis gene sqv-4 encodes a UDP-glucose dehydrogenase that is temporally and spatially regulated. Proc Natl Acad Sci U S A. 2002;99:14224–14229. doi: 10.1073/pnas.172522499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hwang HY, Horvitz HR. The SQV-1 UDP-glucuronic acid decarboxylase and the SQV-7 nucleotide-sugar transporter may act in the Golgi apparatus to affect Caenorhabditis elegans vulval morphogenesis and embryonic development. Proc Natl Acad Sci U S A. 2002;99:14218–14223. doi: 10.1073/pnas.172522199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hwang HY, Olson SK, Brown JR, Esko JD, Horvitz HR. The Caenorhabditis elegans genes sqv-2 and sqv-6, which are required for vulval morphogenesis, encode glycosaminoglycan galactosyltransferase II and xylosyltransferase. J Biol Chem. 2003;278:11735–11738. doi: 10.1074/jbc.C200518200. [DOI] [PubMed] [Google Scholar]
- 87.Hwang HY, Olson SK, Esko JD, Horvitz HR. Caenorhabditis elegans early embryogenesis and vulval morphogenesis require chondroitin biosynthesis. Nature. 2003;423:439–443. doi: 10.1038/nature01634. [DOI] [PubMed] [Google Scholar]
- 88.Bulik DA, Robbins PW. The Caenorhabditis elegans sqv genes and functions of proteoglycans in development. Biochim Biophys Acta. 2002;1573:247–257. doi: 10.1016/s0304-4165(02)00391-4. [DOI] [PubMed] [Google Scholar]
- 89.Bender AM, Kirienko NV, Olson SK, Esko JD, Fay DS. lin-35/Rb and the CoREST ortholog spr-1 coordinately regulate vulval morphogenesis and gonad development in C. elegans. Dev Biol. 2007;302:448–462. doi: 10.1016/j.ydbio.2006.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lane MC, Koehl MA, Wilt F, Keller R. A role for regulated secretion of apical extracellular matrix during epithelial invagination in the sea urchin. Development. 1993;117:1049–1060. doi: 10.1242/dev.117.3.1049. [DOI] [PubMed] [Google Scholar]
- 91.Herman T, Horvitz HR. Mutations that perturb vulval invagination in C. elegans. Cold Spring Harb Symp Quant Biol. 1997;62:353–359. [PubMed] [Google Scholar]
- 92.Newman AP, White JG, Sternberg PW. The Caenorhabditis elegans lin-12 gene mediates induction of ventral uterine specialization by the anchor cell. Development. 1995;121:263–271. doi: 10.1242/dev.121.2.263. [DOI] [PubMed] [Google Scholar]
- 93.Kimble J, Hirsh D. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev Biol. 1979;70:396–417. doi: 10.1016/0012-1606(79)90035-6. [DOI] [PubMed] [Google Scholar]
- 94.Sherwood DR. Cell invasion through basement membranes: an anchor of understanding. Trends Cell Biol. 2006;16:250–256. doi: 10.1016/j.tcb.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 95.Ziel JW, Hagedorn EJ, Audhya A, Sherwood DR. UNC-6 (netrin) orients the invasive membrane of the anchor cell in C. elegans. Nat Cell Biol. 2009;11:183–189. doi: 10.1038/ncb1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hagedorn EJ, Yashiro H, Ziel JW, Ihara S, Wang Z, Sherwood DR. Integrin acts upstream of netrin signaling to regulate formation of the anchor cell’s invasive membrane in C. elegans. Dev Cell. 2009;17:187–198. doi: 10.1016/j.devcel.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ozanne BW, Spence HJ, McGarry LC, Hennigan RF. Transcription factors control invasion: AP-1 the first among equals. Oncogene. 2007;26:1–10. doi: 10.1038/sj.onc.1209759. [DOI] [PubMed] [Google Scholar]
- 98.Sherwood DR, Butler JA, Kramer JM, Sternberg PW. FOS-1 promotes basement-membrane removal during anchor-cell invasion in C. elegans. Cell. 2005;121:951–962. doi: 10.1016/j.cell.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 99.Adiseshaiah P, Vaz M, Machireddy N, Kalvakolanu DV, Reddy SP. A Fra-1-dependent, matrix metalloproteinase driven EGFR activation promotes human lung epithelial cell motility and invasion. J Cell Physiol. 2008;216:405–412. doi: 10.1002/jcp.21410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Perez SE, Cano DA, Dao-Pick T, Rougier JP, Werb Z, Hebrok M. Matrix metalloproteinases 2 and 9 are dispensable for pancreatic islet formation and function in vivo. Diabetes. 2005;54:694–701. doi: 10.2337/diabetes.54.3.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Matus DQ, Li XY, Durbin S, Agarwal D, Chi Q, Weiss SJ, Sherwood DR. In vivo identification of regulators of cell invasion across basement membranes. Sci Signal. 2010;3:ra35. doi: 10.1126/scisignal.2000654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Newman AP, White JG, Sternberg PW. Morphogenesis of the C. elegans hermaphrodite uterus. Development. 1996;122:3617–3626. doi: 10.1242/dev.122.11.3617. [DOI] [PubMed] [Google Scholar]
- 103.Estes KA, Hanna-Rose W. The anchor cell initiates dorsal lumen formation during C. elegans vulval tubulogenesis. Dev Biol. 2009;328:297–304. doi: 10.1016/j.ydbio.2009.01.034. [DOI] [PubMed] [Google Scholar]
- 104.Estes KA, Kalamegham R, Hanna-Rose W. Membrane localization of the NlpC/P60 family protein EGL-26 correlates with regulation of vulval cell morphogenesis in Caenorhabditis elegans. Dev Biol. 2007;308:196–205. doi: 10.1016/j.ydbio.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 105.Hanna-Rose W, Han M. The Caenorhabditis elegans EGL-26 protein mediates vulval cell morphogenesis. Dev Biol. 2002;241:247–258. doi: 10.1006/dbio.2001.0514. [DOI] [PubMed] [Google Scholar]
- 106.Newman AP, Inoue T, Wang M, Sternberg PW. The Caenorhabditis elegans heterochronic gene lin-29 coordinates the vulval-uterine-epidermal connections. Curr Biol. 2000;10:1479–1488. doi: 10.1016/s0960-9822(00)00827-7. [DOI] [PubMed] [Google Scholar]
- 107.Cinar HN, Richards KL, Oommen KS, Newman AP. The EGL-13 SOX domain transcription factor affects the uterine pi cell lineages in Caenorhabditis elegans. Genetics. 2003;165:1623–1628. doi: 10.1093/genetics/165.3.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hanna-Rose W, Han M. COG-2, a sox domain protein necessary for establishing a functional vulval-uterine connection in Caenorhabditis elegans. Development. 1999;126:169–179. doi: 10.1242/dev.126.1.169. [DOI] [PubMed] [Google Scholar]
- 109.Chang C, Newman AP, Sternberg PW. Reciprocal EGF signaling back to the uterus from the induced C. elegans vulva coordinates morphogenesis of epithelia. Curr Biol. 1999;9:237–246. doi: 10.1016/s0960-9822(99)80112-2. [DOI] [PubMed] [Google Scholar]
- 110.Rajakumar V, Chamberlin HM. The Pax2/5/8 gene egl-38 coordinates organogenesis of the C. elegans egg-laying system. Dev Biol. 2007;301:240–253. doi: 10.1016/j.ydbio.2006.08.068. [DOI] [PubMed] [Google Scholar]
- 111.Chamberlin HM, Palmer RE, Newman AP, Sternberg PW, Baillie DL, Thomas JH. The PAX gene egl-38 mediates developmental patterning in Caenorhabditis elegans. Development. 1997;124:3919–3928. doi: 10.1242/dev.124.20.3919. [DOI] [PubMed] [Google Scholar]
- 112.Ihara S, Hagedorn EJ, Morrissey MA, Chi Q, Motegi F, Kramer JM, Sherwood DR. Basement membrane sliding and targeted adhesion remodels tissue boundaries during uterine-vulval attachment in Caenorhabditis elegans. Nat Cell Biol. 2011;13:641–651. doi: 10.1038/ncb2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gurskaya NG, Verkhusha VV, Shcheglov AS, Staroverov DB, Chepurnykh TV, Fradkov AF, Lukyanov S, Lukyanov KA. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat Biotechnol. 2006;24:461–465. doi: 10.1038/nbt1191. [DOI] [PubMed] [Google Scholar]
- 114.Choi J, Richards KL, Cinar HN, Newman AP. N-ethylmaleimide sensitive factor is required for fusion of the C. elegans uterine anchor cell. Dev Biol. 2006;297:87–102. doi: 10.1016/j.ydbio.2006.04.471. [DOI] [PubMed] [Google Scholar]
- 115.Newman AP, Acton GZ, Hartwieg E, Horvitz HR, Sternberg PW. The lin-11 LIM domain transcription factor is necessary for morphogenesis of C. elegans uterine cells. Development. 1999;126:5319–5326. doi: 10.1242/dev.126.23.5319. [DOI] [PubMed] [Google Scholar]
- 116.Broday L, Kolotuev I, Didier C, Bhoumik A, Gupta BP, Sternberg PW, Podbilewicz B, Ronai Z. The small ubiquitin-like modifier (SUMO) is required for gonadal and uterine-vulval morphogenesis in Caenorhabditis elegans. Genes Dev. 2004;18:2380–2391. doi: 10.1101/gad.1227104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bettinger JC, Euling S, Rougvie AE. The terminal differentiation factor LIN-29 is required for proper vulval morphogenesis and egg laying in Caenorhabditis elegans. Development. 1997;124:4333–4342. doi: 10.1242/dev.124.21.4333. [DOI] [PubMed] [Google Scholar]
- 118.White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Vol. 314. London: Phil. Trans. Royal Soc; 1986. [DOI] [PubMed] [Google Scholar]
- 119.Li C, Chalfie M. Organogenesis in C. elegans: positioning of neurons and muscles in the egg-laying system. Neuron. 1990;4:681–695. doi: 10.1016/0896-6273(90)90195-l. [DOI] [PubMed] [Google Scholar]
- 120.Garriga G, Desai C, Horvitz HR. Cell interactions control the direction of outgrowth, branching and fasciculation of the HSN axons of Caenorhabditis elegans. Development. 1993;117:1071–1087. doi: 10.1242/dev.117.3.1071. [DOI] [PubMed] [Google Scholar]
- 121.Shen K, Bargmann CI. The immunoglobulin superfamily protein SYG-1 determines the location of specific synapses in C. elegans. Cell. 2003;112:619–630. doi: 10.1016/s0092-8674(03)00113-2. [DOI] [PubMed] [Google Scholar]
- 122.Colavita A, Tessier-Lavigne M. A Neurexin-related protein, BAM-2, terminates axonal branches in C. elegans. Science. 2003;302:293–296. doi: 10.1126/science.1089163. [DOI] [PubMed] [Google Scholar]
- 123.Asakura T, Ogura K, Goshima Y. UNC-6 expression by the vulval precursor cells of Caenorhabditis elegans is required for the complex axon guidance of the HSN neurons. Dev Biol. 2007;304:800–810. doi: 10.1016/j.ydbio.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 124.Shen K, Fetter RD, Bargmann CI. Synaptic specificity is generated by the synaptic guidepost protein SYG-2 and its receptor, SYG-1. Cell. 2004;116:869–881. doi: 10.1016/s0092-8674(04)00251-x. [DOI] [PubMed] [Google Scholar]
- 125.Christiaen L, Davidson B, Kawashima T, Powell W, Nolla H, Vranizan K, Levine M. The transcription/migration interface in heart precursors of Ciona intestinalis. Science. 2008;320:1349–1352. doi: 10.1126/science.1158170. [DOI] [PubMed] [Google Scholar]
- 126.Maloof JN, Kenyon C. The Hox gene lin-39 is required during C. elegans vulval induction to select the outcome of Ras signaling. Development. 1998;125:181–190. doi: 10.1242/dev.125.2.181. [DOI] [PubMed] [Google Scholar]
- 127.Yang L, Sym M, Kenyon C. The roles of two C. elegans HOX co-factor orthologs in cell migration and vulva development. Development. 2005;132:1413–1428. doi: 10.1242/dev.01569. [DOI] [PubMed] [Google Scholar]
- 128.Wang Z, Sherwood DR. Dissection of genetic pathways in C. elegans. Methods Cell Biol. 2011;106:113–157. doi: 10.1016/B978-0-12-544172-8.00005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Datta A, Bryant DM, Mostov KE. Molecular regulation of lumen morphogenesis. Curr Biol. 2011;21:R126–136. doi: 10.1016/j.cub.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Avinoam O, Podbilewicz B. Eukaryotic cell-cell fusion families. Curr Top Membr. 2011;68:209–234. doi: 10.1016/B978-0-12-385891-7.00009-X. [DOI] [PubMed] [Google Scholar]
- 131.Schindler AJ, Sherwood DR. The transcription factor HLH-2/E/Daughterless regulates anchor cell invasion across basement membrane in C. elegans. Dev Biol. 2011;357:380–391. doi: 10.1016/j.ydbio.2011.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ziel JW, Matus DQ, Sherwood DR. An expression screen for RhoGEF genes involved in C. elegans gonadogenesis. Gene Expr Patterns. 2009;9:397–403. doi: 10.1016/j.gep.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Levine M, Davidson EH. Gene regulatory networks for development. Proc Natl Acad Sci U S A. 2005;102:4936–4942. doi: 10.1073/pnas.0408031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mohler WA, Simske JS, Williams-Masson EM, Hardin JD, White JG. Dynamics and ultrastructure of developmental cell fusions in the Caenorhabditis elegans hypodermis. Curr Biol. 1998;8:1087–1090. doi: 10.1016/s0960-9822(98)70447-6. [DOI] [PubMed] [Google Scholar]
- 135.Kao G, Huang CC, Hedgecock EM, Hall DH, Wadsworth WG. The role of the laminin beta subunit in laminin heterotrimer assembly and basement membrane function and development in C. elegans. Dev Biol. 2006;290:211–219. doi: 10.1016/j.ydbio.2005.11.026. [DOI] [PubMed] [Google Scholar]