Abstract
Purpose
Radiation exposure triggers a complex network of molecular and cellular responses that impacts metabolic processes and alters the levels of metabolites. Such metabolites have potential as biomarkers for radiation dosimetry. This review provides an overview of radiation signalling and metabolism, of metabolomic approaches used in the discovery phase, and of instrumentation with the potential to assess radiation injury in the field.
Approach
Recent developments in fast, high-resolution chromatography and mass spectrometry and new data analysis methods allow the quantitative assessment of thousands of metabolites based on biofluids obtained non-invasively. This complex analysis leads to the discovery-phase identification of groups of metabolites useful for screening and biodosimetry by targeted quantitative measurement. Instrumentation for target analysis can be simpler than that used for discovery, so we examine current technologies based on ion mobility.
Conclusions
Recent published results and ongoing studies examine the complex changes in the levels of many metabolites caused by radiation exposure, and identify groups of small-molecule biomarkers for radiation biodosimetry. Based on results showing separation orthogonal to mass, chemical noise suppression, and high sensitivity, differential mobility mass spectrometry (DMS-MS) ion mobility spectrometry appears highly promising for the development of deployable instrumentation.
Keywords: radiation biodosimetry, metabolomics, metabonomics, mass spectrometry, ion mobility, differential mobility, DMS, DMS-MS
Introduction
As discussed elsewhere in this issue and previously (DiCarlo et al. 2010, Pandey et al. 2010), there is a clear need for more effective and higher throughput biodosimetry suitable to triage and assess potentially large numbers of radiation-exposure casualties. Recent advancements in ‘omics’ technologies offer opportunities to complement current approaches and develop field-deployable instrumentation to assess radiation exposure either alone or in combination with other injuries. Transcriptomics and proteomics are relatively mature omic fields that offer exciting opportunities in this regard, but typically rely on complicated and demanding methodologies requiring isolation of intact ribonucleic acid (RNA) or protein and then analyses. Metabolomics is an emerging field with great potential for radiation biodosimetry. Here metabolites, defined as small molecules typically less than 1 kDa, are assessed with modern analytical approaches that can detect thousands of different molecules, and identify informative subsets of manageable size for use in simpler instrumentation in the field. As will be discussed, easily accessible biofluids can be analysed with metabolomic approaches with the potential for very high throughput. Thus, a metabolomics approach could have utility for initial triage after a radiologic or nuclear event, and could comprise one component of a multi-faceted strategy that also involves further assessment of higher risk individuals using other omics measurements as well as standard clinical evaluation. An understanding of underlying mechanisms provides a basis for determining how molecular responses to radiation exposure can affect metabolism, so an overview of radiation signalling will be given first; then we will review the relatively new field of metabolomics, its application to radiation exposure, and a discussion of the development of instrumentation with potential utility to assess radiation injury during an actual event.
Radiation responses and metabolism
Precedence for radiation effects on metabolites
There is already substantial precedence that radiation exposure can affect the levels of particular small molecules in accessible biofluids. Several decades ago, metabolites including cyclic adenosine monophosphate (cAMP), cyclic guanosine monophosphate (cGMP), isoprostane 8-epi-prostaglandin f2α (PGF2α) and thromboxane B2 and 6-keto prostaglandin f1α (6-keto-PGF1α) were found to be increased in urine after ionising radiation (IR*) exposures (Schneidkraut et al. 1984, Yamaoka et al. 1998). Prostaglandins have frequently been found to increase in various tissues and biofluids after IR (discussed in Stancevic and Kolesnick 2010), such as in saliva after radioiodine therapy (Wolfram et al. 2004). Citrulline in blood has been proposed as a potential biomarker to estimate radiation-induced damage to intestinal epithelium (Lutgens et al. 2003). The sphingolipid ceramide is a well-studied small molecule produced by the hydrolysis of sphingomyelin by acid sphingomyelinase, which can be rapidly activated in cell membranes by IR (Stancevic and Kolesnick 2010). This enzyme can be activated by various stress stimuli including cell surface receptors mediating apoptosis and inflammatory responses. Ceramide is an evolutionarily conserved second messenger known to be involved in biologic processes as diverse as apoptosis, growth arrest, senescence and differentiation, and it has been implicated in the gastrointestinal (GI) syndrome after IR (Stancevic and Kolesnick 2010). Interestingly, ceramide has been shown to affect membrane permeability in model systems and such changes in vivo could have broad effects (Stancevic and Kolesnick 2010). Recent advances in metabolomics now permit global assessments of small molecule changes and the identification of biomarkers with potential application to radiation biodosimetry.
Radiation stress signalling
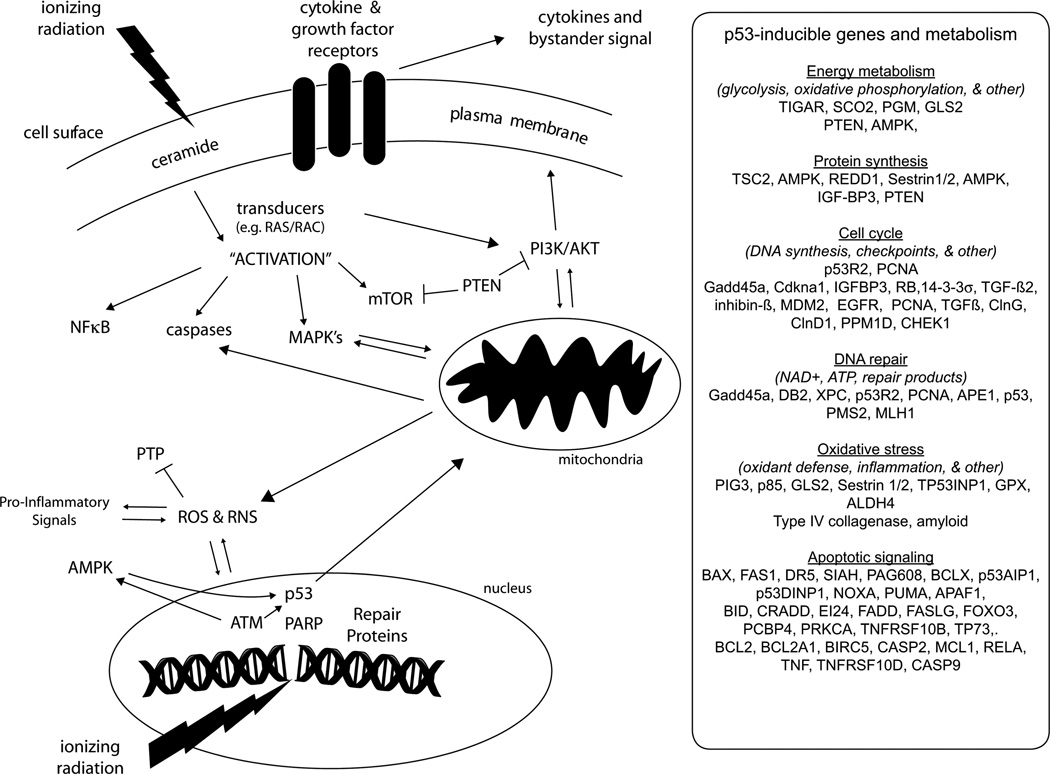
Radiation exposure can trigger a complex series of molecular responses involving many signalling pathways that can affect metabolism either directly, such as by activation of acid sphingomyelinase, or indirectly by altering cell growth. In addition to its well-known effects on deoxyribonucleic acid (DNA), IR directly impacts multiple cellular compartments even at relatively modest doses where only very large macromolecules would be expected to be appreciably ‘hit’ based on target theory (Rohwer 1984, Hall and Giaccia 2006), and ‘hits’ to many of these targets triggers a variety of signalling pathways. An overview of some of the major signalling pathways activated by IR is shown in Figure 1 along with examples of several prominent mediators of these responses. In the case of cellular membranes and particularly the plasma membrane, the signal by IR is probably amplified by radical cascades in lipid membranes with rapid activation of cytokine and growth factor receptors. As reviewed in more detail (Valerie et al. 2007), IR triggers a rapid reactive oxygen species (ROS)-dependent activation of epidermal growth factor receptor (EGFR) family and other tyrosine kinases involved in receptor signalling. Interestingly, both IR and ultraviolet (UV) radiation can trigger ligand-independent activation of cell surface receptors (Rosette and Karin 1996, Valerie et al. 2007). Receptors for growth factors, apoptosis-associated ligands, and inflammatory signalling can show redistribution and clustering after irradiation, and this can facilitate dimerisation with activation in the absence of ligand. In the cytosol, radiation generates ionising events in water, which can also be amplified probably by mitochondria (Valerie et al. 2007), and as mentioned in Figure 1 can enhance protein tyrosine kinase (PTK) activity. With receptor activation, a variety of transducers, such as rat sarcoma (Ras) oncogene, and downstream signalling pathways are activated including pro-growth and potentially protective pathways including the extracellular signal-regulated kinases (ERK) branch of the mitogen-activated protein kinase (MAPK) pathways, nuclear factor kappa B (NFκB), and the interdependent phosphatidylinositol-3-kinase/protein kinase B (PI3K/AKT) and mammalian target of rapamycin (mTOR) pathways. Typically, Insulin-like Growth Factor 1 Receptor (IGF-1R), a tyrosine kinase receptor, signals to PI3K to AKT, a family of serine-threonine kinases, which then signals to downstream transcriptional and other events involved in energy metabolism (Valerie et al. 2007, Vander Heiden et al. 2009). The mTOR pathway upregulates protein synthesis and coordinates with PI3K/AKT signalling to facilitate cell growth. In addition to intracellular events, probably some of the same or related pathways have roles in release of cytokines and other soluble factors that can affect neighbouring cells (i.e., bystander effects) (Hei et al. 2008).
Figure 1.
Overview of radiation stress signalling and metabolism. Examples are shown of some of the important players involved in radiation responses that can affect cellular metabolism and mitochondrial function. As described in the text, actual signal transduction pathways form a complex network and many details have been omitted for clarity. IR triggers responses in cellular membranes, such as activation of ASM and ceramide generation, leading to activation of cell-surface receptors including those for growth factors, cytokines, and apoptosis-related ligands. Cell surface receptors and their transducers then signal to protein kinase cascades, typically involving PTK, leading to activation of a variety of pathways including the examples shown here. Growth factor signalling as well as inhibitory responses can impact the mTOR and PI3K/AKT pathways, which are major controllers of protein synthesis and energy metabolism. Both IR and subsequent mitochondrial events generate ROS and reactive nitrogen species (RNS), which directly inhibit protein tyrosine phosphatase (PTP) activities with consequent enhanced PTK signalling. DNA damage triggers a complex response involving rapid activation of ATM and other Rad3-related proteins as well as a myriad of DNA damaging processing factors such as PARP, which can affect NAD+ and ATP levels as well as generate a variety of small molecule repair products (Hassa and Hottiger 2008). ATM regulates important metabolic effectors such as AMPK, which can down-regulate growth-stimulatory pathways including PI3K/AKT and mTOR. ATM and AMPK contribute to p53 activation, which triggers induction of many genes and also directly impacts mitochondrial function. Some salient examples of p53-inducible genes are shown in the inset on the right side. In addition to up-regulating PTEN and the β subunit of AMPK, p53 also induces a variety of genes with direct roles in energy metabolism and protein synthesis. As shown in the inset, p53 is known to induce many genes that will either directly or indirectly impact the levels of many small molecules by affecting DNA replication, cell cycle control, DNA repair, ROS levels, and apoptosis.
While some of the above anabolic pathways may have pro-survival features, growth delays (cell cycle checkpoints) are a hallmark of radiation responses and allow time for DNA damage processing and other recovery events that maintain genomic stability. For example, ataxia telangiectasia mutated (ATM) is a master signalling protein kinase that orchestrates a complicated network of downstream events involved in the recovery from DNA damage and activation of multiple cell cycle checkpoints (Kitagawa and Kastan, 2005). Compromise of ATM has also been found to worsen features of the metabolic syndrome in mouse models (Schneider et al. 2006). While ATM contributes to the regulation of many signalling events including p53 as described below, it is notable that it positively regulates AMP-activated protein kinase (AMPK), a protein kinase involved in regulation of energy metabolism (Sanli et al. 2010). Typically, activated AMPK stimulates catabolic pathways controlling glycolysis, fatty acid oxidation, and mitochondrial biogenesis, and inhibits anabolic pathways involved in gluconeogenesis, glycogen, fatty acid and protein synthesis including the mTOR pathway (Vander Heiden et al. 2009, Feng and Levine 2010, Luo et al. 2010). Cell cycle delays will indirectly impact overall metabolism, and IR triggers a variety of both protein 53 (p53)-dependent and independent pathways controlling these delays. In the case of the latter, transcriptomics analysis of a large panel of p53 wild-type (wt) and p53-deficient human cell lines analysis showed rapid repression of many genes associated with G2/M cell cycle transit by a pathway involving the transcription factor E2F4 (Amundson et al. 2008).
p53 signalling and metabolism
The tumour suppressor p53 is a prominent mediator of radiation responses including many that either directly or indirectly affect metabolism. Considering that there are more than 6000 Pubmed citations for the search terms p53 plus radiation, only a limited overview is provided here and more comprehensive reviews are available (e.g., see Hofseth et al. 2004, Kastan and Bartek 2004, Meek and Anderson 2009 and references therein). While p53 can have direct effects on mitochondria and DNA repair as previously reviewed (Smith and Fornace 1996, Schuler and Green 2005), it primarily is a stress-inducible transcription factor that directly regulates more than a hundred genes in vivo (http://www-p53.iarc.fr/targetgenes.html), and functions as a central node in a complex stress signalling network with many upstream regulatory components as well (Meek and Anderson 2009). Representative p53-inducible genes are shown in the inset of Figure 1 with functions in general categories that may affect metabolism either directly or indirectly. p53 directly impacts metabolism by upregulation of the phosphatase and tensin homolog (PTEN) tumour suppressor that inhibits PI3K/AKT and mTOR signalling as well as being both downstream and upstream of AMPK (Vander Heiden et al. 2009, Vousden and Ryan 2009, Feng and Levine 2010).
These metabolism-related p53-regulated genes make up a portion of a complex inter-pathway network that permits irradiated cells to shut down cell growth. p53 directly regulates additional genes encoding proteins involved in energy metabolism such as tumour protein 53 (TP53)-induced glycolysis and apoptosis regulator (TIGAR), which decreases glycolytic activity by dephosphorylating fructose-2,6-bisphosphate and by switching glucose to the pentose phosphate pathway (PPP), along with deceasing ROS generation by promoting glutathione production (see Vander Heiden et al. 2009, Vousden and Ryan 2009, Feng and Levine 2010, Zhang et al. 2010 for more details). p53 as well as AMPK and ATM can thus function as ‘metabolic tumour suppressors’ to control cell growth after stress (Vousden and Ryan 2009, Feng and Levine 2010, Luo et al. 2010, Zhang et al. 2010).
p53 has additional roles in mediating stress signalling related to metabolism by dampening some of the growth stimulatory pathways shown in Figure 1. For example, nuclear factor kappa-light-chain-enhancer of activated B cells (NFκB) and p53 can negatively affect each other at several levels (Feng and Levine 2010, Lowe et al. 2010, Salminen and Kaarniranta 2010). In addition to the growth stimulatory ERK branch of the MAPK pathway, IR can lead to activation of members of the c-Jun N-terminal kinase (JNK) and p38 stress MAPK branch often in a tissue-specific manner (Bulavin and Fornace 2004, Valerie et al. 2007). Both p38 and JNK signalling can contribute to p53 activation and particularly p38 can oppose high ERK signalling by triggering pro-apoptotic and growth inhibitory responses (Bulavin and Fornace 2004, Meek and Anderson 2009). Interestingly, p38 is a major mediator of inflammatory responses (Schieven 2005), and IR is known to trigger a variety of both short-and long-term responses associated with inflammation (Lorimore et al. 2003). The pro-inflammatory features of radiation exposure can also contribute to prolonged elevations of ROS levels and oxidative stress (Lorimore et al. 2003).
While not shown in Figure 1, p53 can also repress many genes after IR. For example, p53 can repress Myc (similarity to myelocytomatosis viral oncogene [v-Myc]) oncogene expression, and Myc is known to contribute to the transcriptional activation of genes encoding glycolytic enzymes (discussed in Feng and Levine 2010). p53 can also impact energy metabolism by inhibiting the expression of the glucose transporters GLUT1 and GLUT4, and can decrease the levels of phosphoglycerate mutase (Vousden and Ryan 2009). The mechanisms responsible for the specificity of suppression of transcription by p53 have been somewhat unclear since many p53-repressed genes do not have typical p53-binding sites. Recently, p53 has been reported to be involved in the transcription of at least one of the large intergenic noncoding RNA (lincRNA) namely lincRNA-p21 that can function as a repressor in p53-dependent transcriptional responses (Huarte et al. 2010). This RNA affected the expression of hundreds of gene targets normally repressed by p53.
The actual number of p53-regulated human genes remains to be determined but could approach a thousand. In one study, more than 500 loci for p53 binding sites in the human genome were identified (Wei et al. 2006). Transcriptomic studies indicate many p53-regulated genes can be induced by IR based on reduced responsiveness in isogenic p53-deficient cells compared to p53 wt (Amundson et al. 2005). When the responses in spleen and liver from irradiated wt mice compared to p53-null mice were analysed by microarrays, the total number of potentially p53-regulated genes also exceeded 500 (Burns and El-Deiry 2003). In another array study (Kannan et al. 2001), p53 expression affected a large number of genes including many involved in cellular metabolism such as Lysyl oxidase-like protein, uridine diphophate (UDP)-Galactose 4 epimerase, cAMP activated Protein Kinase (PKA), Lysosomal Mannosidase alpha B, Carboxylesterase, ABC3, Apolipoprotein C-I, Human cocaine and amphetamine regulated transcript (CART), Lecithin-cholesterol acyltransferase, Rhodanese, and Glutamine phosphoribosylpyrophosphate (PRPP) amidotransferase. Thus, p53 either directly or via downstream effectors, such as lincRNA-p21, can have a very broad effect on global gene expression and consequently metabolism after IR.
Metabolomics: Context and methods
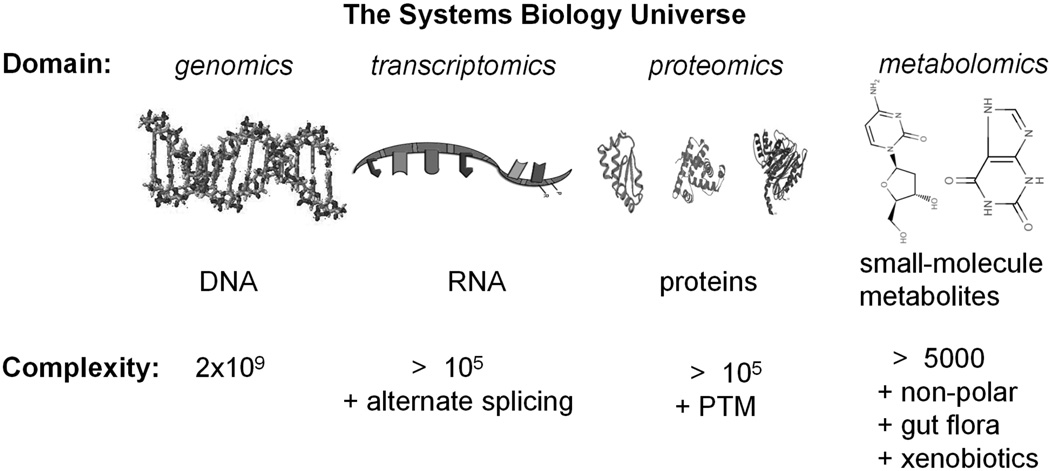
Living systems contain a complex network of interactions that, until recently, have been studied reductively and analytically rather than with an integrative systems approach. The field of systems biology takes up this systemic approach, combining information from different domains to find new insights into the operation of biological systems. As shown in Figure 2, information from genomics, transcriptomics, and proteomics are now supplemented by information provided by the relatively new field of metabolomics.
Figure 2.
Living systems are a complex network of interacting elements. Systems biology views these elements in an integrative way. Metabolomics is a recent addition to the domains of genomics, transcriptomics, and proteomics that qualitatively or quantitatively analyse small-molecule species and related pathways. Metabolomic responses result from proteomic and transcriptomic events and may be sensitive indicators of system health. Complexity is highlighted by estimates of the number of nucleotides in genomic DNA, approximate number of mammalian proteins, which does not include a vast number of post-translational modifications, and metabolites.
The goal of metabolomics is to provide a qualitative and quantitative description of metabolites and their connected pathways. The metabolites synthesised in a biological system are known collectively as the metabolome, and the goal of metabolomics is the ‘unbiased simultaneous identification and quantification’ of the elements and pathways in the metabolome (Fiehn 2002). The definitions for metabolomics and metabonomics have some subtle distinctions but are used interchangeably here (http://en.wikipedia.org/wiki/Metabolomics). Metabolites are generally in the small molecule class, with current data showing a bimodal distribution in size with more than 30% in the 100–400 Da range and a similar number in the 700–900 Da range, and with physico-chemical properties that are generally distinct from xenobiotics such as drugs and toxins (Khanna and Ranganathan 2009). An overview of the metabolomics of toxins and xenobiotics is provided in two recent reviews (Patterson and Idle 2009, Patterson et al. 2010a), while a third review provides a perspective on the use of mass spectrometry (Patterson et al. 2010b). To minimise overlap with these publications, we provide here only a brief overview of the metabolomics field, data acquisition methods and data handling, and focus in later sections on current results related to radiation response.
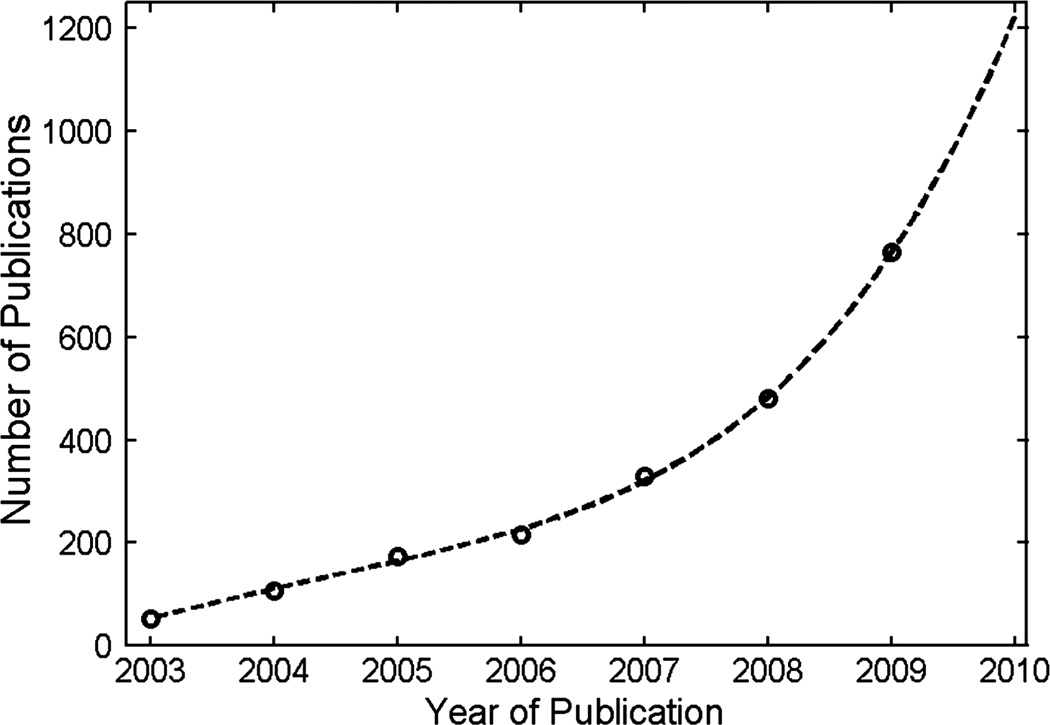
The goals of metabolomics for identification and quantification of the metabolome are challenging, but the promise of the new science is great, largely because, as noted by (Fiehn 2002) “Metabolites are the end products of cellular regulatory processes, and their levels can be regarded as the ultimate response of biological systems to genetic or environmental changes”. It is this suggestion that metabolomics may be among the most sensitive and accessible windows into biology that fuels current interest. Metabolomics is now one of the most rapidly evolving fields in science, as illustrated by the nearly-exponential growth in the number of peer-reviewed papers shown in Figure 3.
Figure 3.
Metabolomics is a rapidly growing field with a rapidly increasing number of publications appearing in the peer-reviewed literature.
The comprehensive characterisation of the metabolome, however, is a daunting task since the endogenous metabolites vary widely in their physical and chemical properties, occurring across the range of chemical classes, which in turn makes their concurrent extraction, separation, and detection a major challenge (Dettmer and Hammock 2004, Dettmer et al. 2007). The explosive growth of new results is important from a comprehensive scientific perspective; however, an ultimate goal of metabolomics in a systems biology context is to be of use in a clinical and screening setting, in our case to identify exposed individuals so that informed decisions can be made on treatment for radiation exposure. Reaching this goal requires a two-step process. In the discovery phase, key metabolite signatures are determined and validated by pathway analysis. This is a top-down, non-targeted, analysis that acquires and interprets huge data sets using sophisticated mathematical techniques and pathway analysis. Some of the recent promising results in the area of radiation exposure are described in the following section. When the identities of biomarkers or related groups of biomarkers are validated and in hand, it is possible to design targeted assays for diagnostic purposes, in our case, for radiation biodosimetry based on minimally-invasive sampling. These bottom-up, targeted analyses can use new technologies and instrumentation that can be simpler and lower in cost than the state of the art complex instrumentation used in the discovery phase. We discuss promising developments for targeted analysis in the section entitled Large-scale screening for radiation exposure based on metabolomic biomarkers.
Recent technology advancements have led to the development of several analytical platforms for qualitative and quantitative metabolomics experimentation. While the nuclear magnetic resonance (NMR)-based approach has traditionally made important contributions in the development of the metabolomics field, it is relatively insensitive and slow and typically measures relatively abundant metabolites in given biofluids (Want et al. 2005). In some recent work, microdroplet microcoil NMR has been combined with liquid chromatography (LC) mass spectrometry (MS) (Lin et al. 2008) to form a LC-MS-NMR platform. The slower NMR analysis achieves high sensitivity by being performed off-line in this case. Another technique, capillary electrophoresis (CE) is a fast and high resolution method which has been employed for targeted analysis of metabolites because the separation is based on the difference in their mass-to-charge (m/z) ratio in solution (Schauer et al. 2005), but cannot easily be used in discovery. The workhorse of platforms for discovery remains techniques involving gas chromatography or liquid chromatography and high-resolution and/or multiple-reaction-monitoring (MRM) mass spectrometry, both for discovery and in designs for diagnostic instrumentation.
Gas chromatography coupled with mass spectrometry (GC-MS) is ideal for thermally stable metabolites including compounds such as sugars, fatty acids, amino acids, aromatic amines, sugar alcohols, as well as for many lipids. This technology has an advantage of unambiguous identification of compounds because of the availability of a comprehensive spectral library for small molecule metabolites (Schauer et al. 2005, Issaq et al. 2008). However, the samples need to be initially derivatised to be amenable for ionisation and detection by GC-MS, and this chemical process can prevent the detection of some metabolites.
LC-MS provides versatility and high selectivity by combining different chromatographic techniques including reverse-phase, normal phase, size exclusion and hydrophilic interaction chromatography, with different ionisation methods, unit or accurate mass resolution and fragmentation analysis. The use of liquid chromatography makes chemical derivatisation unnecessary (enabling the ‘dilute and shoot’ approach) and is compatible with compounds of varying polarity such as lipids, peptides, nucleotides, etc., with respect to ionisation and detection and hence is a good method of choice for global metabolomics. When combined with ultra performance liquid chromatography (UPLC), MS instrumentation can typically yield 5,000 ions or more in both positive and negative electrospray ionisation (ESI) modes. Each of these ions will have a retention time value on the LC column, a mass-to-charge ratio (m/z), and an intensity value. Each biological sample may therefore generate 30,000 data points. A typical metabolomic experiment with such methodologies can easily generate a million data points, so informatics approaches including multivariate data analysis (MDA) methods, are utilised for data reduction.
For complete access to metabolic constituents and pathways, it is necessary to use a combination of techniques. The platform used by the analytical services company Metabolon is detailed in (Evans et al. 2009). They broaden the range of detected metabolites by using three separate protocols on each sample: GC-MS with derivatisation, LC-MS for positive ions, and LC-MS for negative ions. The three sets of results are then critically evaluated and combined prior to data analysis.
First, pre-processing methods are used to process the spectral data through peak filtering, detection, alignment, and normalisation, into discrete peak list where each peak is represented as a function of m/z, retention time, and ion abundance (peak intensity). This augments noise and data reduction as well as removal of systematic bias for downstream analysis. Several software tools have been developed for LC-MS metabolomic data preprocessing. These include MarkerLynx, Met-Align, XCMS, and MZmine. Other packages, some of them specific for LC-MS-based metabolomics, have been reviewed (Katajamaa and Oresic 2007).
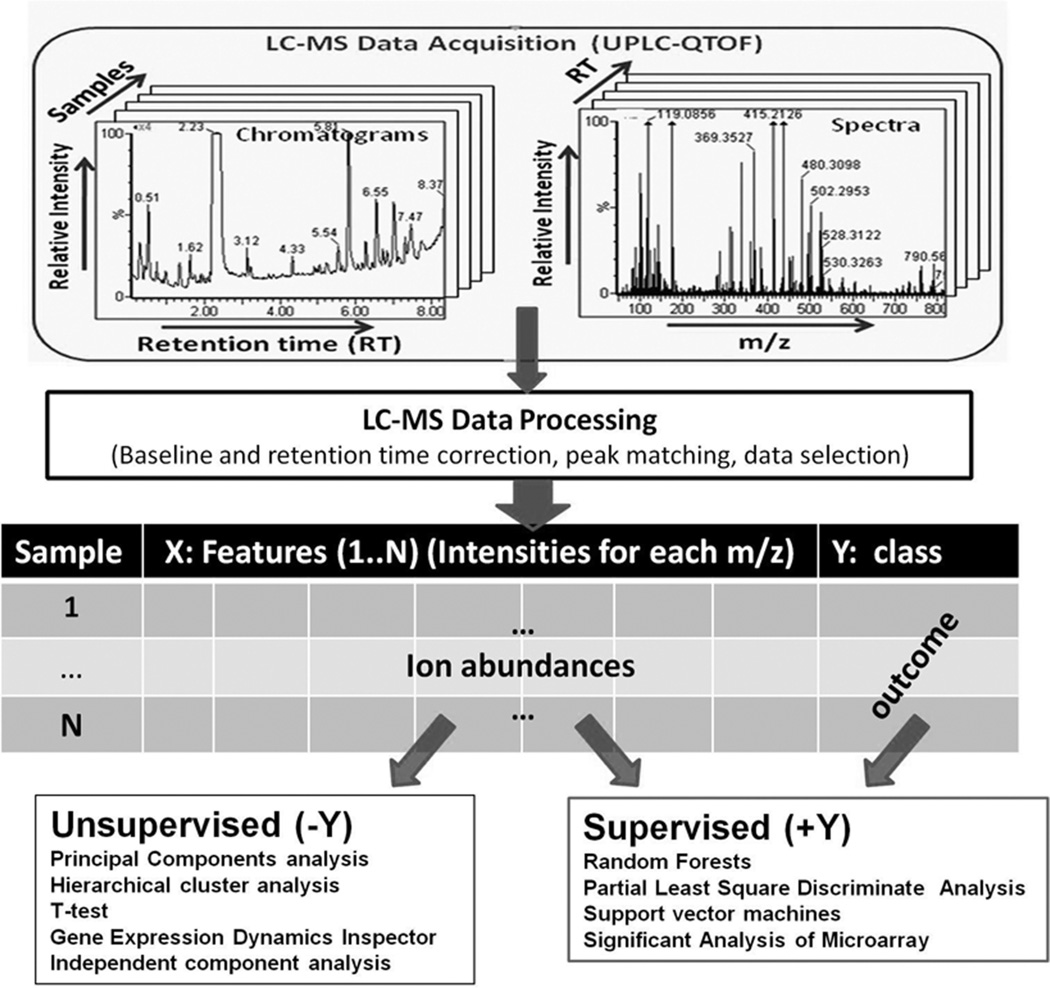
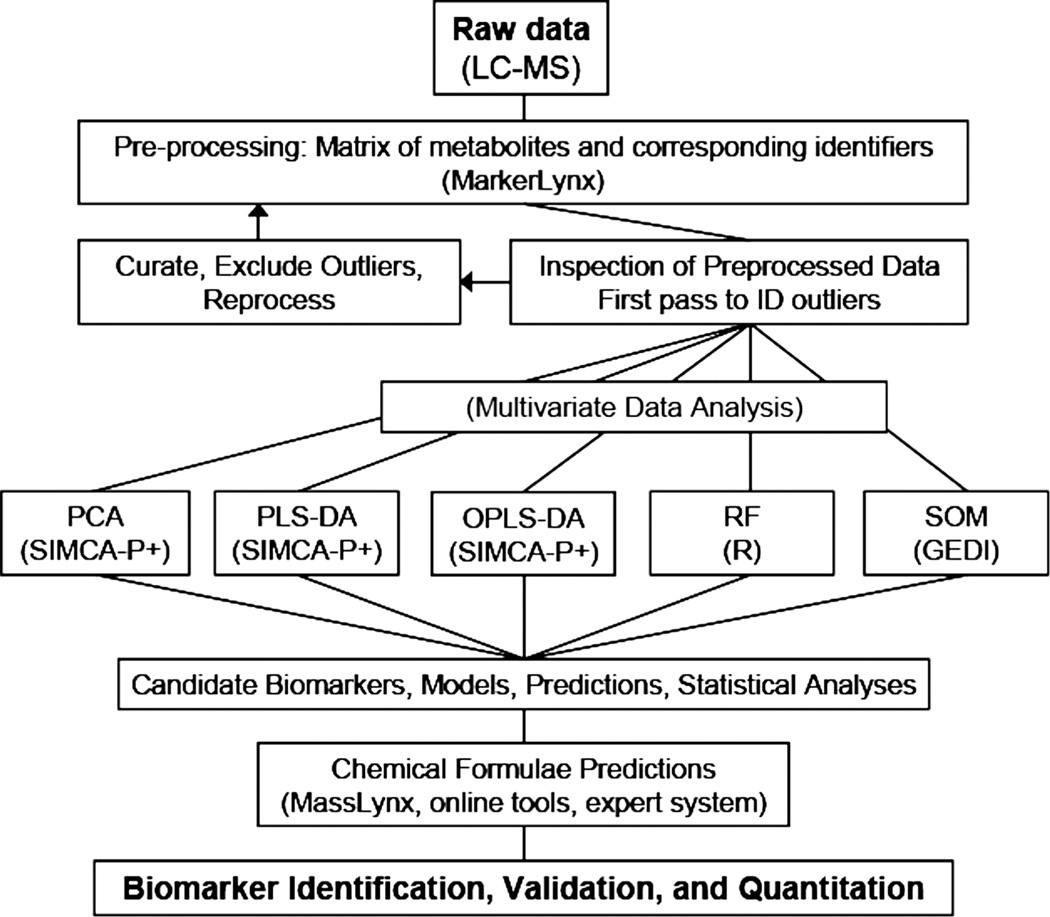
Following data preprocessing, multivariate data analysis and statistical methods are generally used to identify significant differences in metabolic changes between different biological groups. Since preprocessed LC-MS and microarray data share several common features, statistical methods previously developed for microarray data analysis have been utilised for difference detection. An overview of our methods for LC-MS data is presented in Varghese et al. (2010). Notable techniques for data interpretation and visualisation include the Gene Expression Dynamics Inspector (GEDI) (Eichler et al. 2003) originally developed for microarray data analysis, which has been used for UPLC time-of-flight mass spectrometry (TOFMS) radiation studies in (Patterson et al. 2008) to identify subsets of dose-dependent metabolites in the cellular response to radiation. Other methods, such as Significant Analysis of Microarray (SAM) or Empirical Bayesian Analysis of Microarray (EBAM), MeltDB30, t-test, principle component analysis (PCA), independent component analysis (ICA), (orthogonal) projection to latent structures discriminant analysis ((O)PLS-DA), soft-independent analysis of class analogy (SIMCA) methods are widely used to reduce the dimensions of metabolomics data and identify relevant metabolites (Yin et al. 2006, Bao et al. 2009, Kim et al. 2009, Ramautar et al. 2009). Use of these techniques in discovery requires a comparative evaluation of performance, as is done in (Bylesjo et al. 2006) for OPLS, partial least squares (PLS), OPLS-DA, partial least squares discriminant analysis (PLS-DA), and SIMCA. Each of these techniques has its relative strengths, resulting in a sometimes confusing landscape, but one in which results are supported by views from different perspectives. The overview in Figure 4 shows the role of these techniques in discovery. The techniques bifurcate into supervised and unsupervised classes. Supervised analysis makes use of the information about the treatment, which generated each sample (dose, cohort properties, etc.), while unsupervised analysis searches for clusters and discriminating properties using only the data itself. Supervised and unsupervised methods support each other in reducing the list of thousands of ions to a much smaller list with a dose response characteristic for radiation exposure.
Figure 4.
LC-MS (or GC-MS) are frequently used to obtain metabolic profile data for different populations and levels of radiation exposure because of their high sensitivity and peak capacity. Thousands of positively and negatively charged metabolites are identified in this way by mass/charge ratio and ion abundance. To identify combinations of metabolites most directly related to the radiation dose, the data is clustered both with methods that make use of the exposure level for each individual (supervised) and those that examine the data without that class information (unsupervised). Combining results from the different techniques, and validation by chemical pathways information results in a consistent picture of the metabolic response.
The final step in the metabolomics workflow involves mass-based metabolite identification and validation. Several databases are available to retrieve putative identifications (Smith et al. 2005, Wishart et al. 2007, 2009, Cui et al. 2008). However, the identification is insufficient for unambiguous metabolite identification. Therefore, further validation needs to be performed which involves comparison of fragmentation spectra and retention time with an authentic standard combined with an understanding of metabolic pathways to select the correct chemical identity and to eliminate variable responses such as those related to gut flora and diet.
Because of the explosion of results in metabolomics shown in Figure 3, it would be counter-productive to attempt a survey of the entire field. Some notable examples of successful use of metabolomics are given in (Spratlin et al. 2009) that range from discovery to clinical applications in oncology. They indicate that metabolomic analyses can often be performed non-invasively in vivo and usefully imaged by techniques such as [18F] 2-fluoro-2-deoxy-D-glucose positron emission tomography (FDG-PET) and magnetic resonance spectroscopy. We believe that the same utility is in reach for radiation exposure biodosimetry.
Radiation metabolomics
Metabolomics approach in radiobiology
The current metabolomics methodology to obtain global metabolic profiles and subsequently biomarkers has been described elsewhere (Patterson et al. 2008, Tyburski et al. 2008, 2009, Laiakis et al. 2010). Briefly, protein contents in samples are precipitated with an organic solvent and the supernatant is analysed by UPLC-TOFMS (Waters, Milford, MA, USA) in positive and negative ionisation modes to obtain global metabolic profiles. Deconvolution, noise reduction, peak alignment, and normalisation are performed with the software MarkerLynx (Waters, Milford, MA, USA) or alternatively MZmine or XCMS, leading to the extraction of ions, each with a descriptive identity of m/z and retention time.
A number of informatics approaches for multivariate data analysis allow for the identification of ions that show unique and statistically significant differences between groups of choice. The software SIMCA-P+ vs. 12.0 (Umetrics, Umea, Sweden) allows for unsupervised and supervised analysis of the data following Pareto scaling to control the influence of ion abundance (van den Berg et al. 2006). PCA, PLS-DA, and OPLS are the methods of choice. Additionally, data are analysed with the supervised machine-learning algorithm Random Forests (RF), generating lists of characteristic ions based on their importance and abundance through the construction of a large number of classification trees. Trends of ion changes between groups or time courses can also be visualised with the construction of heatmaps or self-organising maps through GEDI. A general strategy is shown in Figure 5 utilising many of the above approaches.
Figure 5.
Analytical framework for metabolomics data analyses and biomarker identification. An overview of some approaches used in radiation metabolomics are shown; see text for more detail.
Based on their m/z, potential ion identities can be searched through a number of online databases such as Madison Metabolomics Consortium Database (MMCD), Lipid Maps, Biological Magnetic Resonance Data Bank (BMRB), and the Human Metabolome Database (HMDB). Validation of the markers is conducted with tandem mass spectrometry (MS/MS) and spectra generated following collision with various energies are compared to spectra generated from pure chemicals. Quantitation of the ions is thereafter performed on a AB SCIEX 4000 QTRAP (Foster City, CA, USA) mass spectrometer coupled to a UPLC with the generation of a standard curve for each molecule based on the pure chemical standard and analysis with the software Analyst (Applied Biosystems, Carlsbad, CA, USA). Alternatively, quantitation is performed on the UPLC-TOFMS with the simultaneous generation of a standard curve, and concentrations are extracted with the software QuanLynx (Waters, Milford, MA, USA). Of the number of biologically important metabolites originally tested, only a subset of 20–30% can typically be positively identified due to the limitations of online databases.
Model studies in culture human cells
Our initial studies focused on radiation responses in human cell lines with well-characterised stress signalling properties (Patterson et al. 2008). The TK6 human lymphoblastoid cell line, as well as other p53 wt hematopoietic lines, show robust transcriptional responses even at low doses where there is little cytotoxicity, as well as at either high or low dose rates (Amundson et al. 1999, 2003, 2005, 2008, Li et al. 2007). In addition, the magnitude of these stress gene responses is often proportional to dose at physiologically relevant doses. The TK6 line has been a mainstay in cellular and molecular toxicology and has been used to assess in vitro toxicological effects for both conventional cytological endpoints and more recently for gene expression (Ku et al. 2007, Goodsaid et al. 2010). With a transcriptomics approach, we have found that overall signalling responses to IR show some qualitative differences from other classes of genotoxic agents and can be clearly distinguished from responses triggered by non-genotoxic agents (Amundson et al. 2005, Li et al. 2007). Using an UPLC-TOFMS approach to assess primarily aqueous metabolites from TK6 cell lysates, both dose as well as time dependent changes were seen at the metabolomics level (Patterson et al. 2008). Representative results are shown for 1 h after γ-irradiation in Figure 6 where overall responses for a variety of metabolites showed clustering of replicate samples by dose. At this time many of the metabolites contributing to this separation were down-regulated, and included nicotinamide and nicotinamide adenine dinucleotide (NAD+), proline and 5-oxoproline, phosphocholine, uridine monophosphate (UMP), adenosine monophosphate (AMP), and spermine and are discussed in detail in the primary report (Patterson et al. 2008). In the same study, this approach was also taken in BJ cells, a human p53 wt fibroblast line, where similar discrimination between unirradiated and irradiated cells was observed. While many non-polar metabolites as well some very small highly polar metabolites were not analysed in this study, this approach clearly demonstrates that a robust cellular response to IR occurs at the metabolic level. We expect that use of approaches to assess additional non-polar metabolites, e.g., GC-MS, will further expand the scope of small molecule changes triggered by IR. As discussed previously (Patterson et al. 2008), some but not all metabolite changes after IR may be attributable to known signalling pathways, and future studies using genetic approaches should facilitate elucidation of the pathways from genes to small molecules.
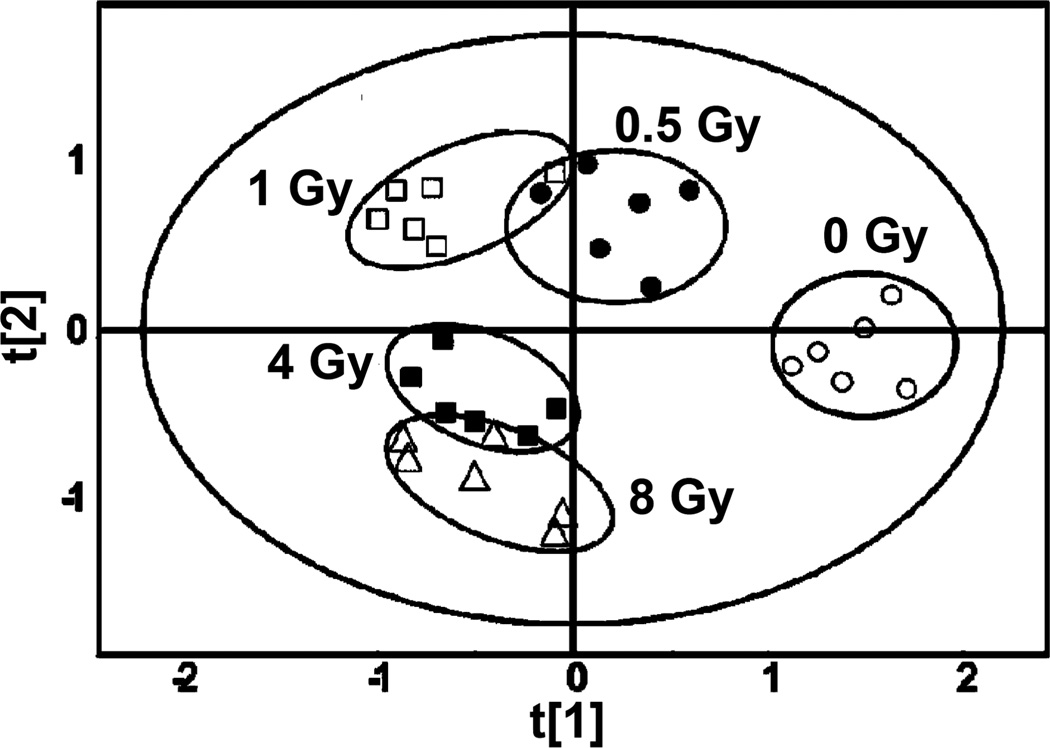
Figure 6.
Clustering of metabolomic responses by γ-ray dose in human cells. Results are taken fromPatterson et al. (2008) where the similarity of responses for individual samples are shown in this dose response experiment using samples of TK6 cells collected 1 h after IR. Multivariate data analysis of UPLC-TOFMS results was carried out as described using PLS-DA (Patterson et al. 2008), proximity of the data points indicate relative similarity of responses.
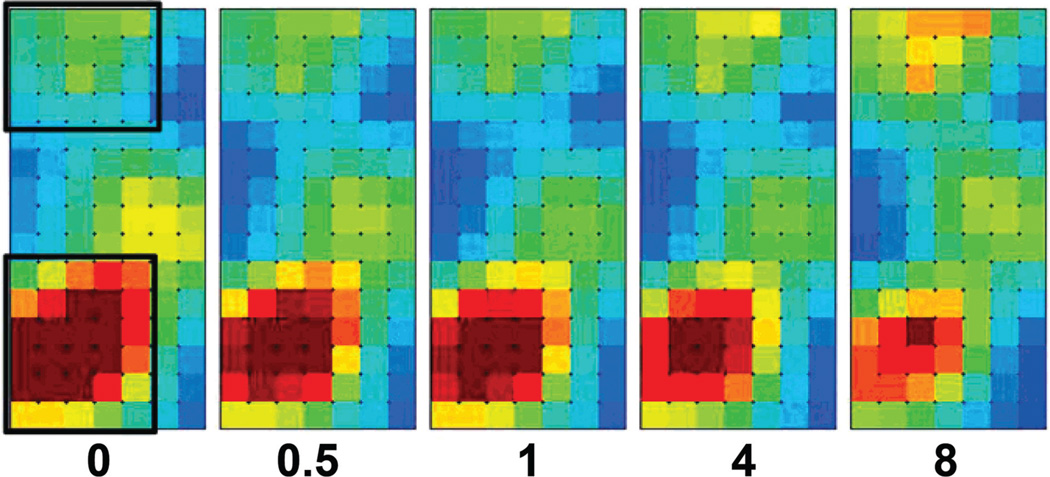
A variety of approaches has been employed to visualise overall ‘omic’ changes for metabolites (Patterson et al. 2010b). One interesting approach has been the adaptation of a transcriptomics program called Gene Expression Dynamics Inspector (GEDI) where self-organising maps (SOM) of tiles containing similar patterns of responses, whether transcripts (Eichler et al. 2003) or metabolites (Patterson et al. 2008, Tyburski et al. 2008), are used to translate high-dimensional profiles of sample classes into animated, coherent and robust montages of images in a two-dimensional heatmap display for the various tiles. This approach was used in the TK6 study and results are reproduced in Figure 7 from the 16 h time point where there is a clear trend suggestive of a dose-response relationship. Lymphoid lines such as TK6 show appreciable apoptosis by this time particularly after the two higher doses, and how apoptosis events affect the overall changes observed can be addressed in subsequent studies, such as where apoptosis could be blocked by either a chemical or genetic approach.
Figure 7.
Self-organising maps using GEDI software provide an overview of cellular responses to various doses of IR. Results are taken from Patterson et al (Patterson et al. 2008), where tiles containing similarly responsive metabolites are displayed with a heat map approach; red indicates clusters of ions with the highest abundance and blue the lowest. The values below each panel indicate the dose (Gy). Dose-dependent trends can be seen for tiles of metabolites that decrease in abundance with dose 16 h after IR in the lower left quadrants of these figures, and for tiles of other metabolites showing a trend to increase with dose in the upper portion.
In vivo metabolomic responses
Recent LC-MS developments have greatly expanded our capability to assess mammalian metabolomes. Our efforts were initiated five years ago using a new Waters Acquity UPLC and QTOF Premier TOFMS. The priority was to use an easily accessible biofluid so we focused our efforts on urine, an access point for the water-soluble metabolome. Our initial studies employed relatively high physiologic doses near the LD50 where we expected a robust response based on that seen at the mRNA and protein levels as discussed earlier. Our rationale was that if no response could be detected after a potentially lethal dose of 8 Gy, a level that results in a robust transcriptional response in mice, then a metabolomics approach with this biofluid would not be a suitable platform. However, this was not the case. We found significant and consistent metabolic changes at this dose. A loading plot analysis demonstrated that the levels of up to approximately 100 potential metabolites or metabolite fragments could be appreciably changed after IR and promising markers were confirmed and validated. As described in more detail (Tyburski et al. 2008), visualisation with the GEDI approach showed dose-dependent overall changes in the levels of many metabolites, and promising biomarkers included N-hexanoylglycine and β-thymidine after 8 or a lower dose of 3 Gy, 3-hydroxy-2-methylbenzoic acid 3-O-sulfate after 3 Gy, and taurine after 8 Gy. Such results established that our approach with modern instrumentation could be effectively employed for radiation metabolomics. Subsequent studies focused on expanding the dose range and timing of these metabolomic responses.
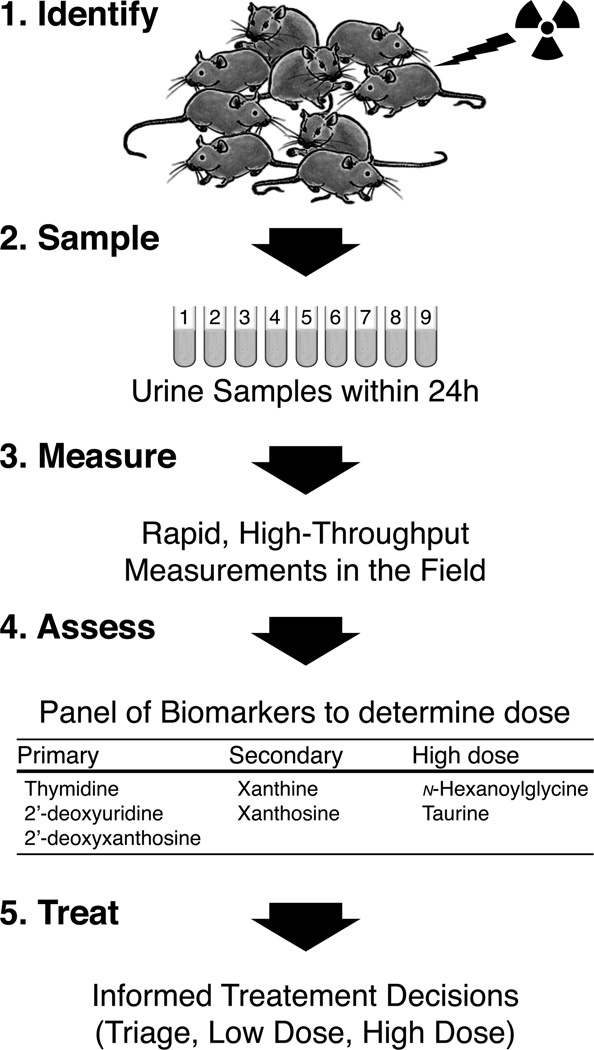
Mouse model for radiation biodosimetry
Studies carried out using metabolomics to discover urine biomarkers for radiation exposures in the mouse provides us with a proof of principle that the same may be accomplished in humans. A general biodosimetry paradigm based on current state-of-the-art biomarker discovery in the mouse is proposed in Figure 8 with some representative biomarkers identified in the course of our studies (Tyburski et al. 2008, 2009). Once a suspected radiation exposure or release has been confirmed, the first step is to identify the population at-risk for exposures that have the potential to cause adverse health effects, most importantly acute radiation syndrome. Urine samples are collected from members of the at-risk population for rapid, high-throughput measurements of biomarkers like thymidine, deoxyuridine, and deoxyxanthosine. These biomarkers can be envisioned to represent a set that can be used to determine doses up to about 3 Gy in the mouse with strong correlation to dose and each other (Tyburski et al. 2008, 2009). Beyond sublethal doses of 3 Gy or so, the response in these markers saturates, and the linear dose-response characteristics are lost. Secondary and high-dose markers are used to further confirm the primary biodosimetry results and expand the sensitivity for detecting higher doses. The results from the panel measurements can then be used to estimate the biologically effective dose received by each individual and inform the course of treatment as necessary. Just as important is the triage of individuals not receiving an appreciable radiation dose and that require no medical attention. This information will enhance the efficiency with which post-exposure medical countermeasures are utilised.
Figure 8.
Preliminary paradigm for radiation biodosimetry established using small molecule biomarkers in mouse urine. Radiation metabolomics has been used to establish a panel of urine biomarkers for identifying within a mixed-exposure population of mice the ones that received low or high doses of gamma radiation (Tyburski et al. 2008, 2009). The first step is to: (1) Identify the at-risk population and then (2) collect urine samples within the first 24 h post-exposure for (3) targeted quantitative measurement using a field deployable device of (4) a panel of validated biomarkers that can be used to screen out unexposed individuals and to identify individuals that received either a low/sublethal or high/lethal dose. The information obtained within these first 24 h will (5) inform treatment decisions and enhance the efficiency with which post-exposure medical countermeasures are implemented. Examples of potential representative biomarkers are included.
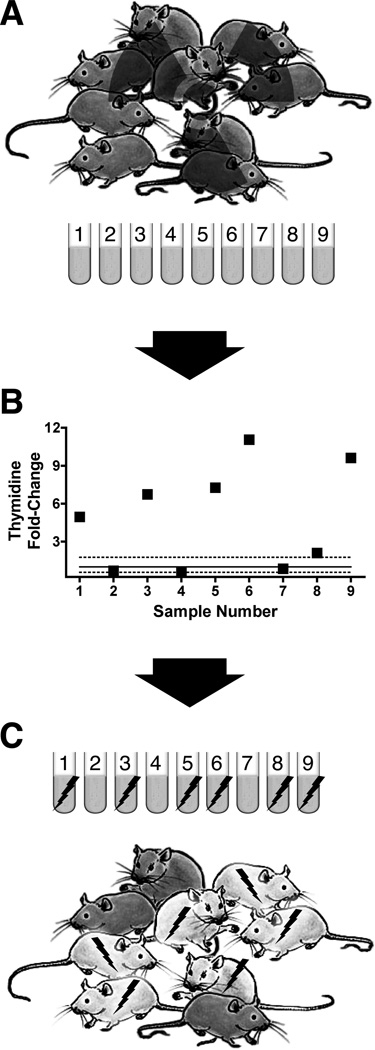
A simplified scheme is shown in Figures 9 to 11 for triaging individuals (mice in this example) receiving detectable radiation exposures, and then using select biomarker subsets to help distinguish low from higher, potentially lethal, doses. As for transcriptomics (discussed in Paul and Amundson 2008), a battery of biomarker sets will probably be required, but simplified examples with a few known radio-responsive metabolites are shown in these figures for illustrative purposes only. For example, in irradiated mice (Tyburski et al. 2008, 2009) changes in urinary thymidine were found to be a relatively reliable indicator of exposure. Therefore, we envision that biomarkers such as thymidine for mice and comparable urine biomarkers for humans can serve as the ‘first-pass’ criteria measurement for identifying exposed individuals among a mixed-exposure population (Figure 9). Elevated thymidine excretion in the urine correlates with dose up through 3 Gy γ radiation at high dose rate (~1 Gy/min), above which a saturation point is exhibited. What is more, the mouse excretes a detectable baseline normal level of thymidine in the urine that remains stable over time and appears to be independent of ageing (unpublished data). Shown in panel B of Figure 9 are the fold-increases in thymidine concentration over control baseline level in 24-h urine samples collected from unexposed and mixed-dose mice. The elevated thymidine levels in these representative samples can be used to reliably predict exposures of ≥1 Gy. Shown are just a few representative sample results.
Figure 9.
Radiation biodosimetry in mice using urine samples. While the levels of numerous metabolites change after IR, a simplified scheme is shown for thymidine which could be used as one of the first-pass biomarkers for identifying exposed individuals and triaging the unexposed (panel A) (Tyburski et al. 2008). Urine samples collected within the first 24 h post-exposure are analysed for thymidine concentration. Concentrations elevated above the upper limit of the baseline normal range indicate radiation exposure and will guide further testing. The magnitude of the elevated concentrations correlates with dose (panel B): mouse 1 received 2 Gy; mice 3 and 5 received 3 Gy; mice 6 and 9 received 8 Gy; mouse 8 received 1 Gy; mice 2, 4, and 7 were not exposed. Individual data are shown as fold-change compared with the mean urine thymidine concentration, normalised by creatinine concentration, of six healthy control mice not exposed to gamma radiation. A solid line indicates baseline, and dotted lines represent the lower and upper bounds of the 95% confidence interval of the baseline mean/baseline mean quotient with an alpha of 0.05. Results were calculated from data presented in by Tyburski et al. (2008, 2009). Once exposed mice have been found within the population (panel C), the findings can be validated and enhanced through further testing. These are representative results representing many experiments carried out under these conditions.
Figure 11.
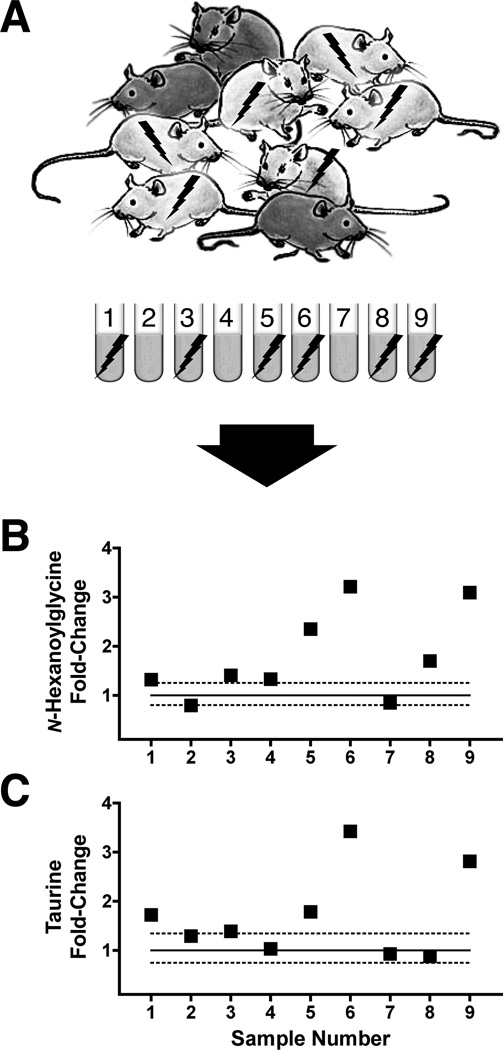
Mouse biomarkers for high-dose radiation biodosimetry. After identification and confirmation of exposed individuals within a mixed-exposure population (panel A) using radiation biomarkers that are sensitive enough to detect as little as 1 Gy but saturate in their response level with even sublethal doses around 3 Gy, attention may be turned toward measurements of the biomarkers of higher doses and two examples, n-hexanoylglycine and taurine (Tyburski et al. 2009), are shown. The elevation above baseline of n-hexanoylglycine (panel B) is reliably observed at doses of 3 Gy or higher. The response threshold for taurine concentration is similar (panel C). Sample 5 is from a mouse exposed to 3 Gy, and samples 6 and 9 are from mice exposed to 8 Gy. Sample 8 was only exposed to 1 Gy so the elevated n-hexanoylglycine is artifact. These markers tend to be a bit less reliable than the purine and pyrimidine markers but when used together suggest exposure to doses above 3 Gy when elevated above normal limits. The plots shown are fold-change over control with lower and upper 95% confidence limits of the baseline (1, solid line) indicated by dotted lines. Representative results were calculated from data in Tyburski et al. (2008, 2009).
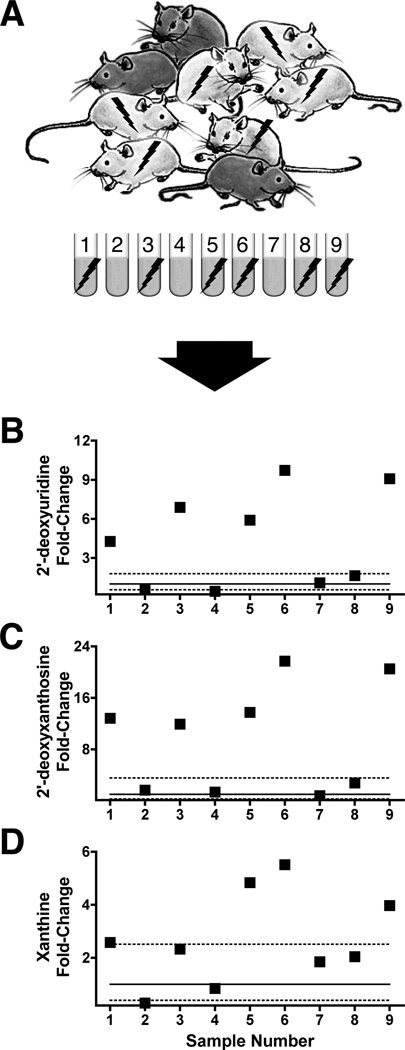
In this idealised scenario, additional sets of metabolites would be used to further confirm radiation exposure. Figure 10 shows the respective fold-increases over baseline of 2′-deoxyuridine, 2′-deoxyxanthosine, and xanthine in our representative sample set (panel A). The covariation of 2′-deoxyuridine (panel B) and 2′-deoxyxanthosine (panel C) with thymidine levels are such that linear regression yields a coefficient of determination (R2) for each of >0.98. Xanthine (panel D) as a marker is less sensitive but still provides a confirming dimension for biodosimetry. Current capabilities for biodosimetry in the mouse are limited in terms of dose-determination if purines and pyrimidines alone are used. However,Tyburski et al. (2008) reported that both taurine and n-hexanoylglycine excretion in the urine of mice exposed to ≥3Gy is elevated. We show here in Figure 11 that both n-hexanoylglycine (panel B) and taurine (panel C) can be used to identify individuals exposed to 8 Gy among a mixed-dose population. In the plots shown, mice 6 and 9 received 8 Gy, and these exhibit the largest fold-increase over baseline of both high-dose markers. In contrast, lower doses elicit only modest fold-increases in these markers over baseline at best.
Figure 10.
Biomarkers for validation of preliminary radiation biodosimetry results. Initial identification of exposed individuals among a mixed-exposure population (panel A) may be further confirmed using a set of radiation biomarkers including 2′-deoxyuridine, 2′-deoxyxanthosine, and xanthine among others. Fold-changes above baseline normal level (solid line) for 2′-deoxyuridine (panel B) and 2′-deoxyxanthosine (panel C) correlate very well with those of thymidine (R2 > 0.98, regression not shown). Xanthine (panel D) as a radiation biomarker is not quite as sensitive but can still be used to indicate exposures of 2 Gy or above. Dotted lines represent the lower and upper limits of the 95% confidence intervals for baseline mean/baseline mean quotients (1-fold) using alpha of 0.05. Representative results were calculated from data in Tyburski et al. (2008, 2009).
In the case of our examples with elevated urinary pyrimidines or purines after irradiation, we are not aware of any reports prior to (Tyburski et al. 2008, 2009) that describe this in mice, although there are some older reports in other species dating back to the late 1950s through the early 1970s. For example, Parizek et al. (1958) reported their observations that urinary deoxycytidine was elevated in a dose-dependent manner in the first 24 h after irradiation with as little as 0.5 Gy exposures to adult rats. These investigators did not, however, observe changes in urinary thymidine excretion. Interestingly, Tyburski and colleagues observed attenuated urinary excretion of deoxycytidine and elevated excretion of thymidine in irradiated mice. The elevated deoxycytidine excretion shown by Parizek et al. exhibits a similar dose-response range and saturation level as do the purines and pyrimidines measured by (Tyburski et al. 2008, 2009).
Ongoing studies in animal models for radiation biodosimetry
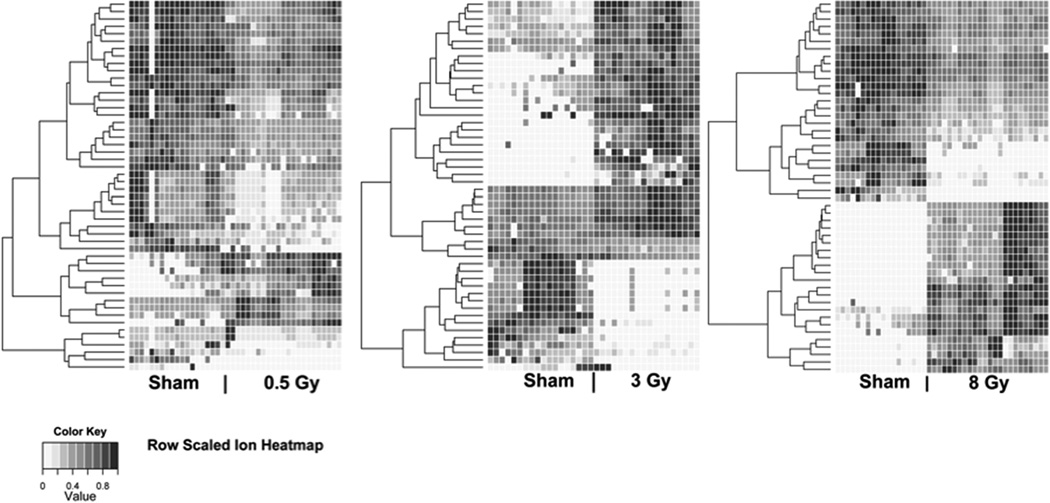
Efforts on radiation metabolomics and biodosimetry have been extended beyond the mouse urinary metabolome and various studies are in the process of being prepared for publication. For example, serum metabolomics have shown promising results with detectable responses, which persist beyond a week after irradiation. The scope of changes triggered by IR can be seen in Figure 12 where at least 50 potential ions are responsive. This further highlights that metabolomics radiation responses, like those at the transcriptomics and proteomics levels (as already discussed), are complex and probably represent the net result of perturbations in a variety of signalling pathways. Efforts are also underway with saliva, another easily accessible biofluid, from mice (and human patients). In addition to wild type C57Bl/6 mice, mutant mouse models are being utilised in order to elucidate the effect of radiation on various metabolic pathways. Another high priority will be to assess the metabolomic effects of low dose rate, partial body irradiations, exposures to inhaled and other internal emitters, and early responses that may be predictive of radiation lethality. In search of common metabolomic signatures between different species, studies are being carried out in a variety of species. For example, we have observed robust responses in rabbit biofluids after irradiation. An important priority will be to compare such responses to those in humans.
Figure 12.
Heatmaps of serum metabolites (ESI+ mode) from γ-irradiated mice. Each heatmap was constructed from the top 50 metabolites of importance generated through analysis with the machine-learning algorithm Random Forests. Each mouse is represented on the heatmap by triplicate runs of its serum sample. The same data from sham-irradiated mice were utilised for analysis in each of the three comparison runs. The metabolites were hierarchically clustered by complete linkage using the Euclidian distance. Differences in trends of metabolites between doses become increasingly apparent with increasing dose. Specifically, in panel A (sham vs. 0.5 Gy) most of the metabolites show less striking differences between the two groups; however, panels B and C (sham vs. 3 or 8 Gy, respectively) display more prominent metabolite changes. This Figure is reproduced in colour in the online version of the International Journal of Radiation Biology.
Towards the development of radiation biomarkers in humans
The atomic bombs of Hiroshima and Nagasaki, nuclear accidents such as Chernobyl and the Three Mile Island incident, exposure to radioactive material from abandoned medical devices such as in Goiania Brazil, and the threat of nuclear terrorism that has developed in recent years, constitute imperatives for the development of radiation biomarkers based on human samples that will provide effective high-throughput biodosimetry.
In order to address issues of exposure to radiation and identification of early biomarkers, we initiated human population studies at Georgetown University Hospital based on partial body irradiation. One cohort included prostate cancer patients undergoing hypofractionated stereotactic body radiation therapy with the CyberKnife system, which consists of a small linear accelerator mounted on a robotic arm (Accuray, Sunnyvale, CA, USA). Each patient completed a course of five 8 Gy fractions (a total dose of 40 Gy) to the prostate over 1–2 weeks. Urine, serum, and saliva were collected prior to the first treatment (control group) and thereafter at 1 h, 48 h and 3 months post irradiation, with each patient serving as his own control. Global metabolic profiles of small molecules were obtained with the UPLC-TOFMS platform at Georgetown University. Initial analysis of urine samples from 18 patients in positive ionisation mode and analysis of the data with the machine learning algorithm Random Forests (RF) revealed a 91.2% classification accuracy of the patient samples into the correct group (Figure 13) when assessing the top five hundred metabolites determined by RF. These promising initial results signify that metabolic differences between preirradiated and post-irradiated populations can lead to discrimination of one group from the other.
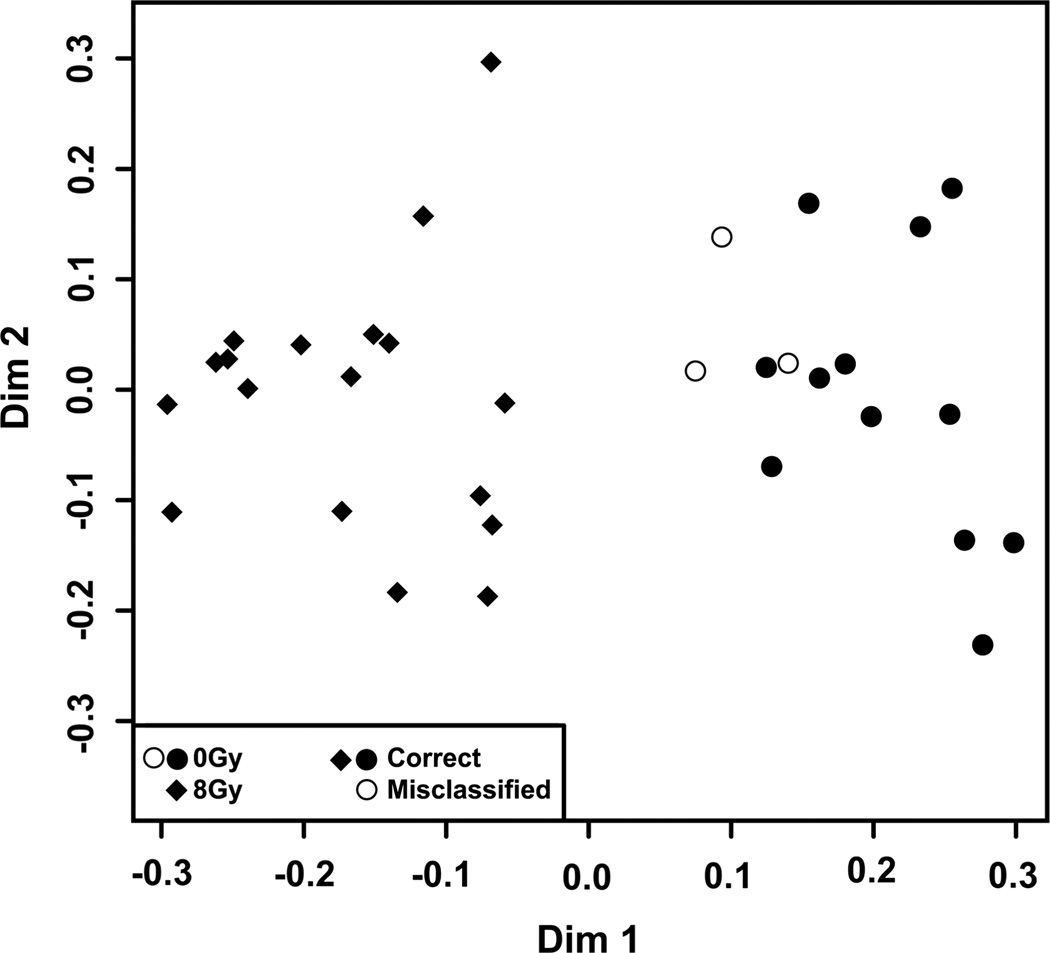
Figure 13.
MDS plot of the top 500 ions from human urines analysed in positive ionisation mode. Urines were collected from 18 prostate cancer patients undergoing hypofractionated stereotactic body radiation therapy with the CyberKnife system at Georgetown University Hospital before irradiation and 48 h post prostate irradiation (8 Gy). Global metabolic profiles were obtained by analysis with the UPLC-TOFMS and the software MarkerLynx. Each sample was normalised to its respective creatinine levels. Further analysis with the machine-learning algorithm Random Forests revealed a 91.2% classification accuracy of the samples in the correct group sample when the top 500 metabolites were utilised for construction of the MDS plot. Only three samples were misclassified, but still group overall with other unirradiated samples. Certain aspects of these samples based on the Random Forests criteria do not fit with the overall control group, which may be due to variability in metabolite profiles unrelated to irradiation.
Additional studies currently underway involve individuals undergoing total body irradiation (TBI) of ~1.25 Gy per fraction prior to bone marrow transplantation. Samples being collected include urine and serum prior to irradiation, 6 h and 24 h after the start of TBI. Transcriptomic analysis of blood from similarly treated patients was initiated by Amundson et al. (2004) and is ongoing in parallel with metabolomic studies, the latter showing robust small molecule responses so far. These concurrent studies will lead to developing connections and correlations between gene expression and metabolite changes. Recruitment of a larger number of patients in both of the human studies in order to increase statistical significance of the results is an ongoing effort, as well as initiation of collaborations to obtain irradiated non-human primate samples. Biomarker elucidation based on the limited number of patient samples that have been collected is underway and validation will be conducted on new incoming samples as they become available.
Large-scale screening for radiation exposure based on metabolomic biomarkers
Rapid screening of a large population for radiation exposure presents new challenges in addition to the discovery effort described above. Several different approaches to rapid screening are being developed as part of three projects under the Center for High-throughput Minimally-invasive Radiation Biodosimetry (http://www.cmcr.columbia.edu/), including cytomics (Garty et al. 2010), functional genetics (Brengues et al. 2010), and metabolomics (Coy et al. 2010). In addition, other approaches have been reported including breath analysis (Fedrigo et al. 2010) and NMR of lipids (Santini et al. 2006). Our discussion will focus on detection of validated small-molecule metabolic biomarkers that have already been identified and reported. Biomarkers from metabolomic studies are an especially promising avenue for detection of responses to insults like exposure to IR. Variation in metabolic products is a result of a chain of amplifying proteomic and transcriptomic events, and becomes, as a result, a sensitive indicator of cellular response.
Identification of the metabolic markers of acute radiation exposure was accomplished through the use of controlled experimental design, protocols, and lab-scale instrumentation to acquire large data sets. This work has included UPLC-TOFMS with electrospray ionisation in mouse models (Tyburski et al. 2008, 2009) and in TK6 cells (Patterson et al. 2008), as well as derivatisation gas chromatography mass spectrometry with electron-impact ionisation (GC-EI-MS) in rat model experiments (Lanz et al. 2009). These several experimental approaches are among those most frequently used in metabolomics and have been reviewed (Halket et al. 2005). These experimental methods detected thousands of compounds in both positive and negative ion mode. These data sets were then analysed with a variety of bioinformatic techniques to provide the reported biomarker metabolites. Several bioinformatic techniques were used, including RF (Lanz et al. 2009), SOM (Patterson et al. 2008, Tyburski et al. 2008), and OPLS (Tyburski et al. 2008). For reliable identification of metabolites truly associated with IR, these analytical techniques must be used in combination with a deep knowledge of the biology and biochemistry of metabolic pathways (Varghese et al. 2010). This process results in the validation of a small number of metabolic markers for radiation exposure, with known dose-response behaviour, whether up or down regulated.
Analytical diagnostic screening, regardless of the underlying technique requires attention to additional performance issues that include:
Sample preparation time;
Background noise levels;
Speed of analysis;
Interference from isobaric compounds and fragments and from charged species of the same m/z;
Quantitative accuracy and ionisation efficiency/ion suppression;
Instrumental cost and complexity;
Online validation of instrumental performance.
Many of these concerns have been evaluated for proteomics marker analysis (Addona et al. 2009, Carr 2009, Abbatiello et al. 2010). The platform of choice for analytical measurements of biomarkers is isotope dilution mass spectrometry coupled with MRM for additional selectivity in the mass spectrometer (Carr 2009). MRM is intended in this case as method of eliminating interferences based on collision-induced fragmentation patterns, while controlled addition of a stable isotope of the target provides an intensity calibration point. For complex biological samples, this level of selectivity may not be enough. (Abbatiello et al. 2010) note that MRM is not free of interferences: “MRM-MS with stable isotope-labeled internal standards (SIS) is increasingly being used to develop quantitative assays in complex biological matrices. These assays can be highly precise and quantitative, but the frequent occurrence of interferences requires that MRM-MS data be critically reviewed”. In addition, chemical noise generated in the ion source, or in the mass spectrometer inlet can prevent the quantitation of low levels of small molecule biomarkers. (Covey et al. 2009) state “At low mass, ions exist at nearly every nominal mass. With the improved sensitivity of atmospheric pressure ionisation (API) sources, chemical noise has appeared even under tandem MS conditions, confounding spectra interpretation and increasing baseline noise under multiple reaction monitoring conditions”.
In order to reduce the complexity and cost of the instrumentation, but at the same time achieve performance that matches, or exceeds, that of a standard or high-resolution lab mass spectrometer, we have investigated the use of ion-mobility separations between the ion source and the mass analyser, both ion mobility spectrometry (IMS) and differential ion mobility spectrometry (DMS). As has been observed experimentally (Dwivedi et al. 2008) and reviewed (McLean 2009), IMS is rather strongly correlated with m/z, especially for smaller molecules in the range of metabolic biomarkers. In contrast, DMS is much less correlated with mass because behaviour arises from a range of chemical properties like clustering affinity, polarity, charge distribution and others. DMS separations are due to attractive interactions between ion and neutral drift gas molecule. These interactions result in ion-neutral clustering that varies dynamically during the DMS separation field, creating a difference between ion mobility in high and low fields (Krylov and Nazarov 2009, Schneider et al. 2010c). DMS separations are due to dynamic interactions between an ionic analyte and the neutral gas as substrate, analogous to chromatography, and thus can vary strongly among ions of the same m/z but different structure. Even ions as similar as ephedrine and pseudoephedrine, differing in the orientation of a single hydroxyl group, can be separated (Schneider et al. 2010c).
Because of space constraints, we cannot provide a complete discussion of the system configuration and performance of DMS ion filtration used with API mass spectrometers. These details are given in a recent series of publications (Coy et al. 2010, Krylov et al. 2010, Schneider et al. 2010a, 2010b, 2010c), and the references therein. The characteristics of DMS-MS that make it valuable for analytical measurements in a targeted screening application on biological samples include the following:
Selectivity
Chemical noise reduction. Ions generated by charge transfer and fragmentation in the ion source are suppressed by 30X or more;
Complementary to m/z separation of the mass spectrometer. Ions of the same m/z but different chemical structure are usually separated, because DMS separation is chemical-affinity based, like LC or GC. Separations can be further enhanced by modifiers (Levin et al. 2006, Krylov and Nazarov 2009, Schneider et al. 2010a, 2010c);
Separation of charge states. Ions of the same m/z but different charge states are always separated by DMS.
Sensitivity
Filtration of the intact molecular ion. DMS-MS provides selectivity, which is equivalent to MS/MS but produces the intact molecular ion so that signal is not reduced;
Short residence time. Because the residence time in the DMS filter is about 5 msec, data acquisitions which target multiple species, even with MRM transitions, are not slowed by DMS response time. In Schneider et al. (2010c), 70 MRM transitions were recorded with 30 msec dwell time, for a total analysis time of hardly more than 2 sec.
Ease of use
Intensity independent of DMS tuning. Transmission efficiency for a DMS interface is weakly and smoothly independent of DMS tuning parameters, making quantitative calibration straight-forward;
Transparent mode. When DMS tuning voltages are off, the DMS interface operates with low loss. This makes instrument quality control rapid and setup of analytical methods easier.
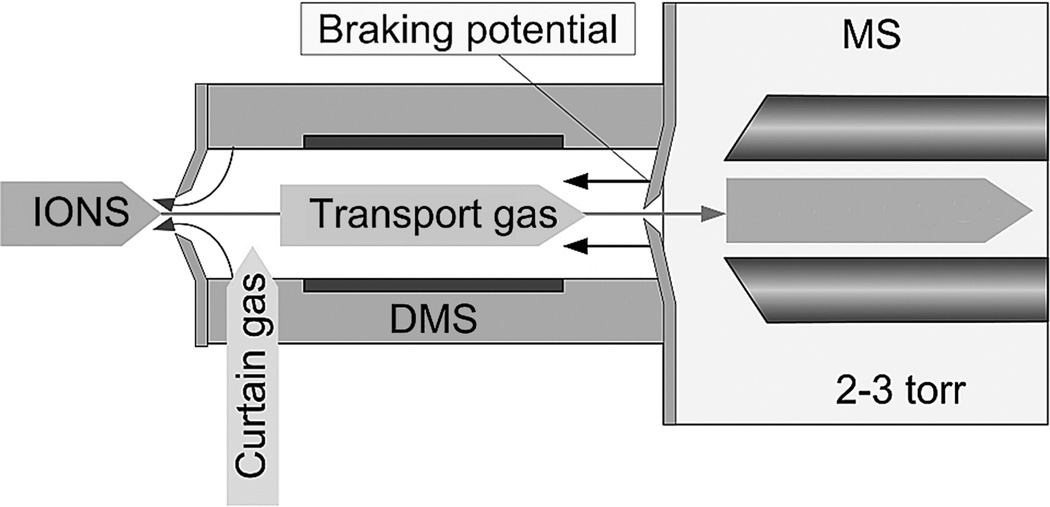
A DMS interface can be implemented for almost any ion source and any API MS inlet configuration. A typical schematic is shown in Figure 14. Planar DMS electrodes are placed in the atmospheric pressure region between the ion source and desolvating curtain gas interface and the orifice (or capillary) that transfers the ions into the first MS region. DMS electrodes are typically 1–2 cm in length with a residence time of 2–6 msec. The short residence time prevents any impact on the data acquisition time for methods that include large sets of MRM transitions. This speed is critical for targeted metabolomic biomarker instrumentation because the loadings in discriminating components or the dimensions in MDS (Figure 12) typically include multiple metabolite ions which will need to be determined simultaneously.
Figure 14.
DMS-MS interface schematic. Planar DMS electrodes are placed in the atmospheric pressure region between the ion source and desolvating curtain gas interface and the orifice (or capillary) that transfers the ions into the first vacuum region in the mass spectrometer. DMS electrodes are typically 1–2 cm in length with an ion residence time of 2–6 msec. The short residence time in the DMS analytical region prevents any impact on the analysis time for data acquisition methods, which include large MRM groups.
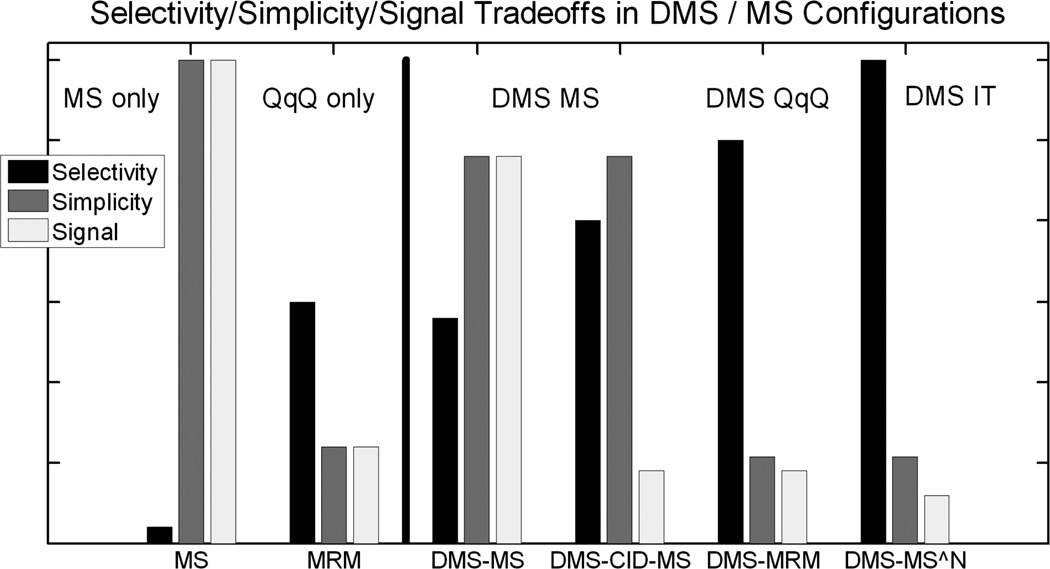
Figure 15 provides a qualitative view of the tradeoffs between selectivity, instrumental simplicity and signal levels for standard MS configurations and for those configurations augmented by a DMS interface, with the groups with and without DMS separated by a vertical bar. The two configurations on the left do not implement DMS and represent a single stage mass detector like a single quadrupole (insufficiently selective for biological applications), and an MRM triple-quad system (QqQ) (more selective, but lossy and still subject to interferences) (Covey et al. 2009, Abbatiello et al. 2010). The four configurations at right illustrate the effect of adding DMS to standard configurations. The first two use a single-stage mass analyser like a single-quadrupole or a TOF. DMS can approach the selectivity of MRM with better signal levels, and the selectivity can be further increased by using inlet voltages to activate the selected ions (DMS-CID-MS). And finally, DMS can be used with MRM or with MSN for even more selectivity at some cost in signal intensity.
Figure 15.
Qualitative comparison of MS and DMS-MS configurations. Selectivity is improved by DMS ion filtration with little cost in signal intensity or in device complexity. MS indicates the use of a single stage mass detector like a single quadrupole, while MRM indicates reaction monitoring in triple quadrupole (QqQ) system, and MS^N indicates a system capable of multiple fragmentation steps, such as an ion trap (IT).
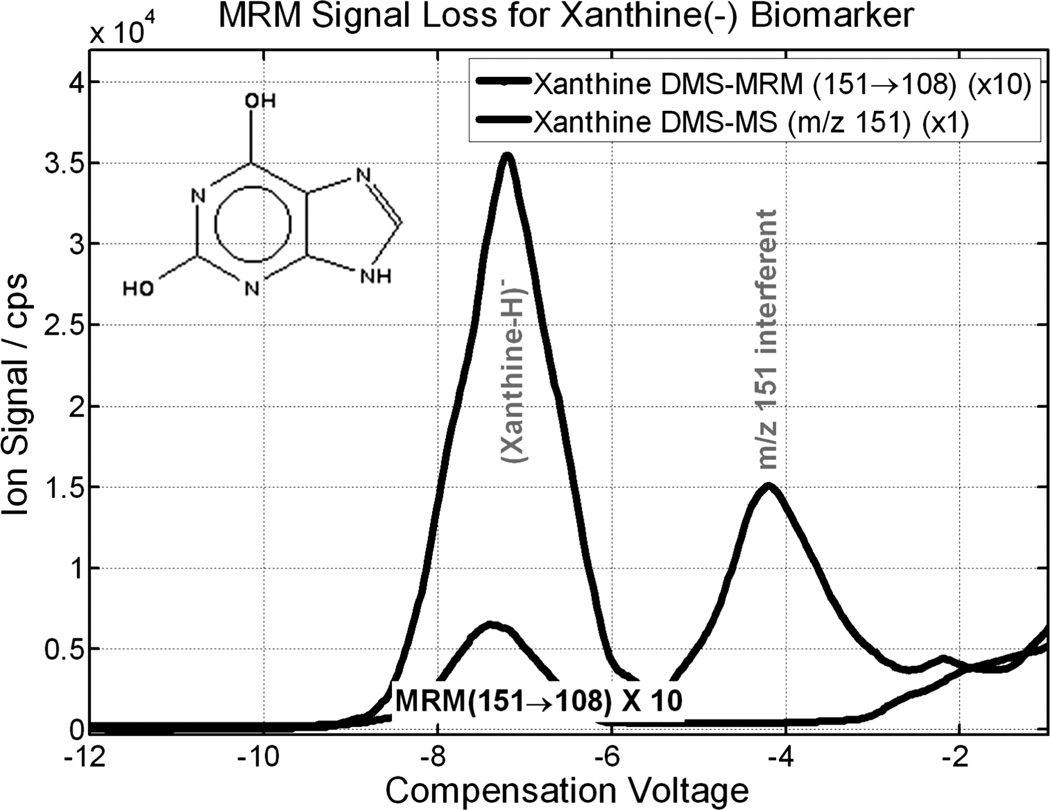
To illustrate the performance benefits, two examples are given, one showing the signal level improvement over MRM, and the second showing the reduction in chemical noise. Figure 16 compares DMS-MS with MRM for xanthine, a metabolic biomarker identified in the current project. Xanthine performs poorly in MRM, with charge loss dominating the strongest MRM transition. DMS-MS separates xanthine without signal loss. It also reveals and separates an isobaric interferent at the molecular ion m/z. For this target ion, the signal comparison is about 50 × in favour of DMS-MS.
Figure 16.
DMS-MS detection avoids signal loss in fragmentation. DMS-MS detection efficiency for biomarker xanthine is about a factor of 50 higher than with detection through MRM (151 m/z → 108 m/z). DMS separation of isobars is also evident. An interferent is present at m/z 151, which is suppressed by both DMS-MS and MRM.
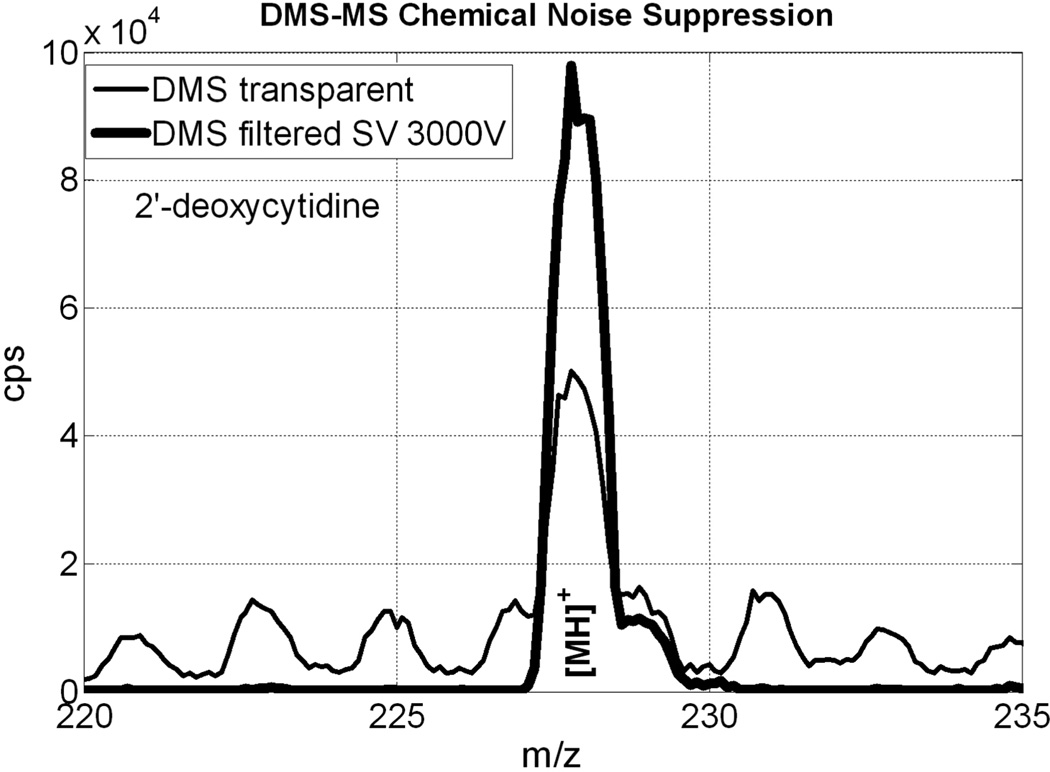
Figure 17 tests suppression of chemical noise on a sample of 2′-deoxycytidine (2′-dC), another identified biomarker. 2′-dC fragments efficiently in MRM, so DMS-MS is not superior in that respect, but, as noted in (Covey et al. 2009), m/z values in this mass range are usually populated at a low level by fragments, clusters, and other species. The Figure compares a DMS-transparent mass spectrum with the same region DMS-filtered for 2′-dC. A close examination of the data behind these two curves shows that chemical noise is suppressed by about a factor of 30.
Figure 17.
DMS-MS chemical noise suppression. In this example, chemical noise is strongly suppressed by DMS filtration optimised for 2′-dC [MH]+. Close examination of the data shows chemical noise to be reduced by a factor of 30 from the DMS-transparent case. The data was acquired on an AB SCIEX DMS-API 3000 prototype instrument operated in DMS-Q1 mode, without MRM.
Since virtually any API mass spectrometer can be fitted with a DMS interface, some of the smaller commercially available instruments with restricted mass range could be fitted with a DMS interface and used as a basis for screening protocols and instrumentation. Nonetheless, these instruments are still relatively large and costly. Development of smaller prototype instruments usable with DMS is proceeding rapidly. We will list two examples. In Tadjimukhamedov et al. (2010) from Prof. R. Graham Cooks lab at Purdue U., a standard Sionex DMS interface has been fitted to their mini-10.5 iontrap mass spectrometer (10 kg weight). Excellent results have been obtained for separation of morphine from diazepam (differing by 1 mass unit), and for measurements from biological samples. DMS-MRM results were also reported for this system. Also,Manard et al. (2010) report the design and testing of a somewhat larger single quadrupole system, fitted with a similar planar DMS ion filter.
Sample handling procedures and ion source configurations are also of great importance. Recent advances in this area include several new technologies. Sample handling is greatly simplified by the use of paper substrates to support the sample, as is now being done with dried blood spots (DBS). DBS sampling is now used routinely in pharmacokinetic and toxicokinetic studies (http://www.whatman.com/dmpk.aspx). In this form, samples can be shipped or stored at ambient temperature without degradation of analytes and metabolites sensitive to plasma enzymes. For the ionisation step, new technologies are also available Liquid Extraction Surface Analysis (LESA) has recently been announced by Advion (http://www.advion.com/news-events/documents/LESA_Prod_Note_FINAL_lr.pdf). This sampling method originated at Oak Ridge National Laboratory (Van Berkel and Kertesz 2009, Kertesz and Van Berkel 2010) and allows stabilised samples to be extracted from an array of samples, ionised, and analysed by DMS-MS.
In order to create low-cost, deployable instrumentation for analytical testing of small-molecule biomarkers, more is necessary than the simple creation of a small mass spectrometer. Too much performance is lost in mass spectrometer size and cost reduction. Based on current experimental results, DMS technology is able to restore the missing selectivity, while maintaining sensitivity and adding little to instrument complexity.
Conclusion
In this review, we have provided an overview of radiation metabolomics and its potential for biodosimetry. Sections have been included on the myriad biological effects of radiation on metabolism, giving a brief summary of the science of metabolomics and the methods used to identify metabolic effects in the discovery phase, a discussion of current results on metabolic radiation response from cell lines, animal models, and efforts to obtain data on human populations, and a discussion of faster, simpler technologies for targeted analysis.
This is an exciting time to be working in the field of metabolomics because of advances on every front. Improvements are legion, from instrumentation for biomarker discovery, to computer methods for data analysis, to understanding of pathways, to instrumentation for targeted analysis. These advances, and the advances in genomics and transcriptomics that open up connections from gene to metabolite, have made work in metabolomics more fruitful, as is evident from the rapid growth of the field seen in Figure 3.
The recent published results, and ongoing studies in cell lines and animal models, indicate dose response, and show clustering and discrimination based on combined detections of groups of metabolites derived from samples such as urine that can be obtained non-invasively. Data from these studies analysed by bioinformatic methods leads to the discovery-phase identification of groups of metabolites useful for screening and biodosimetry. This is extremely encouraging, and makes extension of the work to human populations important, even with the limitations inherent in such a study.
Finally, the power of the results from discovery is realised if they can be utilised to streamline targeted analysis and to provide new kinds of information in discovery. Ion mobility methods, such as DMS-MS, provide simplified quantitative detection of biomarkers, but also allow the elimination or simplification of sample cleanup and pre-separation. With the reduction in overall analysis time from 30 min or more to a minute or two, it is possible to follow kinetics or responses at a rate that is otherwise impossible, and it is possible to process data from larger cohorts in discovery.
The future of metabolomics in understanding radiation response is an exciting prospect. The result of this new understanding will be both a deeper knowledge of pathways and affected systems, and new technologies that enhance discovery and provide clinical tools.
Acknowledgements
The authors would like to acknowledge Dr David J. Brenner and Dr Frank J. Gonzalez for their support. Some of the authors, SLC, JBT, ECL, and AJF, were supported as part of the Columbia Center for High-Throughput Minimally Invasive Radiation Biodosimetry (P.I. David Brenner) and funded by NIH (NIAID) grant U19 AI067773. Efforts by AJF were also supported in part by grant R31-10069 (WCU program) through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology.
Abbreviations
- AMP
adenosine monophosphate
- AMPK
AMP-activated protein kinase
- API
atmospheric-pressure ionisation
- ATM
ataxia telangiectasia mutated
- BMRB
Biological Magnetic Resonance Data Bank
- DBS
dried blood spots
- cAMP
cyclic adenosine monophosphate
- CART
Human cocaine and amphetamine regulated transcript
- CE
capillary electrophoresis
- DNA
deoxyribonucleic acid
- cGMP
Cyclic Guanosine Monophosphate
- DBS
Dried Blood Spots
- DMS
differential ion mobility spectrometry
- EBAM
Empirical Bayesian Analysis of Microarray
- EGFR
epidermal growth factor receptor
- ERBB
epidermal growth factor receptor
- ERK
extracellular signal-regulated kinases
- ESI
electrospray ionisation
- FDG-PET
[18F]2-fluoro-2-deoxy-D-glucose positron emission tomography
- GC-MS
gas chromatography coupled with mass spectrometry
- GC-EI-MS
gas chromatography mass spectrometry with electron-impact ionisation
- GEDI
Gene Expression Dynamics Inspector
- GI
gastrointestinal
- HMDB
Human Metabolome Database
- ICA
independent component analysis
- IGF-1R
Insulin-like Growth Factor 1 Receptor
- IMS
ion mobility spectrometry
- IR
ionising radiation
- IT
ion trap
- JNK
c-Jun N-terminal kinase
- LC
liquid chromatography
- LESA
liquid extraction surface analysis
- MAPK
mitogen-activated protein kinase(s)
- MDA
multivariate data analysis
- MDS
multi-dimensional scaling
- MMCD
Madison Metabolomics Consortium Database
- MRM
multiple-reaction-monitoring
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- mTOR
mammalian target of rapamycin
- Myc
similarity to myelocytomatosis viral oncogene [v-Myc]
- m/z
mass-to-charge
- NAD+
nicotinamide adenine dinucleotide
- NFκB
nuclear factor kappa-light-chain-enhancer of activated B cells
- NMR
nuclear magnetic resonance
- OPLS-DA
orthogonal projection to latent structures discriminant analysis
- p53
protein 53
- PCA
principal component analysis
- PI3K/AKT
phosphatidylinositol-3-kinase/protein kinase B
- 6-keto-PGF1α
6-keto prostaglandin f1α
- PGF2α
isoprostane 8-epi-prostaglandin f2α
- PKA
cAMP activated Protein Kinase
- PLS
partial least squares
- PLS-DA
partial least squares discriminant analysis
- PPP
the pentose phosphate pathway
- PI3K
phosphatidylinositol-3-kinase
- PRPP
phosphoribosylpyrophosphate
- PTEN
phosphatase and tensin homolog
- PTK
protein tyrosine kinase(s)
- PTM
post-translational modifications
- Ras
rat sarcoma
- RF
Random Forests
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
- SAM
Significant Analysis of Microarray
- SIMCA
Soft-independent analysis of class analogy
- SIS
isotope-labeled internal standard(s)
- SOM
self-organising map(s)
- TBI
total body irradiation
- TIGAR
TP53-induced glycolysis and apoptosis regulator
- TOFMS
time-of-flight mass spectrometry
- wt
wild-type
- UPLC
ultra performance liquid chromatography
- UDP
uridine diphophate
- UMP
uridine monophosphate
- UV
ultraviolet
- 2′-dC
2′-deoxycytidine
Footnotes
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Abbatiello SE, Mani DR, Keshishian H, Carr SA. Automated detection of inaccurate and imprecise transitions in peptide quantification by multiple reaction monitoring mass spectrometry. Clin Chem. 2010;56:291–305. doi: 10.1373/clinchem.2009.138420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addona TA, Abbatiello SE, Schilling B, Skates SJ, Mani DR, Bunk DM, Spiegelman CH, Zimmerman LJ, Ham AJ, Keshishian H, et al. Multi-site assessment of the precision and reproducibility of multiple reaction monitoringbased measurements of proteins in plasma. Nature Biotechnology. 2009;27:633–641. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amundson SA, Do KT, Fornace AJ., Jr Induction of Stress Genes by Low Doses of Gamma Rays. Radiation Research. 1999;152:225–231. [PubMed] [Google Scholar]
- Amundson SA, Do KT, Vinikoor L, Koch-Paiz CA, Bittner ML, Trent JM, Meltzer P, Fornace AJJ. Stress-specific signatures: expression profiling of p53 wild-type and -null human cells. Oncogene. 2005;24:4572–4579. doi: 10.1038/sj.onc.1208653. [DOI] [PubMed] [Google Scholar]
- Amundson SA, Do KT, Vinikoor LC, Lee RA, Koch-Paiz CA, Ahn J, Reimers M, Chen Y, Scudiero DA, Weinstein JN, et al. Integrating global gene expression and radiation survival parameters across the 60 cell lines of the National Cancer Institute Anticancer Drug Screen. Cancer Research. 2008;68:415–424. doi: 10.1158/0008-5472.CAN-07-2120. [DOI] [PubMed] [Google Scholar]
- Amundson SA, Grace MB, McLeland CB, Epperly MW, Yeager A, Zhan Q, Greenberger JS, Fornace AJ., Jr Human in vivo radiation-induced biomarkers: gene expression changes in radiotherapy patients. Cancer Research. 2004;64:6368–6371. doi: 10.1158/0008-5472.CAN-04-1883. [DOI] [PubMed] [Google Scholar]
- Amundson SA, Lee A, Koch-Paiz C, Bittner ML, Meltzer P, Trent JM, Fornace AJ., Jr Differential responses of stress genes to low dose-rate gamma-irradiation. Mol Cancer Res. 2003;1:445–452. [PubMed] [Google Scholar]
- Bao Y, Zhao T, Wang X, Qiu Y, Su M, Jia W, Jia W. Metabonomic variations in the drug-treated type 2 diabetes mellitus patients and healthy volunteers. J Proteome Res. 2009;8:1623–1630. doi: 10.1021/pr800643w. [DOI] [PubMed] [Google Scholar]
- Brengues M, Paap B, Bittner M, Amundson S, Seligmann B, Korn R, Lenigk R, Zenhausern F. Biodosimetry on small blood volume using gene expression assay. Health Physics. 2010;98:179–185. doi: 10.1097/01.HP.0000346706.44253.5c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulavin DV, Fornace AJ., Jr p38 MAP kinase’s emerging role as a tumor suppressor. Advances in Cancer Research. 2004;92:95–118. doi: 10.1016/S0065-230X(04)92005-2. [DOI] [PubMed] [Google Scholar]
- Burns TF, El-Deiry WS. Microarray analysis of p53 target gene expression patterns in the spleen and thymus in response to ionizing radiation. Cancer Biol Ther. 2003;2:431–443. doi: 10.4161/cbt.2.4.478. [DOI] [PubMed] [Google Scholar]
- Bylesjo M, Rantalainen M, Cloarec O, Nicholson JK, Holmes E, Trygg J. OPLS discriminant analysis: combining the strengths of PLS-DA and SIMCA classification. J Chemometrics. 2006;20:341–351. [Google Scholar]
- Carr SA. Protein Quantification through Targeted Mass Spectrometry: The Way Out of Biomarker Purgatory? Molecular & Cellular Proteomics. 2009:S16–S16. [Google Scholar]
- Covey TR, Thomson BA, Schneider BB. Atmospheric pressure ion sources. Mass Spectrom Rev. 2009;28:870–897. doi: 10.1002/mas.20246. [DOI] [PubMed] [Google Scholar]
- Coy SL, Krylov EV, Schneider BB, Covey TR, Brenner DJ, Tyburski JB, Patterson AD, Krausz KW, Fornace AJ, Nazarov EG. Detection of Radiation-Exposure Biomarkers by Differential Mobility Prefiltered Mass Spectrometry (DMSMS) Int J Mass Spectrom. 2010;291:108–117. doi: 10.1016/j.ijms.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q, Lewis IA, Hegeman AD, Anderson ME, Li J, Schulte CF, Westler WM, Eghbalnia HR, Sussman MR, Markley JL. Metabolite identification via the Madison Metabolomics Consortium Database. Nat Biotechnol. 2008:162–164. doi: 10.1038/nbt0208-162. [DOI] [PubMed] [Google Scholar]
- Dettmer K, Aronov PA, Hammock BD. Mass spectrometry-based metabolomics. Mass Spectrom Rev. 2007;26:51–78. doi: 10.1002/mas.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer K, Hammock BD. Metabolomics–a new exciting field within the “omics” sciences. Environ Health Perspect. 2004;112:A396–A397. doi: 10.1289/ehp.112-1241997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo AL, Ramakrishnan N, Hatchett RJ. Radiation combined injury: overview of NIAID research. Health Phys. 2010;98:863–867. doi: 10.1097/HP.0b013e3181a6ee32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi P, Wu P, Klopsch SJ, Puzon GJ, Xun L, Hill HH. Metabolic profiling by ion mobility mass spectrometry (IMMS) Metabolomics. 2008;4:63–80. [Google Scholar]
- Eichler GS, Huang S, Ingber DE. Gene Expression Dynamics Inspector (GEDI): for integrative analysis of expression profiles. Bioinformatics. 2003;19:2321–2322. doi: 10.1093/bioinformatics/btg307. [DOI] [PubMed] [Google Scholar]
- Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Analytical Chemistry. 2009;81:6656–6667. doi: 10.1021/ac901536h. [DOI] [PubMed] [Google Scholar]
- Fedrigo M, Hoeschen C, Oeh U. Multidimensional statistical analysis of PTR-MS breath samples: A test study on irradiation detection. International J Mass Spectrometry. 2010;295:13–20. [Google Scholar]
- Feng Z, Levine AJ. The regulation of energy metabolism and the IGF-1/mTOR pathways by the p53 protein. Trends Cell Biol. 2010;20:427–434. doi: 10.1016/j.tcb.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O. Metabolomics–the link between genotypes and phenotypes. Plant Mol Biol. 2002;48:155–171. [PubMed] [Google Scholar]
- Garty G, Chen YH, Salerno A, Turner H, Zhang J, Lyulko O, Bertucci A, Xu YP, Wang HL, Simaan N, et al. The Rabit: A Rapid Automated Biodosimetry Tool for Radiological Triage. Health Physics. 2010;98:209–217. doi: 10.1097/HP.0b013e3181ab3cb6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodsaid FM, Amur S, Aubrecht J, Burczynski ME, Carl K, Catalano J, Charlab R, Close S, Cornu-Artis C, Essioux L, et al. Voluntary exploratory data submissions to the US FDA and the EMA: experience and impact. Nat Rev Drug Discov. 2010;9:435–445. doi: 10.1038/nrd3116. [DOI] [PubMed] [Google Scholar]
- Halket JM, Waterman D, Przyborowska AM, Patel RK, Fraser PD, Bramley PM. Chemical derivatization and mass spectral libraries in metabolic profiling by GC/MS and LC/MS/ MS. J Exp Bot. 2005;56:219–243. doi: 10.1093/jxb/eri069. [DOI] [PubMed] [Google Scholar]
- Hall EJ, Giaccia AJ. Radiobiology for the Radiologist. 2006 [Google Scholar]
- Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci. 2008;13:3046–3082. doi: 10.2741/2909. [DOI] [PubMed] [Google Scholar]
- Hei TK, Zhou H, Ivanov VN, Hong M, Lieberman HB, Brenner DJ, Amundson SA, Geard CR. Mechanism of radiationinduced bystander effects: a unifying model. Journal of Pharmacy and Pharmacology. 2008;60:943–950. doi: 10.1211/jpp.60.8.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofseth LJ, Hussain SP, Harris CC. p53: 25 years after its discovery. Trends Pharmacol Sci. 2004;25:177–181. doi: 10.1016/j.tips.2004.02.009. [DOI] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A Large Intergenic Noncoding RNA Induced by p53 Mediates Global Gene Repression in the p53 Response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issaq HJ, Abbott E, Veenstra TD. Utility of separation science in metabolomic studies. Journal of Separation Science. 2008;31:1936–1947. doi: 10.1002/jssc.200700601. [DOI] [PubMed] [Google Scholar]
- Kannan K, Amariglio N, Rechavi G, Jakob-Hirsch J, Kela I, Kaminski N, Getz G, Domany E, Givol D. DNA microarrays identification of primary and secondary target genes regulated by p53. Oncogene. 2001;20:2225–2234. doi: 10.1038/sj.onc.1204319. [DOI] [PubMed] [Google Scholar]
- Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- Katajamaa M, Oresic M. Data processing for mass spectrometry-based metabolomics. Journal of Chromatography A. 2007:318–328. doi: 10.1016/j.chroma.2007.04.021. [DOI] [PubMed] [Google Scholar]
- Kertesz V, Van Berkel GJ. Liquid microjunction surface sampling coupled with high-pressure liquid chromatographyelectrospray ionization-mass spectrometry for analysis of drugs and metabolites in whole-body thin tissue sections. Analytical Chemistry. 2010;82:5917–5921. doi: 10.1021/ac100954p. [DOI] [PubMed] [Google Scholar]
- Khanna V, Ranganathan S. Physiochemical property space distribution among human metabolites, drugs and toxins. BMC Bioinformatics. 2009;10(Suppl 15):S10. doi: 10.1186/1471-2105-10-S15-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Aronov P, Zakharkin SO, Anderson D, Perroud B, Thompson IM, Weiss RH. Urine metabolomics analysis for kidney cancer detection and biomarker discovery. Mol Cell Proteomics. 2009;8:558–570. doi: 10.1074/mcp.M800165-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa R, Kastan MB. The ATM-dependent DNA damage signaling pathway. Cold Spring Harbor Symposia on Quantitative Biology. 2005;70:99–109. doi: 10.1101/sqb.2005.70.002. [DOI] [PubMed] [Google Scholar]
- Krylov EV, Coy SL, Vandermey J, Schneider BB, Covey TR, Nazarov EG. Selection and generation of waveforms for differential mobility spectrometry. Rev Sci Instrum. 2010;81:024101. doi: 10.1063/1.3284507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylov EV, Nazarov EG. Electric field dependence of the ion mobility. International J of Mass Spectrometry. 2009;285:149–156. [Google Scholar]
- Ku WW, Aubrecht J, Mauthe RJ, Schiestl RH, Fornace AJ., Jr Why Not Start With a Single Test: A Transformational Alternative to Genotoxicity Hazard and Risk Assessment. Toxicol Sci. 2007;99:20–25. doi: 10.1093/toxsci/kfm147. [DOI] [PubMed] [Google Scholar]
- Laiakis EC, Morris GA, Fornace AJ, Jr, Howie SR. Metabolomic analysis in severe childhood pneumonia in the gambia, west Africa: findings from a pilot study. PLoS ONE. 2010;5:e12655. doi: 10.1371/journal.pone.0012655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz C, Patterson AD, Slavik J, Krausz KW, Ledermann M, Gonzalez FJ, Idle JR. Radiation metabolomics. 3. Biomarker discovery in the urine of gamma-irradiated rats using a simplified metabolomics protocol of gas chromatography- mass spectrometry combined with random forests machine learning algorithm. Radiation Research. 2009;172:198–212. doi: 10.1667/RR1796.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DS, Vouros P, Miller RA, Nazarov EG, Morris JC. Characterization of gas-phase molecular interactions on differential mobility ion behavior utilizing an electrospray ionization-differential mobility-mass spectrometer system. Analytical Chemistry. 2006;78:96–106. doi: 10.1021/ac051217k. [DOI] [PubMed] [Google Scholar]
- Li HH, Aubrecht J, Fornace AJ., Jr Toxicogenomics: overview and potential applications for the study of non-covalent DNA interacting chemicals. Mutat Res. 2007;623:98–108. doi: 10.1016/j.mrfmmm.2007.03.013. [DOI] [PubMed] [Google Scholar]
- Lin Y, Schiavo S, Orjala J, Vouros P, Kautz R. Microscale LC-MS-NMR platform applied to the identification of active cyanobacterial metabolites. Analytical Chemistry. 2008;80:8045–8054. doi: 10.1021/ac801049k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorimore SA, Coates PJ, Wright EG. Radiation-induced genomic instability and bystander effects: inter-related non-targeted effects of exposure to ionizing radiation. Oncogene. 2003;22:7058–7069. doi: 10.1038/sj.onc.1207044. [DOI] [PubMed] [Google Scholar]
- Lowe JM, Cha H, Yang Q, Fornace AJ., Jr Nuclear factor-kappaB (NF-kappaB) is a novel positive transcriptional regulator of the oncogenic Wip1 phosphatase. Journal of Biological Chemistry. 2010;285:5249–5257. doi: 10.1074/jbc.M109.034579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z, Zang M, Guo W. AMPK as a metabolic tumor suppressor: control of metabolism and cell growth. Future Oncol. 2010;6:457–470. doi: 10.2217/fon.09.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutgens LC, Deutz NE, Gueulette J, Cleutjens JP, Berger MP, Wouters BG, von Meyenfeldt MF, Lambin P. Citrulline: a physiologic marker enabling quantitation and monitoring of epithelial radiation-induced small bowel damage. International Journal of Radiation Oncology, Biology, Physics. 2003;57:1067–1074. doi: 10.1016/s0360-3016(03)00781-8. [DOI] [PubMed] [Google Scholar]
- Manard MJ, Trainham R, Weeks S, Coy SL, Krylov EV, Nazarov EG. Differential mobility spectrometry/mass spectrometry: The design of a new mass spectrometer for real-time chemical analysis in the field. International J Mass Spectrometry. 2010;295:138–144. [Google Scholar]
- McLean JA. The mass-mobility correlation redux: the conformational landscape of anhydrous biomolecules. J Am Soc Mass Spectrom. 2009;20:1775–1781. doi: 10.1016/j.jasms.2009.06.016. [DOI] [PubMed] [Google Scholar]
- Meek DW, Anderson CW. Posttranslational modification of p53: cooperative integrators of function. Cold Spring Harb Perspect Biol. 2009;1:a000950. doi: 10.1101/cshperspect.a000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey BN, Kumar A, Tiwari P, Mishra KP. Radiobiological basis in management of accidental radiation exposure. International Journal of Radiation Biology. 2010;86:613–635. doi: 10.3109/09553001003746059. [DOI] [PubMed] [Google Scholar]
- Parizek J, Arient M, Dienstbier Z, Skoda J. Deoxycytidine in urine as an indicator of changes after irradiation. Nature. 1958;182:721–722. doi: 10.1038/182721a0. [DOI] [PubMed] [Google Scholar]
- Patterson AD, Gonzalez FJ, Idle JR. Xenobiotic Metabolism: A View through the Metabolometer. Chemical Research in Toxicology. 2010a;23:851–860. doi: 10.1021/tx100020p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson AD, Idle JR. A metabolomic perspective of small molecule toxicity. In: Ballantyne TCMB, Syversen T, editors. General and Applied Toxicology. Chicheste, UK: John Wiley & Sons; 2009. pp. 18–44. [Google Scholar]
- Patterson AD, Lanz C, Gonzalez FJ, Idle JR. The role of mass spectrometry-based metabolomics in medical counter-measures against radiation. Mass Spectrom Rev. 2010b;29:503–521. doi: 10.1002/mas.20272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson AD, Li H, Eichler GS, Krausz KW, Weinstein JN, Fornace AJ, Jr, Gonzalez FJ, Idle JR. UPLC-ESI-TOFMS- based metabolomics and gene expression dynamics inspector self-organizing metabolomic maps as tools for understanding the cellular response to ionizing radiation. Analytical Chemistry. 2008;80:665–674. doi: 10.1021/ac701807v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Amundson SA. Development of gene expression signatures for practical radiation biodosimetry. International Journal of Radiation Oncology, Biology, Physics. 2008;71:1236–1244. doi: 10.1016/j.ijrobp.2008.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramautar R, van der Plas AA, Nevedomskaya E, Derks RJ, Somsen GW, de Jong GJ, van Hilten JJ, Deelder AM, Mayboroda OA. Explorative analysis of urine by capillary electrophoresis-mass spectrometry in chronic patients with complex regional pain syndrome. J Proteome Res. 2009;8:5559–5567. doi: 10.1021/pr900651k. [DOI] [PubMed] [Google Scholar]
- Rohwer RG. Scrapie infectious agent is virus-like in size and susceptibility to inactivation. Nature. 1984;308:658–662. doi: 10.1038/308658a0. [DOI] [PubMed] [Google Scholar]
- Rosette C, Karin M. Ultraviolet light and osmotic stress: activation of the JNK cascade through multiple growth factor and cytokine receptors. Science. 1996;274:1194–1197. doi: 10.1126/science.274.5290.1194. [DOI] [PubMed] [Google Scholar]
- Salminen A, Kaarniranta K. Glycolysis links p53 function with NF-kappaB signaling: impact on cancer and aging process. Journal of Cellular Physiology. 2010;224:1–6. doi: 10.1002/jcp.22119. [DOI] [PubMed] [Google Scholar]
- Sanli T, Rashid A, Liu C, Harding S, Bristow RG, Cutz JC, Singh G, Wright J, Tsakiridis T. Ionizing radiation activates AMP-activated kinase (AMPK): a target for radiosensitization of human cancer cells. International Journal of Radiation Oncology, Biology, Physics. 2010;78:221–229. doi: 10.1016/j.ijrobp.2010.03.005. [DOI] [PubMed] [Google Scholar]
- Santini MT, Romano R, Rainaldi G, Ferrante A, Motta A , Indovina PL. Increases in 1H-NMR mobile lipids are not always associated with overt apoptosis: evidence from MG-63 human osteosarcoma three-dimensional spheroids exposed to a low dose (2 Gy) of ionizing radiation. Radiation Research. 2006;165:131–141. doi: 10.1667/rr3500.1. [DOI] [PubMed] [Google Scholar]
- Schauer N, Steinhauser D, Strelkov S, Schomburg D, Allison G, Moritz T, Lundgren K, Roessner-Tunali U, Forbes MG, Willmitzer L, et al. GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Letters. 2005;579:1332–1337. doi: 10.1016/j.febslet.2005.01.029. [DOI] [PubMed] [Google Scholar]
- Schieven GL. The biology of p38 kinase: a central role in inflammation. Curr Top Med Chem. 2005;5:921–928. doi: 10.2174/1568026054985902. [DOI] [PubMed] [Google Scholar]
- Schneider BB, Covey TR, Coy SL, Krylov EV, Nazarov EG. Control of chemical effects in the separation process of a differential mobility mass spectrometer system. Eur J Mass Spectrom (Chichester, Eng) 2010a;16:57–71. doi: 10.1255/ejms.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider BB, Covey TR, Coy SL, Krylov EV, Nazarov EG. Planar differential mobility spectrometer as a pre-filter for atmospheric pressure ionization mass spectrometry. Int J Mass Spectrom. 2010b;298:45–54. doi: 10.1016/j.ijms.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider BB, Covey TR, Coy SL, Krylov EV, Nazarov EG. Chemical effects in the separation process of a differential mobility/mass spectrometer system. Analytical Chemistry. 2010c;82:1867–1880. doi: 10.1021/ac902571u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JG, Finck BN, Ren J, Standley KN, Takagi M, Maclean KH, Bernal-Mizrachi C, Muslin AJ, Kastan MB, Semenkovich CF. ATM-dependent suppression of stress signaling reduces vascular disease in metabolic syndrome. Cell Metab. 2006;4:377–389. doi: 10.1016/j.cmet.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Schneidkraut MJ, Kot PA, Ramwell PW, Rose JC. Thromboxane and prostacyclin synthesis following whole body irradiation in rats. J Appl Physiol. 1984;57:833–838. doi: 10.1152/jappl.1984.57.3.833. [DOI] [PubMed] [Google Scholar]
- Schuler M, Green DR. Transcription, apoptosis and p53: catch-22. Trends in Genetics. 2005;21:182–187. doi: 10.1016/j.tig.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Smith CA, O’Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- Smith ML, Fornace AJ., Jr The two faces of tumor suppressor p53. American Journal of Pathology. 1996;148:1019–1022. [PMC free article] [PubMed] [Google Scholar]
- Spratlin JL, Serkova NJ, Eckhardt SG. Clinical applications of metabolomics in oncology: a review. Clin Cancer Res. 2009;15:431–440. doi: 10.1158/1078-0432.CCR-08-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stancevic B, Kolesnick R. Ceramide-rich platforms in transmembrane signaling. FEBS Letters. 2010;584:1728–1740. doi: 10.1016/j.febslet.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadjimukhamedov FK, Jackson AU, Nazarov EG, Ouyang Z, Cooks RG. Evaluation of a differential mobility spectrometer/ miniature mass spectrometer system. J Am Soc Mass Spectrom. 2010;21:1477–1481. doi: 10.1016/j.jasms.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Tyburski JB, Patterson AD, Krausz KW, Slavik J, Fornace AJJ, Gonzalez FJ, Idle JR. Radiation metabolomics. 1. Identification of minimally invasive urine biomarkers for gamma- radiation exposure in mice. Radiation Research. 2008;170:1–14. doi: 10.1667/RR1265.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyburski JB, Patterson AD, Krausz KW, Slavik J, Fornace AJ, Jr, Gonzalez FJ, Idle JR. Radiation metabolomics. 2. Dose- and time-dependent urinary excretion of deaminated purines and pyrimidines after sublethal gamma-radiation exposure in mice. Radiation Research. 2009;172:42–57. doi: 10.1667/RR1703.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valerie K, Yacoub A, Hagan MP, Curiel DT, Fisher PB, Grant S, Dent P. Radiation-induced cell signaling: inside-out and outside-in. Mol Cancer Ther. 2007;6:789–801. doi: 10.1158/1535-7163.MCT-06-0596. [DOI] [PubMed] [Google Scholar]
- Van Berkel GJ, Kertesz V. Application of a Liquid Extraction Based Sealing Surface Sampling Probe for Mass Spectrometric Analysis of Dried Blood Spots and Mouse Whole-Body Thin Tissue Sections. Analytical Chemistry. 2009;81:9146–9152. doi: 10.1021/ac901712b. [DOI] [PubMed] [Google Scholar]
- van den Berg RA, Hoefsloot HC, Westerhuis JA, Smilde AK, van der Werf MJ. Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics. 2006;7:142. doi: 10.1186/1471-2164-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese RS, Cheema A, Cheema P, Bourbeau M, Tuli L, Zhou B, Jung M, Dritschilo A, Ressom HW. Analysis of LC-MS data for characterizing the metabolic changes in response to radiation. J Proteome Res. 2010;9:2786–2793. doi: 10.1021/pr100185b. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer. 2009;9:691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- Want EJ, Cravatt BF, Siuzdak G. The expanding role of mass spectrometry in metabolite profiling and characterization. Chembiochem. 2005;6:1941–1951. doi: 10.1002/cbic.200500151. [DOI] [PubMed] [Google Scholar]
- Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, et al. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Research. 2009;37:D603–D610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S, et al. HMDB: the Human Metabolome Database. Nucleic Acids Research. 2007;35:D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfram RM, Palumbo B, Chehne F, Palumbo R, Budinsky AC, Sinzinger H. (Iso) Prostaglandins in saliva indicate oxidation injury after radioiodine therapy. Rev Esp Med Nucl. 2004;23:183–188. doi: 10.1016/s0212-6982(04)72279-x. [DOI] [PubMed] [Google Scholar]
- Yamaoka K, Obata T, Iriyama K, Iwasaki T, Hoshi Y. Simultaneous quantitative analysis of prostaglandins and thromboxane after low-dose X irradiation. Radiation Research. 1998;149:103–106. [PubMed] [Google Scholar]
- Yin P, Zhao X, Li Q, Wang J, Li J, Xu G. Metabonomics study of intestinal fistulas based on ultraperformance liquid chromatography coupled with Q-TOF mass spectrometry (UPLC/Q-TOF MS) J Proteome Res. 2006;5:2135–2143. doi: 10.1021/pr060256p. [DOI] [PubMed] [Google Scholar]
- Zhang XD, Qin ZH, Wang J. The role of p53 in cell metabolism. Acta Pharmacol Sin. 2010;31:1208–1212. doi: 10.1038/aps.2010.151. [DOI] [PMC free article] [PubMed] [Google Scholar]