Abstract
Isothermal Titration Calorimetry, ITC, is a powerful technique that can be used to estimate a complete set of thermodynamic parameters (e.g. Keq (or ΔG), ΔH, ΔS, and n) for a ligand binding interaction described by a thermodynamic model. Thermodynamic models are constructed by combination of equilibrium constant, mass balance, and charge balance equations for the system under study. Commercial ITC instruments are supplied with software that includes a number of simple interaction models, for example one binding site, two binding sites, sequential sites, and n-independent binding sites. More complex models for example, three or more binding sites, one site with multiple binding mechanisms, linked equilibria, or equilibria involving macromolecular conformational selection through ligand binding need to be developed on a case by case basis by the ITC user. In this paper we provide an algorithm (and a link to our MATLAB program) for the non-linear regression analysis of a multiple binding site model with up to four overlapping binding equilibria. Error analysis demonstrates that fitting ITC data for multiple parameters (e.g. up to nine parameters in the three binding site model) yields thermodynamic parameters with acceptable accuracy.
Keywords: ITC, calorimetry, fitting, analysis, non-linear regression, three binding site model, multiple binding sites
Introduction
Titration calorimetry has been used for the simultaneous determination of K and ΔH for more than 40 years [1-4]. Isothermal titration calorimetry is now routinely used to directly characterize the thermodynamics of biopolymer binding interactions [5-13]. Knowledge of the thermodynamic profiles for drug-receptor binding interactions greatly enhances drug design and development [14-17]. ITC instruments (available from GE Healthcare (Microcal) and TA Instruments (Calorimetry Sciences)) have adequate sensitivity to measure heat effects as small as 0.1 μcal making it possible to directly determine binding constants as large as 108 to 109 M−1. Even larger values for K may be estimated from competitive binding experiments [18, 19].
To take full advantage of the powerful ITC technique, the user must be able to design the optimum experiment, understand the nonlinear fitting process, and appreciate the uncertainties in the fitting parameters K, ΔH, and n. ITC experiment design and data analysis have been the subject of numerous publications [5, 6, 14-24]. Recent reviews of isothermal titration calorimetry describe the ease of use of modern microcalorimeters [18, 19, 25]. Several papers have described modern uses of isothermal titration calorimetry to study a broad range of chemical equilibria in numerous ways [26-28]. For example, ITC studies are now being used to identify possible binding mechanisms for ligand/DNA complexes based on their thermodynamic signatures [29]. ITC experiments exploring iron binding to E. coli Ferritin were accompanied by a model describing the equations for three independent binding sites [30] while ITC studies of histone nucleoplasm interactions were best fit with a site-specific cooperative model including four equilibrium constants and four enthalpy changes [31]. Examples of the use of ITC experiments to unravel the complicated binding equilibria often occurring in biology are limited since the analysis tools provided by the ITC industry cover only the simplest cases.
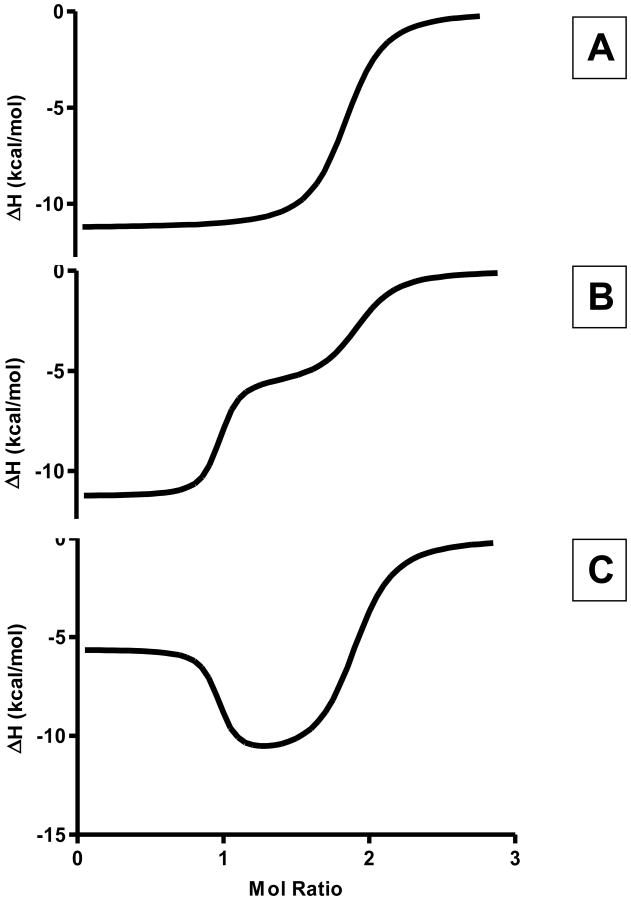
The improved sensitivity of the current ITC instruments has resulted in the ability to accurately estimate thermodynamic parameters for multiple overlapping binding equilibria. Figure 1 shows three unique thermograms that might result from the titration of a system exhibiting two overlapping binding processes. Figures 1B and 1C were simulated from a model for a system having two binding processes, wherein K1 is much greater than K2 and ΔH1 is not equal to ΔH2. These two thermograms show a clear distinction between the first and second binding process, which can be differentiated when fitting data based on the differences in enthalpy change. However, Figure 1A also demonstrates a simulation for a system exhibiting two binding sites in which the binding affinities have the same relative magnitude as the previous cases, K1 ≫ K2, but with ΔH1 equal to ΔH2. In this case, it is not possible to distinguish the two overlapping equilibria and only the weaker binding process (K2) can be modeled. The equivalence of the binding enthalpy changes for the two overlapping processes causes this system to be modeled as a single binding process with n=2.
Figure 1.
Representative two-site ITC titration experiment thermograms. In all three cases, the stoichiometry demonstrates that two ligand molecules are binding to the receptor molecule. The top panel represents a system wherein the enthalpy change for binding of both ligands are experimentally indistinguishable. The middle panel represents a system where the binding site with higher affinity is accompanied by a more exothermic enthalpy change. The bottom panel represents a system where the higher binding affinity site demonstrates a less exothermic enthalpy change than the lower binding affinity site.
The three plots shown in Figure 1 demonstrate the increased complexity that is often seen in ITC thermograms as the number of binding process is increased. ITC thermograms must be understood prior to modeling the data. Understanding the binding profiles ensures that the models being applied to the data give an accurate representation of the chemical equilibria being observed. Furthermore, error analysis of algorithms being applied to analyze ITC thermograms ensures best-fit solutions are accurate. Several papers have been published on methods for evaluating the error introduced into ITC results from either experimental or data fitting considerations [33-40]. In general, the potential for using the ITC method to estimate the number of parameters needed to describe more complex thermodynamic binding models is underappreciated.
In this work, we describe the construction of algorithms that can be used to model ITC data obtained on systems exhibiting three or more binding sites described with overlapping equilibrium constants. We have also used simulated data and the Monte Carlo method [32, 37, 40] to evaluate the uncertainties and cross correlation in the binding parameters (K, ΔH, and n). One and two site systems are used to compare our non-linear fitting routines to those in the commercially available program from GE Healthcare (Microcal, Northampton, MA, Origin 7.0). Simulated data are used to evaluate the three binding site algorithm implemented in MATLAB (MATLAB version R2012a, Natick, MA: The MathWorks Inc., 2012.) for the accuracy of multiple parameters (K1-3, ΔH1-3, and n1-3) determined in an ITC experiment. The analysis techniques described in this paper can be extended to ITC data obtained on other complex systems if the user can construct the appropriate thermodynamic model for the binding interactions and/or linked equilibria. One novel aspect of this work is that our n-sites program for the analysis of systems having up to 4 binding sites is described in detail in the supplementary information section and the fully functional analysis program can be downloaded from our website (http://lewis.chemistry.msstate.edu/download.html)
Materials and Methods
Multiple Binding Site
Algorithms for modeling ITC thermograms demonstrating up to four overlapping binding processes were developed using MATLAB software. The algorithms model the binding affinity, Kj, the molar enthalpy change, ΔHj, and the total stoichiometric ratio, nj, for each of the j = 1 to 4 binding process using non-linear regression techniques. The mass balance and equilibrium equations were manipulated to produce an (n+1)th degree polynomial for the n-binding sites, with free ligand as the indeterminate.
General Binding Site Model
The thermodynamic model algorithms were developed from combination of the appropriate mass balance and equilibrium constant expressions. Equations 1 and 2 are the generalized mass balance and binding equilibrium equations, respectively. Each equation is written in a simplified form that can be expanded to include n-binding site.
| (1) |
| (2) |
Equation 1 establishes that at any point in the titration, the total ligand present in the reaction vessel (Lt) must be either bound to one of the n-binding sites, ( ), or 1 free in the reaction vessel ([L]). The equilibrium constants in Equation 2 have been rewritten to express the fraction of process j bound (Θj) as a function of the binding affinity, Kj, and the presence of free ligand. Substitution of Θj into Equation 1 and expanding yields a (n+1)th degree polynomial where [L] is the indeterminate.
| (3) |
Equation 3 represents a simplified form of this polynomial, where αi represents the ith coefficient. The coefficients, αi, are derived from ligand concentration, macromolecule concentration, stoichiometric ratios, and binding affinities. Calculating the roots of the polynomial shown in Equation 3 determines the concentration of free ligand present in the reaction vessel after the ith injection.
Once the concentration of free ligand is determined, substitution of [L] into Equation 2 yields the fraction of site n bound after the ith injection. The total heat produced from the start of the titration through the ith injection, Qi, can be calculated using Equation 4,
| (4) |
| (5) |
where, Vo is the active calorimeter cell volume, and where is the total reaction heat calculated as the sum of the heat produced in all of the overlapping binding reactions from the start of the titration through the ith injection. Equation 5 is then used to calculate the differential heat produced during the ith injection, ΔQi, where Qi and Qi−1 represent the total heats produced from the start of the titration through injection points i and i−1, and Mt is the macromolecule concentration in the reaction vessel (calorimeter cell) as corrected for dilution and for displacement from the calorimeter cell as titrant is added. Equation 5 assumes that all reaction heat is sensed by the calorimeter and that no heat is lost due to heat losses up the fill tube or to reactions occurring outside of the calorimeter cell and thus incompletely sensed by the calorimeter. ΔQ can be changed to include heat produced from reactions occurring outside of the reaction vessel. One such assumption is that the heat produced outside of the calorimeter cell (i.e. in the fill tube) is only measured with half the efficiency as that inside of the reaction vessel. A correction term for the heat being produced outside of the reaction vessel can be easily introduced into the fitting algorithms.
The algorithms used here introduced a correction term for the heat being produced outside of the reaction vessel by restricting the concentration of macromolecule (Mt) and the concentration of the ligand (Lt) after each injection. Equations 6 and 7 are the iterative functions used to determine the concentration of macromolecule and ligand after the ith injection. Mt,i and Lt,i are the concentration of macromolecule and ligand, respectively, after the ith injection, Li is the concentration of ligand being injected into the cell, Vi is the injection volume, Vo is the active cell volume. The initial conditions for Mt and Xt are Mt,0 = Mi and Xt,0 = 0, where Mi is the concentration of macromolecule loaded into the cell initially.
| (6) |
| (7) |
Equations 6 and 7 were constructed to accurately mimic the concentrations of ligand and macromolecule present in the active cell volume of the calorimeter. After each injection, the macromolecule concentration is lowered by the amount of macromolecule leaving the active cell volume. Similarly, the concentration of ligand is increased after each injection, but after the initial injection, a correction is also made for the amount of ligand that leaves the active cell volume. These equations assume that the stirring rate is efficient enough that the ligand is accurately mixed into the active cell volume directly after the injection.
The difference between the heats calculated for ΔQi (Equation 5) and those measured during an ITC experiment were used to determine the goodness-of-fit. A Levenberg-Marquardt nonlinear regression model common to MATLAB, was used to determine best-fit parameters. (No other minimization routines were tried.) By allowing the model to iterate over the parameters (K1 …, Kn, n1 …, nn, ΔH1 …, ΔHn), a solution is found such that the χ2 merit function is minimized by steepest descent and quadratic minimization.
Algorithm Comparison
To compare the MATLAB and Origin 7.0 non-linear regression analysis programs, the same simulated data sets were fit with both programs. Each set of simulated data was generated using 25 injections of 5 μL with a ligand concentration of 1 mM and a macromolecule concentration of 40 μM at 298 K. Randomly generated normally distributed noise, 0.1 μcal, was added to the simulated thermograms prior to fitting with either Origin 7.0 or MATLAB routines. Solutions resulting from the MATLAB algorithms developed two binding site model were compared to solutions obtained using Microcal Origin 7.0 to ensure that the resulting best-fit parameters were in agreement between the two non-linear regression analysis programs. Simulated ITC thermograms were created for two different cases: two binding sites, and three binding sites. The parameters used to simulate ITC thermograms and the resulting best-fit parameters for the two binding site model are given in Table 1. The parameters used to simulate ITC test data for the three binding site case are given in Table 2, where the best-fit parameters are only listed for the MATLAB program. (Origin 7.0 is limited to solving equilibria for two or fewer reactions although the sequential model in Origin could be transformed and used to simulate thermograms for three independent sites.)
Table 1.
Parameters used for the generation of two-competitive processes simulated ITC titration data. Best-fit parameters (K1, ΔH1, n1, K2, ΔH2, and n2 values) are listed for the Origin 7.0 and MATLAB solutions for the non-linear regression analysis of the three two-competitive processes test cases. The comparison shows that both programs result in the same best-fit parameters. Average best fit parameters are also listed for 100 Monte Carlo analyses of the Case 2 two site test data using both algorithms. The standard deviations are also listed for the six average parameters for both methods.
| Two-Competitive-Site Model: ITC Test Data Simulation Parameters | |||
|---|---|---|---|
| Parameter | Case 1 | Case 2 | Case 3 |
| K1 | 2.00×106 | 2.00×108 | 2.00×108 |
| ΔH1 (kcal/mol) | −9.000 | −12.000 | −3.000 |
| n1 | 1.000 | 1.000 | 1.000 |
| K2 | 1.00×105 | 1.00×106 | 1.00×106 |
| ΔH2 (kcal/mol) | −3.000 | −5.000 | −12.000 |
| n2 | 1.000 | 1.000 | 1.000 |
| Two-Competitive-Site Model: Best Fit Parameters | ||||||
|---|---|---|---|---|---|---|
| Case 1 | Case 2 | Case 3 | ||||
| Parameter | ORIGIN | MATLAB | ORIGIN | MATLAB | ORIGIN | MATLAB |
| K | 2.00×106 | 1.98×106 | 2.02×108 | 1.99×108 | 2.04×108 | 2.02×108 |
| ΔH1 (kcal/mol) | −8.99 | −8.94 | −12.01 | −11.93 | −2.99 | −2.98 |
| n1 | 0.99 | 1.00 | 0.99 | 1.00 | 0.99 | 1.00 |
| K2 | 0.99×105 | 0.98×105 | 0.94×106 | 0.98×106 | 1.02×106 | 1.00×106 |
| ΔH2 (kcal/mol) | −3.12 | −3.09 | −5.01 | −4.97 | −12.03 | −11.93 |
| n2 | 0.98 | 0.99 | 0.99 | 1.00 | 0.99 | 1.00 |
| Case 2: Two-Competitive-Site Model: Average Monte Carlo Parameters & Std. Dev.s | ||||
|---|---|---|---|---|
| ORIGIN | MATLAB | |||
| Parameter | Average | Std. Dev. | Average | Std. Dev. |
| K1 | 1.991×108 | 0.068 ×108 | 1.944×108 | 0.066 ×108 |
| ΔH1 (kcal/mol) | −12.008 | ±0.001 | −11.950 | ±0.001 |
| n1 | 1.005 | ±0.001 | 1.010 | ±0.001 |
| K2 | 0.995×106 | ±0.003×106 | 0.990×106 | ±0.003×106 |
| ΔH2 (kcal/mol) | −5.004 | ±0.002 | −4.950 | ±0.002 |
| n2 | 1.005 | ±0.001 | 1.013 | ±0.001 |
Table 2.
Parameters used for the generation of simulated ITC titration data for the three-competitive processes model. The simulated data were used in the Monte Carlo analysis of the three-competitive processes fitting algorithm. Resulting Monte Carlo mean parameter values and 95% confidence intervals determined for each of the nine parameters (K1-3, ΔH1-3, and n1-3) obtained as a solution for the three-competitive processes model with the MATLAB code given in the supplemental materials. They Monte Carlo analysis demonstrates that all of the three-competitive processes model parameters are well defined and have acceptable error. A correlation between n and ΔH was observed. The 95% confidence intervals show that the equilibrium constants, molar enthalpy changes, and stoichiometric ratios are well determined for all three processes. The binding affinities for each process are slightly overestimated and the third-process molar enthalpy change, ΔH3, and stoichiometry, n3, exhibit the largest uncertainties.
| Three Competitive Sites Model: Simulation Parameters | |||
|---|---|---|---|
| Model Parameter | Site 1 | Site 2 | Site 3 |
| Ki (M−1) | 1.0×108 | 1.0×106 | 1.0×104 |
| ΔHi (kcal/mol) | −12.0 | −8.0 | −4.0 |
| ni | 1.0 | 1.0 | 1.0 |
| Three Sites Binding Algorithm: Monte Carlo Analysis “Best Fit” Parameters | |||
| 95 % Confidence Intervals | |||
| Model Parameter | Monte Carlo Mean | minimum | maximum |
| K1 (M−1) | 1.06×108 | 1.02×108 | 1.09×108 |
| K2 (M−1) | 1.06×106 | 1.02×106 | 1.10×106 |
| K3 (M−1) | 1.11×104 | 1.03×104 | 1.19×104 |
| ΔH1 (kcal/mol) | −12.00 | −11.999 | −12.001 |
| ΔH2 (kcal/mol) | −7.99 | −7.993 | −8.002 |
| ΔH3 (kcal/mol) | −4.24 | −4.147 | −4.339 |
| n1 | 0.99 | 0.999 | 1.000 |
| n2 | 0.99 | 0.999 | 1.000 |
| n3 | 1.08 | 1.052 | 1.101 |
Monte Carlo Analysis
Monte Carlo analysis [32, 37, 40] was used to estimate the uncertainties for each of the nine parameters (K1-3, ΔH1-3, n1-3) required to fit the three binding site model. The Monte Carlo analysis involved the creation of a perfect data set with the MATLAB algorithm. This perfect data set was then used to create 1000 virtual data sets by adding random normally distributed noise to each data point in the perfect data set. The ITC instrument noise was taken to be ± 0.1 μcal per injection [18]. The MATLAB algorithm was then used to fit each of the 1000 virtual ITC experiments and to obtain a set of best-fit parameters for each of the virtual experiments. The mean value for each of the nine fitting parameters was then calculated from the 1000 sets of best-fit values obtained in the Monte Carlo procedure.
Error Analysis
Two-dimensional error plots for each of the nine parameters involved in modeling the three binding sites were generated. The error plot for parameter X was established by fixing the value of X and allowing all other parameters to be iterated until a best-fit solution was found. Fixed values for parameter X were chosen such that Δ Log (X) = Log (X) - Log (perfect value) had a range of −3 to 3. The error plot was produced by plotting Δ Log (Parameter) versus Log (Error). Transformation of the data using logs condenses the data and exaggerates the local and global minima being observed. These error plots were used to evaluate the interdependence of the parameters and to establish the range of acceptable initial parameter guess values required in modeling three binding site.
Results
Algorithm Comparisons
ITC experiments were simulated for multiple two site models. These simulated data (including random noise) were fit using both Origin 7.0 and the MATLAB algorithms. This comparison was done to ensure that our MATLAB algorithms yielded a solution that was in agreement with the solution obtained with Origin 7.0. The best-fit parameters obtained with both programs, for all of the simulated ITC experiments, are in excellent agreement with each other and returned parameters close to the parameters used to create the simulated ITC data sets.
Monte Carlo Analysis
A Monte Carlo analysis was used to estimate the uncertainty in each of the thermodynamic parameters determined in modeling simulated ITC data for a system exhibiting three binding sites. Because modeling a three-process system is not currently implemented in any of the commercially available ITC data analysis programs, a rigorous determination of the uncertainties in the best-fit parameters was conducted in lieu of a direct comparison with other approaches or solutions.
Table 2 gives the mean values and 95% confidence intervals for each parameter estimated by the Monte Carlo method. Comparison of the mean Monte Carlo parameters with the test data simulation parameters (shown at the top of Table 2), demonstrates that the MATLAB algorithm finds a solution minimum in which all nine of the binding parameters are well determined. As seen from Table 2, each of the binding constants (K1, K2, K3) have 95% confidence intervals that are slightly skewed to higher values than the values used to simulate the ITC data. Overestimating the binding constants is typical for binding algorithms, and can be attributed to the number of data points in the slope of the thermogram and the interdependence of the binding affinity on the other parameters. The molar enthalpy changes and stoichiometric ratios for processes 1 and 2 have 95% confidence intervals that are equally distributed around the noiseless values. This indicates that ΔH1, n1, ΔH2, and n2 are determined with the greatest accuracy. Because the stoichiometric ratios are well determined for the two processes with the higher affinities and larger heats, the algorithms correctly identify the solutions for accurate estimates of ΔH1, n1, ΔH2, and n2. However, the 95% confidence interval for ΔH3 (and/or n3) demonstrates that these best-fit parameters contain the largest error of the nine model parameters. ΔH3 is skewed toward more exothermic values. This error results from the inability of the binding algorithm to determine an accurate endpoint for the third process where the heat signal and integrated heats between successive injections is small.
Error Surfaces
A two-dimensional error plot was created for each of the nine parameters required to fit the three binding sites model. MATLAB was used to fix the value of the parameter of interest while allowing all other parameters to be optimized by nonlinear regression. The error plots were constructed by plotting Δ Log (X) versus Log (error), where Δ Log (X) = Log (X) - Log (noiseless value). The error plots demonstrate the interdependence of each parameter, shown by the presence of multiple local minima in several of the parameters. However, the parameters are all found to have global minima at Δ Log (X) equal to zero. This implies that the algorithm finds the correct minima when the starting parameters are adequately set.
A typical parameter 2-D error plot is shown in Figure 2. This figure shows the plot of Δ Log (K2) versus Log (error), where Δ Log (K2) = Log (K2) − Log(1×106). As can be seen from the K2 error plot, at least one local minimum exists that would result in a solution not corresponding to the global minimum. Local minima may be the result of the interdependence of one or more of the fitting parameters. For K2, the shallow local minimum occurring at binding affinities two orders of magnitude larger than the correct value results from the algorithm finding a two site solution wherein K2 and K3 are indistinguishable. This indicates the codependence of the binding affinity values. Hence, it is up to the individual user applying these algorithms to use an appropriate number of binding processes to arrive at an optimal thermodynamic solution that is consistent with the ITC thermograms (i.e. fitting the experimental data within the expected or observed experimental error). Furthermore, as seen from Figure 3 which shows a 3-D error plot for the two parameters ΔH3 and n3, it may be necessary for the individual user to enter appropriate starting values for each parameter being evaluated, and to verify that the minimum solution found corresponds to a global minimum and represents a plausible solution for the actual chemical equilibria occurring in the system.
Figure 2.
Error plot produced by holding the value of K2 constant while all other three-site parameters were optimized using nonlinear regression. The value of K2 ranged from 101 to 108 to cover a large enough range of values (i.e. −3 < ΔLog(K2) < 2). Error plots were produced for all nine parameters, each showing several local minima and a global minimum at ΔLog(X) = 0. The error plot shown illustrates the interdependence of parameters by the local minimum found at -1.5. Error plots for nk and ΔHk show similar local minima due to their interdependence. The error plot demonstrates that appropriate starting positions are necessary to determine an optimal best-fit solution.
Figure 3.
Error plot produced by holding the values of n3 and ΔH3 constant while all other three-site parameters were optimized using nonlinear regression. The value of ΔH3 ranged from 0 to -10 kcal/mol and the value of n3 ranged from 0 to 2, to include the best fit values for each of these parameters. Panel A shows the error plot for the whole range of values for the two parameters while Panel B magnifies the error surface near the best fit values for the parameters ΔH3 and n3 at the global minimum.
Discussion
Multiple Binding Site Models
The theory used for developing the three-processes binding model and fitting algorithm can be extended to model multiple binding site models of almost any order. The challenge in modeling such complex systems can be attributed to two factors. First, the ability to determine the roots of the polynomial (Equation 3) used for calculating the concentration of free ligand within the reaction vessel. For modeling simpler binding models including one or two binding sites, the polynomial expands only to a quadratic or cubic equation, respectively. Although closed form solutions can be obtained for both quadratic and cubic equations, the solution for the cubic equation can be quite cumbersome. For more complex cases, such as three or more binding sites, a closed form solution for the polynomial does not exist. Software such as MATLAB can provide numerical methods for efficient and accurate means of determining the correct root of Equation 3. It is possible to simplify the determination of roots from Equation 3 based on chemical restrictions. The restriction that the concentration of free ligand in the cell cannot fall below zero or exceed the injected ligand concentration allows for the root finding processes to efficiently determine the correct root of the polynomial.
Second, using the smallest correct number of reactions, and introducing restrictions to the binding algorithm can greatly increase the efficiency of complex algorithms. Knowing that the thermodynamic model accurately describes the system under study, (i.e. includes only chemical equilibria actually observed in the system), allows for the minimum number of fitting parameters to be determined by the non-linear regression analysis. Similarly, if any restrictions can be placed on the binding algorithm to reduce the number of modeled parameters, (e.g. fixing any of the parameters using experimentally determined values), the uncertainty in the best-fit parameters can be dramatically reduced. In particular, restrictions placed on the binding stoichiometry, n, are generally helpful for improving fitting efficiency.
The stoichiometric ratio, n, can also greatly impact the enthalpy change, ΔH, of the reaction because these two variables are found to be indirectly proportional. That is, if the ΔH of the reaction is increased by an order of magnitude the stoichiometric ratio will decrease proportionally. Caution must be taken when using these complex algorithms to ensure that the proper minimum is being approached. Due to the compensation of stoichiometries and enthalpy changes, it is possible for the algorithm to enter a local minimum where the enthalpy change becomes increasingly large to compensate for underestimated binding affinities and/or stoichiometric ratios. The relationship between ΔH and n can be seen in Equation 4, which is the only equation in which ΔH appears. Because ΔH enters linearly into the binding models, it is possible to further improve the binding algorithms through linearization of the ΔH parameter.
Monte Carlo Evaluation
Monte Carlo evaluation is a valuable technique for determining the statistical uncertainties in best-fit binding parameters. The uncertainties estimated from a Monte Carlo analysis for one possible three binding site model in which ΔH1 < ΔH2 < ΔH3 is discussed below. The Monte Carlo analysis demonstrated a strong correlation between ΔH and n when determining best-fit parameters. Other combinations of molar enthalpy changes not simulated in this work may result in different parameter uncertainties. However, the largest factor when evaluating uncertainties in the ITC method is the size of the raw heats produced from the reactions occurring in the calorimeter. Because the standard deviation in each injection interval heat, ΔQi, is ±0.1 μcal, larger heats are less affected by experimental error in the integrated injection heats. The results given here from the Monte Carlo analysis of the fit parameter uncertainties are expected to be typical for modeling a system with three binding sites. The parameters with the most uncertainty are typically n3 and ΔH3 because these parameters are sensitive to the end-point of the titration. Thermodynamic parameters obtained from titrations exhibiting a well-defined end-point (i.e. having a large value for Kn) are well determined with the algorithms developed here. Furthermore, the above algorithms, if extended beyond three binding sites, can be used to evaluate even more complex systems. The Monte Carlo method provides a rigorous test of the reliability of the best-fit parameters obtained in the non-linear regression modeling of the three binding site system. The Monte Carlo method can be used to estimate the uncertainties in parameters obtained in the non-linear regression modeling of experimental data in the same manner as used here for testing the non- linear fitting of simulated data but should incorporate experimentally determined error.
Error Analysis
The error plots produced for all parameters involved in modeling three binding sites clearly demonstrated the interdependence of binding parameters (Figure 2). Because several local minima are observed for each parameter, stress is placed on the user to accurately enter reasonable starting parameters to avoid getting trapped in a local minimum. It is clear that several parameters have strong interdependence (i.e. K2 vs. K3, ΔH3 vs. n3) and must be given starting values that will ensure that the true global minimum is found. The user can check the validity of the solution obtained from nonlinear regression analysis by entering several starting locations for parameters that demonstrate large variability. If the same minimum is reached regardless of starting positions, it is more likely that the solution represents a global minimum. The user must take care to ensure that the solution is both descriptive of the system, and represents a plausible thermodynamic and chemical result. However, it must be remembered that the binding model chosen strongly influences the best-fit parameters obtained in the nonlinear regression analysis of ITC data, and binding parameters will only accurately reflect the system when the correct model is chosen.
Example Systems Exhibiting Three Binding Sites
Biological systems exhibiting multiple overlapping may be more common than previously thought. Several ITC studies of these more complex systems have been recently reported [30, 31, 41, 43]. We present three examples of quadruplex DNA binding either a small ligand or a protein that further illustrate the utility of our MATLAB n-binding sites ITC data analysis program.
ITC data for the titration of a 27-mer c-MYC promoter sequence G-quadruplex construct (having the sequence: 5′-TGGGGAGGGTGGGGAGGGTGGGGAAGG) at low ionic strength, 20mM K+, with the cationic porphyrin ligand TMPyP4 (5, 10, 15, 20-meso-tetra (N-methyl-4-pyridyl) porphine) are shown in Figure 4. This WT c-MYC promoter sequence quadruplex binds four ligands at saturation. At physiologic ionic strength (150 mM KCl), the four binding sites in the model are represented by two sets of two sites with each set having a characteristic affinity. In effect, two ligands bind to the two high affinity sites and two ligands bind to the low affinity sites [41]. We have assigned the high affinity sites to end stacking on the two ends of the G-tetrad stack, while the two lower affinity sites have been assigned to the two intercalation sites located between the first and second and the second and third G-tetrads. At low ionic strength (20 mM KCl), a more complicated model is required having three sites exhibiting three different affinities. We have suggested that at the lower ionic strength the two ends of the G-quadruplex are no longer equivalent and the overhanging single strand is stiffened due to electrostatic repulsion. Based on the three binding site fit of the ITC data, the first TMPyP4 binds to one end, the next two ligands bind in the two intercalation sites, and the fourth TMPyP4 binds to the sterically hindered low affinity end of the G-quadruplex. Whether this structural model is correct or not, the ITC thermogram observed in low salt conditions is well fit with the three binding site model and cannot be fit with any simpler model.
Figure 4.
ITC data for the reaction of a cationic porphyrin, TMPyP4, with the WT c-MYC 27-mer polypurine P1 promoter sequence G-quadruplex at low ionic strength (20 mM K+ in BPES buffer at pH=7.0. The lower ionic strength data must be fit to the three-binding site model. The best fit parameters are: K1 = 3.9 × 108 M−1, K2 = 4.2 × 107 M−1, K3 = 2.9 × 106 M−1, ΔH1 = −0.2 kcal/mol, ΔH2 = −12.6 kcal/mol, ΔH3 = -5.9 kcal/mol, and n1 = 1, n2 = 2, n3 = 1.
G-quadruplex ligand studies have typically been limited to single stranded constructs. In order to better model the G-quadruplex motif in the presence of its complementary strand we have constructed capped G-quadruplex constructs in which the c-rich strand has been replaced with a shorter partially complementary strand having the ability to form short flanking duplex regions on either side of the quadruplex and with a short non-interaction region of 4, 5, or 6 Ts bridging the quadruplex in the capped construct. ITC data for the TMPyP4 titration of one such construct is shown in Figure 5. The interaction of TMPyP4 with the “capped” quadruplex is more complicated than the interaction with the singled stranded quadruplex having the same sequence. In effect, the cap results in a small amount of unfolding of the G-quadruplex and in the case of the WT sequence results in a shift in the equilibrium between the 1:6:1 and the 1:2:1 foldamers. The TMPyP4 titration of the 5-T capped 1:6:1 mutant c-MYC 32-mer promoter sequence is fit to a three binding site model after the data have been corrected for some ligand induced quadruplex folding and dilution effects. The three binding events must be the result of mixed end binding and intercalation interactions. We don't currently understand the stoichiometry 1.5:1.0:1.5, however we are continuing to search for structural models that are consistent with the titration results and the best fit parameters shown in Figure 5. (The ITC data shown here for the titration of the capped quadruplex with TMPyP4 are unpublished and provided here as an example of the need for the analysis of ITC data for systems exhibiting multiple overlapping equilibria.)
Figure 5.
ITC data for the reaction of a cationic porphyrin, TMPyP4, with the 1:6:1 c-MYC 32-mer polypurine P1 promoter sequence G-quadruplex capped with a partially complementary 17-mer 5-T capping oligonucleotide in100 mM K+/BPES buffer at pH=7.0. These ITC data have been corrected for ligand induced folding and dilution heat effects and fit to the three binding site model. The best fit parameters are: K1 = 7.4 × 109 M−1, K2 = 5.2 × 107 M−1, K3 = 7.0 × 105 M−1, ΔH1 = -7.0 kcal/mol, ΔH2 = −13.7 kcal/mol, ΔH3 = -9.9 kcal/mol, and n1 = 1.5, n2 = 1, n3 = 1.5.
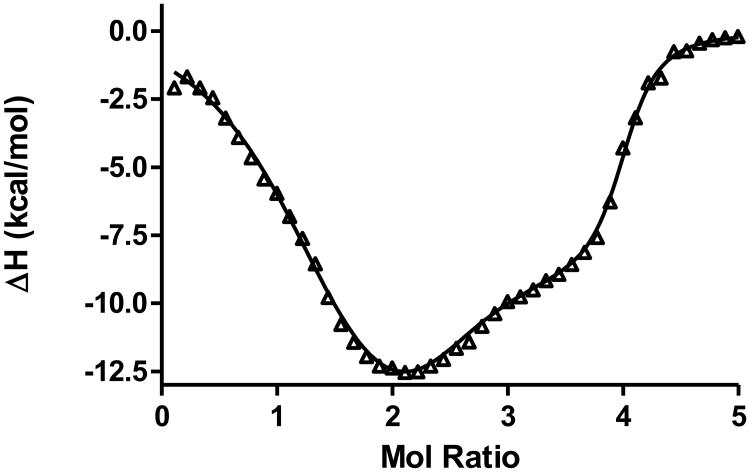
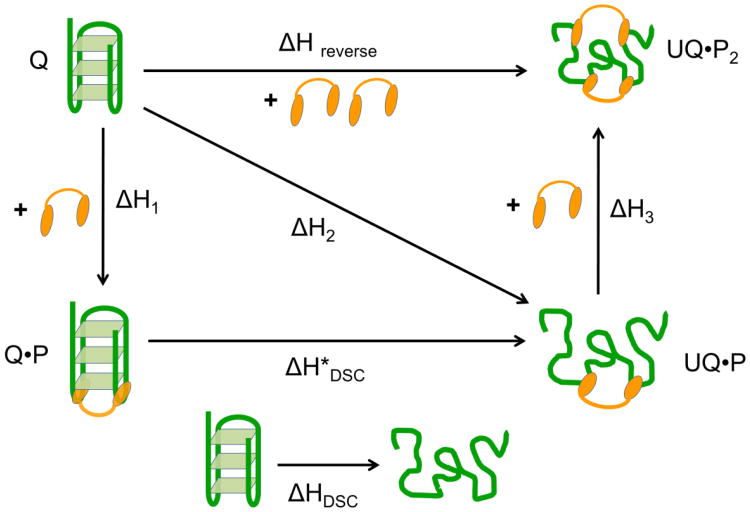
Human telomeric DNA contains tandem repeats of the DNA sequence (TTAGGG) and is capable of forming G-quadruplexes. Unfolding of the telomeric G- quadruplex DNA leads to elongation of the telomere by TERT in vitro. UP1 is the proteolytic fragment of the hnRNP A1 protein that is responsible for several gene regulatory processes at the post-transcriptional level including mRNA alternative splicing, transportation and pri-miRNA processing. A crystal structure of UP1 complexed with a partial human telomeric sequence d(TTAGGG)2 revealed that the interaction between UP1 and the telomeric DNA is established trough three nucleobases “TAG”. The “TAG” binding motif located at several corners of G-quartets was found to be critical for UP1 binding and initiating unfolding of the G-quadruplex structure. ITC experiments, including both forward and reverse titrations and DSC experiments were used to probe how UP1 interacts with human telomeric G-quadruplex DNA in Na+ buffer conditions. ITC data and the three binding sites fit from a forward titration of Tel-22 with UP1 are shown in Figure 6. These studies reveal that UP1 binds and unfolds the Tel-22 G-quadruplex DNA at 2:1 molar ratio. The binding involves loop recognition plus unfolding plus single strand binding for a second UP1 molecule. This binding, unfolding, binding process is described by the thermochemical cycle shown in Figure 7. (The UP1/Tel-22 data are unpublished and are provided by Dr. David Graves at UAB, Department of Chemistry.)
Figure 6.
Screen capture of the MATLAB user interface showing the three binding sites fit of the ITC data obtained for the addition of UP1 to the human telomere quadruplex, Tel-22. Experimental parameters, [L], [M], calorimeter cell volume, injection volume, number of injections, and temperature are shown in the upper left quadrant of the screen. Starting parameters ni, Ki, and ΔHi are entered in the table shown mid-left on the screen capture. Best Fit parameters and the standard deviations are shown at the bottom left of the screen. The right side of the screen shows the ITC data (corrected for heat of dilution) and the best fit line through the data. The MATLAB program can be downloaded at http://lewis.chemistry.msstate.edu/download.html.
Figure 7.
Thermochemical cycle (Hess' Law diagram) for the binding of 2 mols of UP1 to the Tel-22 G-quadruplex. The values of ΔH1, ΔH2, and ΔH3 are obtained directly from the three binding sites model fit to the forward titration data for the addition of UP1 to Tel-22. The value for ΔHreverse is obtained from the “Model Free” reverse titration. The value of ΔH*DSC is obtained as the difference between ΔH1 and ΔH2 and compared to the ΔHDSC value obtained for the thermal denaturation of the naked Tel-22 G -quadruplex. The value of ΔH3 is obtained directly from the three binding sites model and compared to the difference between ΔH2 and ΔHreverse
Further Models
The models developed here assume that the only chemical equilibria being observed are reactions involving the binding of a ligand molecule to one or more specific binding sites in a macromolecule (Equation 2). In these systems, each type of binding site competes with any other binding sites present in the macromolecule and the binding at any site occurs with unitary stoichiometry
We have shown that a large number of thermodynamic parameters may be accurately estimated from an ITC experiment, e.g. the nine parameters in the three binding site model were all well determined. The larger the number of parameters that need to be determined the more uncertainty will be introduced in parameter determination. There will always be some benefit in designing the experiments or in combining data from complimentary studies in order to limit the number of parameters that are fit. For example The enthalpy change for the strongest binding reaction may be best determined in so called “model free” experiments [42] while the thermodynamic parameters for the weakest binding process may be best determined in additional reverse titration experiments where the low affinity complexes are more highly populated [31]. In conclusion, we would like to state that ITC data can be fit with acceptable accuracy to complex thermodynamic models having as many as three or more overlapping binding equilibria.
Supplementary Material
Acknowledgments
Supported in part by grant CA35635 from the National Cancer Institute (to JBC) and by the James Graham Brown Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hansen LD, Christensen JJ, Izatt RM. Entropy titration. A calorimetric method for determination of ΔG° (k), ΔH° and ΔS°. Chem Comm. 1965;36 [Google Scholar]
- 2.Christensen JJ, Izatt RM, Hansen LD, Partridge JA. Entropy Titration. A Calorimetric Method for the Determination of ΔG, ΔH, and ΔS from a Single Thermometric Titration. J Phys Chem. 1966;70:2003–10. [Google Scholar]
- 3.Christensen JJ, Wrathall DP, Oscarson JO, Izatt RM. Theoretical Evaluation of Entropy Titration Method for Calorimetric Determination of Equilibrium Constants in Aqueous Solution. Anal Chem. 1968;40:1713–17. [Google Scholar]
- 4.Eatough DJ, Lewis EA, Hansen LD. In: Determination of ΔHr and Keq Values Chapter 5 in Analytical Solution Calorimetry. Grime K, editor. John Wiley & Sons; New York: 1985. pp. 137–16130. [Google Scholar]
- 5.Wiseman T, Williston S, Brandts JF, Lin LN. Rapid Measurement of Binding Constants and Heats of Binding Using a New Titration Calorimeter. Anal Biochem. 1989;179:131–7. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 6.Freire E, Mayorga OL, Straume M. Isothermal Titration Calorimetry. Anal Chem. 1990;62:950A–959A. [Google Scholar]
- 7.Doyle ML. Characterization of binding interactions by isothermal titration calorimetry. Curr Opin Biotechnol. 1997;8:31–5. doi: 10.1016/s0958-1669(97)80154-1. [DOI] [PubMed] [Google Scholar]
- 8.Holgate GA. Making cool drugs hot: Isothermal titration calorimetry as a tool to study binding energetics. BioTechniques. 2001;30:164–6. [PubMed] [Google Scholar]
- 9.Ladbury JE. Isothermal titration calorimetry: application to structure-based drug design. Thermochim Acta. 2001;380:209–15. [Google Scholar]
- 10.Ladbury JE. Application of isothermal titration calorimetry in the biological sciences: things are heating up! Biotechniques. 2004;37:885–7. doi: 10.2144/04376TE01. [DOI] [PubMed] [Google Scholar]
- 11.Ababou A, Ladbury JE. Survey of the year 2005: literature on applications of isothermal titration calorimetry. J Mol Recognit. 2007;20:4–14. doi: 10.1002/jmr.803. [DOI] [PubMed] [Google Scholar]
- 12.Ladbury JE, Klebe G, Freire E. Adding calorimetric data to decision making in lead discovery: a hot tip. Nat Rev Drug Discov. 2010;9:23–7. doi: 10.1038/nrd3054. [DOI] [PubMed] [Google Scholar]
- 13.Ladbury JE. Calorimetry as a tool for understanding biomolecular interactions and an aid to drug design. Biochem Soc Trans. 2010;38:888–93. doi: 10.1042/BST0380888. [DOI] [PubMed] [Google Scholar]
- 14.Chaires JB. Calorimetry and Thermodynamics in Drug Design. Annu Rev Biophys. 2008;37:135–151. doi: 10.1146/annurev.biophys.36.040306.132812. [DOI] [PubMed] [Google Scholar]
- 15.Freire E. Do enthalpy and entropy distinguish first in class from best in class? Drug Discov Today. 2008;13:869–874. doi: 10.1016/j.drudis2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freire E. A thermodynamic approach to the affinity optimization of drug candidates. Chem Biol Drug Des. 2009;74:468–472. doi: 10.1111/j.1747-0285.2009.00880.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garbett NC, Chaires JB. Thermodynamic studies for drug design and screening. Expert Opin Drug Discov. 2012;7:299–314. doi: 10.1517/17460441.2012.666235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freyer MW, Lewis EA. Isothermal titration calorimetry: experimental design, data analysis, and probing macromolecule/ligand binding and kinetic interactions. Methods Cell Biol. 2008;84:79–113. doi: 10.1016/S0091-679X(07)84004-0. [DOI] [PubMed] [Google Scholar]
- 19.Velazquez-Campoy A, Freire E. Isothermal titration calorimetry to determine association constants for high-affinity ligands. Nat Protoc. 2006;1:186–91. doi: 10.1038/nprot.2006.28. [DOI] [PubMed] [Google Scholar]
- 20.Hansen LD, Fellingham GW, Russell DJ. Simultaneous determination of equilibrium constants and enthalpy changes by titration calorimetry: Methods, instruments, and uncertainties. Anal Biochem. 2010;409:220–9. doi: 10.1016/j.ab.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 21.Lewis EA, Murphy KP. Isothermal titration calorimetry. Methods Mol Biol. 2005;305:1–16. doi: 10.1385/1-59259-912-5:001. [DOI] [PubMed] [Google Scholar]
- 22.Freire E, Schon A, Velazquez-Campoy A. Isothermal titration calorimetry: gener al formalism using binding polynomials. Methods Enzymol. 2009;455:127–55. doi: 10.1016/S0076-6879(08)04205-5. [DOI] [PubMed] [Google Scholar]
- 23.Velazquez-Campoy A, Ohtaka H, Nezami A, Muzammil S, Freire E. Isothermal titration calorimetry. Curr Protoc Cell Biol Chapter. 2004;17:8. doi: 10.1002/0471143030.cb1708s23. Unit 17. [DOI] [PubMed] [Google Scholar]
- 24.Velazquez-Campoy A, Freire E. Isothermal titration calorimetry to determine association constants for high-affinity ligands. Nat Protoc. 2006;1:186–91. doi: 10.1038/nprot.2006.28. [DOI] [PubMed] [Google Scholar]
- 25.Lewis EA. eLS. John Wiley & Sons Ltd; Chichester: 2009. Calorimetry. http://www.els.net. [DOI] [Google Scholar]
- 26.Breslauer KJ, Freire E, Straume M. Calorimetry: a tool for DNA and ligand- DNA studies. Methods Enzymol. 1992;211:533–67. doi: 10.1016/0076-6879(92)11030-m. [DOI] [PubMed] [Google Scholar]
- 27.Bundle DR, Sigurskjold BW. Determination of accurate thermodynamics of binding by titration microcalorimetry. Methods Enzymol. 1994;247:288–305. doi: 10.1016/s0076-6879(94)47022-7. [DOI] [PubMed] [Google Scholar]
- 28.Arnaud A, Bouteiller L. Isothermal titration calorimetry of supramolecular polymers. Langmuir. 2004;20:6858–63. doi: 10.1021/la049365d. [DOI] [PubMed] [Google Scholar]
- 29.Chaires JB. A thermodynamic signature for drug-DNA binding mode. Arch Biochem Biophys. 2006;453:26–31. doi: 10.1016/j.abb.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 30.Bou-Abdallah F, Woodhall MR, Velázquez-Campoy A, Andrews SC, Chasteen ND. Thermodynamic analysis of ferrous ion binding to Escherichia coli ferritin EcFtnA. Biochemistry. 2005;44:13837–46. doi: 10.1021/bi0514212. [DOI] [PubMed] [Google Scholar]
- 31.Taneva SG, Banuelos S, Falces J, Arregi I, Muga A, Konarev PV, Svergun DI, Velazquez-Campoy A, Urbaneja MA. A mechanism for histone chaperoning activity of nucleoplasmin: thermodynamic and structural models. J Mol Biol. 2009;393:448–63. doi: 10.1016/j.jmb.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 32.Chaires JB. Possible origin of differences between van't Hoff and calorimetric enthalpy estimates. Biophys Chem. 1997;64:15–23. doi: 10.1016/s0301-4622(96)02205-3. [DOI] [PubMed] [Google Scholar]
- 33.Horn JR, Russell D, Lewis EA, Murphy KP. Van't Hoff and calorimetric enthalpies from isothermal titration calorimetry: are there significant discrepancies? Biochemistry. 2001;40:1774–8. doi: 10.1021/bi002408e. [DOI] [PubMed] [Google Scholar]
- 34.Baker BM, Murphy KP. Evaluation of linked protonation effects in protein binding reactions using isothermal titration calorimetry. Biophys J. 1996;71:2049–55. doi: 10.1016/S0006-3495(96)79403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tellinghuisen J. Statistical error in isothermal titration calorimetry. Methods Enzymol. 2004;383:245–82. doi: 10.1016/S0076-6879(04)83011-8. [DOI] [PubMed] [Google Scholar]
- 36.Straume M, Johnson ML. Monte Carlo method for determining complete confidence probability distributions of estimated model parameters. Methods Enzymol. 1992;210:117–29. doi: 10.1016/0076-6879(92)10009-3. [DOI] [PubMed] [Google Scholar]
- 37.Tellinghuisen J. A study of statistical error in isothermal titration calorimetry. Anal Biochem. 2003;321:79–88. doi: 10.1016/s0003-2697(03)00406-8. [DOI] [PubMed] [Google Scholar]
- 38.Tellinghuisen J. Statistical error in isothermal titration calorimetry: variance function estimation from generalized least squares. Anal Biochem. 2005;343:106–15. doi: 10.1016/j.ab.2005.04.026. [DOI] [PubMed] [Google Scholar]
- 39.Correia JJ, Chaires JB. Analysis of drug-DNA binding isotherms: a Monte Carlo approach. Methods Enzymol. 1994;240:593–614. doi: 10.1016/s0076-6879(94)40065-2. [DOI] [PubMed] [Google Scholar]
- 40.Saroff HA. Evaluation of uncertainties for parameters in binding studies: the sum- of-squares profile and Monte Carlo estimation. Anal Biochem. 1989;176:161–9. doi: 10.1016/0003-2697(89)90287-x. [DOI] [PubMed] [Google Scholar]
- 41.Freyer MW, Buscaglia R, Kaplan K, Cashman D, Hurley LH, Lewis EA. Biophysical Studies of the c-MYC NHE III1 Promoter: Model Quadruplex Interactions with a Cationic Porphyrin. Biophys J. 2007;92:2007–15. doi: 10.1529/biophysj.106.097246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chaires JB. Human telomeric G-quadruplex: thermodynamic and kinetic studies of telomeric quadruplex stability. FEBS J. 2010;277:1098–1106. doi: 10.1111/j.1742-4658.2009.07462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lewis E, Munde M, Wang S, Rettig M, Le V, Machha V, Wilson WD. Complexity in the binding of minor groove agents: Netropsin has two thermodynamically different DNA binding modes at a single site. Nucleic Acids Res. 2011;39:9649–58. doi: 10.1093/nar/gkr699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.