Abstract
The molecular structures and enthalpy release during binding of poly(amidoamine) (PAMAM) dendrimers to 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) bilayers were explored through atomistic molecular dynamics. Three PAMAM dendrimer terminations were examined: protonated primary amine, neutral acetamide, and deprotonated carboxylic acid. Fluid and gel lipid phases were examined to extract the effects of lipid tail mobility on the binding of generation-3 dendrimers, which are directly relevant to the nanoparticle interactions involving lipid rafts, endocytosis, lipid removal, and/or membrane pores. Upon binding to gel phase lipids, dendrimers remained spherical, had a constant radius of gyration, and approximately one-quarter of the terminal groups were in close proximity to the lipids. In contrast, upon binding to fluid phase bilayers, dendrimers flattened out with a large increase in their asphericity and radii of gyration. Although over twice as many dendrimer–lipid contacts were formed on fluid versus gel phase lipids, the dendrimer–lipid interaction energy was only 20% stronger. The greatest enthalpy release upon binding was between the charged dendrimers and the lipid bilayer. However, the stronger binding to fluid versus gel phase lipids was driven by the hydrophobic interactions between the inner dendrimer and lipid tails.
Introduction
Dendrimers are a major class of synthetic polymers currently in development for applications such as gene delivery, targeted drug delivery, and enhanced in vivo imaging.1–5 Poly(amidoamine) (PAMAM) dendrimers, in particular, have shown to be very promising in these areas (Figure 1).6–10 Although attempts have been made to elucidate the mechanisms of polymer binding and internalization on living cells,11–22 there is still much work to be done before fully capable nanodevices can be designed and created due to the remaining unknown mechanisms of nanoparticle-membrane interaction.
Figure 1.
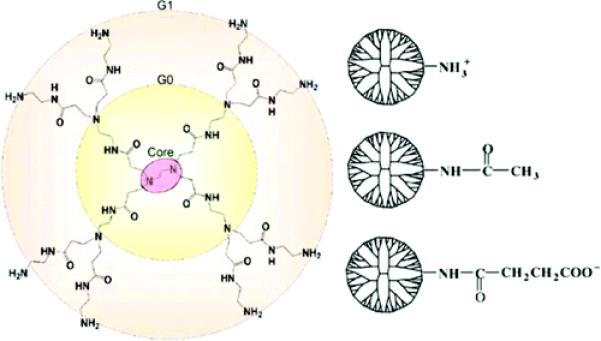
Atomic structure of first-generation (G1) PAMAM dendrimer and terminal groups: protonated primary amine (–NH3+), uncharged acetamide (–Ac), and deprotonated carboxylic acid (–COO−). There are 32 terminal groups per G3 dendrimer, and they are all modified to become +32e charged G3–NH3+, uncharged G3–AC, and the –32e charged G3–COO−. Note that the G1 PAMAM dendrimer illustrated has eight terminal groups (the –NH2 groups on the largest concentric circle). The remaining functional groups are referred to in the text as internal dendrimer moieties as defined in Figure 4.
To address complicated, multicomponent biological systems, models often dissect which components are most relevant to the observed or desired behavior. Lipid molecules are the most prevalent component of the plasma membrane, and a more thorough understanding of the interaction of nanoparticles with lipid molecules will address many relevant issues pertaining to real cells. Lipid bilayer models have been shown to give qualitatively accurate predictions for in vitro cell studies in regardstomembranepermeabilityandpolymerinternalization.12,18–20,23,24
PAMAM dendrimers are used in medical applications for many of the same reasons that also make them good models for nanotoxicity and polymer internalization studies; dendrimers have high homogeneity (polydispersity index ≈ 1.01), high water solubility, numerous modifiable end groups, low immunogenicity, small diameter (<10 nm), and are highly deformable for multivalent interactions.8,9,21,25 The heterogeneous nature of other polymer samples often prevent researchers from determining which particle properties are inducing the observed cellular response. By contrast, because of their homogeneity, PAMAM dendrimers are prime study cases and are utilized in a focused approach as models to address many specific issues in cell biology, nanotoxicity, and medicine.
A companion paper addressing the binding free energy profile along the interaction coordinate and the morphology of generation-3 (G3) PAMAM dendrimers on DMPC bilayers is also found within this publication.17 The purpose of the present paper is to address, in a comparative manner, the differences between the dendrimer interaction with a fluid phase and with a gel phase of the lipid bilayer. This is important because experimental studies have shown that fluid phase lipids exposed to polymers have increased propensity to yield membrane pore formation, membrane leakage, and particle internalization, whereas the gel phase lipids tend to be more resistive to permeation and degradation.20,24 Our study will aid in achieving a better understanding of the binding and internalization mechanisms of polymers into living cells. Particularly, the means by which polymers interact with the fluid versus gel phases of the lipid bilayers are important to cellular mechanisms involving lipid rafts and endocytosis.26–28
The polymer’s surface greatly influences its interaction with cells. Active polymer targeting with specific receptor binding can be hindered by charged polymer moieties, such as protonated primary amine or deprotonated carboxylic acid. This is due to the highly energetic nonspecific binding of these polymers to both targeted and nontargeted cells alike.7,8 Moreover, model membranes are heavily degraded by charged polymers, whereas neutral polymers cause less membrane disruption.12,18,19
Atomic force microscopy (AFM) has yielded nanometer resolution images of supported DMPC bilayers affected by PAMAM dendrimers of various generations (G3, G5, and G7) and terminations (primary amine, acetamide, and carboxylic acid).19,23 Experimental studies have shown that increasing dendrimer size or charge magnitude yields greater bilayer disturbance, as consistent with previous observations of a surface area dependence.18,29 It was also observed that no disruption of gel phase bilayers occurred, whereas neighboring fluid phase bilayers were highly degraded by primary amine-terminated, G7 PAMAM dendrimers.20 These studies demonstrated a strong dependence on dendrimer and lipid properties for the resulting dendrimer–bilayer interaction.
The binding of dendrimers to lipid bilayers has been previously explored computationally via coarse-grained molecular dynamics by Lee and Larson.22 That study approximated the PAMAM dendrimers and 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) bilayer through the motion of computer-modeled beads designed to each represent approximately four heavy atoms and associated hydrogen atoms. That coarse graining procedure was an extension of the coarse-grained force field developed by Marrink et al. for proteins.30 Even though the simulations involved no dendrimer–dendrimer interactions and only 0.5 μs simulation time, observations of G3 and G5 PAMAM dendrimers of varying acetylation on bilayers in the gel and fluid phases were roughly consistent with those from AFM. Coarse-grained simulations yielded significant morphology differences for the dendrimers bound to gel versus fluid phase bilayers. The primary amine-terminated dendrimers bound to the fluid phase lipids, deformed, intercalated into the bilayer, and interacted with the lipid tails. In contrast, the dendrimers bound to the gel phase were confined to the top of the bilayer in a compact shape.
Lee and Larson22 observed that the morphology of the bound dendrimer varied greatly with dendrimer termination such that the amine-terminated dendrimers caused bilayer pores, whereas the acetylated dendrimers avoided contact with the lipid tails, regardless of lipid phase. This observation is somewhat different from the AFM conclusions, which postulated that the acetylated G5 dendrimers rested fully within the lipid tail region and were not seen on the surface of intact bilayers. These differences could easily be attributed to the short simulation time, approximate simulation force-field parameters, experimental artifacts from the mica supporting the lipids, and/or incorrect interpretation of the AFM data which only provided topographic and elastic information.
Our work presented in this manuscript uses an all-atom simulation to examine the binding of G3 PAMAM dendrimers of multiple terminations to lipids of varying phase for a more thorough understanding of the atomistic differences in dendrimer–membrane binding. Three hypotheses are put forth to elucidate the mechanisms of dendrimer–lipid binding. They demonstrate our expected dependence of the resulting interaction on the lipid phase and the dendrimer termination:
Hypothesis 1
The fluid phase lipid bilayer deforms to accommodate dendrimer–lipid interactions with more of the dendrimer and lipids in contact than the gel phase lipids.
Hypothesis 2
The dendrimers achieve a stronger interaction energy with fluid phase lipids than gel phase lipids due to the mobility of the fluid phase lipids allowing the dendrimers’ terminal groups to obtain more favorable interactions.
Hypothesis 3
The charged dendrimers (G3–NH3+ and G3–COO−) release more enthalpy upon binding than the uncharged dendrimer (G3–Ac) and, as such, the charged dendrimers are more morphologically altered upon binding.
The results presented here support the first hypothesis while providing counter evidence to the second and third hypotheses. In comparison to the hydrophobic dendrimer moieties, the dendrimer terminal groups make a relatively small contribution to the fluid versus gel phase lipid energetics and deformation upon binding. Within this manuscript the morphology and the mechanisms of dendrimer bound to lipid bilayers are examined quantitatively. Through these atomistic simulations, greater detail is obtained about the particular atomic configuration and morphology of the bound structures, as well as the enthalpic components to binding. The dendrimer radius of gyration, asphericity, and elastic energy of deformation are examined to quantitatively describe the dendrimer during binding. Further, the number and type of dendrimer–lipid contacts are described both morphologically and enthalpically. Through this analysis we directly address our three hypotheses.
Methods
Atomistic simulations of G3 PAMAM dendrimers and DMPC lipids were performed. Dendrimer parameters were constructed from the CHARMM parameters for generic proteins (para22), whereas the lipid (DMPC) parameters came directly from CHARMM27.31,32 Simulations were run at 300 K with nonbonded interactions cutoff at 13 Å and switched from 8 Å. Time steps of 2 fs were taken with implementation of the SHAKE routine to remove variation in hydrogen atom bond lengths.
Initial dendrimer configurations were made with a recursive script in CHARMM.33 Both the dendrimer and the lipids were separately equilibrated for 2 ns before being were combined into nine different simulations (three dendrimers with three lipid states). All simulations ran until dendrimers stopped moving along the direction perpendicular to the lipid bilayer for more than 5 ns; the total simulation time for all nine systems was 90 ns. The atomic configurations were analyzed every 2 ps over the final 4 ns. All systems required greater than 10 ns to transition from initially contacting the fluid phase bilayer to fully bind. All images of the molecular structure within this manuscript were created with the software VMD.34
The effects of the solvent were modeled implicitly by a distance-dependent dielectric (ε = 4r) to mimic solvent screening effects and to make the atomistic simulations of macromolecules computationally feasible. This solvent approximation has been used previously in molecular dynamics simulations of macromolecules, such as dendrimers, and yielded reasonable agreement with experimental results.17,21,35,36 An explicit solvent representation in this large (3600 nm3) system would require an exceedingly long sampling time and, as such, was deemed unfeasible for the present purpose. A more thorough discussion of the implications and rational behind this approximation is given in the companion manuscript also in this journal.17 Generalized Born solvation models were examined but not pursued due to the lack of appropriate parametrization.37,38
The lipid phase was imposed on the bilayer by varying the amount of restriction on the mobility of the lipid tails. The boundary condition for the fluid phase bilayer was a cylinder of fixed lipid molecules with an inner diameter of 10 nm. This setup maintains experimentally determined lipid surface density and bilayer thickness39,40 while permitting a circular lipid bilayer which drastically decreased computational time by reducing the number of lipid molecules as compared to a rectangular patch with periodic boundary conditions (although the latter conditions tend to be generally considered more accurate because they eliminate end effects, they are not free of artifacts, either41). Membrane undulations may be an important aspect of nanoparticle–membrane interactions and are limited to length scales of the simulated system. However, the small size of membrane simulations does not contribute to the disagreements in experimental and simulated membrane structural properties.42
This membrane model of a 10 nm lipid disk provided sufficient surroundings for the dendrimer to not sense the end of the lipids directly (i.e., the smallest distance between any dendrimer atom and the lipid edge was smaller than the cutoff of the pairwise nonbonded interactions). However, the hard wall boundary conditions imposed here are expected to limit both lipid spreading and long-range membrane effects, such as induced curvature. Unfortunately, these two membrane properties are particularly difficult to model simultaneously with reasonable computational resources. To better model lipid spreading, periodic boundary conditions would be preferred. However, this would require significantly more particles in the corners of the simulation box that are not influential to the binding process. To better model long-range membrane effects, a bilayer patch several orders of magnitude larger is required, which would increase prohibitively the computational cost. In compromise of these computational limitations, a large bilayer disk with hard, cylindrical boundary was modeled.
The gel phase bilayer was created through the complete immobilization of the lipid tails of the equilibrated fluid phase bilayer, maintaining the same surface roughness, lipid density, and bilayer thickness in both bilayer phases. The gel versus fluid bilayer differences in lipid density were not incorporated into this model so that the only difference between these simulated phases was the lipid tail mobility. These models best examine the effects of lipid tail mobility on the structure of bound nanoparticles.
The interactions and deformations of the dendrimer have been examined in terms of the enthalpic contributions to binding. These were calculated as averages over the production simulation run of the CHARMM potential energy terms corresponding to the atomic bonds (distance, angles, and torsions), van der Waals, and electrostatic energies. The all-atom trajectory of thermal fluctuations around equilibrium was also analyzed in separate regions of the system to determine the potential energy stored in the various parts. The dendrimer self-energy and the dendrimer–lipid interaction energies were examined. This analysis did not incorporate the effects of entropy and thus is not a free energy calculation; it quantifies solely the enthalpic component (or more accurately for the canonical ensemble in which the simulations were run, the energy component).
Results
G3 PAMAM dendrimers of varying termination were allowed to freely interact with DMPC bilayers of either fluid or gel phase. The resulting equilibrated, bound configuration was analyzed in terms of the atomistic morphology and the enthalpic contributions to binding. The dendrimers were roughly spherical both when far from the lipid bilayers and when bound to the gel phase lipid bilayer (Figure 2). The gel phase lipid tails were restricted from moving, and the gel phase bilayer retains its planar shape upon dendrimer binding. On the contrary, the fluid phase lipids tails were free to move and did so substantially upon dendrimer binding. The dendrimer flattened and elongated upon binding to the fluid phase lipids due to interactions of hydrophobic dendrimer components to the lipid tails (Figures 2 and 3).
Figure 2.
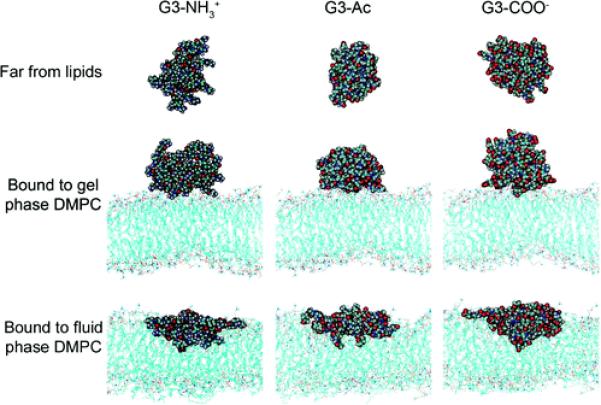
Representative images of the G3 dendrimer with varying terminations in equilibrated states: far from the lipids, bound to the gel phase DMPC bilayer, or bound to the fluid phase DMPC bilayer. These structures have been quantified in terms of dendrimer number and type of dendrimer–lipid contacts (Figures 4 and 5), radius of gyration (Figure 6A), asphericity (Figure 6B), and self-energy (Figure 7). Animations of the equilibrated dendrimers on the DMPC bilayers are found in the Supporting Information.
Figure 3.
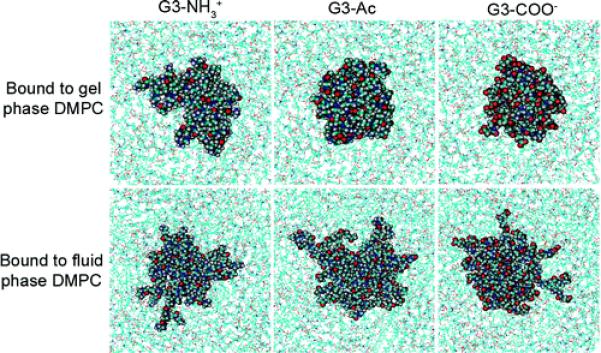
Top view of three different dendrimers on gel and fluid phase DMPC bilayers. The dendrimers increased both radius of gyration and asphericity as they bind to the fluid phase more so than upon binding to gel phase lipids, as represented here by the greater spreading of the dendrimers upon binding to fluid vs gel phase. This increased interaction coincides with the availability of the hydrophobic lipid tails to interact with the hydrophobic dendrimer moieties.
The inner dendrimer components include hydrophobic and hydrophilic moieties. These moieties have been analyzed to determine their individual enthalpic contribution to binding. To facilitate the discussion of the internal dendrimer moieties, each has been given a unique name, as shown in Figure 4. Similarly, each moiety within a termination has been named with the same convention utilized for the inner dendrimer components. The amides found throughout the dendrimer have been broken into carbonyl and secondary amine components for more detailed analysis than the traditional grouping as a single amide would permit.
Figure 4.
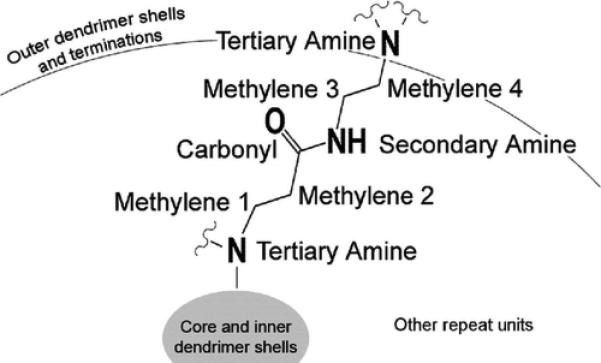
Internal structure of the G3 PAMAM dendrimers includes 60 of the repeating units shown here. Within each dendrimer, each methylene, secondary amine, and carbonyl is found 60 different times, while the tertiary amines occur only 30 different times per dendrimer. These internal dendrimer moieties are identical between all dendrimers. A similar naming convention has been implemented for the moieties within the dendrimer terminations.
The fluid phase bilayer formed a concave depression that accommodated a greater area for dendrimer–bilayer interaction. This dimple in the bilayer permitted more of the dendrimer to interact with the bilayer as well as more lipid molecules to interact with the dendrimer. This result is quantified by counting the number of dendrimer moieties within 3 Å of the lipids and the number of lipid molecules within 3 Å of the dendrimer (Figure 5). In most situations, there were over twice as many interactions between dendrimer binding to fluid phase lipids than the gel phase lipids. The differences in deformability of the fluid versus gel phase lipid bilayer resulted in a significantly different dendrimer binding morphology.
Figure 5.
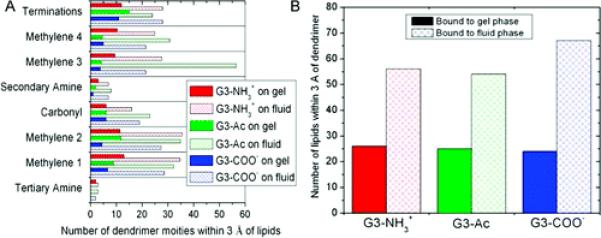
(A) Number of dendrimer moieties within 3 Å of the lipid molecules and (B) the number of lipid molecules within 3 Å of the dendrimer. The fluid phase bilayer permits a greater number of dendrimer–lipid interactions.
When either far from lipids or bound to gel phase lipids, the individual charged terminal groups of G3–NH3+ and G3–COO− frequently extended away from the dendrimer core, whereas the G3–Ac maintained a highly compact structure. However, upon binding to fluid phase lipids, G3–Ac becomes most spread out as the hydrophobic dendrimer moieties reach to interact with the lipid tails. The morphological differences of G3–Ac with varied lipid environments are much greater than those of either G3–NH+ or G3–COO−.
Dendrimer morphology is quantified in terms of radius of gyration (RG) and asphericity (A). The radius of gyration calculates the size of the dendrimer while incorporating density distributions:
| (1) |
with the mass and location of the whole dendrimer (M, x0) and each dendrimer atom (mi, xi).
The dendrimer radius of gyration is only slightly increased (<2%) upon binding to the gel phase lipids; however, upon binding to the fluid phase lipids, the radius of gyration increased between 10% and 40% (Figure 6A). In comparing dendrimers with differing termination, G3–Ac was the smallest dendrimer in both the isolated and gel phase lipid environments but obtained the largest radius of gyration on the fluid phase lipids. The trend is observed that, upon binding to fluid phase lipids, a dendrimer’s radius of gyration increased more if it was hydrophobic and less if it was charged. G3–COO− represented the middle ground in this regard between G3–Ac and G3–NH3+ due to the partially hydrophobic spacer present between the terminal secondary amine and the carboxylic acid.
Figure 6.
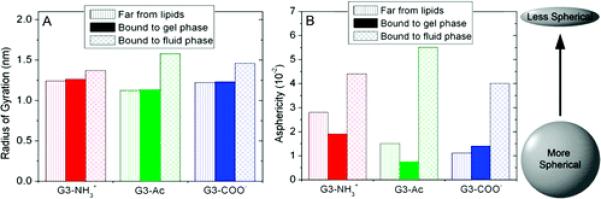
Dendrimer structure quantified in terms of its (A) radius of gyration and (B) asphericity, as defined in eqs 1 and 2. Upon binding to the fluid phase lipids, the dendrimers both increase in size and flatten considerably; however, they do not change shape significantly upon binding to gel phase lipids. The standard deviation of the dendrimer asphericity over time is 17%, 13%, and 24% when far from the lipids, bound to gel phase lipids, or bound to fluid phase lipids, respectively. The standard deviation of the dendrimer radius of gyration over time is less than 1% for all systems.
The morphological deviation of the dendrimer from that of a uniform density sphere was quantified by the dendrimer asphericity (A):43
| (2) |
where Ix,y,z are the eigenvectors of the dendrimer’s moment of inertia matrix. Upon binding to the gel phase bilayer, the asphericity of the amine and acetamide dendrimers decreased by 32% and 50%, respectively, while that of the carboxyl dendrimer increased by 23% (Figure 6B). Upon binding to fluid phase bilayers, the dendrimer asphericity increased by 50–360% as the dendrimers flatten and increase the number of lipid–dendrimer contacts.
The changes in dendrimer shape and binding were analyzed in terms of the contributions to intradendrimer internal energy, i.e., the energy incorporated into the dendrimer which allowed for dendrimer deformation and enhanced binding. Quantification of this self-energy represents the extent to which the dendrimer deformed as a result of the dendrimer–lipid interaction (Figure 7). Dendrimers increased their self-energy by 20–50% upon binding to the gel phase lipids and 60–440% upon binding to fluid phase lipids, with large variations depending on dendrimer termination.
Figure 7.
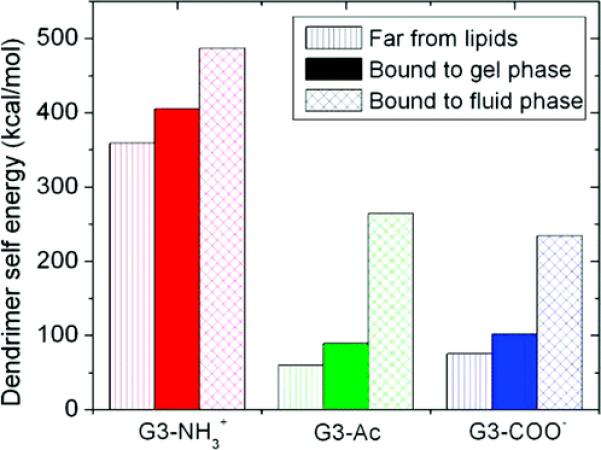
Dendrimer deformation quantified as the macromolecular self-energy. The self-energy of G3 PAMAM dendrimers is equal to the energy of the bonding, electrostatic, and van der Waals terms of the CHARMM force field between atoms in the dendrimer; this is a quantitative description of the dendrimer deformation upon binding. The dendrimers are in a higher energy, more deformed state when bound to the lipids, and more so for the fluid vs gel phase lipids. G3–NH3+ begins in a more energetic state and deforms less to mediate binding as compared to G3–Ac or G3–COO−.
The self-energy landscape of each system was approximately normally distributed in the equilibrium configuration. The differences between the means of each distribution are much greater than the standard error of the means for all data presented. The enthalpies of the lipid conformations are not presented since the differences between the mean enthalpy of various conformations was not significant due to the large number of lipid atoms not directly involved in the dendrimer binding and contributing to greater relative widths of the observed distributions.
Similarly, the strength of the dendrimer–membrane interaction was analyzed in terms of enthalpy. The binding of the dendrimer to the lipids is encouraged by an enthalpy release. This interaction enthalpy was calculated as the average, over the simulated trajectory, of the interaction energy between the whole dendrimer and the lipids as well as between different components of the dendrimer and the lipids (Figure 8). The restriction of the dendrimer to the bilayer plane has an entropic cost; however, binding may allow a greater number of conformations of lipid or dendrimer atoms. Therefore it is not clear whether or not binding was entropically favorable. Since these unbiased simulations demonstrated favorable binding between the dendrimer and the lipids, the binding process is favorable by free energy, and the magnitude of this free energy (calculated using a rigorous umbrella sampling formalism) is the subject of a companion article also in this journal.17
Figure 8.
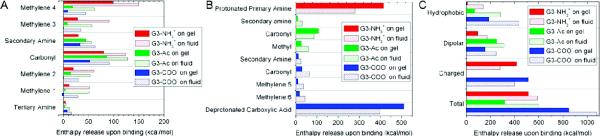
Enthalpy release from the interaction of the dendrimer with the lipids is shown for various parts of the dendrimer. For the inner dendrimer moieties shown in Figure 4, the enthalpies of interaction for each moiety type are shown in (A). For each moiety found within the dendrimer terminations, as shown in Figure 1, the enthalpies of interaction are shown in (B). The results of (A) and (B) are combined into (C) as the dendrimer moieties are categorized as hydrophobic, dipolar, or charged. The charged moieties less strongly bound to the fluid vs gel phase lipids where as the hydrophobic moieties bind over twice as strongly. The majority of the fluid vs gel phase lipid binding differences is mediated by the hydrophobic dendrimer components.
The enthalpy of interaction was calculated for all six dendrimer–lipid configurations and for many dendrimer moieties within both the inner dendrimer shells (Figure 8A) and the terminations (Figure 8B). Figure 8C combines all the dendrimer moieties into the categories hydrophobic, dipolar, or charged where the enthalpic contribution is shown for each type of moiety.
Within this sizable data set, there are multiple interesting observations:
The inner dendrimer shells did not interact strongly with the gel phase lipids. There were stronger interactions between the inner dendrimer and the fluid phase lipids where the dendrimer and lipids deform to mediate binding.
The hydrophobic moieties displayed increased binding enthalpy on fluid versus gel phase lipids as they favorably interact with the hydrophobic lipid tails in fluid phase lipids.
The charged moieties decreased in their net enthalpic interaction on fluid versus gel phase lipids even though over twice as many were within 3 Å of the lipids.
G3–NH3+ showed the least increase in total interaction energy on fluid versus gel phase lipids and G3–NH3+ contains the fewest hydrophobic moieties.
Discussion
The most energetically influential moieties in binding were the charged moieties; however, their interactions were remarkably similar to the sum of the hydrophobic moieties on the fluid lipids. These results highlight the importance of internal dendrimer structure and hydrophobic/hydrophilic composition to biological activity.
The results reported in the present work provide specific energetic underpinnings for improving dendrimer design for enhanced targeting of particular membrane components. Initial binding is enhanced greatly by the addition of charged moieties to the dendrimer by strong dendrimer–lipid headgroup interactions. However, the dendrimers are more likely to penetrate into the lipid tails, deform, and prefer fluid phase bilayer if the dendrimer contains hydrophobic regions accessible to the lipid tails without diminishing the hydrophilic interactions.
These simulations provide further evidence regarding the location of dendrimers bound to lipids. In all systems, the dendrimers bound to the lipids. On gel phase lipids, the dendrimers remained atop of the bilayer and did not significantly deform to accommodate binding. On fluid phase lipids, however, the dendrimers did not remain atop the bilayer. The observed intercalation of the dendrimer into the hydrophobic bilayer regions is consistent with continuum models44 and partially consistent with coarse-grained models.22 Coarse-grained models suggest that only the charged dendrimers become secluded in the bilayer, whereas both the continuum models and atomistic models presented here suggest all dendrimers intercalate into the bilayer.
As the dendrimers bound to the fluid phase lipids and the lipids formed a divot to accommodate binding, the lipid head groups partially lined the divot. The lipids were arranged such that the hydrophobic dendrimer moieties were accessible to the hydrophobic lipid tails and the polar dendrimer moieties had some polar lipid head groups nearby also.
This paper addresses two central questions: (1) What are the atomistic causes and effects of dendrimers binding to gel versus fluid bilayers? (2) How does the variation in dendrimer termination and lipid phase affect binding? The numerical results presented answer both questions via atomistic molecular dynamics simulations. Three hypotheses have been developed and tested while providing insight into the importance of dendrimer structure and lipid phase in biological functionality.
Hypothesis 1
The fluid phase lipid bilayer deforms to accommodate dendrimer–lipid interactions with more of the dendrimer and lipids in contact than the gel phase lipids.
These simulations are consistent with this hypothesis. The images for the simulations (Figures 2 and 3), show the dendrimers penetrated into the fluid phase lipids and the lipid head groups have formed a depression accepting the dendrimer. Many of the dendrimer moieties were in close contact with the fluid phase lipid tails, which did not occur with gel phase lipids. As shown in Figure 5, approximately three times as many dendrimer moieties are within 3 Å of the lipids and approximately twice as many lipid molecules are within 3 Å of the dendrimer for fluid versus gel lipids.
Hypothesis 2
The dendrimers achieve stronger interaction energy with fluid phase lipids than gel phase lipids due to the mobility of the fluid phase lipids allowing the dendrimers’ terminal groups to obtain more favorable interactions.
Although the dendrimers as a whole obtain a stronger interaction with fluid versus gel phase lipids, it is not mediated by the terminal groups. Rather, the difference in binding is driven primarily by the inner dendrimer moieties, especially the hydrophobic parts (Figure 8). The net enthalpy of binding is 14% stronger for G3–NH3+, 27% stronger for G3–COO−, and 88% stronger for G3–Ac for the fluid versus gel phase lipids. However, the enthalpy of binding for terminal groups of G3–NH3+ and G3–COO− are weaker on fluid versus gel phase lipids, by 48% and 2%, respectively, with the carboxyl terminal group including both the deprotonated carboxylic acid in addition to the hydrophobic spacer (Figure 8B). The uncharged terminal groups on G3–Ac bind more strongly to fluid versus gel phase lipids, but much less so than the internal dendrimer moieties. This result contradicts previously held perceptions that the dendrimer binding is dominated by the dendrimer terminal groups and emphasizes the importance of the inner dendrimer structure on the fluid versus gel phase interactions.
Hypothesis 3
The charged dendrimers (G3–NH3+ and G3–COO−) bind enthalpically stronger than the uncharged dendrimer (G3–Ac) and, as such, the charged dendrimers become more morphologically changed upon binding.
The charged dendrimers bind more strongly than the uncharged dendrimer (Figure 8). However, contrary to this hypothesis, G3–Ac becomes significantly more deformed upon binding to the lipids than does either of the charged dendrimers (Figure 6). The morphological changes of binding for all three dendrimers are similar upon the gel phase lipids. Yet on the fluid phase lipids, greater deformation occurs for dendrimers that are more hydrophobic, regardless of dendrimer charge.
The approximations in this manuscript for fluid versus gel phase lipids directly model the effects of lipid tail mobility on dendrimer binding morphology, binding energetics, and lipid rearrangement. Absent from this calculation are experimental differences between gel and fluid phase lipids such as area per lipid, bilayer thickness, and temperature. The conditions used in this manuscript would be appropriate for a fluid phase lipid, and the gel phase lipids were constructed solely by limiting lipid tail movement. Interestingly, the trends observed in this model would likely be enhanced further upon incorporation lower temperatures and greater lipid densities for the gel phase lipids, namely, reduced interaction between the dendrimer and the lipid tails as well as diminished dendrimer deformation upon binding.
Conclusions
This manuscript addressed the mechanisms of dendrimer binding to lipid bilayers, specifically the role of the terminations of G3 PAMAM dendrimers and the phase of DMPC lipid bilayers. The variations in dendrimer termination demonstrate the strong effect of charged terminal groups on binding and overall dendrimer morphology. For example, the charged terminal groups on average release 28% less enthalpy upon interacting with fluid versus gel phase lipids, even though over twice as many were within 3 Å of the lipids on fluid versus gel lipids.
The inner-dendrimer hydrophobic components are most influential for enhancing binding to fluid versus gel phase lipids. Similar to the charged terminals, over twice as many of the hydrophobic moieties were within 3 Å of the fluid versus gel phase lipids. Contrary to the charged terminals, the hydrophobic moieties released twice as much enthalpy upon binding to fluid versus gel lipids. All dendrimers bind more strongly to the fluid versus gel phase lipids, but this is most pronounced for the uncharged dendrimer (G3–Ac), with net 88% greater enthalpy release on fluid versus gel phase lipids. It is concluded that the hydrophobic dendrimer moieties are the key to enhanced binding to fluid versus gel phase lipids.
Supplementary Material
Acknowledgment
C.V.K. received fellowship support from the NIH Michigan Molecular Biophysics Training Program (T32 GM008270-20), the Applied Physics Program, and the Graham Environmental Sustainability Institute. Computational time was provided by the Center for Advanced Computing at the University of Michigan and Lawrence Livermore National Laboratory. The authors thank Christine Orme, Timothy Sullivan, James R. Baker, Jr., Elizabeth Janus, and Jeffery Wereszczynski. This research was supported by a Grant from the National Institute of Biomedical Imaging and BioEngineering (R01-EB005028). I.A. gratefully acknowledges support from the NSF CAREER award program (CHE-0548047).
Footnotes
Supporting Information Available: Animations of equilibrated G3–NH3+, G3–Ac, and G3–COO− on gel and fluid phase DMPC bilayers. This material is available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- (1).Sahoo SK, Labhasetwar V. Nanotech approaches to delivery and imaging drug. Drug DiscoVery Today. 2003;8(24):1112–1120. doi: 10.1016/s1359-6446(03)02903-9. [DOI] [PubMed] [Google Scholar]
- (2).Yang H, Kao WYJ. Dendrimers for pharmaceutical and biomedical applications. J. Biomater. Sci., Polym. Ed. 2006;17(1–2):3–19. doi: 10.1163/156856206774879171. [DOI] [PubMed] [Google Scholar]
- (3).Bosman AW, Janssen HM, Meijer EW. About dendrimers: Structure, physical properties, and applications. Chem. Rev. 1999;99(7):1665–1688. doi: 10.1021/cr970069y. [DOI] [PubMed] [Google Scholar]
- (4).Tang MX, Redemann CT, Szoka FC. In vitro gene delivery by degraded polyamidoamine dendrimers. Bioconjugate Chem. 1996;7(6):703–714. doi: 10.1021/bc9600630. [DOI] [PubMed] [Google Scholar]
- (5).Stiriba SE, Frey H, Haag R. Dendritic polymers in biomedical applications: From potential to clinical use in diagnostics and therapy. Angew. Chem., Int. Ed. 2002;41(8):1329–1334. doi: 10.1002/1521-3773(20020415)41:8<1329::aid-anie1329>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- (6).Tomalia DA, Baker H, Dewald J, Hall M, Kallos G, Martin S, Roeck J, Ryder J, Smith P. A new class of polymers—starburstdendritic macromolecules. Polym. J. 1985;17(1):117–132. [Google Scholar]
- (7).Quintana A, Raczka E, Piehler L, Lee I, Myc A, Majoros I, Patri AK, Thomas T, Mule J, Baker JR. Design and function of a dendrimer-based therapeutic nanodevice targeted to tumor cells through the folate receptor. Pharm. Res. 2002;19(9):1310–1316. doi: 10.1023/a:1020398624602. [DOI] [PubMed] [Google Scholar]
- (8).Majoros IJ, Thomas TP, Mehta CB, Baker JR. Poly(amidoamine) dendrimer-based multifunctional engineered nanodevice for cancer therapy. J. Med. Chem. 2005;48(19):5892–5899. doi: 10.1021/jm0401863. [DOI] [PubMed] [Google Scholar]
- (9).Patri AK, Majoros IJ, Baker JR. Dendritic polymer macromolecular carriers for drug delivery. Curr. Opin. Chem. Biol. 2002;6(4):466–471. doi: 10.1016/s1367-5931(02)00347-2. [DOI] [PubMed] [Google Scholar]
- (10).Landmark KJ, DiMaggio S, Ward J, Kelly CV, Vogt S, Hong S, Kotlyar A, Penner-Hahn JE, James R, Baker J, Holl MMB, Orr BG. Synthesis, characterization, and in vitro testing of superparamagnetic iron oxide nanoparticles targeted using folic acid-conjugated dendrimers. ACS Nano. 2008;2(4):773–783. doi: 10.1021/nn800034w. [DOI] [PubMed] [Google Scholar]
- (11).Hubbell JA. Multifunctional polyplexes as locally triggerable nonviral vectors. Gene Ther. 2006;13:1371–1372. doi: 10.1038/sj.gt.3302763. [DOI] [PubMed] [Google Scholar]
- (12).Hong SP, Leroueil PR, Janus EK, Peters JL, Kober MM, Islam MT, Orr BG, Baker JR, Holl MMB. Interaction of polycationic polymers with supported lipid bilayers and cells: Nanoscale hole formation and enhanced membrane permeability. Bioconjugate Chem. 2006;17(3):728–734. doi: 10.1021/bc060077y. [DOI] [PubMed] [Google Scholar]
- (13).Lai JC, Yuan CL, Thomas JL. Single-cell measurements of polyamidoamine dendrimer binding. Ann. Biomed. Eng. 2002;30(3):409–416. doi: 10.1114/1.1470180. [DOI] [PubMed] [Google Scholar]
- (14).Fischer D, Li YX, Ahlemeyer B, Krieglstein J, Kissel T. In vitro cytotoxicity testing of polycations: influence of polymer structure on cell viability and hemolysis. Biomaterials. 2003;24(7):1121–1131. doi: 10.1016/s0142-9612(02)00445-3. [DOI] [PubMed] [Google Scholar]
- (15).Manunta M, Nichols BJ, Tan PH, Sagoo P, Harper J, George AJT. Gene delivery by dendrimers operates via different pathways in different cells, but is enhanced by the presence of caveolin. J. Immunol. Methods. 2006;314(1–2):134–146. doi: 10.1016/j.jim.2006.06.007. [DOI] [PubMed] [Google Scholar]
- (16).Manunta M, Tan PH, Sagoo P, Kashefi K, George AJT. Gene delivery by dendrimers operates via a cholesterol dependent pathway. Nucleic Acids Res. 2004;32(9):2730–2739. doi: 10.1093/nar/gkh595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Kelly CV, Leroueil PR, Janus EK, Wereszczynski JM, Baker JR, Orr BG, Holl MMB, Andricioaei I. Poly(amidoamine) dendrimers on lipid bilayers I: free energy and conformation of binding. J. Phys. Chem. B. 2008;112:xxxx. doi: 10.1021/jp801377a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Leroueil PR, Hong SY, Mecke A, Baker JR, Orr BG, Holl MMB. Nanoparticle interaction with biological membranes: Does nanotechnology present a janus face. Acc. Chem. Res. 2007;40(5):335–342. doi: 10.1021/ar600012y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Mecke A, Uppuluri S, Sassanella TM, Lee DK, Ramamoorthy A, Baker JR, Orr BG, Holl MMB. Direct observation of lipid bilayer disruption by poly(amidoamine) dendrimers. Chem.Phys. Lipids. 2004;132(1):3–14. doi: 10.1016/j.chemphyslip.2004.09.001. [DOI] [PubMed] [Google Scholar]
- (20).Mecke A, Lee DK, Ramamoorthy A, Orr BG, Holl MMB. Synthetic and natural polycationic polymer nanoparticles interact selectively with fluid-phase domains of dmpc lipid bilayers. Langmuir. 2005;21:8588–8590. doi: 10.1021/la051800w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Mecke A, Lee I, Baker JR, Holl MMB, Orr BG. Deformability of poly(amidoamine) dendrimers. Eur. Phys. J. E. 2004;14(1):7–16. doi: 10.1140/epje/i2003-10087-5. [DOI] [PubMed] [Google Scholar]
- (22).Lee H, Larson RG. Molecular dynamics simulations of PAMAM dendrimer-induced pore formation in DPPC bilayers with a coarse-grained model. J. Phys. Chem. B. 2006;110:18204–18211. doi: 10.1021/jp0630830. [DOI] [PubMed] [Google Scholar]
- (23).Mecke A, Majoros IJ, Patri AK, Baker JR, Holl MMB, Orr BG. Lipid bilayer disruption by polycationic polymers: The roles of size and chemical functional group. Langmuir. 2005;21(23):10348–10354. doi: 10.1021/la050629l. [DOI] [PubMed] [Google Scholar]
- (24).Hong SP, Bielinska AU, Mecke A, Keszler B, Beals JL, Shi XY, Balogh L, Orr BG, Baker JR, Holl MMB. Interaction of poly(amidoamine) dendrimers with supported lipid bilayers and cells: Hole formation and the relation to transport. Bioconjugate Chem. 2004;15(4):774–782. doi: 10.1021/bc049962b. [DOI] [PubMed] [Google Scholar]
- (25).Hong S, Leroueil PR, Majoros IJ, Orr BG, Baker JR, Holl MMB. The binding avidity of a nanoparticle-based multivalent targeted drug delivery platform. Chem. Biol. 2007;14(1):105–113. doi: 10.1016/j.chembiol.2006.11.015. [DOI] [PubMed] [Google Scholar]
- (26).Brown DA, London E. Functions of lipid rafts in biological membranes. Annu. Rev. Cell Dev. Biol. 1998;14:111–136. doi: 10.1146/annurev.cellbio.14.1.111. [DOI] [PubMed] [Google Scholar]
- (27).Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387(6633):569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- (28).Holl MMB. Cell plasma membranes and phase transitions. In press.
- (29).Leroueil PR, Berry SA, Duthie K, Han G, Rotello VM, McNerny DQ, Baker JR, Orr BG, Holl MMB. Wide varieties of cationic nanoparticles induce defects in supported lipid bilayers. Nano Lett. 2008;8(2):420–424. doi: 10.1021/nl0722929. [DOI] [PubMed] [Google Scholar]
- (30).Marrink SJ, de Vries AH, Mark AE. Coarse grained model for semiquantitative lipid simulations. J. Phys. Chem. B. 2004;108(2):750–760. [Google Scholar]
- (31).Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. Charmm—a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4(2):187–217. [Google Scholar]
- (32).MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102(18):3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- (33).Mercier GA. Dendrimer Builder. 1996 http://server.ccl.net/chemistry/resources/messages/1996/05/20.009-dir/index.html. [Google Scholar]
- (34).Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J. Mol. Graphics. 1996;14(1):33. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- (35).Lee I, Athey BD, Wetzel AW, Meixner W, Baker JR. Structural molecular dynamics studies on polyamidoamine dendrimers for a therapeutic application: Effects of pH and generation. Macromolecules. 2002;35(11):4510–4520. [Google Scholar]
- (36).Paulo PMR, Lopes JNC, Costa SMB. Molecular dynamics simulations of charged dendrimers: low-to-intermediate half-generation PAMAMs. J. Phys. Chem. B. 2007;111(36):10651–10664. doi: 10.1021/jp072211x. [DOI] [PubMed] [Google Scholar]
- (37).Feig M, Brooks CL. Recent advances in the development and application of implicit solvent models in biomolecule simulations. Curr. Opin. Struct. Biol. 2004;14(2):217–224. doi: 10.1016/j.sbi.2004.03.009. [DOI] [PubMed] [Google Scholar]
- (38).Bashford D, Case DA. Generalized born models of macromolecular solvation effects. Annu. Rev. Phys. Chem. 2000;51:129–152. doi: 10.1146/annurev.physchem.51.1.129. [DOI] [PubMed] [Google Scholar]
- (39).Johnson SJ, Bayerl TM, McDermott DC, Adam GW, Rennie AR, Thomas RK, Sackmann E. Structure of an adsorbed dimyristoylphosphatidylcholine bilayer measured with specular reflection of neutrons. Biophys. J. 1991;59(2):289–294. doi: 10.1016/S0006-3495(91)82222-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Kucerka N, Liu YF, Chu NJ, Petrache HI, Tristram-Nagle ST, Nagle JF. Structure of fully hydrated fluid phase DMPC and DLPC lipid bilayers using X-ray scattering from oriented multilamellar arrays and from unilamellar vesicles. Biophys. J. 2005;88(4):2626–2637. doi: 10.1529/biophysj.104.056606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Hunenberger PH, McCammon JA. Effect of artificial periodicity in simulations of biomolecules under Ewald boundary conditions: a continuum electrostatics study. Biophys. Chem. 1999;78(1–2):69–88. doi: 10.1016/s0301-4622(99)00007-1. [DOI] [PubMed] [Google Scholar]
- (42).Castro-Roman F, Benz RW, White SH, Tobias DJ. Investigation of finite system size effects in molecular dynamics simulations of lipid bilayers. J. Phys. Chem. B. 2006;110(47):24157–24164. doi: 10.1021/jp064746g. [DOI] [PubMed] [Google Scholar]
- (43).Rudnick J, Gaspari G. The asphericity of random walks. J. Phys. A: Math. Gen. 1986;19:L191–L193. [Google Scholar]
- (44).Ginzburg VV, Balijepalli S. Modeling the thermodynamics of the interaction of nanoparticles with cell membranes. Nano Lett. 2007;7(12):3716–3722. doi: 10.1021/nl072053l. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


