Abstract
The Protein Arginine Deiminases (PADs) catalyze the hydrolysis of peptidyl-arginine to form peptidylcitrulline. Abnormally high PAD activity is observed in a host of human diseases, however, the exact role of protein citrullination in these diseases, as well as the identities of specific citrullinated disease biomarkers, remain unknown, largely due to the lack of readily available chemical probes to detect protein citrullination. For this reason, we developed a citrulline specific chemical probe, rhodamine-phenylglyoxal (Rh-PG), which we show can be used to investigate protein citrullination. This methodology is superior to existing techniques because it possesses higher throughput and excellent sensitivity. Additionally, we demonstrate that this probe can be used to determine the kinetic parameters for a number of protein substrates, monitor drug efficacy, and identify disease biomarkers in an animal model of ulcerative colitis that displays aberrantly increased PAD activity.
The Protein Arginine Deiminases (PADs) catalyze the posttranslational conversion of peptidyl-arginine to peptidylcitrulline; a process commonly referred to as deimination or citrullination.1,2 PAD activity is aberrantly increased in different diseases,1,2 including rheumatoid arthritis (RA), ulcerative colitis (UC), Alzheimer’s disease (AD), multiple sclerosis (MS), lupus, Parkinson’s disease, and cancer.3-9 For example, increased PAD levels are observed in the RA synovium,10 during the formation of Neutrophil Extracellular Traps (NETs) in colitis and lupus,11 within the brains of patients with MS,12 in the joints of patients with osteoarthritis,10 in UC lesions,13 in the hippocampal extracts of AD patients,5 and in several carcinomas.6 Furthermore, the administration of Cl-amidine, a potent PAD inhibitor,14 or a Cl-amidine analog15, has proven useful in decreasing disease severity in animal models of UC, RA, and cancer.15-17
Although abnormally high PAD activity and protein citrullination is observed in these and other diseases, the exact role(s) and target(s) of these enzymes are still poorly understood, in part due to a lack of chemical tools that can be used to identify the proteins that are citrullinated within a specific disease model. The ability to investigate the specific target and role of the PADs in these diseases will inevitably allow for better, more tailored treatments, and identify biomarkers that can be used to diagnose and monitor the efficacy of treatments for these diseases. Additionally, the ability to rapidly detect changes in protein citrullination in response to the addition of a PAD inhibitor will facilitate the development of such compounds.
Common methods for studying citrullination include the Color Development Reagent (COLDER) assay18-21 and a commercially available Anti-Citrulline (Modified) Detection Kit (ACM kit; Millipore; Billerica, MA).22 Although widely used for studying small molecule and peptide substrates of the PADs, the COLDER assay has a relatively high limit of detection (LOD; ~0.6 nmol) making it difficult to use with less concentrated protein substrates. The ACM kit has also proven useful, but suffers from relatively high cost ($40/assay), a lengthy two day procedure, and specialized analysis. Although other means of detecting citrullinated proteins have been described, these are also antibody based (e.g.,the F95 antibody23 and citrullinated histones H3 and H4). In contrast, the probes described herein are readily synthesized, show excellent sensitivity, will react with any citrulline bearing protein, and can be used by most researchers because detection uses reagents/instruments that are available in virtually all biochemically focused labs.
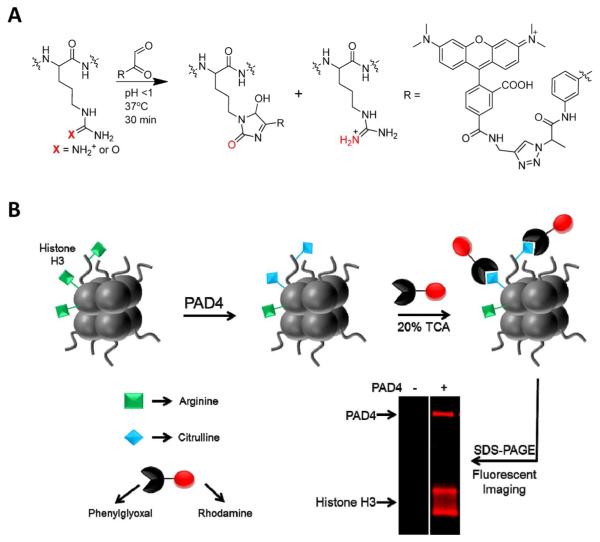
The design of the citrulline specific probe described herein is based on the chemoselective reaction that occurs between glyoxals and either citrulline or arginine under acidic or basic conditions, respectively (Figure 1A and Figure S1).24 Although Fleckenstein et al.,24 reported the derivativation of phenylglyoxal onto the solid phase via the para position, this chemistry was not amenable to the solution phase (Figure S2). Therefore we focused on synthesizing phenylglyoxal compounds with an azido group on the meta position that could be reacted with alkyne modified rhodamine via the copper catalyzed azide-alkyne cycloaddition reaction (Figure S3).25,26 The reaction between citrulline and phenylglyoxal likely proceeds as previously suggested (Figure S4).21,24
Figure 1.
(A) The rhodamine-phenylglyoxal (Rh-PG) probe chemoselectively labels citrulline over arginine at acidic pH. (B) A schematic representation of the method used to monitor citrullination with the Rh-PG probe. Specific arginine residues (green squares) of histone H3 are converted to citrulline (blue diamonds) by PAD4. Treatment of citrullinated protein with the Rh-PG probe under acidic conditions labels citrulline residues selectively. Subsequent separation by SDS-PAGE and fluorescent analysis reveals citrullinated proteins. Note this is an actual gel image, demonstrating the selectivity of Rh-PG.
Probe selectivity was first determined against commonly reactive amino acids, showing that under acidic conditions, Rh-PG reacted selectively with citrulline, homocitrulline, and cysteine (Figure S5). Using a peptide substrate containing both citrulline and cysteine, we showed that the thiohemiacetal generated between Rh-PG and Cys is hydrolyzed at neutral pH, while the cyclic system formed between Rh-PG and citrulline is not (Figure S6,). Since the gel-based screening method includes a neutralization step prior to gel loading, the problem of cysteine cross-reactivity is effectively negated; the selectivity versus Cys under these conditions is ≥650-fold. After demonstrating that our PG probe is capable of selectively reacting with either arginine or citrulline in a pH dependent manner (Figure S1), conditions for probe labeling were optimized, including the ideal probe concentration (i.e., 100 μM; Figure S7), ideal labeling time (i.e., 30 min; Figure S8), and the probe sensitivity/LOD (i.e., ~10 ng (0.67 pmol) of citrullinated histone H3 and ~1 ng (12.7 fmol) of autodeiminated PAD4; Figure S9). The different LODs are due to differences in the citrulline content of the two proteins. Since Rh-PG labeling is performed in 20% trichloroacetic acid (TCA) at 37 °C, it is important to note that increased labeling temperatures and times lead to the formation of SDS insoluble protein aggregates and decreased gel loading (Figure S10). Also, quenching of excess probe with citrulline after labeling reduced background labeling of arginine residues upon resuspension of the protein (data not shown).
To demonstrate that the Rh-PG probe detection method compares favorably to the commercially available ACM kit, we performed a comparative analysis of the two methods using PAD4 autodeimination as the endpoint. PAD4, autodeiminated for various amounts of time, was analyzed with Rh-PG (Figure 2A; top) and the ACM kit (Figure 2A; bottom) according to the manufacturer’s instructions. Silver staining of the fluorescent gel was done to confirm equal protein loading (Figure 2A; middle). Analysis of this data (Figure 2B) indicates that the two techniques provide virtually identical results. The rate of recombinant histone H3 citrullination was also followed and the results of the two methods again show essentially identical results (Figure S11). These data indicate that the Rh-PG method is comparable to commercially available techniques. However, compared to the ACM kit, the Rh-PG labeling method takes significantly less time (~3 h versus ≥25 h), requires fewer steps (6 versus 12), and simpler analysis (fluorescent imaging versus Western blotting).
Figure 2.
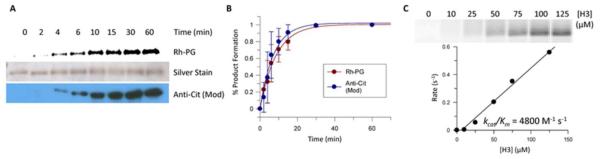
(A and B) Time course of PAD4 autodeimination is similar for the Rh-PG probe and commercially available antibody methods. (A) Rh-PG probed fluorescent image (top) and Anti-Cit (Mod) Kit Western blot analysis (bottom) of autodeiminated PAD4. Silver staining (middle) shows equal protein loading. (B) Plot of percent citrullination over time for each type of analysis, indicating that the two methods are comparable. (C) The Rh-PG labeling method can also be used to determine the kcat/Km value of histone H3 citrullination by PAD4.
Given the high sensitivity of Rh-PG, we next determined whether this methodology could be used to accurately quantify changes in citrullination by determining the steady state kinetic parameters for a protein substrate. Although we and others have used the traditional COLDER assay to do this type of analysis on protein substrates,27 it is difficult to obtain highly accurate rates due to the limited sensitivity of the COLDER assay and the high concentrations of proteins required to accurately measure citrulline production. For the Rh-PG kinetic experiments, we first monitored H3 citrullination as a function of time to ensure that the kinetic parameters were determined under initial velocity conditions (Figure S12). Note that Rh-PG fluorescence was converted to citrulline concentration, using a citrulline standard curve (see supplementary information for details). Having identified a specific time that could be used to obtain the initial rates, we used this method to determine the steady-state kinetic parameters for the citrullination of histone H3 by PAD4 (Figure 2C). The results of this analysis provided a kcat/Km value of 4800 ± 1100 M−1 s−1, which compares favorably with our previously reported value of 3700 M−1 s−1 27. Similar analyses performed on the PAD4 catalyzed deimination of histone H4 gave a value of 2800 ± 200 M−1 s−1 (Figure S13), which is also similar to the previously reported value.27 Given the poor sensitivity and high signal to noise ratios of current methods for determining the rates of protein citrullination (i.e., the COLDER analysis and ammonia detection methods),9,28 the accuracy and excellent sensitivity of our method (note that samples had to be diluted 60 fold before analysis) provides a unique tool to study protein deimination because it permits the determination of Km values for high affinity protein substrates and allows for the study of proteins that are difficult to produce in large quantities.
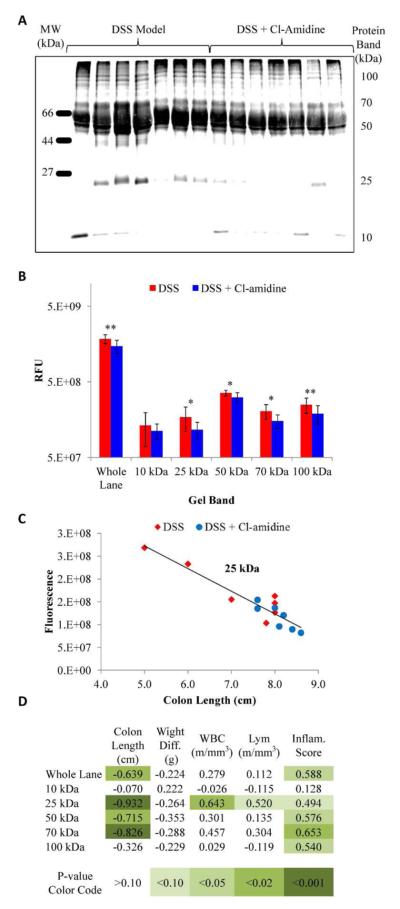
We envisioned that a key advantage of this probe would be its facile ability to visualize target engagement (i.e., decreased citrullination upon inhibitor treatment) as well as the detection/identification of disease-associated biomarkers. To demonstrate its utility for these types of analyses, we evaluated changes in protein citrullination using serum samples from our previous study demonstrating that Cl-amidine, a PAD inhibitor, reduces disease severity in the DSS mouse model of ulcerative colitis.16 Citrullinated proteins present in these samples were labeled with the Rh-PG probe, separated by SDS-PAGE, and fluorescent proteins imaged (Figure 3A). We subsequently analyzed whole lane fluorescence as a function of the addition of Cl-amidine (Figure 3B). Consistent with our previous studies, which used the COLDER assay to measure changes in total protein citrullination, analysis of whole lane fluorescence revealed a significant (P-value <0.01) reduction in fluorescence intensity for the serum samples treated with Cl-amidine. Note that selectivity analysis indicated the Cl-amidine itself does not react with the probe (data not shown). Unlike the previous analysis, which only measures total protein citrullination, the Rh-PG method allowed us to monitor effects on specific proteins, as opposed to global serum citrullination. Based on visual inspection of the fluorescent images, we identified five protein bands (at 10, 25, 50, 70, and 100 kDa) within each lane that appeared to show a marked change in protein citrullination as a function of Cl-amidine. From a comparison of the fluorescent intensities of these bands between the two groups, we observed that 4 out of the 5 protein bands show statistically significant decreases in citrullination in response to Cl-amidine (P-value <0.05; Table S1). The only protein band analyzed that did not show a significant decrease in response to Cl-amidine (P-value 0.44) was the band at 10 kDa.
Figure 3.
(A) Fluorescent image of Rh-PG labeled serum samples from a mouse model of DSS induced UC. (B) Analysis of the UC samples indicates a significant decrease in protein citrullination in response to Cl-amidine (*P-value <0.05; **P-value <0.01). (C) Correlation plot showing good correlation between colon length and fluorescence of the protein band at 25 kDa. (D) Table of correlation coefficients between various disease scores and specific protein band fluorescent intensity.
Given these findings, we hypothesized that direct correlations between the citrullination of these proteins and disease severity may exist. To test this hypothesis, we compared the fluorescence intensity data for the five proteins described above to each of five different disease metrics obtained for the mice in this study (i.e., colon length, weight difference, white blood cell count, lymphocyte count, and inflammation score) (Table S2). Correlation plots (Figure 3C and Figure S14) and correlation coefficients (Figure 3D) were generated, and colon length (4 out of 6 significant correlations) and inflammation scores (5 out of 6 significant correlations) showed the highest correlations between disease severity and citrulline levels. Note that these two endpoints are the key indicators of disease severity.16,29,30
Interestingly, the 10 kDa protein, the only protein with no significant decrease in citrullination in response to Cl-amidine, also has no significant correlation to any of the disease scores, meaning it essentially serves as a negative control. The highest correlation coefficients, and therefore the most relevant correlations, exist between colon length and the levels of the citrullinated 25 kDa and 70 kDa proteins (correlation coefficients = −0.932 and −0.826, respectively; Figure 3C). Important to note is that the average protein levels, as analyzed by coomassie staining, did not change between untreated and Cl-amidine treated samples for either of these protein bands, indicating that the change in fluorescence is indeed due to a change in citrullination. The 25 kDa band also showed the greatest number of significant correlations with disease severity (i.e., 4 out of 5 disease activity scores). The only outlier is weight loss, which also did not correlate with any of the other protein bands analyzed. The strong correlation between citrullination of the 25 kDa protein and common disease activity scores, especially colon length, suggests that this protein is a biomarker of ulcerative colitis.
Although the Rh-PG probe is clearly useful for studying recombinant proteins, which is itself beneficial to the PAD field, this work demonstrates its true utility in studying more complex samples from disease states (e.g., UC, RA, Alzheimer’s disease, Parkinson’s disease, and cancer, to name a few) in which dysregulated PAD activity plays a role. For example, while anti-citrulline protein antibodies (ACPAs) are well established biomarkers for RA, less is understood about the targets of these antibodies. Additionally, we do not know the identities of citrullinated protein biomarkers for the other diseases in which the PADs are involved.
In total, these data demonstrate that the Rh-PG probe is a powerful chemical probe of protein citrullination that will undoubtedly be useful for providing robust and telling insights into the role of particular PAD substrates in PAD related diseases. Further work is currently underway to determine the identity of that 25 kDa protein and identify and characterize unique disease biomarkers in other diseases in which the PADs play a role.
Supplementary Material
ACKNOWLEDGMENT
Financial support for this work provided by NIH grants GM079357 (P.R.T.), CA151304 (P.R.T. and L.J.H), and TSRI.
Footnotes
ASSOCIATED CONTENT
Synthetic procedures, experimental details, and supplemental figures. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interests.
REFERENCES
- (1).Jones JE, Causey CP, Knuckley B, Slack-Noyes JL, Thompson PR. Curr Opin Drug Discov Devel. 2009;12:616. [PMC free article] [PubMed] [Google Scholar]
- (2).Vossenaar ER, Zendman AJ, van Venrooij WJ, Pruijn GJ. Bioessays. 2003;25:1106. doi: 10.1002/bies.10357. [DOI] [PubMed] [Google Scholar]
- (3).Schellekens GA, de Jong BA, van den Hoogen FH, van de Putte LB, van Venrooij WJ. J. Clin. Invest. 1998;101:273. doi: 10.1172/JCI1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Moscarello MA, Mastronardi FG, Wood DD. Neurochem. Res. 2007;32:251. doi: 10.1007/s11064-006-9144-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Ishigami A, Ohsawa T, Hiratsuka M, Taguchi H, Kobayashi S, Saito Y, Murayama S, Asaga H, Toda T, Kumura N, Maruyama N. J. Neurosci. Res. 2005;80:120. doi: 10.1002/jnr.20431. [DOI] [PubMed] [Google Scholar]
- (6).Chang X, Han J, Pang L, Zhao Y, Yang Y, Shen Z. BMC Cancer. 2009;9:40. doi: 10.1186/1471-2407-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Chang X, Han J. Mol. Carcinog. 2006;45:183. doi: 10.1002/mc.20169. [DOI] [PubMed] [Google Scholar]
- (8).Jones JE, Causey CP, Knuckley BE, Slack JL, Thompson PR. Curr. Opin. Drug Discov. Devel. 2009;12:616. [PMC free article] [PubMed] [Google Scholar]
- (9).Kearney PL, Bhatia M, Jones NG, Yuan L, Glascock MC, Catchings KL, Yamada M, Thompson PR. Biochemistry. 2005;44:10570. doi: 10.1021/bi050292m. [DOI] [PubMed] [Google Scholar]
- (10).Kinloch A, Lundberg K, Wait R, Wegner N, Lim NH, Zendman AJ, Saxne T, Malmstrom V, Venables PJ. Arthritis Rheum. 2008;58:2287. doi: 10.1002/art.23618. [DOI] [PubMed] [Google Scholar]
- (11).Wang Y, Li M, Stadler S, Correll S, Li P, Wang D, Hayama R, Leonelli L, Han H, Grigoryev SA, Allis CD, Coonrod SA. J. Cell. Biol. 2009;184:205. doi: 10.1083/jcb.200806072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Wood DD, Ackerley CA, Brand B, Zhang L, Raijmakers R, Mastronardi FG, Moscarello MA. Lab. Invest. 2008;88:354. doi: 10.1038/labinvest.3700748. [DOI] [PubMed] [Google Scholar]
- (13).Chen CC, Isomoto H, Narumi Y, Sato K, Oishi Y, Kobayashi T, Yanagihara K, Mizuta Y, Kohno S, Tsukamoto K. Clin. Immunol. 2008;126:165. doi: 10.1016/j.clim.2007.09.001. [DOI] [PubMed] [Google Scholar]
- (14).Luo Y, Arita K, Bhatia M, Knuckley B, Lee Y-H, Stallcup MR, Sato M, Thompson PR. Biochemistry. 2006;45:11727. doi: 10.1021/bi061180d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Wang Y, Li P, Wang S, Hu J, Chen XA, Wu J, Fisher M, Oshaben K, Zhao N, Gu Y, Wang D, Chen G, Wang Y. Journal of Biological Chemistry. 2012;287:25941. doi: 10.1074/jbc.M112.375725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Chumanevich AA, Causey CP, Knuckley BA, Jones JE, Poudyal D, Chumanevich AP, Davis T, Matesic LE, Thompson PR, Hofseth LJ. American Journal of Physiology - Gastrointestinal and Liver Physiology. 2011;300:G929. doi: 10.1152/ajpgi.00435.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Willis VC, Gizinski AM, Banda NK, Causey CP, Knuckley B, Cordova KN, Luo Y, Levitt B, Glogowska M, Chandra P, Kulik L, Robinson WH, Arend WP, Thompson PR, Holers VM. The Journal of Immunology. 2011;186:4396. doi: 10.4049/jimmunol.1001620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Knipp M, Vasak M. Anal. Biochem. 2000;286:257. doi: 10.1006/abio.2000.4805. [DOI] [PubMed] [Google Scholar]
- (19).Knuckley B, Causey CP, Jones JE, Bhatia M, Dreyton CJ, Osborne TC, Takahara H, Thompson PR. Biochemistry. 2010;49:4852. doi: 10.1021/bi100363t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Knuckley B, Causey CP, Pellechia PJ, Cook PF, Thompson PR. ChemBioChem. 2010;11:161. doi: 10.1002/cbic.200900698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Holm A, Rise F, Sessler N, Sollid LM, Undheim K, Fleckenstein B. Analytical Biochemistry. 2006;352:68. doi: 10.1016/j.ab.2006.02.007. [DOI] [PubMed] [Google Scholar]
- (22).Moelants EAV, Van Damme J, Proost P. PLoS ONE. 2011;6:e28976. doi: 10.1371/journal.pone.0028976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Nicholas AP, Whitaker JN. Glia. 2002;37:328. [PubMed] [Google Scholar]
- (24).Tutturen AEV, Holm A, Jørgensen M, Stadtmüller P, Rise F, Fleckenstein B. Analytical Biochemistry. 2010;403:43. doi: 10.1016/j.ab.2010.04.012. [DOI] [PubMed] [Google Scholar]
- (25).Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. Angew Chem Int Ed Engl. 2002;41:2596. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- (26).Tornoe CW, Christensen C, Meldal M. J Org Chem. 2002;67:3057. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- (27).Slack JL, Jones LE, Bhatia MM, Thompson PR. Biochemistry. 2011;50:3997. doi: 10.1021/bi200309e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Sugawara K, Oyama F. Journal of Biochemistry. 1981;89:771. doi: 10.1093/oxfordjournals.jbchem.a133257. [DOI] [PubMed] [Google Scholar]
- (29).Hou Y-C, Chu C-C, Ko T-L, Yeh C-L, Yeh S-L. European Journal of Nutrition. 2012 doi: 10.1007/s00394-012-0416-3. DOI: 10.1007/s00394-012-0416-3. [DOI] [PubMed] [Google Scholar]
- (30).Poudyal D, Mai Le P, Davis T, Hofseth AB, Chumanevich AP, Chumanevich AA, Wargovich MJ, Nagarkatti M, Nagarkatti PS, Windust A, Hofseth LJ. Cancer Prevention Research. 2012;5:685. doi: 10.1158/1940-6207.CAPR-11-0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.