Abstract
Calcium is a second messenger in virtually all cells and tissues1. Calcium signals in the nucleus have effects on gene transcription and cell growth that are distinct from those of cytosolic calcium signals; however, it is unknown how nuclear calcium signals are regulated. Here we identify a reticular network of nuclear calcium stores that is continuous with the endoplasmic reticulum and the nuclear envelope. This network expresses inositol 1,4,5-trisphosphate (InsP3) receptors, and the nuclear component of InsP3-mediated calcium signals begins in its locality. Stimulation of these receptors with a little InsP3 results in small calcium signals that are initiated in this region of the nucleus. Localized release of calcium in the nucleus causes nuclear protein kinase C (PKC) to translocate to the region of the nuclear envelope, whereas release of calcium in the cytosol induces translocation of cytosolic PKC to the plasma membrane. Our findings show that the nucleus contains a nucleoplasmic reticulum with the capacity to regulate calcium signals in localized subnuclear regions. The presence of such machinery provides a potential mechanism by which calcium can simultaneously regulate many independent processes in the nucleus.
The messenger actions of calcium have been characterized extensively in the cytosol, where this cation regulates such varied processes as contraction, locomotion, morphogenesis, growth, differentiation and secretion1. Calcium can regulate very different cytosolic processes, in part, through local subcellular signals. Calcium in different organelles also has specific roles in cell regulation; for example, the endoplasmic reticulum (ER) is the principal intracellular calcium store1, but the Golgi and mitochondria are also involved in calcium signalling2,3.
Free calcium in the nucleus regulates important functions such as protein transport across the nuclear envelope4,5 and the transcription of some genes6–8. There is controversy, however, about the mechanism by which free calcium in the nucleus is controlled. Nuclear machinery has been identified that would permit calcium to be released from the nuclear envelope directly into the nucleoplasm9,10, but there also is evidence that calcium diffuses into the nucleus from the cytosol through nuclear pores11,12. In either pathway, calcium would reach the interior of the nucleus through diffusion from the nuclear boundary, and the kinetic analysis of nuclear calcium waves is consistent with this interpretation13. But this simple mechanism would allow the nucleus to behave only as a single compartment that is regulated in a uniform fashion by calcium, whereas the nucleus actually is a heterogeneous compartment in which many processes are regulated simultaneously. We therefore examined whether the nucleus contains more intricate calcium signalling machinery that can release calcium locally in discrete regions of the nuclear interior.
We identified a nucleoplasmic reticulum in SKHep1 epithelial cells in several ways. First, we labelled cells with the ER membrane dye ER-Tracker. Because of the small size of these intranuclear membranes, we examined the cells using two-photon excitation to minimize photobleaching14,15. This detected a fine, branching intranuclear network that was continuous with the nuclear envelope and the ER (Fig. 1a, b). This network was less apparent using epifluorescence microscopy (see Supplementary Information Fig. S1). Similar intranuclear extensions of the ER have been described previously and are thought to be widespread among mammalian cell types16. In addition, they may be dynamic structures that become altered during cell proliferation or in some disease states17.
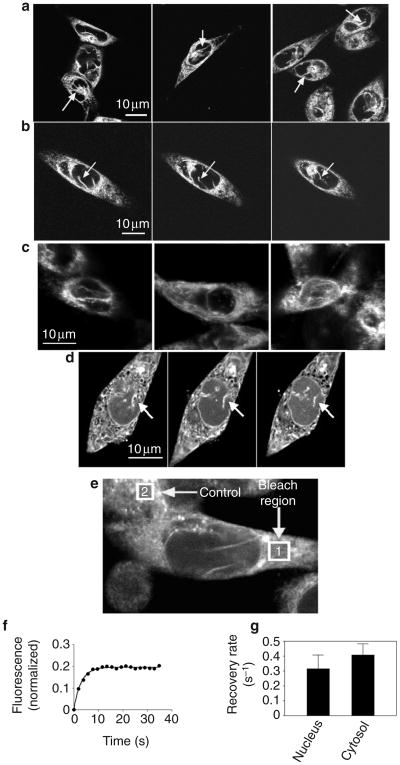
Figure 1. The nucleus of SKHep1 cells contains a nucleoplasmic reticulum.
a, Different fields of SKHep1 cells labelled with the dye ER-Tracker and visualized by two-photon microscopy show the presence of reticular structures in the nucleus (arrows). b, Serial focal planes of a single cell show one of these reticular structures traversing the nucleus (arrows). c, Confocal immunofluorescence images of three different SKHep1 cells labelled with antibodies against calreticulin show the presence of reticular structures in the nucleus. d, Serial focal planes of an SKHep1 cell labelled with the calcium dye fluo-4/AM and visualized by confocal microscopy show that the nucleoplasmic reticulum (arrows) stores calcium. The nucleoplasmic reticulum can be followed from the ER and nuclear envelope into the nuclear interior. e, FRAP was used to examine the kinetics of refilling of mag-fluo-4 in the ER and the nucleoplasmic reticulum. The fluorescence of a bleach region (1) in the ER of a cell is monitored over time and is normalized to the fluorescence of a control region (2) in a nearby cell that is not photobleached. f, FRAP of the bleached region is closely described by a mono-exponential decay equation. Data points and the corresponding non-linear regression curve for a representative experiment are shown. g, Rate constants are similar for FRAP regions in the nucleus and the cytosol (P > 0.45).
To confirm the relationship of these structures to the ER, we examined the intracellular distribution of the ER protein calreticulin by immunofluorescence (Fig. 1c). Images were recorded near the middle of the nucleus, at least 2 μm from either the upper or the lower nuclear boundary. Similarly to ER-Tracker, calreticulin labelling was distributed in a reticular pattern that was not only in the cytosol, but also in the nucleus. The reticular labelling in the nucleus extended from the nuclear envelope to deep within the nuclear interior. These findings identify a nucleoplasmic reticulum in SKHep1 cells, similarly to previous observations in other cell types using other ER markers16.
To determine whether these structures store calcium, we loaded the cells with fluo-4/AM under conditions that favour compartmentalization of dye in the ER18 and examined them by confocal microscopy. This detected a fine, branching intranuclear network that was continuous with the nuclear envelope and the ER (Fig. 1d), similarly to what we observed in cells labelled with ER-Tracker. Discrete intranuclear accumulations of the calcium dye fluo-3 have also been described19. As an alternative approach, we also loaded cells with the calcium dye mag-fluo-4/AM, which preferentially labels ER calcium stores owing to its low affinity for calcium (dissociation constant (Kd) = 22 μM). As with fluo-4, mag-fluo-4 fluorescence also detected a fine, branching intranuclear network that was continuous with the nuclear envelope and the ER (Fig. 1e and Supplementary Information Fig. S2).
We carried out fluorescence recovery after photobleaching (FRAP) to determine whether intranuclear fluorescence represented dye associated with heterogeneously distributed nuclear proteins, rather than dye in a membrane-bound compartment (Fig. 1f, g). FRAP in both cytosolic and nuclear regions was described by a single exponential term according to the equation f(t) = f0(1 − e−kt), where f0 is the initial fluorescence, f(t) is the fluorescence t seconds after photobleaching, and k is the rate constant for fluorescence recovery. The data were closely approximated using this equation (Fig. 1f), with an R2 value of 0.91 ± 0.01 (mean ± s.e.m; R2, measure of goodness of fit for non-linear regression). The recovery rate constant was 0.32 ± 0.09 s−1 (n = 14) for nuclear fluorescence and 0.40 ± 0.08 s−1 for cytosolic fluorescence (n = 19; P > 0.45; Fig. 1g). As the fluorescence recovery of mag-fluo-4 follows similar kinetics in the nucleus and the cytosol, this finding suggests that the nuclear fluorescence of mag-fluo-4 was contained in a membrane-enclosed compartment similar to the ER. Taken together, these findings show that the nuclear interior contains a calcium-storing network that is continuous with the ER and the nuclear envelope.
Calcium signalling in SKHep1 cells is mediated only by the InsP3 receptor because these cells do not express the ryanodine receptor20. We therefore examined the expression and subcellular distribution of InsP3 receptor isoforms in these cells. Immunoblot analysis showed that these cells express the type II and III isoforms of the InsP3 receptor, but not the type I isoform (Fig. 2a). The nuclear and cytosolic fractions were also analysed by immunoblot (Fig. 2b and Supplementary Information Fig. S3). Densitometric analysis showed that the amount of the type II isoform in the nucleus was 51 ± 15% of its amount in the cytosol. By contrast, the nucleus contained only 24 ± 7% of the cytosolic amount of the type III isoform (P < 0.03; n = 3 sets of blots for each isoform). Thus, both the type II and type III InsP3 receptor are expressed in these cells, but the type II receptor is enriched in the nucleus relative to the type III isoform.
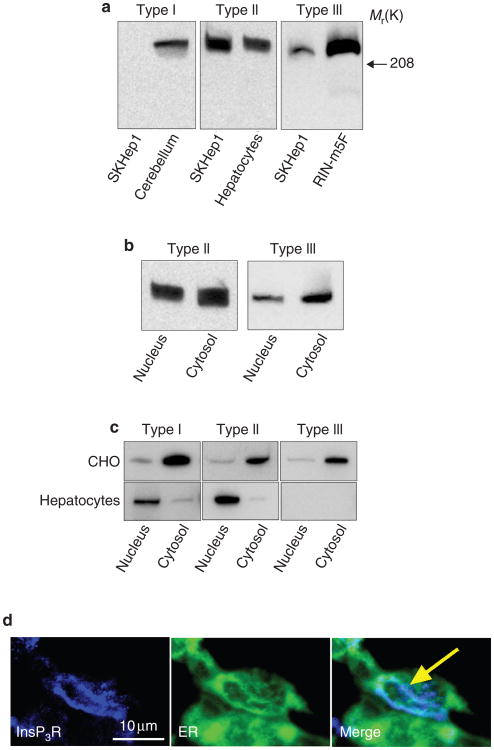
Figure 2. SKHep1 cells express InsP3 receptors in the nucleoplasmic reticulum.
a, Membranes extracted from SKHep1 whole-cell lysates and probed with antibodies specific for the InsP3 receptor isoforms show expression of type II and III receptors but not the type I receptor. Positive controls were rat cerebellum (type I receptor), rat hepatocytes (type II) and RIN-m5F cells (type III)36. b, Nuclear and cytosolic cell fractions probed with antibodies against the type II and type III InsP3 receptors show that each compartment expresses both isoforms. c, Nuclear and cytosolic fractions of CHO cells and hepatocytes probed with antibodies specific for the InsP3 receptor isoforms show cell-to-cell differences in the relative distribution of each isoform in the nucleus and the cytosol. d, Confocal immunofluorescence shows that the type II InsP3 receptor is expressed in both the cytosol and the nucleus of SKHep1 cells, and that it is distributed along the nucleoplasmic reticulum in the nucleus (arrow). The InsP3 receptor was labelled with the same antibodies used for immunoblots. The ER and its nuclear extensions were labelled with the calreticulin antibody used in Fig. 1. No InsP3 receptor labelling was observed in negative controls labelled with secondary antibody alone (not shown).
To determine whether differences in the subcellular distribution of InsP3 receptor isoforms also occur in other types of cell, we analysed nuclear and cytosolic fractions from Chinese hamster ovary (CHO) cells and primary rat hepatocytes by immunoblot (Fig. 2c). We examined these two cell types because they have been shown to contain extensions of the ER into their nuclei16. Densitometric analysis showed that the nuclear/cytosolic ratio of the type I, II and III InsP3 receptors in CHO cells was 0.48 ± 0.17, 0.23 ± 0.12 and 0.43 ± 0.20, respectively. By contrast, the nuclear/cytosolic ratio of the type I and type II InsP3 receptors in hepatocytes was 3.89 ± 0.41 and 19.5 ± 3.9, respectively, whereas the type III isoform was not expressed. Thus, biochemical fractionation studies suggest that SKHep1 cells, CHO cells and primary hepatocytes each have a distinct distribution of InsP3 receptor isoforms in their nucleus and cytosol.
As the above studies do not exclude the possibility that the nuclear fractions may be contaminated with ER (see Supplementary Information Fig. S3), we examined SKHep1 cells by confocal immunofluorescence to confirm and to determine the distribution of the type II InsP3 receptor in the nucleus (Fig. 2d). Cells were double-labelled with an antibody specific for calreticulin to facilitate identification of the ER and its intranuclear extensions. The intranuclear InsP3 receptor was localized not only to the nuclear envelope but also to the nucleoplasmic reticulum. Thus, the nucleus of SKHep1 cells is relatively enriched in the type II InsP3 receptor, and this receptor is expressed, in part, along the nucleoplasmic reticulum.
We examined the function of intranuclear InsP3 receptors in several ways. First, we stimulated cells with hepatocyte growth factor (HGF) because growth factors may preferentially activate calcium signalling machinery that is associated with the nucleus21. Calcium signals induced by HGF (50 ng ml−1) were observed in 51 cells. The nuclear calcium signal preceded the cytosolic calcium signal by 2.63 ± 0.25 s (P < 0.0001) in 21 of these cells (Fig. 3a, b), and began simultaneously (that is, less than 1-second apart) in the nucleus and cytosol in the other 30 cells. This shows that growth factors can preferentially induce nuclear calcium signals. Both the epidermal growth factor (EGF) and fibroblast growth factor (FGF) receptors can translocate to the nucleus22,23, which provides a potential route by which receptor tyrosine kinases may selectively activate calcium signalling pathways in the nucleus.
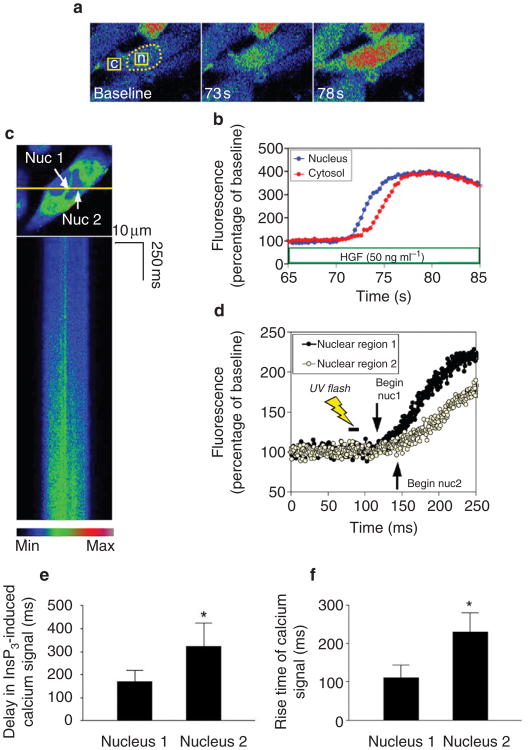
Figure 3. Nuclear calcium signals begin in the nucleoplasmic reticulum.
a, Serial confocal images of an SKHep1 cell stimulated with HGF. Broken line delineates the nucleus; fluorescence was monitored in the nuclear and cytosolic regions of interest indicated by the squares in the left panel. Fluorescence increases first in the nucleus (73 s) and then in the cytosol (78 s). All calcium images are pseudocoloured according to the scale in c. b, Representation of the fluorescence increases in the nuclear and cytosolic regions indicated in a. The nuclear increase in calcium precedes the cytosolic increase. c, High-speed confocal line scanning microscopy in an SKHep1 cell labelled with fluo-4/AM. The nucleus is examined while microinjected NPE-caged InsP3 is released by ultraviolet flash photolysis. Top, the cell and the line used for line scanning (yellow); bottom, the line scan collected as InsP3 is uncaged. The calcium signal in the nucleoplasm begins near the area of the nucleoplasmic reticulum (nuc1) and then spreads to the rest of the nucleoplasm (nuc2). d, Representation of the fluorescence increases in the line scan. The increase in calcium begins in the region of the nucleoplasmic reticulum (nuclear region 1) and then spreads to more distant regions in the nucleoplasm (nuclear region 2). e, Calcium signals in the nucleus begin sooner in the region of the nucleoplasmic reticulum. The latency period between photorelease of InsP3 and the onset of nuclear calcium signalling was shorter in the region of the nucleoplasmic reticulum (*P < 0.05, n = 10). f, Calcium signals in the nucleus rise more quickly in the region of the nucleoplasmic reticulum. The time required for signals to increase from 25% to 75% of their maximum value was shorter in the region of the nucleoplasmic reticulum (*P < 0.002, n = 10).
Next, we examined the function of intranuclear InsP3 receptors more directly through the photorelease of nitrophenylethyl ester (NPE)-caged InsP3 in SKHep1 cells. NPE-caged InsP3 was microinjected into the cells and then released in a controlled fashion by ultraviolet flash photolysis. Confocal line scanning microscopy24 was used to obtain adequate spatial and temporal resolution of the InsP3-induced calcium signals (Fig. 3c, d). Release of InsP3 resulted in calcium signals that began within 170 ± 51 ms in the region of the nucleoplasmic reticulum (mean ± s.e.m., n = 10), but only within 323 ± 102 ms elsewhere in the nucleus (P < 0.05 by paired t-test; Fig. 3e). Similarly, the rise time for calcium signals was 111 ± 33ms near the nucleoplasmic reticulum but was 230 ± 154 ms elsewhere in the nucleus (P < 0.001; Fig. 3f). The observation that calcium signals begin sooner and rise faster in the region of the nucleoplasmic reticulum indicates that InsP3-induced calcium signals in the nucleus may begin in that region.
A previous study in HeLa cells addressed this topic and showed that the calcium ionophore ionomycin caused greater increases in fluo-3 fluorescence in intranuclear ‘hot spots’ than elsewhere in the nucleus19. In that study, however, images were collected only every 10–20 s. In addition, the data were not normalized to account for initial differences in fluorescence distribution in the nucleus; therefore, it was not clear whether calcium increased sooner in the region of these hot spots or even whether the percentage increase in fluorescence was greater than elsewhere in the nucleus. By contrast, our findings provide quantitative evidence through high-resolution spatial and temporal measurements to support the idea that InsP3-induced calcium signals in the nucleus begin at or near the nucleoplasmic reticulum.
To examine directly the effect of InsP3 on subnuclear calcium release, we developed a technique for localized, intranuclear flash photolysis of NPE-caged InsP3 using two-photon excitation (Fig. 4). The single photon absorption of the NPE caging group is maximal at 260 nm, therefore it is difficult to photolyse NPE-caged compounds efficiently by two-photon excitation using even the shortest wavelength (∼690 nm) available from a mode-locked Ti:Sapphire (Ti:S) laser. To achieve spatially localized uncaging using two-photon excitation, we doubled the frequency of the Ti:S laser fundamental in the range of 860–920 nm to produce femtosecond pulses of 430–460 nm, corresponding to single-photon excitation at about 215–230 nm.
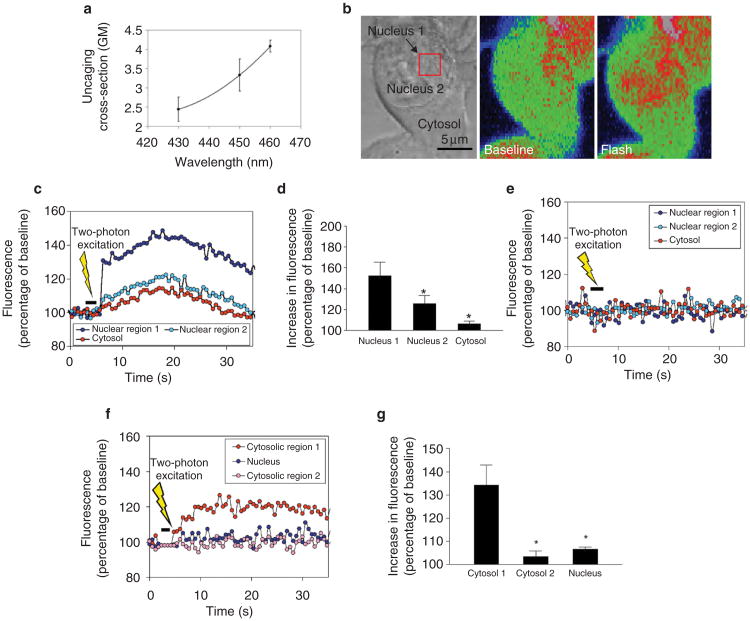
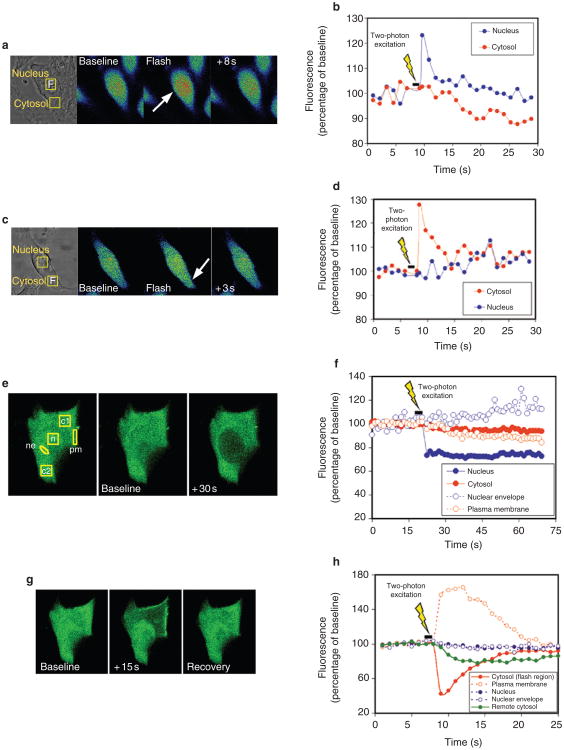
Figure 4. The nucleoplasmic reticulum provides localized release of calcium.
a, Optimal conditions for two-photon photorelease of NPE-caged compounds. The effective two-photon uncaging cross-section of the NPE caging group was measured using NPE-ATP and was maximal at 460 nm. b, Localized two-photon flash photolysis of caged InsP3 in the nucleus. Left, arrow indicates the cell injected with NPE-InsP3; the red square indicates the intranuclear region subjected to two-photon flash photolysis. Middle, pseudocoloured image of the injected cell before two-photon excitation. Right, two-photon photorelease of InsP3 increases calcium predominantly in the nucleus. Although spatial resolution is decreased in the pseudocolour images to maximize collection speed, distinct regions in the nucleus and cytosol can be distinguished. c, Representation of calcium signalling during localized intranuclear photorelease of InsP3. The increase in calcium begins in the region of the nucleoplasmic reticulum (nucleus 1), before spreading throughout the nucleus (nucleus 2) and into the cytosol. The increase in calcium is much smaller and slower in onset than when InsP3 is uncaged throughout the cell (see Fig. 3d). d, Summary of intranuclear uncaging experiments. The main increase in calcium occurs in the region of uncaging near the nucleoplasmic reticulum (nucleus 1). A smaller increase is detected elsewhere in the nucleus (nucleus 2), and a minimal calcium signal is detected in the cytosol. Data are the mean ± s.e.m of six experiments (*P < 0.001 by repeated measures analysis of variance; ANOVA). e, Two-photon excitation in sham-injected cells does not increase calcium. Results are representative of those seen in ten separate cells. f, Localized two-photon uncaging of InsP3 in the cytosol results in localized puffs of cytosolic calcium. An increase in calcium is detected in the region of uncaging (cytosol 1), but no significant increase is detected elsewhere in the cytosol (cytosol 2) or in the nucleus. g, Summary of cytosolic uncaging experiments. The main increase in calcium occurs in the region of uncaging in the cytosol (cytosol 1). A minimal increase is detected elsewhere in the cytosol (cytosol 2) and in the nucleus. Data are the mean ± s.e.m of six experiments (*P < 0.0005 by repeated measures ANOVA).
The effective two-photon uncaging cross-section of the NPE caging group was measured over this range using NPE-ATP and reached a maximum of about 4 Göppert-Mayer (GM; 1 GM = 10−50 cm4 s per photon) at 460 nm (Fig. 4a). To our knowledge, this is the highest two-photon uncaging cross-section that has been measured so far. The photophysical properties of NPE-caged compounds are relatively independent of the caged moiety25, therefore we used these cross-section values to estimate local concentrations of InsP3 after photolysis of NPE-InsP3. Although higher cross-sections might be obtained at wavelengths closer to 520 nm, for stability and reasons of doubling efficiency we worked at 460 nm (corresponding to a 920-nm fundamental). Two-photon uncaging of InsP3 was combined with confocal imaging to ensure that calcium signals were observed in the focal plane in which InsP3 was photoreleased (Supplementary Information Fig. S4).
Photorelease of InsP3 within 1 μm of the nucleoplasmic reticular structures resulted in small increases in calcium that began in the region of the nucleoplasmic reticulum (Fig. 4b). The calcium increase was greatest at the site of photorelease of InsP3, was significantly attenuated elsewhere in the nucleus and was minimal in the cytosol (Fig. 4c, d). Two-photon excitation of the region near the nucleoplasmic reticulum did not increase nuclear or cytosolic calcium in sham-injected cells (Fig. 4e) or in non-injected cells (not shown). In addition, cells could tolerate up to several seconds of pulsed light at 460 nm without bleb formation, spontaneous calcium increases or other evidence of damage. Localized release of caged InsP3 in the cytosol resulted in localized puffs of calcium in the cytosol that did not spread to distant regions of cytosol or to the nucleus (Fig. 4f, g).
Taken together, these findings show that this approach can be used to photorelease NPE-caged InsP3 in a highly localized fashion in individual cells. In fact, this technique should be useful for photorelease from other caging groups that have not been previously amenable to photolysis by two-photon excitation, such as CNB ((α-2-carboxy)-2-nitrobenzyl)26. In addition, these findings show that the nucleoplasmic reticulum is an InsP3-gated calcium store that can give rise to local calcium signals in the nuclear interior.
Cytosolic and nuclear calcium regulate distinct processes in the cell. Most notably, nuclear calcium signals are thought to regulate preferentially the transcription of some genes6–8. But immediate effects of nuclear calcium signals have not been visualized directly. Because protein kinase C (PKC) can translocate to either the plasma membrane or the nuclear envelope27, we examined the effects of cytosolic and nuclear calcium on the cellular distribution of a green fluorescent protein (GFP) fusion protein of PKC-γ encompassing the calcium-sensitive (C2) regulatory domain28,29. For these studies, calcium was transiently increased in discrete regions of 6 × 6 μm in the nucleus (Fig. 5a, b) or the cytosol (Fig. 5c, d) by two-photon flash photolysis of caged calcium. We used caged calcium for these experiments rather than caged InsP3 to maintain more precise control of the amount of free calcium released and the duration of the calcium transient20,30.
Figure 5. Nuclear and cytosolic calcium have distinct effects on PKC translocation.
a, Localized uncaging of calcium in the nucleus of an SKHep1 cell using two-photon flash photolysis. Transmission image shows the rectangular regions in the nucleus and cytosol where calcium was monitored. Two-photon flash photolysis (F) was restricted to a volume of ∼40 fl (<1% of the total cell volume). Serial confocal images show that photorelease of calcium causes a transient increase in calcium that is restricted to the nucleus. b, Representation of a shows the transient increase in calcium in the nucleus but not the cytosol. Data are representative of 17 cells. c, Localized uncaging of calcium in the cytosol of an SKHep1 cell using two-photon flash photolysis as in a. The photorelease of calcium causes a transient increase in calcium that is restricted to the cytosol. d, Representation of c shows the transient increase in calcium in the cytosol but not the nucleus. Data are representative of 12 cells. e, Localized uncaging of calcium in the nucleus of an SKHep1 cell causes translocation of GFP–PKC-γ to the region of the nuclear envelope. Left, GFP–PKC-γ fluorescence (green) is distributed uniformly throughout the nucleus and cytosol before stimulation. The experimental regions of interest are labelled as follows: nucleus (n), nuclear envelope (ne), cytosolic region 1 (c1), cytosolic region 2 (c2), plasma membrane (pm). Middle and right, photorelease of calcium in the rectangular region in the nucleus shifts GFP fluorescence from the nuclear interior to the region of the nuclear envelope. f, Representation of e shows that two-photon uncaging of calcium in the nucleus causes a rapid decrease in nuclear GFP fluorescence plus an associated increase near the nuclear envelope. No change in fluorescence is detected in the cytosol or at the plasma membrane. Data are representative of eight experiments. g, Localized uncaging of calcium in region c1 of the cytosol causes translocation of GFP–PKCγ to the nearby portion of the plasma membrane. h, Representation of g shows that two-photon uncaging of calcium in cytosolic region 1 causes a rapid decrease in GFP fluorescence in that region plus an associated increase near the plasma membrane. A more gradual decrease in fluorescence occurs elsewhere in the cytosol (cytosolic region 2). No change in fluorescence is detected in the nucleus or in the region of the nuclear envelope. Data are representative of five experiments.
As shown previously29, GFP-PKC-γ was distributed uniformly across the nucleus and the cytosol of unstimulated cells. Photorelease of calcium in the nucleus induced loss of nuclear PKC with translocation to the region of the nuclear envelope in most of the cells, but it did not affect the distribution of cytosolic PKC in any of the cells (n = 8 cells; Fig. 5e, f). Photorelease of calcium in the cytosol induced a rapid, transient translocation of cytosolic PKC to the plasma membrane (n = 5 cells). The translocation was most marked in the region of the cytosol where calcium was uncaged, and it was not associated with a change in the distribution of PKC in the nucleus (Fig. 5g, h). In control cells not loaded with caged calcium, two-photon excitation of nuclear or cytosolic regions (n= 8 each) was not associated with a redistribution of GFP–PKC-γ (data not shown). These findings show that localized nuclear calcium signals can have effects on translocation of PKC that are distinct from those of cytosolic calcium signals.
The presence of calcium signalling machinery in the nuclear interior has several implications. First, although calcium signals can be transmitted directly12 or indirectly31 to the nucleus from the cytosol, several intranuclear processes require specific nuclear rather than cytosolic increases in calcium. For example, gene transcription mediated by the cAMP response element (CRE), CRE-binding protein (CREB) or CREB-binding protein (CBP) depends specifically on increases in nuclear calcium6,7. Transcriptional activation of Elk-1 by EGF also depends on nuclear rather than cytosolic calcium8. In addition, calcium can bind to and directly regulate certain nuclear transcription factors32 and can also affect DNA structure33. Our findings provide a cellular mechanism by which such events can be regulated without the coactivation of calcium-dependent events in the cytosol.
Second, both local and global calcium signals occur in the cytosol and, in fact, subcellular calcium signals are responsible for the differential regulation of cytosolic processes by this second messenger1. Although evidence had suggested that calcium signals in the nucleus result simply from diffusion of calcium from either the cytosol12 or the nuclear envelope13,34, the nucleus is divided into functionally distinct subcompartments35. By showing that a nucleoplasmic reticulum can give rise to localized calcium gradients in the nuclear interior, we have identified a potential mechanism by which calcium-dependent events can be regulated differentially in the nucleus, just as they are in the cytosol. The presence of machinery to generate spatially defined calcium signalling patterns in the nucleus, such as calcium puffs and calcium waves, may uncover a new layer of control of nuclear function by this second messenger.
Methods
Cells and materials
We obtained SKHep1 cells from ATCC (Manassas, VA) and examined them 2–3 d after plating them onto glass coverslips. ER-Tracker, fluo-4/AM, mag-fluo-4/AM and DM-nitrophen (DMNP-EDTA) were from Molecular Probes (Eugene, OR) and NPE-InsP3 was from Calbiochem (San Diego, CA). Monoclonal antibodies against calreticulin were from Stressgen (Victoria, BC, Canada). Antibodies specific for the InsP3 receptor isoforms were used for immunoblotting and immunofluorescence. The type I InsP3 receptor antibody24 was from Research Genetics (Huntsville, AL), the type II InsP3 receptor antibody36 was a gift from R. J. Wojcikiewicz (SUNY Syracuse) and the type III InsP3 receptor antibody24 was from BD Transduction Laboratories (Lexington, KY). A gene encoding GFP–PKC-γ containing the C2 (calcium-sensing) but not the C1 (diacylglycerol-sensitive) domain28,29 was a gift from T. Meyer (Stanford University).
Fluorescence imaging
Cells were loaded with ER-Tracker dye (100 nM) for 30 min, fluo-4/AM (6 μM) for 30-60 min or mag-fluo-4/AM (6 μM) for 45 min at room temperature. In some experiments, fluo-4/AM was preferentially loaded into the ER by incubating cells with the dye at 37 °C for 60 min (ref. 18). Coverslips containing the cells were transferred to a custom-built perfusion chamber on the stage of a Bio-Rad MRC-1024 confocal microscope (Hercules, CA), equipped with a Spectra-Physics Tsunami Ti:S laser and a Millenia X pump laser (Mountain View, CA) for two-photon excitation, and were observed using a 63×, 1.4 NA objective lens. ER-Tracker was excited at 790 nm by two-photon excitation or at 350–400 nm using conventional epifluorescence; fluo-4 was excited at 488 nm using a krypton/argon laser. We observed two-photon fluorescence at 500–540 nm by using custom-built external detectors and collected emission signals between 505 and 550 nm to detect the confocal fluorescence of fluo-4. Both confocal and two-photon images were recorded near the middle of the nucleus, at least 2 μm from either the upper or the lower nuclear boundary identified by serial confocal sectioning.
Immunoblots and immunofluorescence
SKHep1 cell immunoblots were done by standard methods20,24. We used a commercially available kit from Pierce (Rockford, IL) to prepare nuclear and cytosolic cell fractions. Blots were visualized by enhanced chemiluminesence, and quantitative analysis of the blots was done on a Bio-Rad GS-700 imaging densitometer (Bio-Rad, Hercules, CA). We carried out immunofluorescence as described20,24. Cells were double-labelled with antibodies against the InsP3 receptor isoforms and against calreticulin, and then incubated with secondary antibodies conjugated to Alexa 594 and Alexa 488 (Molecular Probes) to detect the InsP3 receptor and calreticulin antibodies, respectively. Confocal immunofluorescence images were obtained by excitation at 488 nm with observation at 505–550 nm to detect Alexa 488, followed by excitation at 633 nm with observation at > 650 nm to detect Alexa 594. We used a Zeiss LSM 510 confocal microscope (Thornwood, NY) for immunofluorescence imaging.
Detection of calcium and GFP
For calcium imaging cells were incubated with fluo-4/AM (6 μM), whereas for GFP imaging cells were transfected 48 h beforehand with GFP–PKC-γ using Effectene (Qiagen, Valencia, CA). We used an MRC-1024 confocal microscope (Bio-Rad) to monitor fluo-4 or GFP fluorescence at a rate of either 2–10 frame per second or 6 ms per line during line scanning24. Changes in fluorescence were normalized by the initial fluorescence (F0) and are expressed as (F/F0) × 100%.
NPE-caged InsP3 (1–5 mM) was microinjected into cells as described20,24 and uncaged by ultraviolet or two-photon flash photolysis. Injected cells were allowed to recover for at least 5 min before flash photolysis. For ultraviolet flash photolysis, we released NPE-InsP3 by using a mercury arc lamp coupled to a 1-mm quartz fibre optic cable through a high-speed shutter and filter wheel20,24.
For photorelease of InsP3 by two-photon excitation, the Ti:S laser was tuned to a wavelength of 920 nm with a pulse width of ∼ 100 fs and an output power of 500 mW. The 920-nm fundamental was then doubled in frequency to 460 nm using a 0.5-mm LBO crystal (Super Optronics, Los Angeles, CA). The 460-nm beam was coupled into the confocal optical path by a 470-nm long-pass dichroic (see Supplementary Information Fig. S4) after passing through an 8× eyepiece for beam expansion and parfocality adjustment. The focal point of the doubled Ti:S was adjusted to overlap with the 488-nm focus of the confocal scanner to assure that flash photolysis occurred in the imaging plane of focus. Parfocality was adjusted and verified with a bleachable fluorescent polymer by collecting confocal x–z scans of the bleach spot. We controlled the two-photon flash duration by using a UniBlitz shutter (Vincent Associates, Rochester, NY) and controlled intensity by detuning the frequency doubler.
In some experiments, caged calcium was selectively photoreleased in the nucleus or the cytosol using two-photon excitation as described20,30. In brief, cells were loaded with the cell-permeant form of DM-nitrophen (2 μM), and the Ti:S laser was tuned to a wavelength of 730 nm with a pulse width of 90 fs and an output power of 600 mW. Calcium was then uncaged in predetermined 40-fl regions in the nucleus or the cytosol while fluo-4 or GFP was monitored throughout the cell.
Measurement of the two-photon NPE uncaging cross-section
The two-photon uncaging cross-section is defined as (quantum efficiency of photolysis) × (two-photon absorption cross-section). Measurements for NPE-caged compounds were carried out by raster scanning 10 μl aliquots of NPE-ATP through a 40×/1.2NA water immersion objective lens for 500–3,000 scans at a scan speed of ∼ 1 m s−1. A high raster scan speed was used to ensure minimal depletion of NPE-ATP in the focal volume.
Measurements were done using a doubled Ti:S laser at 430, 450 and 460 nm with an output power of 0.5–3 mW. We quantified photoreleased ATP by high performance liquid chromatography (HPLC); we used caged ATP because ATP is easier to quantify by HPLC than is InsP3. The concentration of photoreleased ATP was quadratically related to the output power over the power range used. The reported values (Fig. 4a) were calculated on the basis of the number of molecules released, the illumination power, the focal volume, the concentration of NPE-ATP and the second order coherence (g). We calculated g on the basis of the laser repetition rate (80 MHz), the pulse width (∼300 fs at the sample) and the pulse shape factor37.
Supplementary Material
Supplementary Information accompanies the paper on www.nature.com/naturecellbiology.
Acknowledgments
We thank B. Ehrlich for comments on the manuscript; T. Meyer for the GFP–PKC-γ construct and R. Wojcikiewicz for providing antibodies against the type II InsP3 receptor. This work was supported by an AASLD–Schering Advanced Hepatology Fellowship Award, a Glaxo Institute for Digestive Health Basic Science Research Award and a Yale Child Health Research Center Award (to W.E.); a grant from Conselho Nacional de Desenvolvimento Científico e Tecnológico (to M.F.L.); and NIH grants (to W.R.Z. and M.H.N).
Footnotes
Competing Financial Interests: The authors declare that they have no competing financial interests.
References
- 1.Berridge MJ, Lipp P, Bootman MD. The versatility and universality of calcium signalling. Nature Rev Mol Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 2.Pinton P, Pozzan T, Rizzuto R. The Golgi apparatus is an inositol 1,4,5-trisphosphate-sensitive Ca2+ store, with functional properties distinct from those of the endoplasmic reticulum. EMBO J. 1998;17:5298–5308. doi: 10.1093/emboj/17.18.5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jouaville LS, Ichas F, Holmuhamedov EL, Camacho P, Lechleiter JD. Synchronization of calcium waves by mitochondrial substrates in Xenopus laevis oocytes. Nature. 1995;377:438–441. doi: 10.1038/377438a0. [DOI] [PubMed] [Google Scholar]
- 4.Stehno-Bittel L, Perez-Terzic C, Clapham DE. Diffusion across the nuclear envelope inhibited by depletion of the nuclear Ca2+ store. Science. 1995;270:1835–1838. doi: 10.1126/science.270.5243.1835. [DOI] [PubMed] [Google Scholar]
- 5.Perez-Terzic C, Pyle J, Jaconi M, Stehno-Bittel L, Clapham DE. Conformational states of the nuclear pore complex induced by depletion of nuclear Ca2+ stores. Science. 1996;273:1875–1877. doi: 10.1126/science.273.5283.1875. [DOI] [PubMed] [Google Scholar]
- 6.Chawla S, Hardingham GE, Quinn DR, Bading H. CBP: a signal-regulated transcriptional coactivator controlled by nuclear calcium and CaM kinase IV. Science. 1998;281:1505–1509. doi: 10.1126/science.281.5382.1505. [DOI] [PubMed] [Google Scholar]
- 7.Hardingham GE, Chawla S, Johnson CM, Bading H. Distinct functions of nuclear and cytoplasmic calcium in the control of gene expression. Nature. 1997;385:260–265. doi: 10.1038/385260a0. [DOI] [PubMed] [Google Scholar]
- 8.Pusl T, et al. Epidermal growth factor-mediated activation of the ETS-domain transcription factor Elk-1 requires nuclear calcium. J Biol Chem. 2002;277:27517–27527. doi: 10.1074/jbc.M203002200. [DOI] [PubMed] [Google Scholar]
- 9.Divecha N, Rhee SG, Letcher AJ, Irvine RF. Phosphoinositide signalling enzymes in rat liver nuclei: phosphoinositidase C isoform β1 is specifically, but not predominantly, located in the nucleus. Biochem J. 1993;289:617–620. doi: 10.1042/bj2890617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malviya AN, Rogue P, Vincendon G. Stereospecific inositol 1,4,5-[32P]trisphosphate binding to isolated rat liver nuclei: evidence for inositol trisphosphate receptor-mediated calcium release from the nucleus. Proc Natl Acad Sci USA. 1990;87:9270–9274. doi: 10.1073/pnas.87.23.9270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allbritton NL, Oancea E, Kuhn MA, Meyer T. Source of nuclear calcium signals. Proc Natl Acad Sci USA. 1994;91:12458–12462. doi: 10.1073/pnas.91.26.12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lipp P, Thomas D, Berridge MJ, Bootman MD. Nuclear calcium signalling by individual cytoplasmic calcium puffs. EMBO J. 1997;16:7166–7173. doi: 10.1093/emboj/16.23.7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox JL, Burgstahler AD, Nathanson MH. Mechanism of long-range Ca2+ signalling in the nucleus of isolated rat hepatocytes. Biochem J. 1997;326:491–495. doi: 10.1042/bj3260491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 15.Williams RM, Zipfel WR, Webb WW. Multiphoton microscopy in biological research. Curr Opin Chem Biol. 2001;5:603–608. doi: 10.1016/s1367-5931(00)00241-6. [DOI] [PubMed] [Google Scholar]
- 16.Fricker M, Hollinshead M, White N, Vaux D. Interphase nuclei of many mammalian cell types contain deep, dynamic, tubular membrane-bound invaginations of the nuclear envelope. J Cell Biol. 1997;136:531–544. doi: 10.1083/jcb.136.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wamhoff BR, Dixon JL, Sturek M. Atorvastatin treatment prevents alterations in coronary smooth muscle nuclear Ca2+ signaling in diabetic dyslipidemia. J Vasc Res. 2002;39:208–220. doi: 10.1159/000063686. [DOI] [PubMed] [Google Scholar]
- 18.Malgaroli A, Milani D, Meldolesi J, Pozzan T. Fura-2 measurement of cytosolic free Ca2+ in monolayers and suspensions of various types of animal cells. J Cell Biol. 1987;105:2145–2155. doi: 10.1083/jcb.105.5.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lui PPY, Kong SK, Kwok TT, Lee CY. The nucleus of HeLa cell contains tubular structures for Ca2+ signalling. Biochem Biophys Res Commun. 1998;247:88–93. doi: 10.1006/bbrc.1998.8649. [DOI] [PubMed] [Google Scholar]
- 20.Leite MF, et al. Molecular basis for pacemaker cells in epithelia. J Biol Chem. 2002;277:16313–16323. doi: 10.1074/jbc.M109207200. [DOI] [PubMed] [Google Scholar]
- 21.Clark EA, Brugge JS. Integrins and signal transduction pathways: the road taken. Science. 1995;268:233–239. doi: 10.1126/science.7716514. [DOI] [PubMed] [Google Scholar]
- 22.Reilly JF, Maher PA. Importin β-mediated nuclear import of fibroblast growth factor receptor: role in cell proliferation. J Cell Biol. 2001;152:1307–1312. doi: 10.1083/jcb.152.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin SY, et al. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nature Cell Biol. 2001;3:802–808. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- 24.Hagar RE, Burgstahler AD, Nathanson MH, Ehrlich BE. Type III InsP3 receptor channel stays open in the presence of increased calcium. Nature. 1998;396:81–84. doi: 10.1038/23954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu QQ, Schnabel WE, Schupp H. Formation and decay of nitronic acid in the photoarrangement of o-nitrobenzyl esters. J Photochem. 1987;36:317–332. [Google Scholar]
- 26.Gee KR, Wieboldt R, Hess GP. Synthesis and photochemistry of a new photolabile derivative of GABA. Neurotransmitter release and receptor activation in the microsecond time region. J Am Chem Soc. 1994;116:8366–8367. [Google Scholar]
- 27.Oancea E, Teruel MN, Quest AFG, Meyer T. Green fluorescent protein (GFP)-tagged cysteine-rich domains from protein kinase C as fluorescent indicators for diacylglycerol signaling in living cells. J Cell Biol. 1998;140:485–498. doi: 10.1083/jcb.140.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teruel MN, Meyer T. Parallel single-cell monitoring of receptor-triggered membrane translocation of a calcium-sensing protein module. Science. 2002;295:1910–1912. doi: 10.1126/science.1065028. [DOI] [PubMed] [Google Scholar]
- 29.Oancea E, Meyer T. Protein kinase C as a molecular machine for decoding calcium and diacylglycerol signals. Cell. 1998;95:307–318. doi: 10.1016/s0092-8674(00)81763-8. [DOI] [PubMed] [Google Scholar]
- 30.Leite MF, Burgstahler AD, Nathanson MH. Ca2+ waves require sequential activation of inositol 1,4,5-trisphosphate receptors and ryanodine receptors in pancreatic acinar cells. Gastroenterology. 2002;122:415–427. doi: 10.1053/gast.2002.30982. [DOI] [PubMed] [Google Scholar]
- 31.Deisseroth K, Heist EK, Tsien RW. Translocation of calmodulin to the nucleus supports CREB phosphorylation in hippocampal neurons. Nature. 1998;392:198–202. doi: 10.1038/32448. [DOI] [PubMed] [Google Scholar]
- 32.Carrion AM, Link WA, Ledo F, Mellstrom B, Naranjo JR. DREAM is a Ca2+-regulated transcriptional repressor. Nature. 1999;398:80–84. doi: 10.1038/18044. [DOI] [PubMed] [Google Scholar]
- 33.Dobi A, Agoston DV. Submillimolar levels of calcium regulates DNA structure at the dinu-cleotide repeat (TG/AC)n. Proc Natl Acad Sci USA. 1998;95:5981–5986. doi: 10.1073/pnas.95.11.5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerasimenko OV, Gerasimenko JV, Tepikin AV, Petersen OH. ATP-dependent accumulation and inositol trisphosphate- or cyclic ADP-ribose-mediated release of Ca2+ from the nuclear envelope. Cell. 1995;80:439–444. doi: 10.1016/0092-8674(95)90494-8. [DOI] [PubMed] [Google Scholar]
- 35.Phair RD, Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- 36.Wojcikiewicz RJH. Type I, II, and III inositol 1,4,5-trisphosphate receptors are unequally susceptible to down-regulation and are expressed in markedly different proportions in different cell types. J Biol Chem. 1995;270:11678–11683. doi: 10.1074/jbc.270.19.11678. [DOI] [PubMed] [Google Scholar]
- 37.Xu C, Zipfel W, Shear JB, Williams RM, Webb WW. Multiphoton fluorescence excitation: new spectral windows for biological nonlinear microscopy. Proc Natl Acad Sci USA. 1996;93:10763–10768. doi: 10.1073/pnas.93.20.10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allbritton NL, Meyer T, Stryer L. Range of messenger action of calcium ion and inositol 1,4,5-trisphosphate. Science. 1992;258:1812–1815. doi: 10.1126/science.1465619. [DOI] [PubMed] [Google Scholar]
- 39.Meyer T, Stryer L. Molecular model for receptor-stimulated calcium spiking. Proc Natl Acad Sci USA. 1988;85:5051–5055. doi: 10.1073/pnas.85.14.5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramos-Franco J, Fill M, Mignery GA. Isoform-specific function of single inositol 1,4,5-trisphosphate receptor channels. Biophys J. 1998;75:834–839. doi: 10.1016/S0006-3495(98)77572-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information accompanies the paper on www.nature.com/naturecellbiology.