Abstract
Using whole-genome microarray datasets of the Immunological Genome Project, we demonstrate a closer transcriptional relationship between NK and T cells than any other leukocytes, distinguished by their expression of similar signaling functions. While resting NK cells were known to share expression of a few genes with cytotoxic CD8+ T cells, transcriptome-wide analysis demonstrates that the commonalities extend to hundreds of genes, many with unknown functions. The NK cell response to viral infection is dampened relative to cytotoxic CD8+ T cells, in part due to their “pre-primed” state. Collectively, the data provide global context for known and novel molecular aspects of NK cell identity and function by delineating the genome-wide repertoire of gene expression of NK cells in various states.
INTRODUCTION
The Immunological Genome Project (ImmGen) is a consortium of laboratories aimed at establishing a comprehensive database of gene expression within the mouse immune system1. As part of this collaboration, we have identified the gene expression programs of Natural Killer (NK) cells and analyzed them under steady-state and during response to a viral infection in order to generate a resource for interrogating NK cell biology.
The immune system of vertebrates is classically divided into innate and adaptive branches. The innate immune system responds rapidly to infectious agents, whereas the adaptive response requires cell division and differentiation of effector cells. NK cells and innate-like lymphocytes, which include γδ T cells, invariant NKT (iNKT) cells, intestinal epithelial lymphocytes (iELs), B1 cells, and marginal zone B cells, exhibit both adaptive and innate features2, 3. For example, these cell types use somatically rearranged genes encoding receptors to recognize specific structures from microbes and self-antigens2, whereas functionally, innate-like lymphocytes mount quick effector responses such as cytolysis and rapid cytokine, chemokine, and antibody secretion.
Since their first description in 19754, 5, the relationship of NK cells to lymphoid and myeloid cells has been a topic of debate. The ability of certain T cell populations, such as γδ T cells and some activated αβ-T cell receptor (TcR)-bearing T cells to mediate “NK-like” cytolysis as well as the shared expression of numerous cell surface antigens and effector molecules between NK cells and T cells (e.g. CD2, CD7, CD90, perforin, granzyme A, and interferon-gamma (IFN-γ)) have led to the proposal that NK cells might simply represent a developmental or differentiation stage of T cells. However, the lack of productive rearrangement of TcR genes in mature NK cells and the development of NK cells in mice lacking a thymus or the recombinase genes required for TcR rearrangement unambiguously distinguish NK cells as a third, distinct lineage of lymphoid cells6. A relationship between NK cells and myeloid cells has been argued based on shared expression of cell surface markers, such as CD11b and CD11c. However, subsequent studies defining properties of hematopoietic progenitor populations have demonstrated that most NK cells are derived from progenitors shared with lymphocytes rather than myeloid cells7.
Global transcriptional analysis is a powerful approach to yield new insights into the biology of specific cellular subsets8, 9. Early studies using this approach to analyze human and mouse NK cellsidentified sets of genes that were specifically expressed in NK cells, as well as transcriptional changes during NK cell activation in vitro10–12. In this study, we have systematically defined the transcriptome of mouse NK cells in several contexts, including activation states and relative to all other lymphocyte and myeloid populations profiled by the ImmGen consortium. The transcriptional profiling technique was multi-dimensional, making this study unique from the previous analyses because of the large number of datasets (e.g. conditions and cell types) that were simultaneously compared. The findings presented herein provide molecular definitions of NK cell identity and function, providing novel insights into the nature of NK cells and a publicly available resource documenting the transcriptome of NK cells in various states.
RESULTS
Transcription-based organization of the major leukocyte subsets
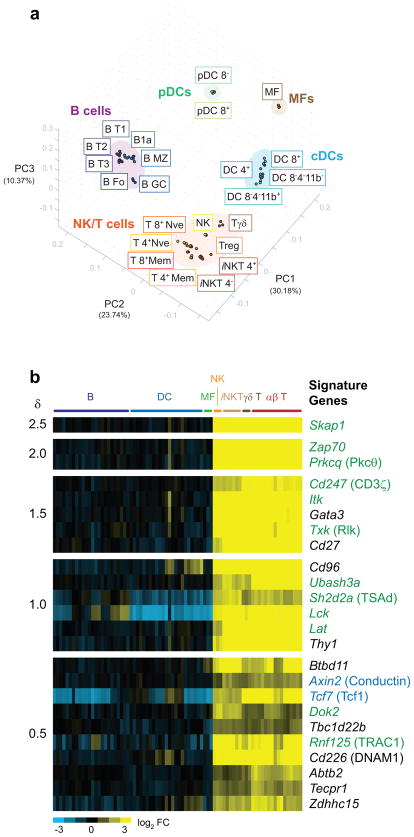
In order to establish a molecular definition of NK cell identity, we investigated the relatedness of naïve NK cells to other leukocyte populations using principal component analysis (PCA), a method that identifies gene expression patterns (principal components) that most strongly explain variance across a dataset. Delineating population relationships using the three most informative principle components, as defined by the 15% most variable genes across all splenic leukocyte populations, revealed a striking segregation of the populations into five discrete clusters (Fig. 1a). Lymphoid cells including B cells, NK cells, iNKT cells, γδ T cells, and αβ T cell subsets formed groups distant from macrophages. While plasmacytoid dendritic cells (pDC) group close to macrophages, conventional CD11c+ dendritic cell populations (cDC) cluster between macrophages and lymphoid cell subsets. Among lymphoid cells, there was a distinct separation of a cluster containing B cell subsets and a cluster containing NK, iNKT cells, γδ T cells, and αβ T cell subsets (hereafter referred to as the “NK/T complex”). Notably, NK cells and other innate-like T lymphocytes did not discernibly segregate from the adaptive T cell populations at this level of resolution.
Figure 1. NK and T cells exhibit close relative similarity at the transcriptome level.
(a) Splenic leukocyte populations were separated into distinct groupings by PCA. The top three PC are displayed with their percentage contributions to inter-sample variation. Individual replicate samples are displayed. (b) Significant genes discriminating the NK/T complex from other leukocyte populations are displayed as a heat map in decreasing order of significance (δ score), with individual replicates shown. Green text = molecules in the ITAM signaling pathway; blue text = molecules in the Wnt/β-catenin pathway. Common names are in parenthesis. Populations abbreviations: B T1, B cells, transitional type 1; B T2, B cells, transitional type 2; B T3, B cells, transitional type 3; B Fo, B cells, follicular; B1a, B1a B cells; B MZ, B cells, marginal zone; B GC, B cells germinal center; pDC 8−, plasmacytoid dendritic cells, CD8−; pDC 8+, plasmacytoid dendritic cells, CD8+; MF, macrophages; DC 4+, dendritic cells, CD4+; DC 8+, dendritic cells, CD8+; DC 8−4−11b+, dendritic cells, CD8−CD4−CD11b+; DC 8− 4− 11b−, dendritic cells, CD8−CD4−CD11b−; NK, natural killer cells; Tγδ, TCRγδ T cells; Treg, T regulatory cells; iNKT 4+, invariant NKT CD4+ cells; iNKT 4−, invariant NKT CD4− cells; T 4+ Nve, CD4+ naïve cells; T 8+ Nve, CD8+ naïve cells; T 4+ Mem, CD4+ memory cells; T 8+ Mem, CD8+ memory cells.
Comprehensive pairwise comparisons of each NK/T population with B cell and myeloid (MF, DC, and pDC) populations, followed by identification of the intersection of all pairwise comparisons, identified a 24-gene signature that distinguishes resting NK, iNKT cells, γδ T cells, and αβ T cell subsets from other leukocyte populations (Fig. 1b). These genes were strongly enriched in the components of the immunoreceptor tyrosine-based activation motif (ITAM) signaling pathway, including signaling molecules known to regulate activation of NK/T cells such as Lck, Zap70, Tec kinases (Itk and Txk), PKC-θ (Prkcq)13, STS-2 (Ubash3a), TRAC1 (Rnf125), and adaptor proteins encoded by Cd247, Lat, Skap1, Sh2d2a, and Dok214. The finding of such a prominent enrichment of these signaling molecules upon unbiased evaluation reflects the shared biology of NK and T cells and is a strong validation that this functional similarity in signaling constitutes a large component of the overall similarity between NK and T cells.
Molecular organization of the NK/T complex
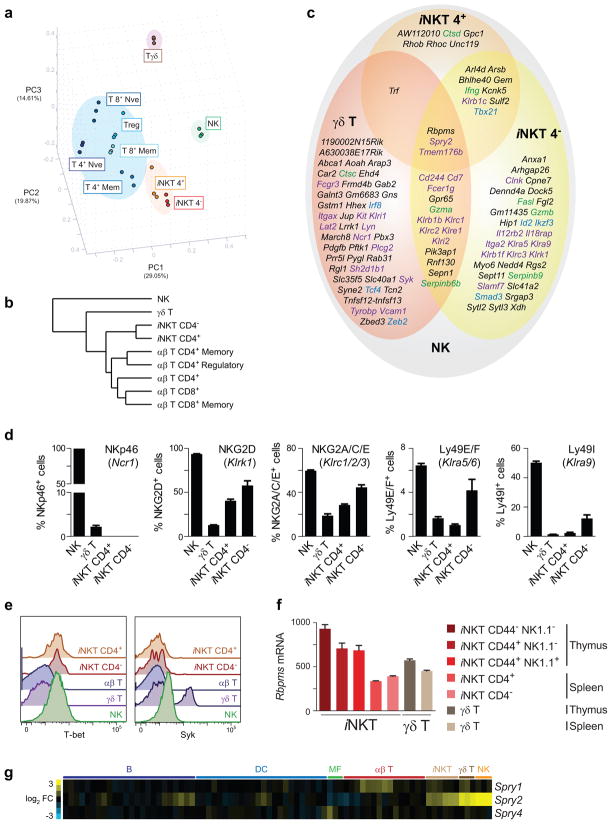
PCA of the 15% most variable genes across the 9 NK/T complex populations revealed grouping of the adaptive T cell populations (Fig. 2a). In contrast, the innate populations failed to group together, with only the iNKT subsets (iNKT CD4+ and iNKT CD4−) showing a close relationship. A similar organization was observed with hierarchical clustering (Fig. 2b).
Figure 2. Organization of the innate and adaptive branches of the NK/T complex.
(a) NK and innate-like T cells form distinct associations from one another and from adaptive αβ T cell populations, which group together. The top three PC are depicted with their percentage contributions to inter-sample variation. Individual replicate samples are shown. (b) Hierarchical clustering of the NK/T complex populations demonstrates close grouping of adaptive CD4+ and CD8+ T cell populations. Clustering is based on Euclidean distances of averaged arrays with a population, with all genes expressed in any of the depicted populations included in the analysis. (c) Overlap of genes expressed in NK, γδ T, and iNKT, but not adaptiveαβ T cell populations. Genes depicted met a δ score threshold of 0.5. Green text = effector molecules; purple = surface molecules; blue = transcription factors. Genes unique to resting NK cells are shown in Figure 3. (d,e) Flow cytometric analysis of NKR, T-bet, and Syk expression by NK and innate-like lymphocytes relative to TCRαβ+ T cells. Data represent n = 3 from 2 independent experiments. (f) Rbpms transcript level in iNKT and γδ T populations isolated from thymus or spleen. (g) Heat map analysis of Spry family member expression by splenic leukocyte populations. Spry3 was not expressed above background levels in any population and is not shown.
We hypothesized that despite the diversity of the innate populations of the NK/T complex, transcriptional commonalities would exist that distinguish innate populations from adaptive T cells. To determine these shared programs, we identified differentially expressed genes for each innate population against adaptive T cells and assessed conservation across the four comparisons. The 112 genes most significantly upregulated by resting NK cells and at least one other innate cell subset as compared with αβ T cell subsets were enriched in genes encoding surface and signaling receptors and molecules (Fig. 2c). These genes included activating and inhibitory NK cell receptors (NKRs) (e.g. Fcgr3 [CD16], Ncr1 [NKp46], Klrc2 [NKG2C], Klrk1 [NKG2D], Slamf7 [CRACC], Klra5 [Ly49E], Klra9 [Ly49I], Klrc1 [NKG2A]), transmembrane proteins and other surface receptors (e.g. Il12rb2, Fasl, Kit, Cd7), integrins (e.g. Itga2 [CD11b], Itgax [CD11c]), kinases (e.g. Syk, Lyn), and adapters (e.g. Fcer1g [FcRγ], Tyrobp [DAP12], Lat2 [NTAL], Sh2d1b1 [EAT-2], and Clnk). Many of these differentially expressed genes were previously shown to have higher expression in innate lymphocytes, providing validation of these data8, 15, 16. In addition, we found a novel set of genes (e.g. Fgl2, Sulf2, Lrrk1, Aoah, Car2) that exhibit a distinctive expression pattern in NK and innate lymphocytes, but whose function in these lineages is currently unknown. Staining for a representative set of cell surface antigens and intracellular molecules revealed that in most cases, the measured levels of transcript were reflected in the frequencies of NK, iNKT or γδ T cells staining positive for these markers (Fig. 2d,e). The exceptions were NKG2D and Ly49E/F, which were expressed on the surface of NK and iNKT CD4− cells and also detected in iNKT CD4+ and γδ T cells, albeit at reduced levels as compared to NK cells.
Given the observations that unprimed NK, iNKT and γδ T cells respond rapidly to stimulation, it was not surprising that resting NK cells express elevated mRNA levels of effector molecules including Ifng, proteases (e.g. Gzmb, Gzma, Ctsc [cathepsin C], and Ctsd [cathepsin D]), and protease inhibitors (Serpinb6b and Serpinb9). Expression of several genes involved in vesicle transport (e.g. Rab31, Sytl2, and Sytl3) and regulation of cytoskeleton (e.g. Sept11 and Myo6) was higher in NK and/or iNKT and γδ T cells, suggesting that changes in vesicle trafficking or cytoskeletal rearrangements may effect the generation or release of lytic granules. In addition, transcription factors Tbx21 (T-bet) and Id2 showed elevated expression in NK and iNKT cell lineages (Fig. 2c,e), consistent with reports indicating their requirement for the development and function of these cells17. Notably, innate genes identified in this analysis do not merely represent genes characteristic of cell activation: only a fraction of genes more highly expressed in NK and innate-like T cell populations relative to αβ T cells corresponded to genes induced during cell activation and/or proliferation.
Consistent with our observation that innate NK/T subsets are heterogeneous, only three genes (Rbpms, Tmem176b, and Spry2) were significantly shared by resting NK, iNKT, and γδ T cell populations (Fig. 2c). RNA-binding protein gene with multiple splicing (Rbpms) regulates Transforming growth factor-β (TGFβ) signaling18. Given that TGFβ is critical for the development of iNKT and γδ T cell lineages19, it is intriguing that Rbpms mRNA is highest in the more immature thymic iNKT and γδ T cell populations (Fig. 2f). While the expression of Tmem176b is specific to innate NK/T subsets, its expression is much lower than in DCs. Sprouty homolog 2 (Spry2) is a member of a sprouty gene family; there are four Spry genes in higher vertebrates with only Spry2 being highly and specifically expressed in NK, iNKT, and γδ T cell populations (Fig. 2g). Given that Sprouty proteins are involved in a negative feedback mechanism to limit antigen receptor-mediated signaling20, SPRY2 might represent a novel regulator of an innate lymphocyte developmental or activation program. These analyses confirm known functions of NK and innate-like T cells and identify novel molecular components of innate-like lymphocyte populations.
A transcriptional signature defining resting NK cell identity
The shared repertoire of surface receptors, signaling molecules, and transcription factors expressed by NK cells and other innate-like T cell populations blurs the distinction between these cells types. We therefore defined a resting NK signature by identifying genes more highly expressed in NK cells than in all other leukocyte populations. Nearly half of the 25 most significant NK cell-enriched genes encoded NKRs, the most specific of which were Klra8 (Ly49H) and Ncr1 (NKp46) (Fig. 3a). Ly49H, while expressed in only 50% of NK cells in C57BL/6 mice, is not detectable in any other leukocyte populations (Fig. 3b). NKp46 has previously been shown to have selective expression in NK cells with two exceptions: rare T cell subsets (Fig. 2d)21, 22 and a mucosal population of innate lymphoid cells that express retinoid-related orphan receptor (ROR)-γt23. Additional genes preferentially expressed by NK cells that were previously identified include sphingosine 1-phosphate receptor 5 (S1pr5)24, adhesion molecules Mcam25 and Itga2 (CD49b)26, and effector molecules Gzma, Gzmb, and Prf127, 28. Among the genes whose expression was uniquely increased in NK cells, Adamts14, Serpinb9b, and Styk1 have not yet been reported to be expressed by this subset of lymphocytes. A disintegrin and metalloproteinase with thrombospondin motifs 14 (Adamts14) processes extracellular matrix proteins29, which we speculate may be important for the migratory ability of NK cells. Serine peptidase inhibitor, clade B, member 9 (Serpinb9b) inactivates granzyme B in an irreversible manner30 and is required to protect cytotoxic lymphocytes from granzyme B-mediated cell death31 and may protect NK cells from being killed by their own cytolytic molecules. Serine/threonine/tyrosine kinase 1 (Styk1) shares homology with platelet-derived growth factor/fibroblast growth factor receptors and has been suggested to regulate cell proliferation and survival by activating both MAP kinase and phosphatidylinositol 3′-kinase32, its function in NK cells is currently unknown and it has not previously been identified as a potentially NK cell-restricted gene expressed in the hematopoietic system. These molecules represent novel intracellular proteins that distinguish NK cells from other resting leukocytes.
Figure 3. Molecular uniqueness of resting NK cells.
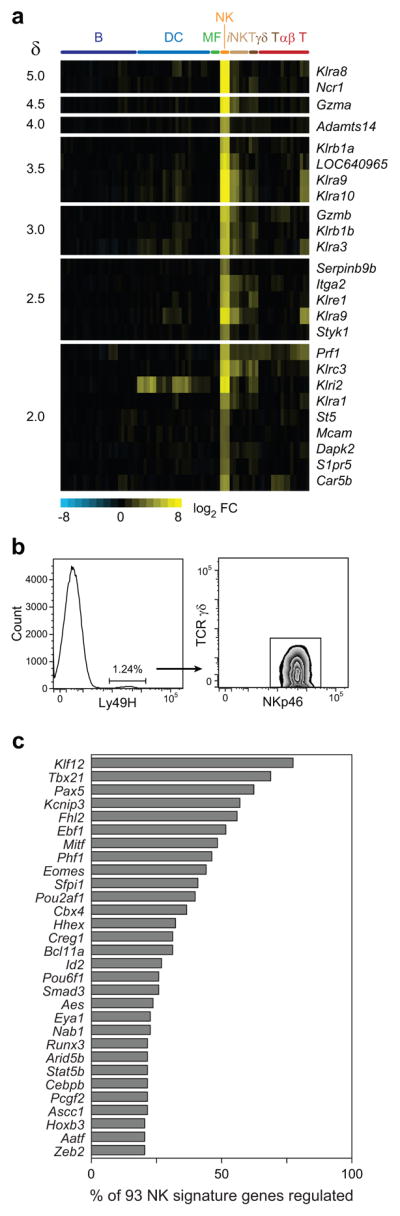
(a) Heat map of genes most strongly enriched in NK cells over all other populations, shown in decreasing order of significance. A full list of genes is available in Supplementary Table 1. (b) Flow cytometric analysis of Ly49H expression in splenic leukocytes. Cells staining positive for Ly49H co-express NKp46, but not TCRγδ. Data are representative of three independent experiments. (c) Predicted regulators of resting NK cell signature genes, in decreasing order of predicted influence on the NK signature genes shown in (a).
We further sought to understand the regulatory control of NK cell “uniqueness” and identified putative transcriptional regulators of the resting NK cell signature genes using a network modeling approach (described in Supplementary Note 1). Of the predicted regulators, a number of genes known to influence the development or function of NK cells, such as Tbx21, Eomes, Mitf, Sfpi1, Id2, Smad3, Runx3, and Stat5b were identified17 (Fig. 3c). However, the majority of the identified regulators have no known role in NK cell development, despite strong associations with genes enriched in NK cells. For example, Klf12 is predicted to regulate 80% of the identified NK fingerprint, however, the role of this Kruppel-like zinc finger protein in NK cells is unknown. These data suggest that a rich biology related to transcriptional definition of NK cell identity remains undiscovered.
Transcriptional priming of effector functions in NK cells
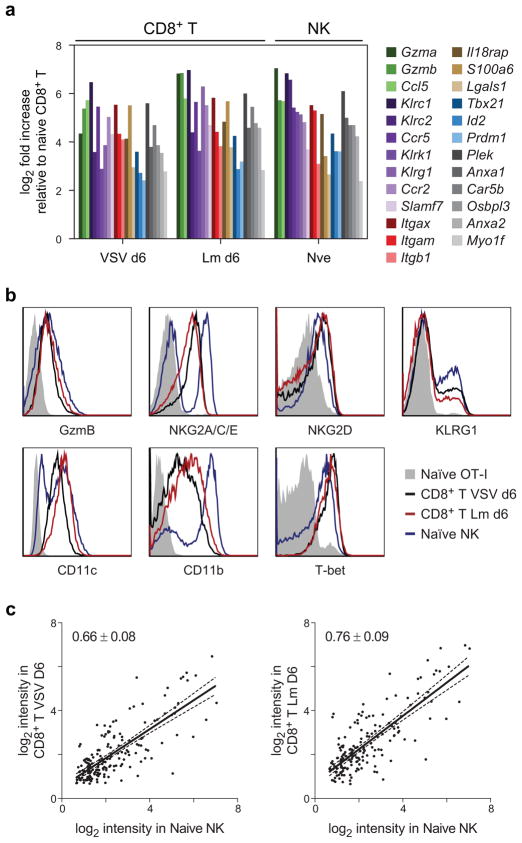
NK cells are pre-primed to allow rapid activation of some effector functions. We explored this concept at the genome level by identifying genes highly expressed in naive NK cells and induced in effector CD8+ T cells after either Vesicular Stomatitis Virus (VSV) or Listeria monocytogenes (Lm) infections, as compared to naïve CD8+ T cells.
Expression programs shared by naïve NK cells and effector CD8+ T cells include effector functions, NKRs, adhesion and homing, transcription factors, and signaling molecules (Fig. 4a, b). Killing of infected cells is a critical effector function of both NK cells and effector CD8+ T cells and is mediated by the release of perforin and granzymes; accordingly, expression of Gzma and Gzmb was high in both naïve NK cells and effector CD8+ T cells. Naïve NK cells and effector CD8+ T cells also shared expression of other effector molecules such as Ifng and Ccl5 (RANTES) (Fig. 4a, Supplementary Table 2). Expression of several activating (e.g. Klrc2 [NKG2C], Klrk1 [NKG2D], Slamf7 [CRACC] and inhibitory (e.g. Klrc1 [NKG2A], Klrg1) NKRs were also shared between naïve NK cells and effector CD8+ T cells, consistent with prior observations33 (Fig. 4a, b). For effector cells to respond against foreign invaders, they acquire the ability to migrate to sites of infection, and this is largely attributed to the increased surface expression of chemokine receptors and adhesion molecules. We observed that genes encoding chemotactic proteins Ccr2 and Ccr5 and the cell adhesion proteins Itgam (CD11b), Itgax (CD11c), and Itgb1 (CD29) were already highly expressed by naïve NK cells, in contrast to requiring induction in effector CD8+ T cells. Thus, the concerted action of these molecules may influence the appropriate tissue distribution of NK and effector CD8+ T cells.
Figure 4. Naïve NK cells are primed for effector responses.
(a) Genes significantly expressed in resting NK cells as compared to resting CD8+ T cells, and also induced in effector CD8+ T cells during responses to VSV and Lm are shown. Color groups represent secreted effector molecules (green), receptors (purple), integrins (red), regulatory molecules (brown), transcription factors (blue), and others (grey). Only the most significant genes are depicted (δ score > 2.0); a complete list is available in Supplementary Table 2. (b) Antibody staining of genes identified in (a) as being expressed in naïve NK cells and effector CD8+ T cells, but not naïve CD8+ T cells. Data are representative of at least two independent experiments. (c) Normalized log intensities for naïve NK cells and CD8+ T cells responding to VSV or Lm are plotted for each significant gene and subjected to linear regression. Slopes are shown with 95% confidence bands.
The transcription factors (e.g. Tbx21 [T-bet], Id2, and Prdm1 [Blimp-1]) were also expressed in naïve NK cells and effector CD8+ T cells, suggesting a common differentiation program. The elevated expression of Blimp-1 in naïve NK cells is intriguing, given the role of this transcription factor in regulating the differentiation of effector CD8+ T cells34, 35.
A comparison of expression levels of the effector genes revealed that, on average, they are expressed at higher levels in naïve NK cells than in effector CD8+ T cells (Fig. 4c). This suggests that with regard to transcriptional pre-priming, NK cells are maximally expressing levels of these effector molecules as part of their persistently “alerted” state. These findings demonstrate that the pre-primed state previously described for Ifng and granzymes also applies transcriptome-wide to many additional putative effector genes.
Transcriptional profile of NK cells during MCMV infection
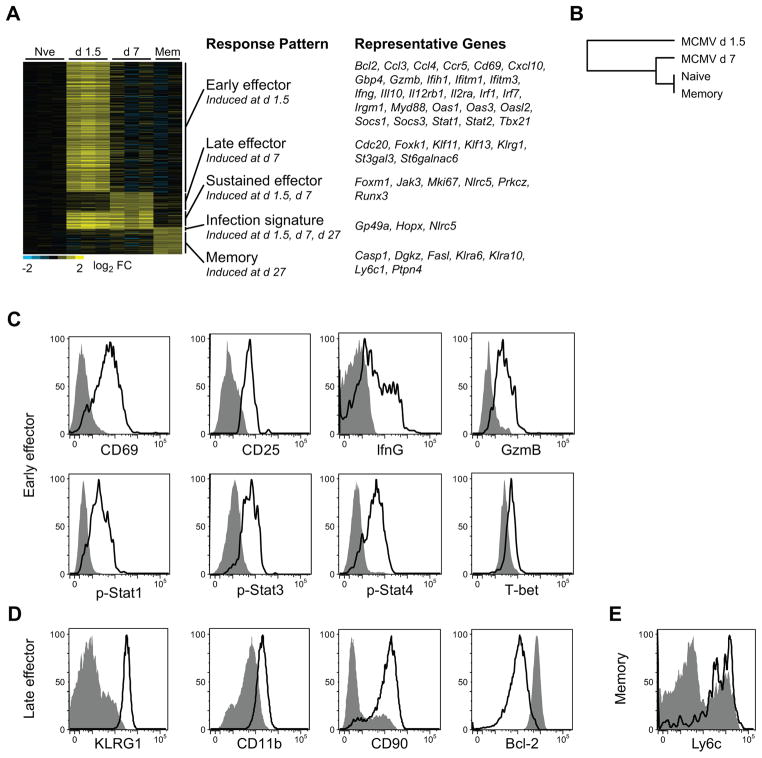
The transcriptional baseline defined above represents a single state – the resting state -of an NK cell’s existence. To explore NK cell changes during pathogen-specific activation, we generated a kinetic portrait of gene expression by profiling naïve, early effector (day 1.5 post-infection (PI), late effector (day 7 PI) and memory (day 27 PI) Ly49H+ NK cells following MCMV infection.
The largest changes occurred early during infection as revealed by a number of genes differentially expressed in activated Ly49H+ NK cells (875 genes upregulated) (Fig. 5a, Supplementary Table 3). The majority of this response was diminished by day 7 PI, although some expression was sustained. On the whole, day 7 PI late effector and day 27 PI memory NK cells were more similar to one another in their gene expression patterns than any other population pair (Fig. 5b). Additionally, a number of genes were specifically upregulated in memory NK cells, supporting the notion that NK cell memory reflects a unique state compared to that of naïve or effector NK cells36.
Figure 5. The Ly49H+ NK cell response to MCMV infection is dominated by an early activation response, followed by effector and memory responses.
(a) A heat map depicting all genes significantly induced at any time point post-infection relative to naïve Ly49H+ NK cells, grouped by hierarchical clustering. (b) The day 1.5 transcriptome of Ly49H+ NK cells responding to MCMV segregates from NK cells at other times during the response. Averaged populations were organized by hierarchical clustering based on Euclidean distance using all genes expressed at any time point. (c–e) Flow cytometric surface expression of CD69, CD25, and intracellular expression of IFNγ, GzmB, pStat-1, pStat-3, pStat-4, T-bet at day 1.5 PI (c), KLRG1, CD11b, CD90, and intracellular Bcl-2 at day 7 PI (d) and Ly6c at day 27 PI (e) is shown in Ly49H+ NK cells from MCMV-infected (black histogram) or uninfected mice (gray histogram). Data are representative of three independent experiments.
Transcripts upregulated at day 1.5 PI included indicators of inflammation (e.g. Cd69, Ifih1, Ifitm1, Ifitm3), proliferation (e.g. Il2ra encoding CD25), and effector function (e.g. Ifng, GzmB) (Fig. 5a, Supplementary Table 3). Increased expression of a representative set of these molecules was also confirmed by flow cytometry (Fig. 5c). Previous studies have demonstrated that IL-12 signaling through STAT proteins promotes IFN-γ production37. IL-12 receptor (Il12rb1) and STAT (Stat1, Stat2) transcripts increase early after MCMV infection, suggesting that mechanisms are in place for activated NK cells to more easily sense IL-12 and signal via STATs to mediate optimal production of effector cytokines such as IFN-γ (Fig. 5a). Phosphorylated STAT1, STAT3, and STAT4 are all found at elevated amounts at day 1.5 PI (Fig. 5c). Although Tbx21 (T-bet) is expressed in resting NK cells (Fig. 2e), it is further upregulated at the transcript and protein level (Fig. 5a,c); whether the increased expression of this transcription factor influences the effector function of NK cells remains to be determined. Because NK cells are such potent effector cells when activated, these cells must also be immediately regulated so as to not generate uncontrolled inflammation in the environment, evidenced by an increase in suppressor of cytokine signaling (SOCS1 and SOCS3) transcripts also observed at this early time point. Indeed, IFN-γ production by NK cells peaks at day 1.5 PI, but is rapidly abrogated, likely due to the activity of SOCS proteins38. In addition, NK cells transcribe and express IL-10 early after MCMV infection, which serves to regulate the magnitude of the immune response and limit pathology39. Thus, both pro-inflammatory and regulatory molecules exert their influences on NK cell activation and effector function early after MCMV infection.
Day 7 following MCMV infection marks the peak of clonal expansion for Ly49H+ NK cells40. Consistent with the observation that Ly49H+ NK cells are capable of 100–1000-fold expansion during MCMV infection, regulators of cell cycle (Cell Division Cycle, or CDC proteins) and protein associated with cellular proliferation (MKI67) are elevated at this time point (Fig. 5a, Supplementary Table 3). Transcription factors including Foxm141 and Klf1342, shown to regulate proliferation and survival of T lymphocytes, are also increased in Ly49H+ NK cells at day 7. Additionally, enhanced expression of Klrg1 (KLRG1), Itgam (CD11b), and Thy (CD90) are observed at both the transcript and protein level on Ly49H+ NK cells at day 7 PI compared to resting NK cells (Fig. 5a,d). These data demonstrate that the transcriptional changes for these markers represent all Ly49H+ NK cells, as opposed to a subset.
Following the peak of the effector Ly49H+ NK cell response against MCMV, a “contraction” phase occurs in which most effector cells undergo cell death, leaving behind a long-lived “memory” NK cell pool that persists for months following initial infection40, 43. Consistent with the contraction phase of the NK cell response beginning at approximately day 7 PI, expression of Bcl-2 is diminished at this time point relative to earlier times in infection (Fig. 5d). At day 27 PI, expression of Ly6c1 transcripts in memory NK cells was increased compared to resting NK cells (Fig. 5a) and confirmed by cell surface staining (Fig. 5e). In summary, the data explore the transcriptional dynamics of NK cells during MCMV infection, and further suggest that multiple cellular processes are involved in the differentiation of naïve NK cells into effector and memory cells, despite pre-priming of some effector mechanisms.
Effector NK and CD8+ T cell differentiation
The effector response to infection is characterized by migration of effector cells from lymphoid to non-lymphoid tissues, clonal expansion, secretion of anti-viral cytokines, and cytolysis of infected cells. Although some effector functions are pre-primed in NK cells, they rely on migration to secondary lymphoid organs and dendritic cell-derived signals in order to become fully functional24. To define a transcriptional program central to effector differentiation, we examined changes in gene expression common to NK cell and CD8+ T cell differentiation in response to infection. In order to identify appropriate time points for comparison, correlations were calculated between the gene expression changes in NK cells after days 1.5 and 7 after MCMV infection as compared with each CD8+ T cell time point after infection with either VSV or Lm (Fig. 6a), which revealed that NK cells at day 7 PI were, on the whole, transcriptionally more similar to CD8+ effector T cells than NK cells at day 1.5 PI. Furthermore, this response most closely resembled the CD8+ T cell response after day 6 of infection with either VSV or Lm. We therefore used the three most strongly correlated time points (VSV at days 6, 8, and 15 PI and Lm at days 8, 10, and 15) for the comparison with NK cells at day 7 PI.
Figure 6. Common effector mechanisms of NK cells and CD8+ T cells.
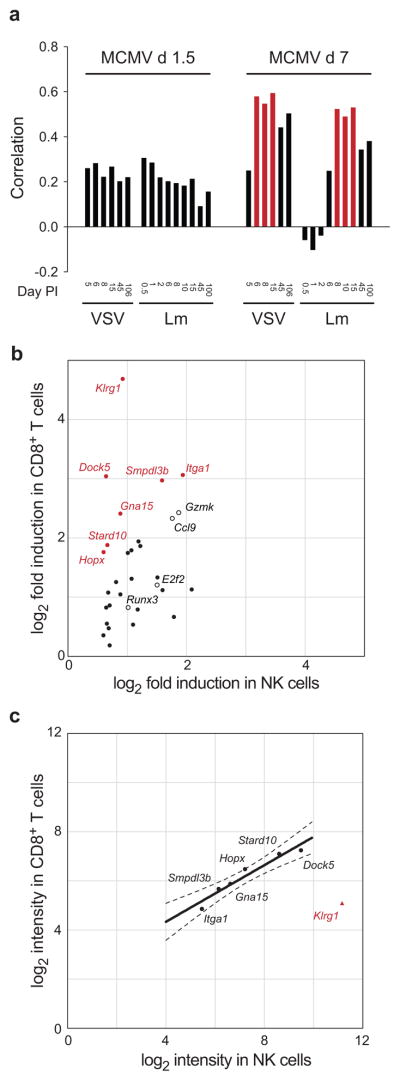
(a) Pearson correlations of the day 1 and day 7 NK cell responses to MCMV infection as compared to CD8+ T cell responses to VSV and Lm. The three most strongly related responses between NK cells and CD8+ T cells responding to VSV and Lm are shown in red. (b) Induction of effector genes in NK cells is muted as compared with CD8+ effector cells. Genes exhibiting greater than two-fold difference in regulation are shown in red text; additional selected representative genes are shown in black text. (c) Muted gene induction in NK cells is at least in part due to higher levels of basal expression in naïve NK cells.
We identified 32 genes with significantly altered expression in effector NK and CD8+ T cells compared with their naïve counterparts (Fig. 6b, Supplementary Table 4). These included genes encoding transcription factors (e.g. Runx3, E2f2, Hmgb2, Zmiz1, and HopX), migration and adhesion molecules (e.g. Itga1 and Ccl9), and effector molecule Gzmk. KLRG1 is found on the surface of both Ly49H+ NK cells (Fig. 5d) and antigen-specific CD8+ T cells undergoing clonal expansion (Fig. 4b) and may provide a means to regulate cells undergoing rapid cell division and limit collateral damage to host tissues.
Comparison of NK and CD8+ T cell effector responses on a gene-by-gene basis revealed that the magnitude of gene induction in NK cells was lower than in CD8+ T cells (Fig. 6b); the median gene induction in NK cells was 85% of that in CD8+ T cells (p = 0.039). Whereas 7 genes displayed greater than two-fold higher induction in CD8+ T cells, no gene reached higher than two-fold induction in NK cells (Fig. 6b; red). The smaller magnitude of induction was largely due to a higher baseline expression of these genes in NK cells, which on average were expressed in naïve CD8+ T cells at 58% of the levels measured in naïve NK cells (Fig. 6c; slope=0.58, 95% CI 0.36–0.79). Klrg1 also followed this trend but to a greater degree, with the high levels of expression in naïve NK cells (67-fold higher than in naïve CD8+ T cells) (Fig. 6c; red). We conclude that because NK cells are naturally primed for rapid responses to pathogens, the magnitude of their transcriptome-wide induction is generally smaller than the specific responses of effector CD8+ T cells, as previously observed for Ifng and Gzmb44.
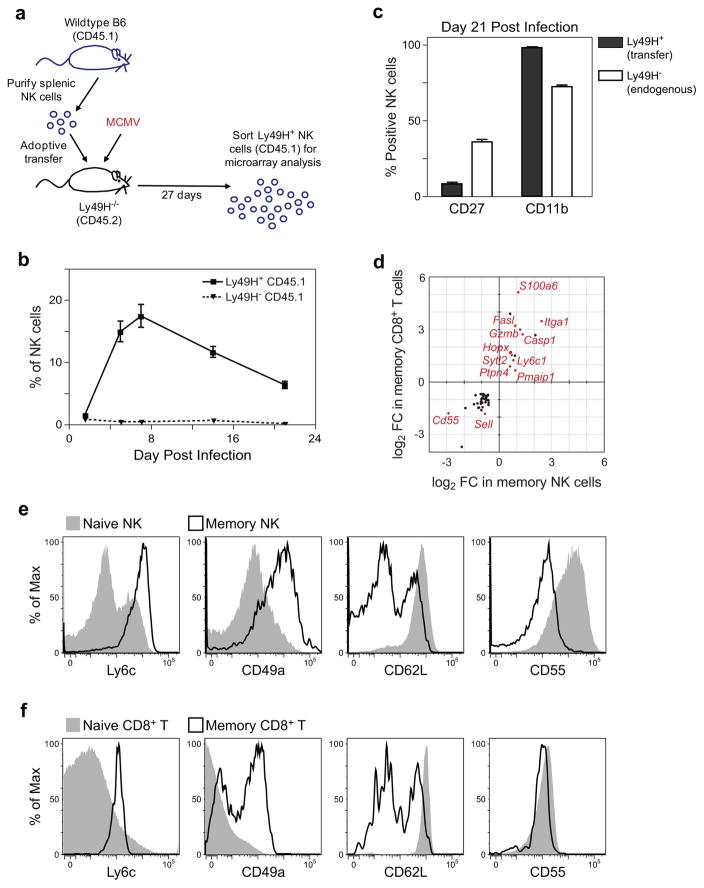
A conserved program underlying memory
During the second phase of both NK and CD8+ T cell immune responses, the majority of the effector cells die, but those that survive go on to seed a pool of long-lived memory cells that can subsequently acquire effector functions much more rapidly upon re-exposure to antigen40, 45. To better understand the underlying program that establishes these functions, we identified the transcriptional changes that accompany memory cell differentiation. Using a previously described adoptive transfer system40, we transferred purified NK cells from wild-type mice into Ly49H-deficient hosts (Fig. 7a). After MCMV infection, Ly49H+ NK cells underwent massive expansion over the course of 7 days, followed by a contraction phase and the generation of long-lived memory NK cells isolated at day 27 PI (Fig. 7b). In contrast to naïve Ly49H+ NK cells or to endogenous Ly49H− NK cells, memory NK cells showed increased expression of CD11b and reduced expression of CD27, consistent with prior observations (Fig. 7c)40.
Figure 7. Common memory responses of NK cells and CD8+ T cells.
(a) Wild-type NK cells (105) from C57BL/6 (CD45.1) mice were transferred into Ly49H-deficient C57BL/6 mice (CD45.2) and infected with MCMV. (b) Percentages of transferred Ly49H+ CD45.1+ and Ly49H− CD45.1+ NK cells within the total NK1.1+ TCRβ− population after MCMV infection. (c) Transferred Ly49H+ CD45.1+ and endogenous Ly49H− CD45.1+ NK cells were analyzed for CD27 and CD11b expression. (d) Genes commonly induced are shown with fold changes (FC) in each population. Selected representative genes are shown in red text. (e,f) The expression of Ly6c, CD49a, CD62L, and CD55 was assessed in (e) Ly49H+ NK cells from naïve or MCMV-infected mice (day 28 PI) or (f) OTI CD8+ T cells from naïve or VSV-infected mice (day 60 PI, except for CD62L which was measured at day 100 PI). Data are representative of at least two independent experiments.
We compared gene expression profiles of memory Ly49H+ NK cells (day 27 post MCMV) and memory CD8+ T cells (generated following either VSV or Lm) to naïve Ly49H+ NK and CD8+ T cells, respectively, and identified a common set of 47 genes that were coordinately regulated during the differentiation of naïve into memory cells. (Fig. 7d, Supplementary Table 5). Memory specific transcripts include genes involved in signaling potential (e.g. s100a6 and Ptpn4), effector function (e.g. Gzmb, Fasl, and Sytl2), migration (e.g. Sell [CD62L] and Itga1 [CD49a]), and apoptosis (e.g. Casp1 and Pmaip1). A subset of these identified genes including s100a6, Casp1, Itga1, Ly6c1, and Gzmb was also upregulated in memory CD8+ T cells from the lymphocytic choriomeningitis virus (LCMV) model of CD8+ T cell memory differentiation46. Flow cytometric analysis confirmed upregulation of Ly6C and CD49a and downregulation of CD62L and CD55 in both memory NK (Fig. 7e) and memory CD8+ T (Fig. 7f) cells with CD49a and CD55 being the novel cell surface makers of memory NK and T cells. In addition, we identified a transcription factor, homeobox only protein (Hopx) that was upregulated in memory cells. Previous reports that Hopx is upregulated in iTregs and effector memory T cells and was critical for the survival of activated mouse TH1 effector/memory cells47, 48 suggest that this regulator may also promote the persistence of NK memory cells after infection.
Analysis of the expression intensities revealed that similar to gene induction in effector populations, the magnitude of induction was generally greater in memory CD8+ T cells than in memory NK cells (slope = 1.50, 95% CI 1.28 to 1.72). Together, these results demonstrate that there is a common transcriptional program conserved in NK and CD8+ T cell memory differentiation in response to infection.
Discussion
A major goal of this work was to define, from the transcriptome-wide perspective, the NK cells in various states to generate novel insights and a resource describing the specific genes involved with NK cell functions. We used the breadth of cell populations available from the ImmGen project to explore transcripts that define the identity of NK cells in a more robust and in-depth perspective than provided by previous analyses10–12. The results show extensive transcriptional differences between NK cells and other leukocyte populations far beyond the few specific markers commonly used to identify them by flow cytometry. In addition, we found that few transcripts were uniquely specific to NK cells, many being shared with other cell types of the immune system, particularly T cells. These data provide a genome-wide context for interpretation of NK cell functions and will accelerate the discovery of pathways that regulate NK cell activation states.
We demonstrate a close transcriptional relatedness between naïve NK cells and innate-like T cells, suggesting that their gene expression profiles reflect a functional rather than developmental similarity. The abundance of cell surface receptors and signaling molecules expressed in NK and innate-like T cells is consistent with their role as a primary sentinel and reveals a wealth of novel signaling mechanisms for further targeted exploration.
Although NK cells are distinct in their innate properties, they also exhibit properties associated with adaptive CD8+ T cells including cytotoxicity and memory. Our work provides the first systematic identification of the genes associated with these common behaviors and identifies hundreds of genes not previously known to be associated with these functions. We found elements of the effector and memory NK cell differentiation signature that were shared by effector and memory CD8+ T cells, suggesting conservation of some activation mechanisms between NK and CD8+ T cell lineages. However, this must be interpreted in the context of dynamic changes in activation state, as evidenced by distinct NK cell transcriptomes at each stage of differentiation from naïve to activated to late effector to memory cell. Although understanding the function of NK cell memory is in its infancy, one implication of a defined gene expression signature corresponding to memory differentiation is that specific genes could be useful as surrogate markers for memory NK cells with the greatest potential to confer immunologic potential.
As for αβ T cells, the memory NK cell differentiation signature contains genes that are both unique to the memory state (e.g. Casp1, Fasl, and Ly6c1) and genes that are initially expressed in effector cells and are maintained in memory cells (e.g. Itga1 and Hopx) – genes that have not been previously appreciated for a potential role in NK cell memory and worthy of further study. This suggests that the transcriptome of memory NK cells represents a composite of genes uniquely expressed by these long-lived cells and those maintained from prior stages of differentiation, possibly to allow quick re-expression upon secondary exposure to antigen. We speculate that the expression pattern of the memory “repertoire”, rather than the individual genes themselves, is required to define the memory NK cell state.
This study provides a comprehensive transcriptome perspective on various stages of NK cell function in the context of closely related T lymphocytes. The data simultaneously support and extend previous findings, while providing a unique resource for future investigation of NK cell biology.
METHODS
Mice and infections
In accordance with ImmGen standard operating protocol (SOP) (www.immgen.org), six-to-eight week-old male C57BL/6 and B6.SJL-Ptprca Pepcb/BoyJ (congenic CD45.1) mice were received from the Jackson Laboratory and maintained under specific pathogen free conditions. C57BL/6 mice were infected by intraperitoneal injections of Smith strain MCMV (5×104 plaque-forming units (pfu). An adoptive transfer system was used to generate memory NK cells as described40. In brief, purified NK cells from B6.SJL-Ptprca Pepcb/BoyJ mice were adoptively transferred into Ly49H-deficient C57BL/6 mice one day before viral infection. In the case of CD8+ T cells, 5 × 103 CD45.1+ OTI TcR transgenic cells were transferred into C57BL/6 recipients. Mice were infected 1 day after transfer with either 5 × 103 cfu Lm-OVA or 5 × 103 pfu VSV-OVA. To obtain naïve OTI cells, 5 × 106 CD45.1+ OTI cells were transferred into C57BL/6 mice and were purified from mice 2 days post transfer. Experiments were done according to the UCSF Institutional Animal Care and Use Committee guidelines.
Cell Sorting
Cells were prepared according to ImmGen SOP (www.immgen.org). Naïve NK or Ly49H+ NK cells were isolated from spleens of uninfected C57BL/6 mice. Effector Ly49H+ NK cells were isolated from C57BL/6 mice on day 1.5 and 7 after MCMV infection. Memory Ly49H+ NK cells were isolated from spleens of reconstituted Ly49H-deficient mice on day 27 after MCMV infection. All samples were pooled from 3 mice, stained for cell surface markers and ~1–3 × 104 cells (>99% pure) were double-sorted directly into Trizol (Invitrogen) using a FACSAria cell sorter (BD). For each population, independent triplicate samples were sorted, except for memory Ly49H+ NK cells, which independent duplicate samples were sorted from 16 mice.
Microarray hybridization and analysis
Isolated RNA was amplified and prepared for hybridization to the Affymetrix MoGene 1.0 ST array using the GeneChip Whole Transcript Sense Target Labeling Assay in accordance with manufacturer’s instructions. Raw data were normalized using the robust multichip average algorithm in the “Expression File Creator” module (Gene Pattern). Raw data from all ImmGen samples are available in the NCBI GEO database (GSE 15907); processed data are available on ImmGen’s data browsers (www.immgen.org). The consortium-standardized post-normalization threshold of 120 was taken to indicate expression above background, and probes were included in comparisons only if they were expressed in all replicates of at least one population. Additional details49 are provided as Supplementary Note 1 and Supplementary Note 2.
Flow cytometry
Fc receptors were blocked with anti-CD16 and CD32 mAb (2.4G2) (UCSF Antibody Core) at 10μg/ml prior to surface staining. Antibodies to the following cell surface markers were purchased or generously provided to ImmGen by eBioscience: NK1.1 (clone PK136), NKp46 (29A1.4), TCRβ (H57-597), CD4 (RM4–5 or GK1.5), CD8 (53-6.7), CD3 (145-2C11 or eBio500A2), CD5 (53-7.3), CD19 (MB19-1), CD25 (PC61.5), GR-1 (RB6-8C5), B220 (RA3-6B2), Ter119 (Ter119), Ly49E/F (CM4), Ly49H (3D10), Ly49I (YLI-90), NKG2D (MI-6), NKG2A/C/E (20d5), CD11b (M1/70), CD69 (H1.2F3), KLRG1 (2F1), CD45.1 (A20), CD45.2 (104), IFNγ (XMG1.2), Bcl2 (10C4), CD45.1 (A20), CD45.2 (104); or other vendors: CD11c (N418), CD55 (RIKO-3), CD69 (H1.2F3), CD90 (30-H12), T-bet (4B10) by BioLegend; CD49a (Ha31/8), CD62L (MEL-14), Ly6C (AL-21), TCRγδ (GL3), GzmB (GB11) by BD Pharmingen; p-STAT1, p-STAT3, p-STAT4, Live/Dead fixable near-IR dye by Invitrogen; and Syk (provided by A. Weiss, UCSF). Intracellular staining was performed according to manufacturer’s instructions (BD). All cells were analyzed on an LSRII (BD) with FloJo software (Tree Star).
Supplementary Material
Acknowledgments
We thank the members of the ImmGen Consortium and M. Dozmorov (Oklahoma Medical Research Foundation) for discussions, the ImmGen core team (M. Painter, J. Ericson, and S. Davis) for data generation and processing, J. Jarjoura and J. Arakawa-Hoyt for assistance in cell sorting, A. Beaulieu, J. Karo, and S. Madera for data from MCMV infection experiments, and eBioscience, Affymetrix, and Expression Analysis for support of the ImmGen Project. This work is supported by R24 AI072073 and AI068129 from NIH/NIAID, ACS fellowship (to N.A.B.), CIHR fellowship (to G.M.), T32AI060537 (to D.W.H.), T32AI060536 (to J.A.B.), and A1072117 (to A.W.G.). J.C.S. is a Searle Scholar, and L.L.L. is an American Cancer Society Professor.
The Immunological Genome Project Consortium
Gautier, E.L.1,2, Jakubzick, C.1, Randolph, G.J.1,2, Best, J.A.3, Knell, J.3, Goldrath, A.3, Miller, J.1, Brown, B.1, Merad, M.1, Jojic, V.4, Koller, D.4, Cohen, N.5, Brennan, P.5, Brenner, M.5, Shay, T.6, Regev, A.6, Fletcher, A.7, Elpek, K.7, Bellemare-Pelletier, A.7, Malhotra, D.7, Turley, S.7, Jianu, R.8, Laidlaw, D.8, Collins, J.J.9, Narayan, K.10, Sylvia, K.10, Kang, J.10, Gazit, R.11, Rossi, D.J.11, Kim, F.12, Rao, T.N.12, Wagers, A.12, Shinton, S.A.13, Hardy, R.R13, Monach, P.14, Bezman, N.A.15, Sun, J.C.15, Kim, C.C.15, Lanier, L.L.15, Heng, T.16, Kreslavsky, T.4, Painter, M.16, Ericson, J. 16 Davis, S. 16, Mathis, D.16, Benoist, C.16
Footnotes
Icahn Medical Institute, Mount Sinai Hospital 1425 Madison Ave New York, NY
Department of Pathology & Immunology, Washington University, 660 South Euclid Ave, St. Louis, MO
Division of Biological Sciences, University of California San Diego, 9500 Gilman Dr., La Jolla, CA
Computer Science Department, Stanford University, 353 Serra Mall, Stanford, CA
Rheumatology, Immunology and Allergy, Brigham and Women’s Hospital, 75 Francis St, Boston, MA
Broad Institute, 7 Cambridge Center, Cambridge, MA
Dana-Farber Cancer Institute and Harvard Medical School, 450 Brookline Ave, Boston, MA
Computer Science Department, Brown University, 115 Waterman St, Providence, RI
HHMI, Department of Biomedical Engineering, Boston University, 44 Cummington St, Boston, MA
University of Massachusetts Medical School 55 Lake Ave North Worcester, MA
Department of Stem Cell and Regenerative Biology, Harvard University, and Program in Cellular and Molecular Medicine, Children’s Hospital, 200 Longwood Ave, Boston, MA
Joslin Diabetes Center, 1 Joslin Place, Boston, MA
Fox Chase Cancer Center, 333 Cottman Ave, Philadelphia, PA
Department of Medicine, Boston University, 72 East Concord St, Boston MA
Department of Microbiology & Immunology, University of California San Francisco, 513 Parnassus Ave, San Francisco
Division of Immunology, Dept of Microbiology & Immunobiology, Harvard Medical School, 77 Ave Louis Pasteur, Boston, MA.
Author contributions
C.C.K. analyzed data; N.A.B. and J.C.S. sorted cell subsets, performed follow-up experiments and analyzed data; G.M., D.W.H., Y.K. performed experiments; J.A.B. and A.W.G. designed and carried out the T cell studies; N.A.B., C.C.K., J.C.S., L.L.L. designed studies and wrote the paper.
References
- 1.Heng TS, Painter MW. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 2.Bendelac A, Bonneville M, Kearney JF. Autoreactivity by design: innate B and T lymphocytes. Nat Rev Immunol. 2001;1:177–186. doi: 10.1038/35105052. [DOI] [PubMed] [Google Scholar]
- 3.Vivier E, et al. Innate or adaptive immunity? The example of natural killer cells. Science. 331:44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herberman RB, Nunn ME, Holden HT, Lavrin DH. Natural cytotoxic reactivity of mouse lymphoid cells against syngeneic and allogeneic tumors. II. Characterization of effector cells. Int J Cancer. 1975;16:230–239. doi: 10.1002/ijc.2910160205. [DOI] [PubMed] [Google Scholar]
- 5.Kiessling R, Klein E, Pross H, Wigzell H. “Natural” killer cellsin the mouse. II. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Characteristics of the killer cell. Eur J Immunol. 1975;5:117–121. doi: 10.1002/eji.1830050209. [DOI] [PubMed] [Google Scholar]
- 6.Lanier LL, Phillips JH. Ontogeny of natural killer cells. Nature. 1986;319:269–270. doi: 10.1038/319269b0. [DOI] [PubMed] [Google Scholar]
- 7.Di Santo JP. Natural killer cell developmental pathways: a question of balance. Annu Rev Immunol. 2006;24:257–286. doi: 10.1146/annurev.immunol.24.021605.090700. [DOI] [PubMed] [Google Scholar]
- 8.Yamagata T, Benoist C, Mathis D. A shared gene-expression signature in innate-like lymphocytes. Immunol Rev. 2006;210:52–66. doi: 10.1111/j.0105-2896.2006.00371.x. [DOI] [PubMed] [Google Scholar]
- 9.Chambers SM, et al. Hematopoietic fingerprints: an expression database of stem cells and their progeny. Cell Stem Cell. 2007;1:578–591. doi: 10.1016/j.stem.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Obata-Onai A, et al. Comprehensive gene expression analysis of human NK cells and CD8(+) T lymphocytes. Int Immunol. 2002;14:1085–1098. doi: 10.1093/intimm/dxf086. [DOI] [PubMed] [Google Scholar]
- 11.Hanna J, et al. Novel insights on human NK cells’ immunological modalities revealed by gene expression profiling. J Immunol. 2004;173:6547–6563. doi: 10.4049/jimmunol.173.11.6547. [DOI] [PubMed] [Google Scholar]
- 12.Dybkaer K, et al. Genome wide transcriptional analysis of resting and IL2activated human natural killer cells: gene expression signatures indicative of novel molecular signaling pathways. BMC Genomics. 2007;8:230. doi: 10.1186/1471-2164-8-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol. 2008;9:495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bezman N, Koretzky GA. Compartmentalization of ITAM and integrin signaling by adapter molecules. Immunol Rev. 2007;218:9–28. doi: 10.1111/j.1600-065X.2007.00541.x. [DOI] [PubMed] [Google Scholar]
- 15.Pont F, et al. The gene expression profile of phosphoantigen-specific human gammadelta T lymphocytes is a blend of alphabeta T-cell and NK-cell signatures. Eur J Immunol. 42:228–240. doi: 10.1002/eji.201141870. [DOI] [PubMed] [Google Scholar]
- 16.Wilson SB, Byrne MC. Gene expression in NKT cells: defining a functionally distinct CD1d-restricted T cell subset. Curr Opin Immunol. 2001;13:555–561. doi: 10.1016/s0952-7915(00)00258-2. [DOI] [PubMed] [Google Scholar]
- 17.Hesslein DG, Lanier LL. Transcriptional control of natural killer cell development and function. Adv Immunol. 109:45–85. doi: 10.1016/B978-0-12-387664-5.00002-9. [DOI] [PubMed] [Google Scholar]
- 18.Sun Y, et al. Potentiation of Smad-mediated transcriptional activation by the RNA-binding protein RBPMS. Nucleic Acids Res. 2006;34:6314–6326. doi: 10.1093/nar/gkl914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li MO, Flavell RA. TGF-beta: a master of all T cell trades. Cell. 2008;134:392–404. doi: 10.1016/j.cell.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akbulut S, et al. Sprouty proteins inhibit receptor-mediated activation of phosphatidylinositol-specific phospholipase C. Mol Biol Cell. 21:3487–3496. doi: 10.1091/mbc.E10-02-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Narni-Mancinelli E, et al. Fate mapping analysis of lymphoid cells expressing the NKp46 cell surface receptor. Proc Natl Acad Sci U S A. 108:18324–18329. doi: 10.1073/pnas.1112064108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu J, et al. NKp46 identifies an NKT cell subset susceptible to leukemic transformation in mouse and human. J Clin Invest. 121:1456–1470. doi: 10.1172/JCI43242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Satoh-Takayama N, et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29:958–970. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 24.Walzer T, et al. Natural killer cell trafficking in vivo requires a dedicated sphingosine 1-phosphate receptor. Nat Immunol. 2007;8:1337–1344. doi: 10.1038/ni1523. [DOI] [PubMed] [Google Scholar]
- 25.Despoix N, et al. Mouse CD146/MCAM is a marker of natural killer cell maturation. Eur J Immunol. 2008;38:2855–2864. doi: 10.1002/eji.200838469. [DOI] [PubMed] [Google Scholar]
- 26.Arase H, Saito T, Phillips JH, Lanier LL. Cutting edge: the mouse NK cell-associated antigen recognized by DX5 monoclonal antibody is CD49b (alpha 2 integrin, very late antigen-2) J Immunol. 2001;167:1141–1144. doi: 10.4049/jimmunol.167.3.1141. [DOI] [PubMed] [Google Scholar]
- 27.Ewen CL, Kane KP, Bleackley RC. A quarter century of granzymes. Cell Death Differ. 19:28–35. doi: 10.1038/cdd.2011.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Young JD, Hengartner H, Podack ER, Cohn ZA. Purification and characterization of a cytolytic pore-forming protein from granules of cloned lymphocytes with natural killer activity. Cell. 1986;44:849–859. doi: 10.1016/0092-8674(86)90007-3. [DOI] [PubMed] [Google Scholar]
- 29.Colige A, et al. Cloning and characterization of ADAMTS-14, a novel ADAMTS displaying high homology with ADAMTS-2 and ADAMTS-3. J Biol Chem. 2002;277:5756–5766. doi: 10.1074/jbc.M105601200. [DOI] [PubMed] [Google Scholar]
- 30.Hirst CE, et al. The intracellular granzyme B inhibitor, proteinase inhibitor 9, is up-regulated during accessory cell maturation and effector cell degranulation, and its overexpression enhances CTL potency. J Immunol. 2003;170:805–815. doi: 10.4049/jimmunol.170.2.805. [DOI] [PubMed] [Google Scholar]
- 31.Zhang M, et al. Serine protease inhibitor 6 protectscytotoxic T cells from self-inflicted injury by ensuring the integrity of cytotoxic granules. Immunity. 2006;24:451–461. doi: 10.1016/j.immuni.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 32.Liu L, et al. A novel protein tyrosine kinase NOK that shares homology with platelet-derived growth factor/fibroblast growth factor receptors induces tumorigenesis and metastasis in nude mice. Cancer Res. 2004;64:3491–3499. doi: 10.1158/0008-5472.CAN-03-2106. [DOI] [PubMed] [Google Scholar]
- 33.Kaech SM, Hemby S, Kersh E, Ahmed R. Molecular and functional profiling of memory CD8 T cell differentiation. Cell. 2002;111:837–851. doi: 10.1016/s0092-8674(02)01139-x. [DOI] [PubMed] [Google Scholar]
- 34.Kallies A, Xin A, Belz GT, Nutt SL. Blimp-1 transcription factor is required for the differentiation of effector CD8(+) T cells and memory responses. Immunity. 2009;31:283–295. doi: 10.1016/j.immuni.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 35.Shin H, et al. A role for the transcriptional repressor Blimp-1 in CD8(+) T cell exhaustion during chronic viral infection. Immunity. 2009;31:309–320. doi: 10.1016/j.immuni.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun JC, Lopez-Verges S, Kim CC, DeRisi JL, Lanier LL. NK cells and immune “memory”. J Immunol. 186:1891–1897. doi: 10.4049/jimmunol.1003035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trinchieri G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat Rev Immunol. 2003;3:133–146. doi: 10.1038/nri1001. [DOI] [PubMed] [Google Scholar]
- 38.Palmer DC, Restifo NP. Suppressors of cytokine signaling (SOCS) in T cell differentiation, maturation, and function. Trends Immunol. 2009;30:592–602. doi: 10.1016/j.it.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee SH, Kim KS, Fodil-Cornu N, Vidal SM, Biron CA. Activating receptors promote NK cell expansion for maintenance, IL-10 production, and CD8 T cell regulation during viral infection. J Exp Med. 2009;206:2235–2251. doi: 10.1084/jem.20082387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun JC, Beilke JN, Lanier LL. Adaptive immune features of natural killer cells. Nature. 2009;457:557–561. doi: 10.1038/nature07665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xue L, Chiang L, He B, Zhao YY, Winoto A. FoxM1, a forkhead transcription factor is a master cell cycle regulator for mouse mature T cells but not double positive thymocytes. PLoS One. 5:e9229. doi: 10.1371/journal.pone.0009229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou M, et al. Kruppel-like transcription factor 13 regulates T lymphocyte survival in vivo. J Immunol. 2007;178:5496–5504. doi: 10.4049/jimmunol.178.9.5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Leary JG, Goodarzi M, Drayton DL, von Andrian UH. T cell-and B cell-independent adaptive immunity mediated by natural killer cells. Nat Immunol. 2006;7:507–516. doi: 10.1038/ni1332. [DOI] [PubMed] [Google Scholar]
- 44.Sun JC, Lanier LL. NK cell development, homeostasis and function: parallels with CD8 T cells. Nat Rev Immunol. 11:645–657. doi: 10.1038/nri3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cui W, Kaech SM. Generation of effector CD8+ T cells and their conversion to memory T cells. Immunol Rev. 236:151–166. doi: 10.1111/j.1600-065X.2010.00926.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wherry EJ, et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27:670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 47.Albrecht I, et al. Persistence of effector memory Th1 cells is regulated by Hopx. Eur J Immunol. 40:2993–3006. doi: 10.1002/eji.201040936. [DOI] [PubMed] [Google Scholar]
- 48.Hawiger D, Wan YY, Eynon EE, Flavell RA. The transcription cofactor Hopx is required for regulatory T cell function in dendritic cell-mediated peripheral T cell unresponsiveness. Nat Immunol. 11:962–968. doi: 10.1038/ni.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malhotra D, Fletcher AL, Astarita J, Lukacs-Kornek V, Tayalia P, Gonzalez SF, Elpek KG, Chang SK, Knoblich K, Hemler ME, Brenner MB, Carroll MC, Mooney DJ, Turley SJ. Immunological Genome Project Consortium. Nat Immunol. 13:499–510. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.