As recently as 12 years ago, breakthroughs aided by laboratory neuroscience and modern neuroimaging had made differentiation and classification of malformations of cortical development seem straightforward (Barkovich et al., 2001). Some resulted from abnormal neuronal proliferation (microcephalies, megalencephalies), others from abnormal neuronal migration (heterotopia and lissencephaly), and still others from abnormal cortical organization (mostly polymicrogyrias). The protein products of the few genes known to be associated with these disorders (LIS1 and DCX with lissencephaly; FLNA with heterotopia) support these mechanistic concepts, as the lissencephaly genes are important in the migration of neurons (specifically in the extension of leading processes and nucleokinesis, the process by which the nucleus is able to follow the leading process); and the assumption that mutations of FLNA (which produces an actin cross-linking phosphoprotein important for cell locomotion) in neurons impair their migration is logical. Although details needed clarification, everything seemed to make sense.
The discovery that tubulin mutations cause brain malformations was easily incorporated into existing malformation classifications. That some infants with lissencephaly had mutations of TUBA1A (Poirier et al., 2007) did not dislodge existing concepts, as both LIS1 and DCX were known to be microtubule-associated proteins (MAPs), which complex with microtubules and/or microtubule motor proteins during cell migration. Reports that mutations of TUBB2B are associated with polymicrogyria (Jaglin et al., 2009) posed more difficulties, but caused no great surprise, as polymicrogyria is hard to explain. This malformation of cortical development is associated with many different conditions, including prenatal injury (infection or ischaemia) and many mutations. The causative mechanisms are unknown, and the appearance on imaging varies greatly. Still, it was easy to accept that disruption of the complex (and poorly understood) processes of terminal neuronal migration and cortical organization could result from TUBB2B mutations. Similarly, the report that TUBB3 mutations are associated with congenital fibrosis of the optic nerves (Poirier et al., 2010) was interesting but not altogether surprising. Microtubules play a large part in axonal navigation through the developing brain, particularly at growth cones—the axonal leading process. The attractive signal received by actin-rich filopodia in the tip (peripheral zone) of the growth cone is transduced to the cytoskeleton, causing polymerization of actin and extension of microtubules moving the growth cone in the direction of attraction; simultaneously, actin is depolymerized and filopodia retract from areas expressing neutral or repulsion signals. This process allows growth cones to navigate through the developing brain to their targets. Abnormalities of microtubules thus impair axon navigation and result in diminished size of nerves and white matter bundles, manifesting in patients with tubulin mutations as abnormalities of the corpus callosum, cranial nerves and internal capsule.
The past year has qualified these simple formulations. We now know that mutations of TUBA1A cause not only lissencephaly but also a polymicrogyria-like malformation of cortical development (Poirier et al., 2012), and that mutations of TUBB2B cause congenital fibrosis of the extraocular muscles, axon dysinnervation, polymicrogyria-like malformations of cortical development, pachygyria, as well as more complex developmental disorders (Cederquist et al., 2012; Guerrini et al., 2012; Romaniello et al., 2012). Two reports published in this issue of Brain now reveal that TUBB2B mutations can cause lissencephaly; that both TUBA1A and TUBB2B mutations can cause a spectrum of cortical malformations associated with mutations that affect specific portions of the tubulin heterodimer (Cushion et al., 2013: see pages 534–546); and that specific mutations of TUBB3 (the E410K phenotype) cause a fascinating new phenotype that can largely be interpreted as resulting from an axonal path-finding defect (Chew et al., 2013: see pages 520–533). This new information builds upon earlier work (Poirier et al., 2010; Tischfield et al., 2010) in advancing our understanding of how microtubules function in brain development and how effects of specific mutations of these genes alter the structure and, consequently, the function of their protein products.
Early neuronal development is complex: the neural tube rapidly expands from proliferation of progenitor cells, which differentiate and migrate before organizing into discrete components. While en route to their destination, neurons extend both large leading processes that aid their migration and small processes that develop into neurites (axons and dendrites) that elongate and form synapses, allowing communication with other neurons and the formation of networks. The dramatic changes in the structure of neurons as they migrate and connect with other neurons in distant parts of the nervous system can only be accomplished by active changes in the cytoskeleton, in particular those occurring in and adjacent to cilia, centrosomes and microtubules (Kuijpers and Hoogenraad, 2011). Therefore, it is not surprising that mutations in genes involved in the formation of these structures result in multiple and varied malformations. In addition to knowledge of when the gene is expressed and the degree of gene redundancy, an understanding of these processes underlying malformations requires knowledge of how each gene mutation affects the structure of its protein product; more specifically, how altered structure of the protein product affects its interaction with other molecules in specific molecular pathways has to be determined. By mapping the sites of amino acid substitutions resulting from TUBB2B and TUBA1A mutations onto a predicted 3D model of an α/β-tubulin heterodimer within microtubule polymer structures, Cushion et al. (2013) have correlated the location of several mutation-altered proteins to the sites of the α/β-tubulin interface and the binding site of DCX to tubulin. Sites near the α/β-tubulin interface should be important in formation of the heterodimer and its stability, both of which are crucial for rapid changes in the cytoskeleton (e.g. axonal pathfinding) of the developing neuron. It has been reported that some TUBB3 mutations (p.E205K, p.A302V, p.M388V and p.M323V) impair tubulin heterodimerization (Poirier et al., 2010; Tischfield et al., 2010), while others increase microtubule stability (Poirier et al., 2010); either of these effects could impair axonal pathfinding in which actin in filopodia rapidly polymerizes and microtubules are simultaneously built (through heterodimerization) or deconstructed upon encountering attractive or repulsive signals, respectively. Mutations that predominantly interfere with heterodimerization may not affect cortical development; indeed, E410 and other proteins that cause congenital fibrosis of the extraocular muscles are located on the opposite face of the heterodimer from those affected by malformations of cortical development resulting from TUBB3 mutations (Poirier et al., 2010). In addition, E410 is located at a kinesin motor protein-binding site of β-tubulin, near the α-tubulin interface, and is likely to be involved in heterodimerization (Fig. 1). The exact mechanisms by which the process is altered remain to be determined. Cushion et al. (2013) also note that the TUBB2B R380 site is located at the DCX-binding site of β-tubulin, close to the α-tubulin interface, but on the opposite side from proteins affected in congenital fibrosis of the extraocular muscles. As mutations of DCX are known to cause pachygyria and lissencephaly, it seems likely that these impact neuronal migration, and it is to be expected that mutations affecting R380 cause malformations of cortical development, in this case polymicrogyria-like cortical malformations. It will be interesting to see, in future studies, whether other tubulin-related causes of malformations of cortical development are associated with mutations near binding sites of other microtubule-associated proteins.
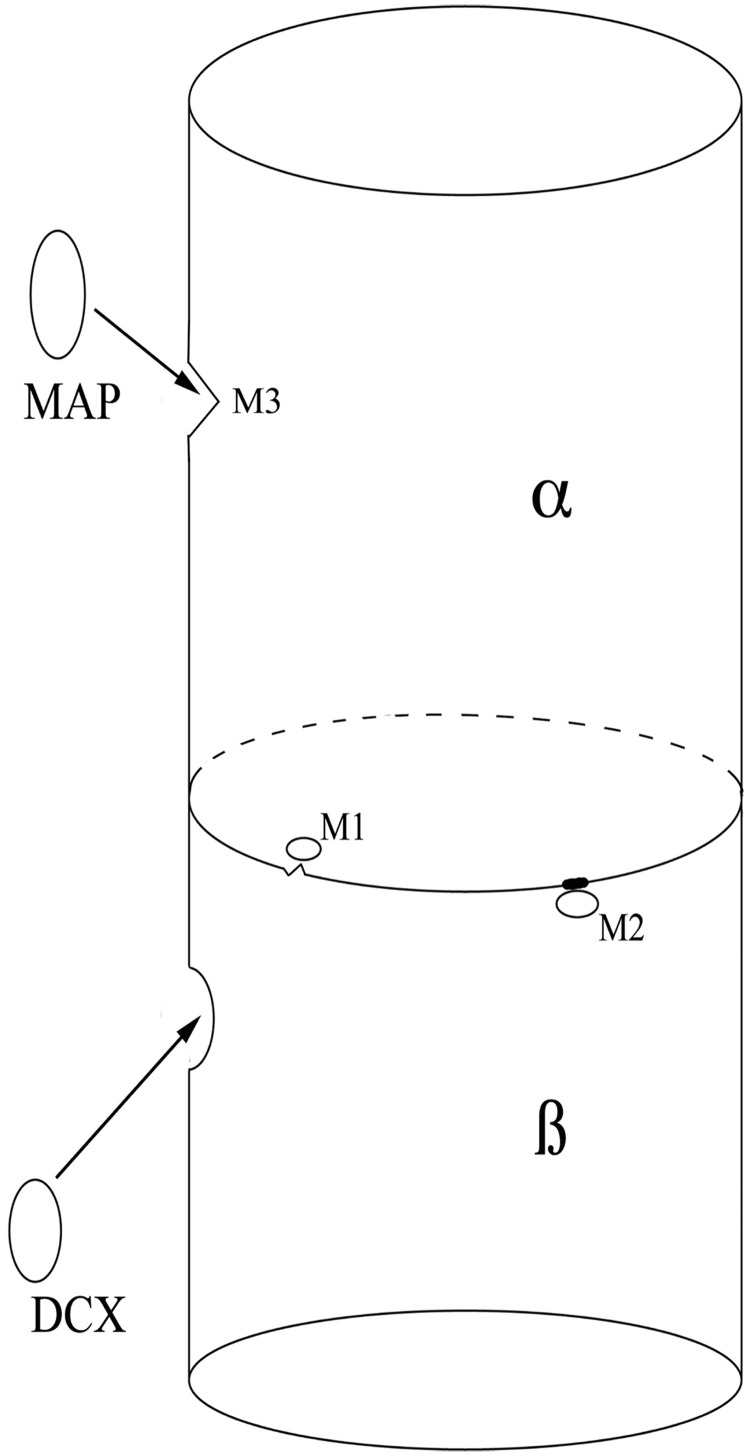
Figure 1.
Schematic showing the effects of mutations on the structure and stability of microtubules. This depicts a microtubule heterodimer, composed of alternating α- and β-subunits. Mutations near the interface of the subunits can interfere both with the dimerization process (owing to unstable regions of the interface that break down, M1) and rapid breakdown of the dimers (owing to impaired breakdown of abnormal regions of interface, M2) that are essential for neuronal migration and axonal pathfinding. This likely leads to hypoplastic axonal structures such as the cortical spinal tracts, corpus callosum and cranial nerves. Interaction with microtubule-associated proteins (MAPs) such as DCX (in β-subunit) is essential for other microtubule functions, such as mitosis and neuronal migration. Mutations near binding sites of the microtubule-associated proteins (M3 in α-subunit) can interfere with microtubule-associated protein binding, disturbing these critical functions in a manner similar to mutations of the microtubule-associated proteins themselves. These mutations are more likely to result in a malformation of cortical development.
Finally, use of the term ‘PMG-like MCD’ (polymicrogyria-like malformations of cortical development) in the article by Cushion et al. (2013) brings to the front an issue that has been simmering for a decade. Physicians who deal frequently with malformations of cortical development have known for a long time that the term ‘polymicrogyria’ includes several, perhaps many, different malformations that have superficial similarities. The so-called ‘cobblestone malformations’, in which neurons migrate into the subarachnoid space through gaps in the pial limiting membrane are often considered to be polymicrogyrias. Other conditions with deep undulations of thin cortex, and those with tiny undulations and rather thick cortex are also put in this group, as are many other disorders with distinct patterns (discussed in Barkovich, 2010). ‘Polymicrogyria’ is also described with infection, ischaemia and many different genetic mutations, not all of which are known directly to affect the brain. The same can be said of lissencephalies and heterotopia and focal cortical dysplasias. Such classifications were acceptable and useful in the past, when the molecular mechanisms could not be determined. Knowledge has now advanced to the point where these fascinating disorders should be classified by the effects of specific mutations on molecular structure and their consequent ramifications upon molecular pathways. It is expected that this will create realistic opportunities for the development of effective therapies.
Funding
A.J.B. is supported NIH R37 NS35129 (Walsh).
References
- Barkovich AJ. MRI analysis of sulcation morphology in polymicrogyria. Epilepsia. 2010;51(Suppl 1):17–22. doi: 10.1111/j.1528-1167.2009.02436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkovich AJ, Kuzniecky RI, Jackson GD, Guerrini R, Dobyns WB. Classification system for malformations of cortical development: update 2001. Neurology. 2001;57:2168–78. doi: 10.1212/wnl.57.12.2168. [DOI] [PubMed] [Google Scholar]
- Cederquist GY, Luchniak A, Tischfield MA, Peeva M, Song Y, Menezes MP, et al. An inherited TUBB2B mutation alters a kinesin-binding site and causes polymicrogyria, CFEOM and axon dysinnervation. Hum Mol Genet. 2012;21:5484–99. doi: 10.1093/hmg/dds393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew S, Balasubramanian R, Chan W-M, Kang PB, Andrews C, Webb BD, et al. A novel syndrome caused by the E410K amino acid substitution in the neuronal β-tubulin isotype 3. Brain. 2013;136:522–535. doi: 10.1093/brain/aws345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushion TD, Dobyns WB, Mullins JGL, Stoodley N, Chung S-K, Fry AE, et al. Overlapping cortical malformations and mutations in TUBB2B and TUBA1A. Brain. 2013;136:536–548. doi: 10.1093/brain/aws338. [DOI] [PubMed] [Google Scholar]
- Guerrini R, Mei D, Cordelli DM, Pucatti D, Franzoni E, Parrini E. Symmetric polymicrogyria and pachygyria associated with TUBB2B mutations. Eur J Hum Genet. 2012;20:995–8. doi: 10.1038/ejhg.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglin XH, Poirier K, Saillour Y, Buhler E, Tian G, Bahi-Buisson N, et al. Mutations in the beta-tubulin gene TUBB2B result in asymmetrical polymicrogyria. Nat Genet. 2009;41:746–52. doi: 10.1038/ng.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijpers M, Hoogenraad CC. Centrosomes, microtubules and neuronal development. Mol Cell Neurosci. 2011;48:349–58. doi: 10.1016/j.mcn.2011.05.004. [DOI] [PubMed] [Google Scholar]
- Poirier K, Keays DA, Francis F, Saillour Y, Bahi N, Manouvrier S, et al. Large spectrum of lissencephaly and pachygyria phenotypes resulting from de novo missense mutations in tubulin alpha 1A (TUBA1A) Hum Mutat. 2007;28:1055–64. doi: 10.1002/humu.20572. [DOI] [PubMed] [Google Scholar]
- Poirier K, Saillour Y, Bahi-Buisson N, Jaglin XH, Fallet-Bianco C, Nabbout R, et al. Mutations in the neuronal beta-tubulin subunit TUBB3 result in malformation of cortical development and neuronal migration defects. Hum Mol Genet. 2010;19:4462–73. doi: 10.1093/hmg/ddq377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier K, Saillour Y, Fourniol F, Francis F, Souville I, Valence S, et al. Expanding the spectrum of TUBA1A-related cortical dysgenesis to Polymicrogyria. Eur J Hum Genet. 2012 doi: 10.1038/ejhg.2012.195. doi:10.1038/ejhg.2012.195. [Epub ahead of print 5 Sept 2012] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romaniello R, Tonelli A, Arrigoni F, Baschirotto C, Triulzi F, Bresolin N, et al. A novel mutation in the β-tubulin gene TUBB2B associated with complex malformation of cortical development and deficits in axonal guidance. Dev Med Child Neurol. 2012;54:765–9. doi: 10.1111/j.1469-8749.2012.04316.x. [DOI] [PubMed] [Google Scholar]
- Tischfield MA, Baris HN, Wu C, Rudolph G, Van Maldergem L, He W, et al. Human TUBB3 mutations perturb microtubule dynamics, kinesin interactions, and axon guidance. Cell. 2010;140:74–87. doi: 10.1016/j.cell.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]