Abstract
Heterotopic ossification (HO) is defined as the formation of ectopic bone in soft tissue outside the skeletal tissue. HO is thought to result from aberrant differentiation of osteogenic progenitors within skeletal muscle. However, the precise origin of HO is still unclear. Skeletal muscle contains two kinds of progenitor cells, myogenic progenitors and mesenchymal progenitors. Myogenic and mesenchymal progenitors in human skeletal muscle can be identified as CD56+ and PDGFRα+ cells, respectively. The purpose of this study was to investigate the osteogenic differentiation potential of human skeletal muscle-derived progenitors. Both CD56+ cells and PDGFRα+ cells showed comparable osteogenic differentiation potential in vitro. However, in an in vivo ectopic bone formation model, PDGFRα+ cells formed bone-like tissue and showed successful engraftment, while CD56+ cells did not form bone-like tissue and did not adapt to an osteogenic environment. Immunohistological analysis of human HO sample revealed that many PDGFRα+ cells were localized in proximity to ectopic bone formed in skeletal muscle. MicroRNAs (miRNAs) are known to regulate many biological processes including osteogenic differentiation. We investigated the participation of miRNAs in the osteogenic differentiation of PDGFRα+ cells by using microarray. We identified miRNAs that had not been known to be involved in osteogenesis but showed dramatic changes during osteogenic differentiation of PDGFRα+ cells. Upregulation of miR-146b-5p and -424 and downregulation of miR-7 during osteogenic differentiation of PDGFRα+ cells were confirmed by quantitative real-time RT-PCR. Inhibition of upregulated miRNAs, miR-146b-5p and -424, resulted in the suppression of osteocyte maturation, suggesting that these two miRNAs have the positive role in the osteogenesis of PDGFRα+ cells. Our results suggest that PDGFRα+ cells may be the major source of HO and that the newly identified miRNAs may regulate osteogenic differentiation process of PDGFRα+ cells.
Introduction
Heterotopic ossification (HO) is defined as the formation of mature lamellar bone in soft tissue sites outside the skeletal periosteum. HO has been recognized to occur in many distinct contexts such as neurologic injury, trauma, and genetic abnormalities. However, the most common site is muscle and soft tissues after surgical trauma, especially total hip arthroplasty (THA) [1]. HO is diagnosed in 0.6% to 90% of patients after THA, with an average incidence of 53%. Ten per cent of patients suffer severe HO with pain in the area of the operated joint combined with a decrease in the range of motion, leading to functional impairment; [2], [3], [4], [5], [6]. Several methods for treatment of HO were reported. Nonsteroidal anti-inflammatory drugs (NSAIDs) reduce the incidence of HO when administered early (3 weeks) after spinal cord injury [7], [8], while etidronate can halt the progression of HO once the diagnosis is made if initiated fairly early (3–6 weeks) [9], [10], [11]. HO is thought to result from inappropriate differentiation of osteogenic progenitor cells that is induced by a pathological imbalance of local or systemic factors. However, the precise origin of HO has not been fully elucidated.
Skeletal muscle contains myogenic stem cells called satellite cells. Satellite cells are suggested to have the ability to differentiate into lineages other than the myogenic lineage, but a lineage-tracing study has demonstrated that they are committed to the myogenic lineage and do not spontaneously adopt nonmyogenic fates [12]. Recent studies revealed the presence of mesenchymal progenitor cells distinct from satellite cells in mouse skeletal muscle. We have identified PDGFRα+ mesenchymal progenitors in mouse muscle interstitium and demonstrated that these cells are responsible for fat infiltration and fibrosis of skeletal muscle [13], [14]. Interestingly, PDGFRα+ mesenchymal progenitors showed osteogenic differentiation in response to bone morphogenetic protein (BMP) stimulation [13], while another report showed that these cells did not differentiate into osteogenic cells when stimulated with dexamethasone, β-glycerophosphate, and ascorbic acid [15]. Wosczyna et al. recently demonstrated that Tie2+PDGFRα+Sca-1+ interstitial progenitors contribute to HO using a BMP2-induced in vivo bone formation model [16]. These results suggest the possibility that HO may result from PDGFRα+ progenitors in skeletal muscle.
MicroRNAs (miRNAs) are short, noncoding RNAs that are involved in the regulation of several biological processes including cell differentiation. It was reported that miRNAs control osteogenic differentiation. miR-138 plays a pivotal role in bone formation in vivo by negatively regulating osteogenic differentiation [17]. BMP2 treatment downregulated the expression of miR-133 and miR-135 that inhibit osteogenic differentiation by targeting Runx2 and Smad5 [18]. miR-206 inhibits osteoblast differentiation by targeting connexin 43 [19]. miR-141 and -200a are involved in pre-osteoblast differentiation in part by regulating the expression of DLX5 [20].
In this study, we isolated CD56+ myogenic progenitors and PDGFRα+ mesenchymal progenitors from human skeletal muscle and examined their osteogenic potentials both in vitro and in vivo. We also investigated miRNA expression by microarray during osteogenic differentiation of human skeletal muscle-derived progenitor cells.
Materials and Methods
Ethics statement
Experiments using human samples were approved by the Ethical Review Board for Clinical Studies at Fujita Health University. Muscle samples were obtained from gluteus medius muscles of patients undergoing total hip arthroplasty. Muscle samples for histological analysis were obtained from 25 patients ranging in age from 30 to 85 years with an average age of 59.5 years. Muscle samples for cell culture were obtained from 33 patients ranging in age from 35 to 82 years with an average age of 64.3 years. All patients gave written informed consent.
HO sample was obtained by thirty five-year-old man who had femoral fracture and subarachnoid hemorrhage by traffic accident and was treated by open reduction and internal fixation. X-ray analysis showed that HO was observed around the left hip joint, especially in rectus femoris muscle and vastus medialis muscle.
Cell preparation
Muscles (typically ∼0.1–0.3 g) were placed in PBS. Nerves, blood vessels, tendons, and fat tissues were carefully removed under a dissecting microscope. Trimmed muscles were minced and digested with 0.2% type II collagenase (Worthington) for 30 min at 37°C. Digested muscles were passed through an 18-gauge needle several times and further digested for 10 min at 37°C. Muscle slurries were filtered through a 100-µm cell strainer (BD Biosciences) and then through a 40-µm cell strainer (BD Biosciences). Erythrocytes were eliminated by treating the cells with Tris-buffered 0.8% NH4Cl. Cells were resuspended in growth medium consisting of DMEM supplemented with 20% FBS, 1% penicillin-streptomycin, and 2.5 ng/ml basic fibroblast growth factor (bFGF) (Invitrogen) and maintained at 37°C in 5% CO2 and 3% O2.
FACS
Cells were trypsinized and resuspended in washing buffer consisting of PBS with 2.5% FBS, and stained with PE-conjugated anti-CD56 (1∶20, Miltenyi Biotec), and biotinylated anti-PDGFRα (1∶200, R&D) for 30 min at 4°C. Cells were then stained with streptavidin-PE/Cy5 (1∶200, BD Pharmingen) for 30 min at 4°C in the dark. Stained cells were analyzed using a FACSCalibur or FACSVantage SE flow cytometer (BD Biosciences). Cell sorting was performed on a FACSVantage SE.
Properties of human skeletal muscle-derived cells
To study the properties of human skeletal muscle-derived cells, we examined viability and population doubling time in vitro. To examine the viability, cells were seeded into 96-well plate (1.3×103 cells/well) and incubated at 37°C in 5% CO2 and 21% O2. After 3 days, ProstoBlue reagent (Invitrogen) was added to the medium. The absorbance at 560 nm of each well was measured with the microplate reader (Thermo Fisher). To examine the population doubling time, cells were seeded into 48-well plate (3.0×103 cells/well) and incubated 37°C in 5% CO2 and 21% O2. After 3 days, cells were counted, and passage into 48-well plate (3.0×103 cells/well). After two more passages, the population doubling time was calculated.
Osteogenic differentiation
Cells were seeded at a density of 4.2×103 cells/cm2 and incubated at 37°C in 5% CO2 and 21% O2. At 50–70% confluency, osteogenesis was induced using osteogenic differentiation medium consisting of 10% FBS-αMEM supplemented with dexamethasone, ascorbate-phosphate, and β-glycerophosphate (R&D). Osteogenic differentiation medium was replaced every 3–4 days.
Alkaline phosphatase staining
Cells were inoculated into 8-well chamber slides (3.5×103 cells/well, Nalge Nunc) and cultured with osteogenic differentiation medium. At different time points, cells were washed with PBS and fixed. Alkaline phosphatase was stained using a Sigma kit #85 according to the manufacturer's protocols.
Alizarin red S staining
For detection of calcification during osteogenic differentiation, cells were washed with PBS and fixed 100% ethanol for 8 minutes. The fixed cells were stained with 1% alizarin red S solution.
Quantification of alkaline phosphatase activity
Cells were inoculated into 96-well plates (8.5×103 cells/well) and cultured with osteogenic differentiation medium. At different time points, cells were washed with 0.9% sodium chloride and lysed in extraction solution (1% NP-40 with 0.9% sodium chloride). Alkaline phosphatase activity was assayed by a spectrophotometric method using a TRACP & ALP Assay Kit (Takara). The absorbance at 405 nm of each well was measured with the microplate reader (Thermo Fisher).
In vivo bone formation model
To study the capacity of human skeletal muscle-derived cells to form bone, cells (1×106 cells/block) were loaded onto 5 mm of PLGA-hydroxyapatite composite blocks (GC R&D Dept) and implanted subcutaneously in C.B-17/lcr-scid/scid Jcl mice (5 wks, male). As a control, the PLGA-hydroxyapatite block alone was transplanted. Implants were removed after 8 wks and fixed with 10% formaldehyde, then embedded in paraffin.
Histochemistry and microscopy
Freshly frozen muscle tissues were sectioned (7-µm thick) using a cryostat. Fresh frozen sections were fixed with 4% PFA for 5 min. HO sample was fixed with 10% formaldehyde, then embedded in paraffin. Paraffin-embedded samples were cut into 5-µm-thick sections. For PDGFRα staining in paraffin-embedded sections, antigen was recovered using Antigen Retrival Reagent (R&D). Sections were blocked with protein-block serum-free reagent (DAKO) for 15 min, and incubated with primary antibodies at 4°C overnight, followed by secondary staining. Primary antibodies used were anti-PDGFRα (1∶200, R&D), anti-CD56 (1∶20, Miltenyi Biotec), anti-Laminin (1∶400, Sigma), and anti-lamin A/C (1∶250, Epitomics). Secondary antibodies used were Alexa Fluor 488 or 568-conjugated anti-Goat IgG (1∶1000, Invitrogen), Dylight 549-conjugated anti-mouse IgG (1∶1000, Jackson ImmunoResearch), Alexa Fluor 488 or 647-conjugated anti-Rabbit IgG (1∶1000, Invitrogen). Specimens were counterstained with DAPI (Invitrogen) and mounted with SlowFade Gold antifade reagent (Invitrogen). In some cases, specimens were subjected sequentially to Hematoxylin-Eosin (H-E) staining as previously described [13]. Stained tissues and cells were photographed with a fluorescence microscope BX51 (Olympus) equipped with a DP70 CCD camera (Olympus) or an inverted fluorescence microscope DMI4000B (Leica) equipped with a DFC350FX CCD camera (Leica). Confocal images of sections were taken using the confocal laser scanning microscope system LSM700 (Carl Zeiss).
Microarray analysis
At different time points of osteogenic differentiation, miRNAs were purified using a miRNeasy Mini Kit (Qiagen). Microarray analysis was performed using a 3D-Gene Human miRNA Oligo chip (Toray). Extracted total RNA was labeled with Hy5 using the miRCURY LNA Array miR labeling kit (Exiqon, Vedbaek, Denmark). The hybridization was performed following the supplier's protocols (www.3d-gene.com). Hybridization signals were scanned using a 3D-Gene Scanner (Toray). The raw date of each spot was normalized by substitution with a mean intensity of the background signal determined by all blank spots' signal intensities of 95% confidence intervals. Measurements of both duplicate spots with the signal intensities greater than 2 standard deviations (SD) of the background signal intensity were considered to be valid. A relative expression level of a given miRNA was calculated by comparing the signal intensities of the averaged valid spots with their mean value throughout the microarray experiments. The microarray data were deposited in the Gene Expression Omnibus database with accession number GSE39361.
qRT-PCR Analysis
miRNAs were extracted using miRNeasy Mini Kit (Qiagen). Reverse transcription reaction was performed using miScript Reverse Transcription Kit (Qiagen). Expression levels of miRNAs were quantified using miScript SYBR® Green PCR Kit (Qiagen). Specific primer sequences are listed below: miR-30a: UGUAAACAUCCUCGACUGGAAG, miR-146b-5p: UGAGAACUGAAUUCCAUAGGCU, miR-199b-5p: CCCAGUGUUUAGACUAUCUGUUC, miR-424: CAGCAGCAAUUCAUGUUUUGAA, miR-7: UGGAAGACUAGUGAUUUUGUUGU, miR-145*: GGAUUCCUGGAAAUACUGUUCU. U6 primer (Qiagen) was used as an internal control.
Prediction of targets of miRNAs
Targets of miRNAs were predicted using TargetScan (http://www.targetscan.org/) and top ten targets of each miRNA were selected.
Inhibition of miRNA during osteogenic differentiation
PDGFRα+ cells from three different patients were inoculated into 8-well chamber slides (3.0×103 cells/well) with 10% FBS-αMEM. At 50–70% confluency, the medium was replaced to osteogenic differentiation medium without antibiotic, and the cells were transfected with 50 nM fluorescein-labeled miRCURY LNA Power Inhibitor (EXIQON) using 0.4 µl Lipofectamine (Invitrogen). For dual inhibition of miRNAs, inhibitors were used at 30 nM each. After 6 hours, the medium was changed to osteogenic differentiation medium and cells were cultured as described above. Second transfection was performed 7 days after osteogenic induction.
Statistical analysis
Statistical significance was assessed by Student's t test. For comparisons of more than two groups, one-way analysis of variance (ANOVA) followed by Steel-Dwass's test was used. A probability of less than 1% or 5% (p<0.01 or p<0.05) were considered statistically significant.
Results
Identification of PDGFRα+ and CD56+ cells in human skeletal muscle
We previously identified PDGFRα+ cells in the interstitial spaces of mouse skeletal muscle and showed that these cells represent unique mesenchymal progenitors distinct from satellite cells [13]. Therefore, we analyzed human skeletal muscle using PDGFRα as a marker for mesenchymal progenitors and CD56 as a marker for satellite cells [21], [22], [23], [24]. PDGFRα+ cells were located in the interstitial spaces of muscle tissue, while CD56+ cells were located beneath the basement membrane of myofibers (Figure 1A). These localizations indicate that the two kinds of progenitors represent distinct populations in skeletal muscle. We next isolated these cells from human muscle. Because limited amounts of human tissue were available, we first cultured human muscle-derived cells, and then the cultured cells were subjected to fluorescence-activated cell sorting (FACS). FACS analysis also revealed that PDGFRα+ cells are different from CD56+ cells (Figure 1B). After cells were sorted, myogenic activity was detected only in CD56+ cells, and adipogenic activity was observed exclusively in PDGFRα+ cells (data not shown). These results suggest that PDGFRα+ cells and CD56+ cells are highly enriched for mesenchymal progenitors and satellite cell-derived myogenic cells, respectively. We examined basic properties of both cell populations. When cell viability was assessed, values of OD 560 nm were 0.804±0.228 for PDGFRα+ cells and 0.708±0.144 for CD56+ cells. Population doubling times were 14.00±0.52 hours for PDGFRα+ cells and 13.61±0.61 hours for CD56+ cells. There were no statistically significant differences in these properties.
Figure 1. Identification of PDGFRα+ cells and CD56+ cells in human skeletal muscle.
(A) Human skeletal muscle sections were stained with antibodies against PDGFRα (green), CD56 (red), and laminin (purple), and counterstained with DAPI (blue). Subsequently, specimens were subjected to H-E staining. Arrows indicate PDGFRα+ cells located in interstitial spaces of muscle tissue. Arrowheads indicate CD56+ cells residing beneath the basement membrane of myofibers. Scale bar: 10 µm. (B) Human muscle-derived cells were analyzed for the expression of PDGFRα and CD56 by FACS.
Osteogenic differentiation potential of human skeletal muscle-derived progenitors in vitro
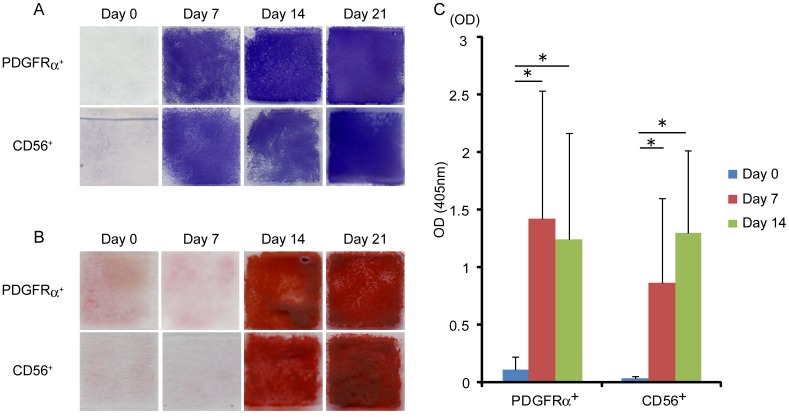
Both PDGFRα+ cells and CD56+ cells were sorted and cultured under osteogenic conditions. To determine the capacity for osteogenic differentiation of human skeletal muscle-derived progenitors in vitro, we performed alkaline phosphatase (ALP) staining and alizarin red S staining. Both PDGFRα+ cells and CD56+ cells became positive for ALP at an early stage of differentiation (Figure 2A), while alizarin red S staining of both cell types did not become positive until 14 days after osteogenic induction (Figure 2B). Next, to compare the osteogenic differentiation potential of PDGFRα+ cells and CD56+ cells, we quantified ALP activity during osteogenic differentiation. After osteogenic induction, both PDGFRα+ cells and CD56+ cells showed much higher ALP activity compared with undifferentiated states. However, both cell types showed similar levels of ALP activity after differentiation, and there was no significant difference between them (Figure 2C).
Figure 2. Osteogenic differentiation potential of human skeletal muscle-derived cells in vitro.
(A) Alkaline phosphatase staining was performed at the time points indicated during osteogenic differentiation of PDGFRα+ cells and CD56+ cells. (B) Alizarin red S staining was performed at the time points indicated during osteogenic differentiation of PDGFRα+ cells and CD56+ cells. (C) Alkaline phosphatase activity of PDGFRα+ cells and CD56+ cells during osteogenic differentiation was quantified. Values are shown as means ± s.d. of ten independent preparations. *P<0.01.
Bone-forming capacity of human skeletal muscle-derived progenitors in vivo
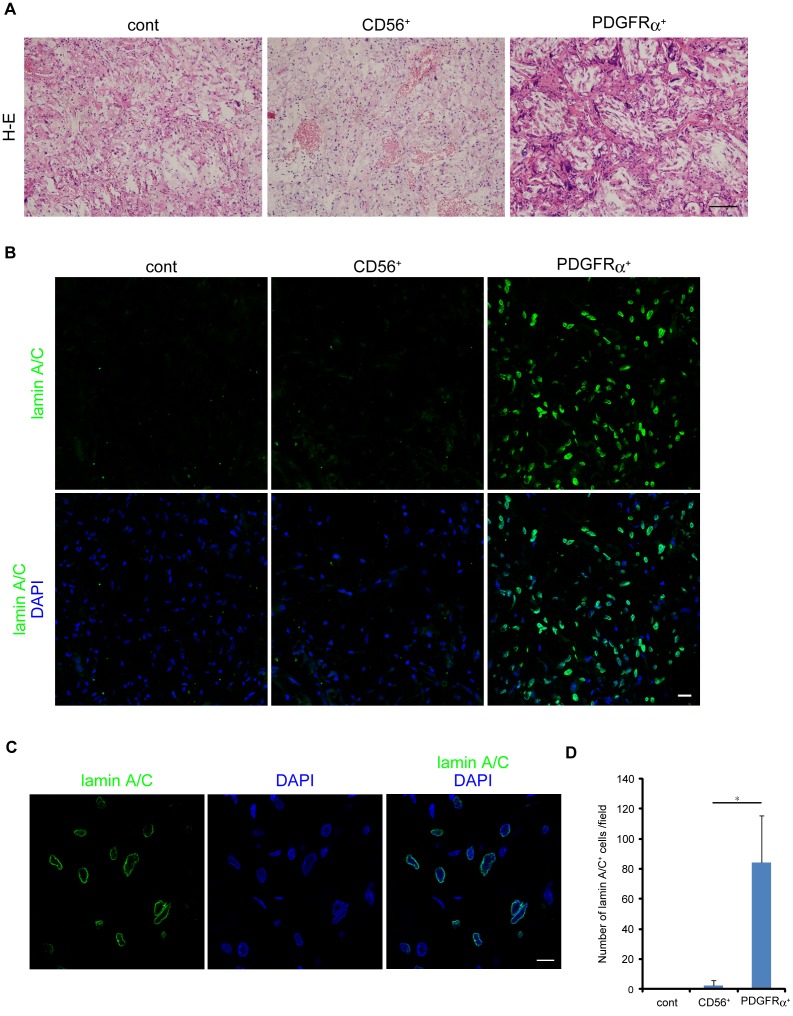
To examine the osteogenic differentiation potential of human skeletal muscle-derived progenitors in vivo, we implanted PLGA-hydroxyapatite scaffolds loaded with 1×106 PDGFRα+ cells or CD56+ cells into mouse backs. This procedure is known to create an osteogenic environment in vivo (unpublished data). After eight weeks, histological analyses were performed. PDGFRα+ cells showed successful engraftment and had formed bone-like tissue. Structures histologically resembling medullary cavities were observed, and multinucleated giant cells were detected at the edges of cavities (Figure 3A). In contrast, CD56+ cells did not create such tissues (Figure 3A). To distinguish transplanted human cells from host mouse cells, we stained them with an antibody that is specific to human lamin A/C. Many human cells were detected in PDGFRα+ cell implants, while only a few human cells were detected in CD56+ cell implants (Figure 3B and D). Confocal laser microscopic analysis with high magnification revealed that nuclear envelope of human cells were clearly stained with this antibody (Figure 3C). These results suggest that PDGFRα+ cells possess far superior ability to adapt to an in vivo osteogenic environment than CD56+ cells. We also examined human skeletal muscle sample with HO in which large ectopic bone was formed within muscle tissue (Figure 4A). Intriguingly, many PDGFRα+ cells were found in proximity to ectopically formed bone (Figure 4A and B). In contrast, we did not observe CD56+ cells around ectopic bone (data not shown). Such aberrant behavior of PDGFRα+ cells were never observed in healthy muscle where only a few PDGFRα+ cells were found to be localized in the interstitial space (Figure 1A).
Figure 3. Bone-forming capacity of human skeletal muscle-derived cells in vivo.
(A) PLGA-hydroxyapatite scaffolds containing PDGFRα+ cells or CD56+ cells were subcutaneously transplanted into immunodeficient mice. Scaffolds containing no cell were used as a control. After 8 wks, implants were removed and subjected to H-E staining. Scale bar: 50 µm. (B) Implants were stained with human lamin A/C specific antibody. Scale bar: 20 µm. (C) High magnification of PDGFRα+-cell transplant. Scale bar: 10 µm. (D) Human lamin-positive nuclei were counted. Number of positive cells is represented as means ± s.d., n = 15 fields from three independent implants. *P<0.01.
Figure 4. Immunohistological analysis of human HO sample.
Human HO sample was subjected to immunofluorescent staining for PDGFRα and subsequently to H-E staining. (A) Image of H-E staining. Scale bar: 100 µm. Right panel shows high magnification image of square region in the left panel. (B) Image of PDGFRα staining. Arrows indicate PDGFRα+ cells surrounding ectopic bone tissue. Scale bar: 10 µm.
Changes of miRNAs during osteogenic differentiation of human skeletal muscle-derived progenitor cells
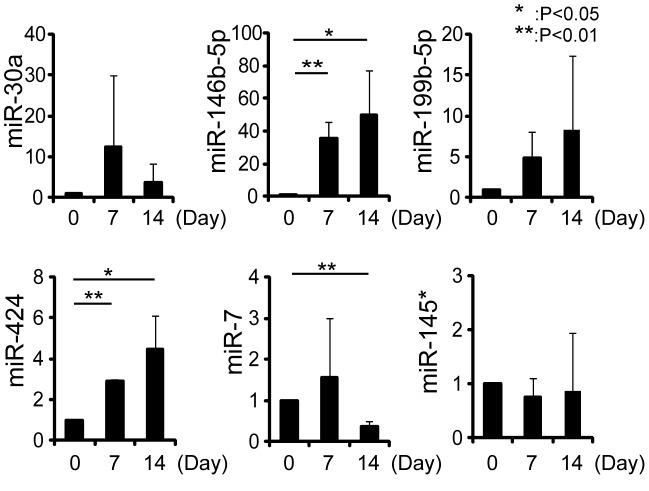
To examine the involvement of miRNAs in osteogenic differentiation of PDGFRα+ cells, we performed microarray analyses. We selected miRNAs that were upregulated more than twofold or downregulated to less than one half at the time points of one and two weeks after osteogenic induction. MiR-19b, -30a, -146b-5p, -199b-5p, and -424 were found to be upregulated, and miR-7, -145*, and -210 were found to be downregulated (Table 1). Interestingly, these miRNAs have not been reported in the context of osteogenesis. We confirmed the changes of these miRNAs during osteogenic differentiation of PDGFRα+ cells of three human different samples using quantitative RT-PCR. Upregulation of miR-146b-5p and -424 and downregulation of miR-7 were confirmed by qRT-PCR (Figure 5). We also examined the changes of these miRNAs during osteogenic differentiation of CD56+ cells from two different patients. miR-146b-5p and -424 tended to be upregulated but fold changes were smaller than that of PDGFRα+ cells, and downregulation of miR-7 were not observed (Table S1). We next predicted potential targets of these miRNAs by TargetScan. Top ten targets of three miRNAs were listed in Table 2.
Table 1. Changes in miRNAs during osteogenic differentiation of PDGFRα+ cells revealed by microarray analysis.
| miRNAs | Day 7 | Day 14 | ||||
| sample1 | sample2 | ave. | sample1 | sample2 | ave. | |
| miR-19b | 2.094 | 2.401 | 2.248 | 4.443 | 5.045 | 4.744 |
| miR-30a | 2.078 | 2.597 | 2.338 | 3.248 | 3.913 | 3.581 |
| miR-146b-5p | 12.255 | 8.533 | 10.394 | 5.692 | 5.161 | 5.427 |
| miR-199b-5p | 3.774 | 4.336 | 4.055 | 2.729 | 3.445 | 3.087 |
| miR-424 | 2.354 | 2.432 | 2.393 | 3.685 | 3.810 | 3.748 |
| miR-7 | 0.299 | 0.347 | 0.323 | 0.101 | 0.167 | 0.134 |
| miR-210 | 0.161 | 0.204 | 0.183 | 0.083 | 0.102 | 0.093 |
| miR-145* | 0.340 | 0.300 | 0.320 | 0.149 | 0.233 | 0.191 |
Values are shown as the relative expression to uninduced cells. Day 7 and Day 14: 7 days and 14 days after osteogenic induction.
Figure 5. Changes in miRNA during osteogenic differentiation of PDGFRα+ cells.
At the different time points during osteogenic differentiation of PDGFRα+ cells, the expressions of miRNAs indicated were quantified by qRT-PCR. Values are represented as the ratio to uninduced cells and shown as means ± s.d. of three independent preparations.
Table 2. Bioinformatics analysis of miRNA targets.
| miRNA | Target gene | Gene name |
| miR-146b-5p | TRAF6 | TNF receptor-associated factor 6 |
| IRAK1 | interleukin-1 receptor-associated kinase 1 | |
| ZBTB2 | zinc finger and BTB domain containing 2 | |
| IGSF1 | immunoglobulin superfamily, member 1 | |
| LCOR | ligand dependent nuclear receptor corepressor | |
| TDRKH | tudor and KH domain containing | |
| WWC2 | WW and C2 domain containing 2 | |
| BCORL1 | BCL6 corepressor-like 1 | |
| ZDHHC7 | zinc finger, DHHC-type containing 7 | |
| NOVA1 | neuro-oncological ventral antigen 1 | |
| miR-424 | FGF2 | fibroblast growth factor 2 (basic) |
| SLC11A2 | solute carrier family 11 (proton-coupled divalent metal ion transporters), member2 | |
| PLAG1 | pleiomorphic adenoma gene 1 | |
| ZBTB34 | zinc finger and BTB domain containing 34 | |
| MGAT4A | Mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A | |
| PLSCR4 | phospholipid scramblase 4 | |
| ANO3 | anoctamin 3 | |
| UNC80 | unc-80 homolog (C. elegans) | |
| TNFSF13B | tumor necrosis factor (ligand) superfamily, member 13b | |
| UBE2Q1 | ubiquitin-conjugating enzyme E2Q family member 1 | |
| miR-7 | UBXN2B | UBX domain protein 2B |
| SPATA2 | spermatogenesis associated 2 | |
| C5orf22 | chromosome 5 open reading frame 22 | |
| ZNF828 | zinc finger protein 828 | |
| POLE4 | polymerase (DNA-directed), epsilon 4 (p12 subunit) | |
| CNO | cappuccino homolog (mouse) | |
| RAF1 | v-raf-1 murine leukemia viral oncogene homolog 1 | |
| IDE | insulin-degrading enzyme | |
| EGFR | epidermal growth factor receptor | |
| RELA | v-rel reticuloendotheliosis oncogene homolog |
Inhibition of miR-146b-5p and -424 lead to suppression of osteogenic maturation of PDGFRα+ cells
To clarify the role of two significantly upregulated miRNAs, miR-146b-5p and -424, in osteogenic differentiation of PDGFRα+ cells, we inhibited these miRNAs using LNA-based miRNA inhibitors. Transfected inhibitors were readily detectable throughout differentiation period (Figure 6A). Inhibition of these miRNAs did not affect early phase of differentiation assessed by ALP staining (Figure 6B). In contrast, alizarin red S staining revealed that inhibition of these miRNAs strongly suppressed bone matrix mineralization of PDGFRα+ cells (Figure 6C). Inhibitory effect of miR-146b-5p inhibitor on matrix mineralization was comparable to that of miR-424 inhibitor, and we did not see synergistic effect when both miRNAs were inhibited (Figure 6C). These results suggest that miR-146b-5p and -424 positively regulate osteogenic maturation of PDGFRα+ cells.
Figure 6. Inhibition of miRNAs during osteogenic differentiation of PDGFRα+ cells.
PDGFRα+ cells were transfected with miRNA inhibitor and osteogenic differentiation was induced. PDGFRα+ cells transfected with control inhibitor were used as a control. Experiments were performed using cells from three independent preparations and we obtained consistent results in each experiment. Data from one representative experiment were shown. (A) Transfected fluorescein-labeled miRNA inhibitor was observed at the time points indicated. Scale bar: 50 µm. (B) Alkaline phosphatase staining was performed at the time points indicated. (C) Alizarin red S staining was performed at the time points indicated.
Discussion
Although HO is thought to derive from osteogenic progenitors, these cells in human skeletal muscle have not been well characterized. Circulating bone marrow-derived osteogenic precursors are present in human peripheral blood [25], and these cells were suggested to participate in ectopic bone formation in mice and humans [26], [27], [28]. In contrast to these studies, it was also reported that there is no evidence of direct contribution of hematopoietic cells to ectopic bone formation [29], [30]. Furthermore, study of a patient with fibrodysplasia ossificans progressiva, a rare genetic disorder of heterotopic skeletogenesis, who had undergone bone marrow transplantation revealed that cells of hematopoietic origin are not sufficient to form ectopic bone [29]. Since the most common cases of HO are in muscle and soft tissues after surgery [1], it is important to understand the osteogenic properties of cells resident in skeletal muscle.
Several groups reported that presence of osteoprogenitors in human skeletal muscle. Levy et al. separated cells from human skeletal muscle into connective tissue and satellite cell fractions and found that human skeletal muscle connective tissue contained osteoprogenitor cells [31]. Mastrogiacomo et al. also isolated osteoprogenitor cells from human skeletal muscle and showed that only a few of them was positive for myogenic markers [32]. Another group isolated mesenchymal progenitor cells from traumatized human skeletal muscle [33], and suggested that these cells are the putative osteoprogenitor cells that initiate ectopic bone formation in HO [34]. In this study, we examined osteogenic properties of PDGFRα+ cells and CD56+ cells derived from human skeletal muscle. PDGFRα+ cells and CD56+ cells showed comparable differentiation potential in vitro with similar levels of ALP activity after osteogenic differentiation. However, PDGFRα+ cells, but not CD56+ cells, showed superior engraftment and bone-like tissue formation in the PLGA-hydroxyapatite scaffold-based an in vivo bone formation model. Many studies of mice showed in vitro osteogenic activity of myogenic cells. Additionally, lineage-tracing experiments have shown that Tie2-expressing cells (presumably endothelial cells) convert to the mesenchymal lineage and contribute to HO [35]. However, recent detailed analyses utilizing Tie2-Cre transgenic mice and a BMP2-induced in vivo HO model demonstrated that myogenic cells (CD31−CD45−PDGFRα−Sca-1− cells) and endothelial cells (Tie2+CD31+ cells) do not contribute to HO, and that CD31−CD45−PDGFRα+Sca-1+ cells (named Tie2+PDGFRα+Sca-1+ progenitors) are the only population capable of generating ectopic bone in vivo [16]. This last study is consistent with our current study using human muscle–derived cells. Several studies have used Sca-1 as a marker to identify the nonmyogenic mesenchymal population in mouse skeletal muscle [15], [36], [37], but Sca-1 is also highly expressed in other cell types, such as endothelial cells. Furthermore, Sca-1 is not applicable to humans because there is no human homologue of Sca-1. We took advantage of the specificity of PDGFRα and identified PDGFRα+ mesenchymal progenitors having a superior in vivo propensity for osteogenesis in the human skeletal muscle interstitium. Taken together, PDGFRα+ mesenchymal progenitors in skeletal muscle can be considered to be one of the best possible candidates for the origin of HO. This notion is further supported by the fact that increased number of PDGFRα+ cells surround the heterotopic bone formed in skeletal muscle in human HO sample.
Interestingly, osteoprogenitor cells isolated from human skeletal muscle [32], [33] and PDGFRα+ mesenchymal progenitors described here possess differentiation potentials toward other mesenchymal lineages such as adipocyte and chondrocytes. Differentiation potentials toward osteogenic, adipogenic and chondrogenic lineages have been generally considered to be hallmarks of so-called mesenchymal stem cells (MSCs). Close relationship between MSCs and pericyte have been reported in several human tissues [38], [39], [40], [41]. Levy et al. also reported similarity between osteoprogenitor cells from human skeletal muscle and pericyte [31]. Crisan et al. reported more direct relationship between MSCs and pericyte. They prospectively isolated perivascular cells, principally pericytes, from multiple human tissues including skeletal muscle as CD146+, CD34−, CD45−, CD56−, ALP+, NG2+, and PDGFRβ+ cells [42]. Purified pericyte have shown to give rise to multipotent cells in culture that fulfill the features of MSCs. They also demonstrated that purified pericytes were readily myogenic in culture and after injection into cardiotoxin-injured or genetically dystrophic immunodeficient mouse skeletal muscle. Dellavalle et al. also reported that pericytes from human skeletal muscle possess high myogenic potential [43]. In contrast, Li et al. showed that non-myogenic cells residing in the fascia of rat or human skeletal muscle contain cells with strong chondrogenic potential [44]. Intriguingly, these chondrogenic progenitor cells lack pericyte marker CD146. Similarly, PDGFRα+ mesenchymal progenitors described here have little myogenic potential and are distinct from pericytes. Lecourt et al. also reported that only CD56+ fraction from human skeletal muscle has myogenic potential and only CD56− fraction show adipogenesis [45]. Further studies are required to completely understand the relationship between MSCs and pericyte, and their myogenic potential.
We identified three miRNAs that show dramatic changes during osteogenic differentiation of PDGFRα+ cells. Although these miRNAs have not been reported in the context of osteogenesis, we showed that matrix mineralization was significantly suppressed by the inhibition of two upregulated miRNAs, miR-146b-5p and -424. Since miR-146b-5p and -424 have distinct sequence in their seed region, they may target different set of mRNAs. However single inhibition of each miRNA and dual inhibition of two miRNAs resulted in comparable suppression of mineralization (Figure 6C). These results suggest that miR-146b-5p and -424 stimulate osteogenic maturation of PDGFRα+ cells by inhibiting different targets lying within the same signaling pathway.
TargetScan predicts TNF receptor-associated factor 6 (TRAF6) and interleukin-1 receptor-associated kinase 1 (IRAK1) as the targets of miR-146b-5p (Table 2). TRAF6 and IRAK1 are key adapter molecules in the NF-κB signaling pathway. NF-κB activation requires the phosphorylation and degradation of inhibitory κB (IκB) proteins, which is induced by IκB kinase α (IKKα) and IKKβ. TRAF6 and IRAK1 mediate IKK activation, thereby lead to NF-κB activation [46]. TRAF6 and IRAK1 have been demonstrated to be the direct targets of miR-146b-5p and therefore miR-146b-5p inhibit NF-κB signaling by targeted repression of TRAF6 and IRAK1 [47], [48], [49]. Interestingly, TargetScan also predicts IRAK2 and IKKβ as the targets of miR-424, although they are not included in top ten targets shown in Table 2. Human IRAK2 has no splice variants, while mouse has four IRAK2 isoforms generated by alternative splicing at the 5′-end of the gene and two of which are inhibitory [50]. Thus IRAK2 in human and mouse may function differently. Importantly, recent study showed that IRAK2 is required for NF-κB activation in human cells [51]. Although NF-κB pathway has been well studied in osteoclasts, recent studies also revealed its importance in osteoblasts as a negative regulator of bone formation [52], [53]. Taken together, miR-146b-5p and -424 may inhibit NF-κB signaling by targeting different signal mediators such as TRAF6, IRAK1, IRAK2 or IKKβ, and exert a stimulatory effect on osteogenesis of PDGFRα+ cells.
In conclusion, we isolated human skeletal muscle-derived progenitor cells and characterized their osteogenic properties. Our results suggest that PDGFRα+ mesenchymal progenitors residing human skeletal muscle are the major origin of HO. In addition, the miRNAs analysis presented here may provide useful information to understand the mechanism whereby miRNAs regulate osteogenic differentiation of PDGFRα+ mesenchymal progenitors.
Supporting Information
(DOC)
Acknowledgments
The authors thank M. Kokubo and M. Nakatani for technical assistance, A. Matsuura for helpful suggestions on histology, and K. Hitachi for advice on miRNA experiments. We also thank K. Ono for proofreading of manuscript.
Funding Statement
This work was supported by a Grant-in-Aid for Scientific Research on Innovative Areas, MEXT (23126527); Intramural Research Grant (23-5) for Neurological and Psychiatric Disorders of NCNP; and MEXT-Supported Program for the Strategic Research Foundation at Private Universities (S1001034). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Nilsson OS, Persson PE (1999) Heterotopic bone formation after joint replacement. Curr Opin Rheumatol 11: 127–131. [DOI] [PubMed] [Google Scholar]
- 2. Brooker AF, Bowerman JW, Robinson RA, Riley LH Jr (1973) Ectopic ossification following total hip replacement. Incidence and a method of classification. J Bone Joint Surg Am 55: 1629–1632. [PubMed] [Google Scholar]
- 3. Ritter MA, Vaughan RB (1977) Ectopic ossification after total hip arthroplasty. Predisposing factors, frequency, and effect on results. J Bone Joint Surg Am 59: 345–351. [PubMed] [Google Scholar]
- 4. Thomas BJ (1992) Heterotopic bone formation after total hip arthroplasty. Orthop Clin North Am 23: 347–358. [PubMed] [Google Scholar]
- 5. Ahrengart L (1991) Periarticular heterotopic ossification after total hip arthroplasty. Risk factors and consequences. Clin Orthop Relat Res 49–58. [PubMed] [Google Scholar]
- 6. Naraghi FF, DeCoster TA, Moneim MS, Miller RA, Rivero D (1996) Heterotopic ossification. Orthopedics 19: 145–151. [DOI] [PubMed] [Google Scholar]
- 7. Banovac K, Sherman AL, Estores IM, Banovac F (2004) Prevention and treatment of heterotopic ossification after spinal cord injury. J Spinal Cord Med 27: 376–382. [DOI] [PubMed] [Google Scholar]
- 8. Banovac K, Williams JM, Patrick LD, Haniff YM (2001) Prevention of heterotopic ossification after spinal cord injury with indomethacin. Spinal Cord 39: 370–374. [DOI] [PubMed] [Google Scholar]
- 9. Banovac K (2000) The effect of etidronate on late development of heterotopic ossification after spinal cord injury. J Spinal Cord Med 23: 40–44. [DOI] [PubMed] [Google Scholar]
- 10. Banovac K, Gonzalez F, Renfree KJ (1997) Treatment of heterotopic ossification after spinal cord injury. J Spinal Cord Med 20: 60–65. [DOI] [PubMed] [Google Scholar]
- 11. Banovac K, Gonzalez F, Wade N, Bowker JJ (1993) Intravenous disodium etidronate therapy in spinal cord injury patients with heterotopic ossification. Paraplegia 31: 660–666. [DOI] [PubMed] [Google Scholar]
- 12. Starkey JD, Yamamoto M, Yamamoto S, Goldhamer DJ (2011) Skeletal muscle satellite cells are committed to myogenesis and do not spontaneously adopt nonmyogenic fates. J Histochem Cytochem 59: 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Uezumi A, Fukada S, Yamamoto N, Takeda S, Tsuchida K (2010) Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nat Cell Biol 12: 143–152. [DOI] [PubMed] [Google Scholar]
- 14. Uezumi A, Ito T, Morikawa D, Shimizu N, Yoneda T, et al. (2011) Fibrosis and adipogenesis originate from a common mesenchymal progenitor in skeletal muscle. J Cell Sci 124: 3654–3664. [DOI] [PubMed] [Google Scholar]
- 15. Favier B, Lemaoult J, Lesport E, Carosella ED (2010) ILT2/HLA-G interaction impairs NK-cell functions through the inhibition of the late but not the early events of the NK-cell activating synapse. FASEB J 24: 689–699. [DOI] [PubMed] [Google Scholar]
- 16. Wosczyna MN, Biswas AA, Cogswell CA, Goldhamer DJ (2012) Multipotent progenitors resident in the skeletal muscle interstitium exhibit robust BMP-dependent osteogenic activity and mediate heterotopic ossification. J Bone Miner Res 27: 1004–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eskildsen T, Taipaleenmaki H, Stenvang J, Abdallah BM, Ditzel N, et al. (2011) MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proc Natl Acad Sci U S A 108: 6139–6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li Z, Hassan MQ, Volinia S, van Wijnen AJ, Stein JL, et al. (2008) A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc Natl Acad Sci U S A 105: 13906–13911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Inose H, Ochi H, Kimura A, Fujita K, Xu R, et al. (2009) A microRNA regulatory mechanism of osteoblast differentiation. Proc Natl Acad Sci U S A 106: 20794–20799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Itoh T, Nozawa Y, Akao Y (2009) MicroRNA-141 and -200a are involved in bone morphogenetic protein-2-induced mouse pre-osteoblast differentiation by targeting distal-less homeobox 5. J Biol Chem 284: 19272–19279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schubert W, Zimmermann K, Cramer M, Starzinski-Powitz A (1989) Lymphocyte antigen Leu-19 as a molecular marker of regeneration in human skeletal muscle. Proc Natl Acad Sci U S A 86: 307–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Illa I, Leon-Monzon M, Dalakas MC (1992) Regenerating and denervated human muscle fibers and satellite cells express neural cell adhesion molecule recognized by monoclonal antibodies to natural killer cells. Ann Neurol 31: 46–52. [DOI] [PubMed] [Google Scholar]
- 23. Fidzianska A, Kaminska A (1995) Neural cell adhesion molecule (N-CAM) as a marker of muscle tissue alternations. Review of the literature and own observations. Folia Neuropathol 33: 125–128. [PubMed] [Google Scholar]
- 24. Lindstrom M, Thornell LE (2009) New multiple labelling method for improved satellite cell identification in human muscle: application to a cohort of power-lifters and sedentary men. Histochem Cell Biol 132: 141–157. [DOI] [PubMed] [Google Scholar]
- 25. Eghbali-Fatourechi GZ, Lamsam J, Fraser D, Nagel D, Riggs BL, et al. (2005) Circulating osteoblast-lineage cells in humans. N Engl J Med 352: 1959–1966. [DOI] [PubMed] [Google Scholar]
- 26. Otsuru S, Tamai K, Yamazaki T, Yoshikawa H, Kaneda Y (2007) Bone marrow-derived osteoblast progenitor cells in circulating blood contribute to ectopic bone formation in mice. Biochem Biophys Res Commun 354: 453–458. [DOI] [PubMed] [Google Scholar]
- 27. Otsuru S, Tamai K, Yamazaki T, Yoshikawa H, Kaneda Y (2008) Circulating bone marrow-derived osteoblast progenitor cells are recruited to the bone-forming site by the CXCR4/stromal cell-derived factor-1 pathway. Stem Cells 26: 223–234. [DOI] [PubMed] [Google Scholar]
- 28. Suda RK, Billings PC, Egan KP, Kim JH, McCarrick-Walmsley R, et al. (2009) Circulating osteogenic precursor cells in heterotopic bone formation. Stem Cells 27: 2209–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kaplan FS, Glaser DL, Shore EM, Pignolo RJ, Xu M, et al. (2007) Hematopoietic stem-cell contribution to ectopic skeletogenesis. J Bone Joint Surg Am 89: 347–357. [DOI] [PubMed] [Google Scholar]
- 30. Kan L, Liu Y, McGuire TL, Berger DM, Awatramani RB, et al. (2009) Dysregulation of local stem/progenitor cells as a common cellular mechanism for heterotopic ossification. Stem Cells 27: 150–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Levy MM, Joyner CJ, Virdi AS, Reed A, Triffitt JT, et al. (2001) Osteoprogenitor cells of mature human skeletal muscle tissue: an in vitro study. Bone 29: 317–322. [DOI] [PubMed] [Google Scholar]
- 32. Mastrogiacomo M, Derubeis AR, Cancedda R (2005) Bone and cartilage formation by skeletal muscle derived cells. J Cell Physiol 204: 594–603. [DOI] [PubMed] [Google Scholar]
- 33. Nesti LJ, Jackson WM, Shanti RM, Koehler SM, Aragon AB, et al. (2008) Differentiation potential of multipotent progenitor cells derived from war-traumatized muscle tissue. J Bone Joint Surg Am 90: 2390–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jackson WM, Aragon AB, Bulken-Hoover JD, Nesti LJ, Tuan RS (2009) Putative heterotopic ossification progenitor cells derived from traumatized muscle. J Orthop Res 27: 1645–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Akao Y, Nakagawa Y, Hirata I, Iio A, Itoh T, et al. (2010) Role of anti-oncomirs miR-143 and -145 in human colorectal tumors. Cancer Gene Ther 17: 398–408. [DOI] [PubMed] [Google Scholar]
- 36. Leblanc E, Trensz F, Haroun S, Drouin G, Bergeron E, et al. (2011) BMP-9-induced muscle heterotopic ossification requires changes to the skeletal muscle microenvironment. J Bone Miner Res 26: 1166–1177. [DOI] [PubMed] [Google Scholar]
- 37. Schulz TJ, Huang TL, Tran TT, Zhang H, Townsend KL, et al. (2011) Identification of inducible brown adipocyte progenitors residing in skeletal muscle and white fat. Proc Natl Acad Sci U S A 108: 143–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shi S, Gronthos S (2003) Perivascular niche of postnatal mesenchymal stem cells in human bone marrow and dental pulp. J Bone Miner Res 18: 696–704. [DOI] [PubMed] [Google Scholar]
- 39. Schwab KE, Gargett CE (2007) Co-expression of two perivascular cell markers isolates mesenchymal stem-like cells from human endometrium. Hum Reprod 22: 2903–2911. [DOI] [PubMed] [Google Scholar]
- 40. Zannettino AC, Paton S, Arthur A, Khor F, Itescu S, et al. (2008) Multipotential human adipose-derived stromal stem cells exhibit a perivascular phenotype in vitro and in vivo. J Cell Physiol 214: 413–421. [DOI] [PubMed] [Google Scholar]
- 41. Covas DT, Panepucci RA, Fontes AM, Silva WA Jr, Orellana MD, et al. (2008) Multipotent mesenchymal stromal cells obtained from diverse human tissues share functional properties and gene-expression profile with CD146+ perivascular cells and fibroblasts. Exp Hematol 36: 642–654. [DOI] [PubMed] [Google Scholar]
- 42. Crisan M, Yap S, Casteilla L, Chen CW, Corselli M, et al. (2008) A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell 3: 301–313. [DOI] [PubMed] [Google Scholar]
- 43. Dellavalle A, Sampaolesi M, Tonlorenzi R, Tagliafico E, Sacchetti B, et al. (2007) Pericytes of human skeletal muscle are myogenic precursors distinct from satellite cells. Nat Cell Biol 9: 255–267. [DOI] [PubMed] [Google Scholar]
- 44. Li G, Zheng B, Meszaros LB, Vella JB, Usas A, et al. (2011) Identification and characterization of chondrogenic progenitor cells in the fascia of postnatal skeletal muscle. J Mol Cell Biol 3: 369–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lecourt S, Marolleau JP, Fromigue O, Vauchez K, Andriamanalijaona R, et al. (2010) Characterization of distinct mesenchymal-like cell populations from human skeletal muscle in situ and in vitro. Exp Cell Res 316: 2513–2526. [DOI] [PubMed] [Google Scholar]
- 46. Vallabhapurapu S, Karin M (2009) Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol 27: 693–733. [DOI] [PubMed] [Google Scholar]
- 47. Taganov KD, Boldin MP, Chang KJ, Baltimore D (2006) NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A 103: 12481–12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. O'Neill LA, Sheedy FJ, McCoy CE (2011) MicroRNAs: the fine-tuners of Toll-like receptor signalling. Nat Rev Immunol 11: 163–175. [DOI] [PubMed] [Google Scholar]
- 49. Hulsmans M, Van Dooren E, Mathieu C, Holvoet P (2012) Decrease of miR-146b-5p in monocytes during obesity is associated with loss of the anti-inflammatory but not insulin signaling action of adiponectin. PLoS One 7: e32794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hardy MP, O'Neill LA (2004) The murine IRAK2 gene encodes four alternatively spliced isoforms, two of which are inhibitory. J Biol Chem 279: 27699–27708. [DOI] [PubMed] [Google Scholar]
- 51. Flannery SM, Keating SE, Szymak J, Bowie AG (2011) Human interleukin-1 receptor-associated kinase-2 is essential for Toll-like receptor-mediated transcriptional and post-transcriptional regulation of tumor necrosis factor alpha. J Biol Chem 286: 23688–23697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chang J, Wang Z, Tang E, Fan Z, McCauley L, et al. (2009) Inhibition of osteoblastic bone formation by nuclear factor-kappaB. Nat Med 15: 682–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Krum SA, Chang J, Miranda-Carboni G, Wang CY (2010) Novel functions for NFkappaB: inhibition of bone formation. Nat Rev Rheumatol 6: 607–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)