Abstract
Recent studies have suggested that plant genomes have undergone potentially rampant horizontal gene transfer (HGT), especially in the mitochondrial genome. Parasitic plants have provided the strongest evidence of HGT, which appears to be facilitated by the intimate physical association between the parasites and their hosts. A recent phylogenomic study demonstrated that in the holoparasite Rafflesia cantleyi (Rafflesiaceae), whose close relatives possess the world's largest flowers, about 2.1% of nuclear gene transcripts were likely acquired from its obligate host. Here, we used next-generation sequencing to obtain the 38 protein-coding and ribosomal RNA genes common to the mitochondrial genomes of angiosperms from R. cantleyi and five additional species, including two of its closest relatives and two host species. Strikingly, our phylogenetic analyses conservatively indicate that 24%–41% of these gene sequences show evidence of HGT in Rafflesiaceae, depending on the species. Most of these transgenic sequences possess intact reading frames and are actively transcribed, indicating that they are potentially functional. Additionally, some of these transgenes maintain synteny with their donor and recipient lineages, suggesting that native genes have likely been displaced via homologous recombination. Our study is the first to comprehensively assess the magnitude of HGT in plants involving a genome (i.e., mitochondria) and a species interaction (i.e., parasitism) where it has been hypothesized to be potentially rampant. Our results establish for the first time that, although the magnitude of HGT involving nuclear genes is appreciable in these parasitic plants, HGT involving mitochondrial genes is substantially higher. This may represent a more general pattern for other parasitic plant clades and perhaps more broadly for angiosperms.
Author Summary
Recent studies have suggested that plant genomes have undergone potentially rampant horizontal gene transfer (HGT), especially in the mitochondrial genome. Here, using phylogenomic approaches, we demonstrate that as much as ∼40% of the mitochondrial genes in the parasitic plant species Rafflesiaceae are acquired from their hosts via HGT. These transgenes are likely functional in their recipient species and in some cases appear to have displaced native copies in the same genomic location. These results establish for the first time that, although the magnitude of HGT involving nuclear genes is appreciable in parasitic plants, HGT involving mitochondrial genes is substantially higher.
Introduction
Recent studies have suggested that plant genomes have undergone potentially rampant horizontal gene transfer (HGT) [1], [2], especially in the mitochondrial genome [3]–[7]. Parasitic plants have provided the strongest evidence of HGT [8]–[12], which appears to be facilitated by the intimate physical association between the parasites and their hosts [8], [10], [12]–[15]. One parasitic plant clade that appears to be prone to HGT is Rafflesiaceae sensu stricto, which belong to the order Malpighiales [8], [16]–[18] and whose members possess the largest flowers in the world. Rafflesiaceae are endophytic holoparasites, which lack leaves and stems. This family includes the genera Rafflesia (∼28 species), Rhizanthes (four species), and Sapria (three species), and provides one of the best opportunities to investigate HGT in plants because (i) the parasites have a very narrow host specialization range on members of the grapevine family (Tetrastigma spp., Vitaceae), (ii) complete genome sequences, including fully annotated mitochondrial and plastid genomes, are available for close relatives of the parasites (Ricinus communis, Euphorbiaceae) [19], [20] and their hosts (Vitis vinifera, Vitaceae) [21]–[23], and (iii) the hosts and parasites are separated by at least 115 million years of evolution (Figure 1A) [24]–[27]. These factors make it easier to distinguish transgenes from native genes in Rafflesiaceae using phylogenomic tools.
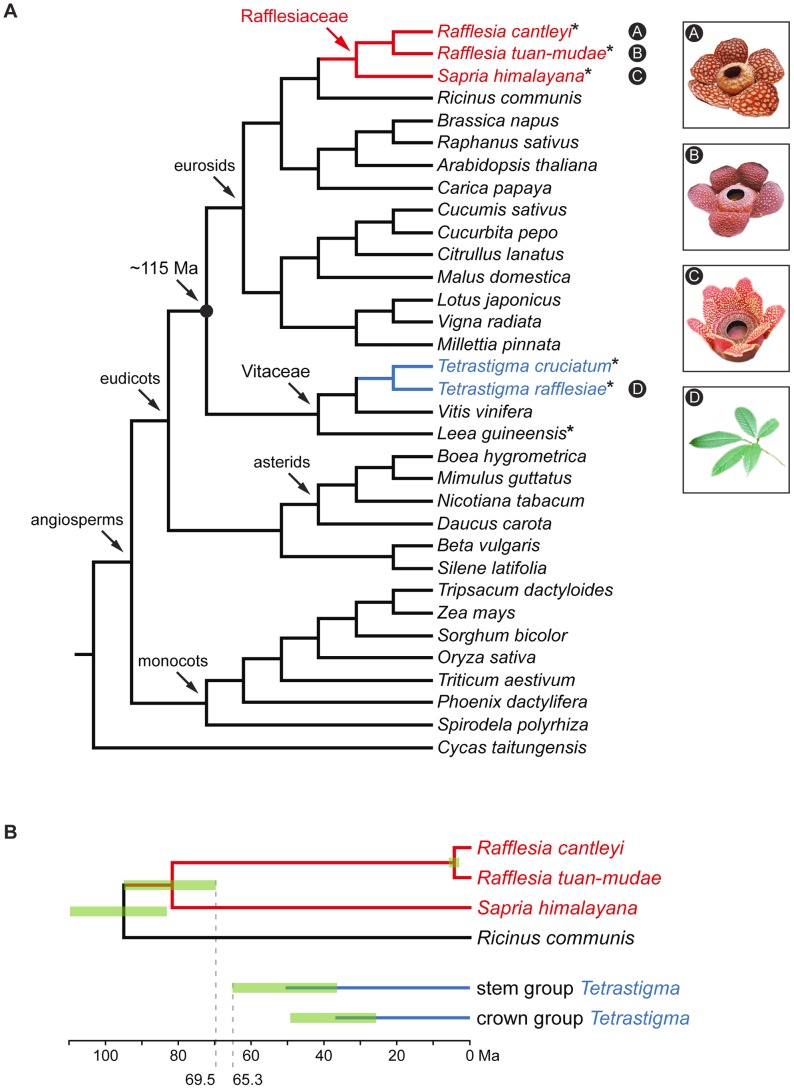
Figure 1. Phylogenetic relationships and divergence times of the three Rafflesiaceae species and two Tetrastigma species included in this study.
Phylogenetic relationships (A) and divergence times (B). Holoparasitic Rafflesiaceae (red) is a member of the order Malpighiales, and its obligate host Tetrastigma (blue) is a member of the Vitaceae family. The approximate divergence time between the parasite and host clade is 115 Ma [24]–[27]. Mitochondrial genome sequences generated in this study are marked with asterisks, and the node age error bars (95% highest posterior density intervals) are shown in green. The accepted phylogenetic relationships are based on APG III [38] and Qiu et al. [40], and the divergence times of Rafflesiaceae and Tetrastigma are based on Bendiksby et al. [48] and Chen et al. [49], respectively.
A recent phylogenomic study demonstrated that in Rafflesia cantleyi, about 2.1% of nuclear gene transcripts were likely acquired from its obligate host [28]. This study, however, did not include a thorough investigation of the mitochondrial genome. Here, we comprehensively sequenced 38 mitochondrial genes from R. cantleyi and five additional species, including two of its closest relatives and two host species. Our results reveal an extraordinarily high degree of HGT in the mitochondrial genome of Rafflesiaceae involving genes that were likely acquired from its host at various time intervals. Most of these transgenic sequences possess intact reading frames and are actively transcribed indicating that they are potentially functional. Additionally, some of these transgenes maintain synteny with their donor and recipient lineages suggesting that native genes in Rafflesiaceae have likely been displaced via homologous recombination. These results establish for the first time that although the magnitude of HGT involving nuclear genes is appreciable in these parasitic plants, HGT involving mitochondrial genes is substantially higher.
Results/Discussion
The mitochondrial provenance of our sequenced genes
We used next-generation sequencing to comprehensively sequence the mitochondrial genomes of three species that span the crown node of Rafflesiaceae: Rafflesia cantleyi, Rafflesia tuan-mudae, and Sapria himalayana (Figure 1A, see also Table S1). We then extracted the 38 mitochondrial genes from our de novo assembled contigs that ranged in size from 2 to 54 kilobases (kb). These 38 protein-coding and ribosomal RNA genes are present in the mitochondrial genomes of both Ricinus and Vitis, and are also common to most angiosperms [29]. We included 35, 33, and 59 gene sequences from R. cantleyi, R. tuan-mudae and S. himalayana, respectively, for further analyses. While repetitive sequences made assembly of the entire chromosome impractical, high sequence coverage (Table S1) ensured that we have sequenced all coding regions in these mitochondrial genomes.
Several lines of evidence suggest that all gene sequences we assembled here are localized to the mitochondrial genome of Rafflesiaceae. First, the genome libraries for two of our three Rafflesiaceae species (i.e., R. cantleyi and S. himalayana) were prepared from fresh tissue using sucrose gradient centrifugation, which are enriched for plant organelles [30]. Since the plastid genome has apparently been lost in Rafflesiaceae [31], our libraries are heavily enriched for mitochondria. Second, plastid, mitochondrial and nuclear genes in plant cells differ widely in copy number: plastid genes are generally present in hundreds to thousands of copies per cell, mitochondrial genes in tens to hundreds of copies per cell, while nuclear genes are usually present in only two copies per cell [7], [32]. To investigate if gene sequences assembled here have copy numbers that correspond with a mitochondrial localization, we compared gene copy number here to 1,305 genes previously determined to be localized to the nuclear genome of R. cantleyi [28] and R. tuan-mudae [18]. Our results demonstrate that copy numbers for all putative mitochondrial gene sequences in Rafflesiaceae are one to two orders of magnitude greater than for nuclear genes (Table S2), with means of 155-, 68-, and 160-fold greater for R. cantleyi, R. tuan-mudae, and S. himalayana, respectively (p-value<2.2×10−16, Welch's t test). These copy numbers are consistent with a mitochondrial localization, but not high enough to suggest localization in, or the existence of, a plastid genome in Rafflesiaceae. Third, when comparing assembled gene sequences from R. cantleyi with our previously published complementary DNA (cDNA) library [28], we identified cytosine-to-uracil (C-to-U) RNA editing in seven genes (i.e., atp1, atp4, atp6, cox2, nad1, rps4, and rps12), which is a common characteristic of mitochondrial genes [33]. These results collectively indicate that these gene sequences are most likely localized to the mitochondrial genome of Rafflesiaceae, although complete assembly of these mitochondrial genomes will be required to definitively confirm our results.
Extraordinarily high, and variable, rates of HGT in the mitochondrial genome of Rafflesiaceae
To estimate the magnitude of HGT in the mitochondrial genome of Rafflesiaceae, we also sequenced the same 38 mitochondrial genes from three species of Vitaceae: Tetrastigma cruciatum, which is the host of S. himalayana [34], Tetrastigma rafflesiae, which is the host of R. cantleyi and R. tuan-mudae [34], [35], and Leea guineensis (Figure 1A, see also Table S1). The latter represents the earliest diverging lineage of Vitaceae [36], [37], which allows us to determine if putative transgenic sequences from Rafflesiaceae are phylogenetically nested within the host clade Vitaceae.
Our newly sequenced mitochondrial gene sequences from the six species were then analyzed using maximum likelihood (ML) with homologous sequences from 27 other seed plants whose mitochondrial genomes have been sequenced and fully annotated (Figure 1A, see also Table S3; Arabidopsis thaliana [Brassicaceae], Beta vulgaris [Amaranthaceae], Boea hygrometrica [Gesneriaceae], Brassica napus [Brassicaceae], Carica papaya [Caricaceae], Citrullus lanatus [Cucurbitaceae], Cucumis sativus [Cucurbitaceae], Cucurbita pepo [Cucurbitaceae], Cycas taitungensis [Cycadaceae], Daucus carota [Apiaceae], Lotus japonicus [Fabaceae], Malus domestica [Rosaceae], Millettia pinnata [Fabaceae], Mimulus guttatus [Phrymaceae], Nicotiana tabacum [Solanaceae], Oryza sativa [Poaceae], Phoenix dactylifera [Arecaceae], Raphanus sativus [Brassicaceae], Ricinus communis, Silene latifolia [Caryophyllaceae], Sorghum bicolor [Poaceae], Spirodela polyrrhiza [Araceae], Tripsacum dactyloides [Poaceae], Triticum aestivum [Poaceae], Vigna radiata [Fabaceae], Vitis vinifera, and Zea mays [Poaceae]). These reference species represent a broad sampling of most major flowering plant clades [38]. Each Rafflesiaceae gene sequence was placed into one of three categories–i.e., VGT, HGT, or unassigned–on the basis of its phylogenetic position and ML bootstrap percentage (BP) support following Xi et al. [28]. We applied two BP thresholds to categorize each gene sequence. Our more conservative estimate applied a 70 BP threshold; this BP threshold has been shown to correspond to a very high probability that the clade is real [39]. Here, gene sequences whose placements were consistent with accepted species' relationships (i.e., Rafflesiaceae gene sequences were sister to their closest relative Ricinus with ≥70 BP; [17], [18]) were scored as VGT; HGT was inferred when gene sequences were placed elsewhere with ≥70 BP; and gene sequences with <70 BP were left unassigned. To explore if our estimates of HGT were sensitive to our thresholds, we also categorized these gene sequences by applying a less conservative threshold using ≥50 BP.
Our phylogenetic analyses of the 38 mitochondrial genes indicated that, for the 30 autotrophic species included here (i.e., 27 reference species, two Tetrastigma species, and Leea; Figure 1A), phylogenetic placements largely agreed with accepted relationships between families [38], [40] using both the 70 and 50 BP thresholds (Figures S1A and S2). The only three exceptions were for atp1 where Brassicaceae (i.e., Arabidopsis+Brassica+Raphanus) was placed sister to the asterids (i.e., Boea+Mimulus+Nicotiana) with 81 BP (a similar topology was also identified by Nickrent et al. [10]); for atp4 where Brassicaceae was placed sister to Fabaceae (i.e., Lotus+Millettia+Vigna) with 93 BP; and for cox1 where Brassicaceae was placed sister to Caryophyllales (i.e., Beta+Silene) with 87 BP (Figure S1A). These results indicate that applying both the 70 and 50 BP thresholds yields very low false positive estimates of HGT in these autotrophic species, for which we expect little or no HGT to occur.
In contrast, in the three holoparasitic Rafflesiaceae species, 11 mitochondrial genes demonstrated evidence for one or more cases of HGT using our more conservative 70 BP threshold: of the 21 gene sequences with ≥70 BP in R. cantleyi, five gene sequences (24%) showed evidence of HGT, 5 of 19 (26%) in R. tuan-mudae, and 11 of 27 (41%) in S. himalayana. Furthermore, vertical placements of these putative transgenic sequences were rejected in 18 of 21 cases using the approximately unbiased (AU) test (Table 1). For the less conservative 50 BP threshold, the number of mitochondrial genes that showed evidence of HGT increased to 16; however, the relative frequencies of HGT are nearly identical with those above: 29% in R. cantleyi (7 of 24), 32% in R. tuan-mudae (7 of 22), and 47% in S. himalayana (16 of 34). This indicates that our less conservative threshold does not increase false positive rates. Thus, given the consistency of our estimates of HGT using both thresholds, we treat these transgenes collectively in the discussion below unless otherwise indicated. Two additional findings support the reliability of our HGT inferences: first, the phylogenetic placements of these transgenic sequences were not obviously biased by C-to-U RNA editing (see Figure S1B for phylograms with RNA editing sites excluded from our alignments); second, seven of our large assembled contigs contained both transgenes and native genes (Figure 2), indicating that these transgenes were clearly integrated into the mitochondrial genome of Rafflesiaceae. Therefore, rates of mitochondrial HGT in Rafflesiaceae appear to be extraordinarily high, and well above the false positive rates established from the 30 autotrophic species included here.
Table 1. Horizontally transferred (HGT) sequences identified in the mitochondrial genomes of Rafflesia cantleyi, Rafflesia tuan-mudae, and Sapria himalayana with associated statistics.
| Gene | Species | Gene identity | HGT donor lineage | BP | AU test | Gene length (base pairs) | Reading frame | RPKM | C-to-U RNA editing site |
| Mitochondrial genes | |||||||||
| atp1 | R. cantleyi | HGT | Vitaceae | 54 | NS | 1520 | I | 2370 | 1039 |
| R. tuan-mudae | HGT | Vitaceae | 54 | NS | 1520 | I | |||
| S. himalayana | HGT | Vitaceae | 54 | NS | 1514 | I | |||
| atp4 | R. cantleyi | HGT | Cucurbitaceae | 83 | NS | 526 | I | 6560 | 168, 337 |
| R. tuan-mudae | HGT | Cucurbitaceae | 83 | NS | 526 | I | |||
| S. himalayana | HGT | Cucurbitaceae | 83 | NS | 520 | I | |||
| atp9 | R. cantleyi | VGT | – | 94 | – | 204 | I | 1225 | |
| R. tuan-mudae | VGT | – | 94 | – | 204 | I | |||
| S. himalayana | VGT | – | 94 | – | 204 | I | |||
| S. himalayana | HGT | Vitaceae | 65 | NS | 195 | I | |||
| cob | R. cantleyi | VGT | – | 96 | – | 1166 | I | 941 | |
| R. tuan-mudae | VGT | – | 96 | – | 1166 | I | |||
| S. himalayana | VGT | – | 96 | – | 1166 | I | |||
| S. himalayana | HGT | Vitaceae | 99 | 0.0004 | 1179 | I | |||
| cox1 | R. cantleyi | HGT | Brassicaceae+Caryophyllales | 71 | 0.0360 | 1542 | I | 449 | |
| R. tuan-mudae | HGT | Brassicaceae+Caryophyllales | 71 | 0.0360 | 1542 | I | |||
| S. himalayana | HGT | Brassicaceae+Caryophyllales | 71 | 0.0360 | 1542 | I | |||
| S. himalayana | HGT | Tetrastigma | 100 | <0.0001 | 1461 | I | |||
| cox2 | R. cantleyi | – | – | <50 | – | 770 | I | 2183 | 14, 61, 153, 233 |
| R. tuan-mudae | – | – | <50 | – | 770 | I | |||
| S. himalayana | – | – | <50 | – | 771 | I | |||
| S. himalayana | HGT | Tetrastigma | 96 | <0.0001 | 246 | I | |||
| cox3 | R. cantleyi | VGT | – | 78 | – | 798 | I | 1810 | |
| S. himalayana | VGT | – | 78 | – | 798 | I | |||
| S. himalayana | HGT | Tetrastigma | 85 | 0.0063 | 337 | I | |||
| rpl2 | R. cantleyi | HGT | Vitaceae | 51 | NS | 408 | I | 44 | |
| R. tuan-mudae | HGT | Vitaceae | 51 | NS | 408 | I | |||
| S. himalayana | HGT | Tetrastigma | 69 | NS | 963 | ψ | |||
| rpl5 | R. cantleyi | VGT | – | 89 | – | 375 | I | 1619 | |
| R. tuan-mudae | VGT | – | 89 | – | 375 | I | |||
| S. himalayana | VGT | – | 89 | – | 528 | I | |||
| S. himalayana | HGT | Tetrastigma | 100 | 0. 0030 | 543 | I | |||
| rps1 | R. cantleyi | HGT | Tetrastigma | 100 | <0.0001 | 342 | I | 155 | |
| R. tuan-mudae | HGT | Tetrastigma | 100 | <0.0001 | 342 | I | |||
| S. himalayana | HGT | Tetrastigma | 100 | <0.0001 | 494 | I | |||
| rps4 | R. cantleyi | VGT | – | 96 | – | 838 | I | 2358 | 50, 61 |
| R. tuan-mudae | VGT | – | 96 | – | 838 | I | |||
| S. himalayana | VGT | – | 96 | – | 1040 | I | |||
| R. cantleyi | HGT | Tetrastigma | 81 | 0.0075 | 143 | I | 99 | ||
| R. tuan-mudae | HGT | Tetrastigma | 81 | 0.0075 | 143 | I | |||
| rps7 | R. cantleyi | HGT | Vitaceae | 96 | 0.0060 | 328 | ψ | 850 | |
| R. tuan-mudae | HGT | Vitaceae | 96 | 0.0060 | 328 | ψ | |||
| S. himalayana | HGT | Vitaceae | 96 | 0.0060 | 186 | ψ | |||
| rps13 | S. himalayana | HGT | Vitaceae | 86 | 0.0033 | 264 | ψ | ||
| rps14 | S. himalayana | HGT | Tetrastigma | 68 | 0.0104 | 303 | I | ||
| sdh3 | S. himalayana | HGT | Vitaceae | 96 | 0.0102 | 272 | I | ||
| sdh4 | R. cantleyi | – | – | <50 | – | 390 | I | 2902 | |
| S. himalayana | – | – | <50 | – | 390 | I | |||
| S. himalayana | HGT | Vitaceae | 63 | NS | 243 | I | |||
| Genes of plastid origin | |||||||||
| atpA | R. tuan-mudae | HGT | Daucus | 94 | 0.0044 | 515 | ψ | ||
| S. himalayana | HGT | Daucus | 94 | 0.0044 | 457 | ψ | |||
| atpB | S. himalayana | HGT | Tetrastigma | 100 | 0.0001 | 477 | ψ | ||
| atpI | S. himalayana | HGT | Tetrastigma | 94 | 0.0003 | 100 | I | ||
| ndhB | S. himalayana | HGT | Tetrastigma | 86 | 0.0133 | 447 | I | ||
| psaB | S. himalayana | HGT | Tetrastigma | 92 | 0.0111 | 362 | ψ | ||
| psbA | S. himalayana | HGT | Tetrastigma | 100 | 0.0001 | 395 | ψ | ||
| psbC | S. himalayana | HGT | Vitaceae | 100 | 0.0004 | 969 | ψ | ||
| psbD | S. himalayana | HGT | Tetrastigma | 100 | <0.0001 | 753 | ψ | ||
| rbcL | S. himalayana | HGT | Vitaceae | 82 | NS | 436 | ψ | ||
| rpoC1 | S. himalayana | HGT | Tetrastigma | 100 | <0.0001 | 1613 | ψ | ||
| rpoC2 | S. himalayana | HGT | Tetrastigma | 100 | 0.0003 | 2000 | ψ | ||
| rps12 | S. himalayana | HGT | Vitaceae | 75 | 0.0304 | 222 | ψ | ||
| rrn16 | S. himalayana | HGT | Tetrastigma | 87 | 0.0297 | 1080 | – | ||
| rrn23 | S. himalayana | HGT | Tetrastigma | 99 | 0.0002 | 1638 | – | ||
Vertically transferred (VGT) sequences are additionally listed when they are present. HGT donor lineages are inferred from our maximum likelihood (ML) phylogenies, which are summarized here with ML bootstrap percentages (BP) from Figure S1A and S1D. AU = approximately unbiased test; I = intact reading frame; NS = not significant (i.e., p-value>0.05); ψ = presence of nonsense mutation. Gene expression level reported here is normalized to reads per kilobase per million reads (RPKM) for R. cantleyi.
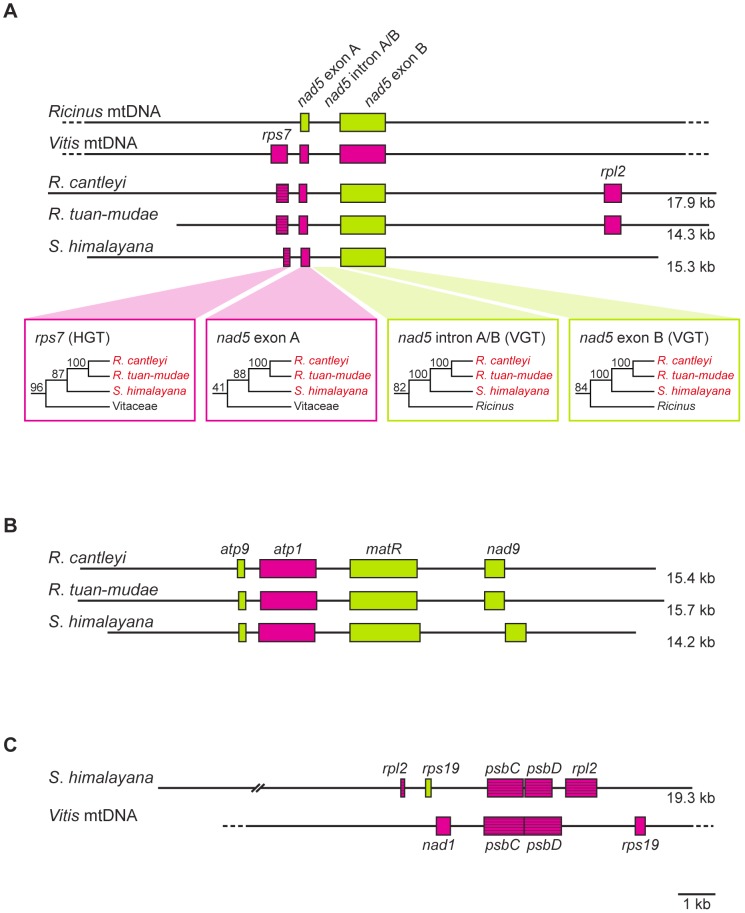
Figure 2. Gene organization of three assembled contigs for Rafflesiaceae (Rafflesia cantleyi, Rafflesia tuan-mudae, and Sapria himalayana), Ricinus communis (Euphorbiaceae), and Vitis vinifera (Vitaceae).
(A–C) The green and red boxes indicate Ricinus-like and Vitis-like genes, respectively. Pseudogenes are represented by striped boxes, and the sequence length (in kilobases [kb]) is indicated to the right of each assembled contig. Gene organization of Ricinus and Vitis mitochondrial genomes (mtDNA) follows Rivarola et al. [20] and Goremykin et al. [23], respectively.
Of the 11 mitochondrial genes that showed evidence of HGT using our more conservative threshold, four (i.e., cob, cox3, rpl5, and rps4) maintained both horizontally and vertically transferred homologs, and seven included only transgenic sequences (i.e., atp4, cox1, cox2, rps1, rps7, rps13, and sdh3) (Table 1). An additional five mitochondrial genes showed evidence of HGT using our less conservative threshold, one of them (atp9) maintained both horizontally and vertically transferred homologs, and four included only transgenic sequences (i.e., atp1, rpl2, rps14, and sdh4) (Table 1). Of those genes that included only transgenic copies in Rafflesiaceae all had homologs present in the mitochondrial genome of Ricinus, which suggests that they were likely present as native copies ancestrally in Rafflesiaceae and were subsequently displaced by transgenic homologs. One example is illustrated by our assembled contig containing the genes nad5 exons A and B and rps7 (Figure 2A). In Rafflesiaceae, nad5 exon B was identified as a native sequence (84 BP) in our phylogenetic analyses, while rps7 was identified as a transgene (96 BP). The phylogenetic placement of nad5 exon A within Vitaceae is also consistent with HGT, but support for this placement is <50 BP (Figure 2A, see also Figure S1C). However, the synteny of nad5 exons A and B is conserved among Ricinus, Vitis, and all three Rafflesiaceae species, suggesting that the native copy of nad5 exon A in Rafflesiaceae may have been displaced by a horizontally transferred DNA fragment via homologous recombination [41]. This hypothesis is further supported by the fact that nad5 exon A is immediately adjacent to the well-placed transgene rps7 in Rafflesiaceae, which exactly matches the synteny of Vitis but not Ricinus. To better locate the recombination breakpoint, we analyzed the intron region between nad5 exons A and B. This ∼1-kb region is highly conserved across angiosperms and can be easily aligned for phylogenetic analysis. We found that nad5 intron A/B was clearly identified as a native sequence (82 BP; Figure 2A, see also Figure S1C), therefore, the breakpoint is likely very close to the junction of nad5 exon A and intron A/B. Although the integration of foreign DNA via homologous recombination is common in bacteria [41], reports of this phenomenon are rare for plants (e.g., atp1 gene [42] and rps11 gene [1], [4]). Such direct homologous recombination, which is likely facilitated by the intimate physical association between Rafflesiaceae and their hosts [14], [15] combined with the frequent fusion of plant mitochondria [1], [43]–[45], may obviate the need to invoke a transposable element, bacterium, or virus for catalyzing the insertion of a DNA fragment from donor to recipient in plants.
Additionally, in the mitochondrial genome of S. himalayana, we found evidence of HGT involving 14 genes that were potentially of plastid origin using our more conservative threshold, only one of which was also identified in R. tuan-mudae (Table 1, see also Figure S1D). Thirteen of these genes support the conclusion that they were acquired via host-to-parasite HGT, because in each case S. himalayana is placed sister to, or nested within, Vitaceae with ≥70 BP. Only atpA from S. himalayana and R. tuan-mudae were placed elsewhere phylogenetically, sister to Daucus with 94 BP. Furthermore, for six of these 14 genes (i.e., atpB, atpI, psaB, psbA, psbC, and psbD), the transgenic sequences from S. himalayana were sister to the mitochondrial and not the plastid homologs from Vitaceae (Figure S1D). These six genes, plus three additional plastid genes (i.e., atpA, ndhB, and rbcL), have been shown to be incorporated into the mitochondrial genome of Vitis [23]. Together, these results suggest that the majority of these plastid genes were likely acquired via HGT from the host mitochondrial genome, instead of from its plastid genome.
Finally, seven of our assembled contigs demonstrated that synteny was maintained between transgenes from Rafflesiaceae and genes from the close relative of their hosts, Vitis, whose mitochondrial and plastid genomes were both fully annotated (i.e., rps7+nad5 exon A, psbC+psbD, sdh3+rpl5+rps14+cob+cox1, rpoC1+rpoC2, and sdh4+cox3; Figure 2 and Figure S3). This, combined with our finding that two transgenes bear introns (i.e., cox1 and rpl2), firmly supports our previous suggestion that transgenes in Rafflesiaceae are likely transferred as larger DNA fragments versus shorter mRNAs [28].
Transgenes are expressed in Rafflesiaceae
Most previously reported mitochondrial transgenes in plants appear to be non-functional, i.e., they have been shown to be either introns (e.g., [3], [6], [8], [46], [47]) or pseudogenes (e.g., [7], [9]). However, among all mitochondrial transgenes identified here, six of seven sequences in R. cantleyi and R. tuan-mudae, and 13 of 16 sequences in S. himalayana maintain their reading frames (Table 1). To further understand if these transgenes are expressed, we re-examined the recently published transcriptome of R. cantleyi [28] to quantify gene expression levels of these mitochondrial genes. Our results indicate that all transgenes in R. cantleyi show evidence of expression (Table 1). Furthermore, although native genes in R. cantleyi show higher overall levels of gene expression than transgenes (Figure S4), this difference is not significant (p-value = 0.19, Welch's t test). Thus, transgenes are actively transcribed in this species, suggesting that they have functional promoters and likely play a role in cellular function.
Timing of HGT events
Our broad phylogenomic assessment of mitochondrial genome provides a unique opportunity to determine if HGT we identified in Rafflesiaceae is relatively ancient or more recent. For five genes that show evidence of HGT (i.e., atp1, atp4, cox1, rps7, and atpA; Figure S1A and S1D), it is most parsimonious to infer that they each result from an ancient HGT event. The more ancient origin is supported by the fact that transgenic sequences from Rafflesia and Sapria form a clade. Furthermore, we found that some of these transgenes maintained synteny between Rafflesia and Sapria (e.g., atp1 and rps7; Figure 2A and 2B). Therefore, these gene transfers appear to have been relatively ancient and likely occurred after the origin of stem group Rafflesiaceae (95% highest posterior density [HPD] interval of 83.1–109.5 Ma; [17], [48]) and before the origin of crown group Rafflesiaceae (69.5–95.9 Ma; [17], [48]). Both of these estimated clade ages, accounting for 95% HPD intervals, are outside the age of stem group Tetrastigma (36.4–65.3 Ma; [49]), and well outside the age of crown group Tetrastigma (25.7–49.3 Ma; [49]) (Figure 1B).
This raises the distinct possibility that Rafflesiaceae has had former host associations with other plant lineages (perhaps within Vitaceae, but also outside of the family), which may have served as past donors of transgenes. We have previously referred to this as the ghost of HGT's past [50]. In support of this possibility, none of the more ancient transgenic sequences we identified grouped with their current hosts Tetrastigma, as would be expected if these species served as hosts. Two genes (i.e., atp1 and rps7) involved in these more ancient HGT events are sister to Vitaceae, suggesting that close relatives of Tetrastigma may have served as past transgenic donors. In three other cases (i.e., atp4, cox1, and atpA), however, transgenic sequences do not group closely to Vitaceae (e.g., Cucurbitaceae and Daucus; Table 1), indicating different transgenic donors (no evidence of gene conversion, which would confound phylogenetic placements, was detected in these genes using the OrgConv package [51] with p-value<0.001). To our knowledge, this is the first evidence that Rafflesiaceae may have previously parasitized different host species, which served as transgenic donors in the past. Further taxon sampling of these genes by co-authors Z.X. and Y.W. is underway and should allow us to determine those previous host donors more precisely.
For the remaining 28 instances of HGT, it is most likely that these were the result of more recent gene transfers. Transgenic sequences in these cases are found exclusively in either Rafflesia (i.e., rps4) or Sapria (e.g., atp9 and cob), or if identified in both Rafflesia and Sapria they do not form a clade (i.e., rpl2 and rps1) (Figure S1A and S1D). Evidence of such recent HGT is especially prevalent in S. himalayana: 17 of its transgenic sequences are sister to, or nested within, Tetrastigma (Table 1). Moreover, our phylogenetic analyses indicate that some of these sequences may have resulted from multiple independent gene transfers involving the same gene because transgenic sequences from Rafflesiaceae do not form a clade. In some cases, these gene transfers appear to involve multiple transgenic sequences within a single species for the same gene (i.e., cox1 in Sapria, which possesses two distinct transgenic sequences that appear to have been transferred independently). In other cases, gene transfers involve multiple transgenic sequences in different species for the same gene (i.e., rpl2 and rps1, which show independent transfer events for Rafflesia and Sapria). These more recent HGT events are further supported by synteny: transgenic sequences involving the same gene from Rafflesia and Sapria are located at different positions in the mitochondrial genome (e.g., rpl2; Figure 2A and 2C). Why some genes exhibit repeated HGT is fertile ground for future investigation.
Conclusion
Our study is the first to comprehensively assess the magnitude of HGT in plants involving a genome (i.e., mitochondria) and a species interaction (i.e., parasitism) where it has been hypothesized to be potentially rampant. These results reveal a high degree of HGT in the mitochondrial genome of Rafflesiaceae involving genes that were likely acquired from its host at various time intervals. We previously established that in R. cantleyi, about 2.1% of nuclear gene transcripts have likely been acquired from its host via HGT [28]. In contrast, our study conservatively indicates that 24–41% of the mitochondrial gene sequences show evidence of HGT in Rafflesiaceae, depending on the species. These results establish for the first time that although the magnitude of HGT involving nuclear genes is appreciable, HGT involving mitochondrial genes in these parasitic plants is an order of magnitude higher. This elevated rate of HGT involving the mitochondrial genome may represent a more general pattern for other parasitic plant clades, and perhaps more broadly for angiosperms.
Materials and Methods
Molecular techniques and next-generation sequencing
For R. cantleyi and S. himalayana, mitochondria were isolated from ∼30 grams of fresh material from flower buds using the sucrose gradient centrifugation protocols of Jansen et al. [30]. DNA extracted from purified mitochondria was amplified with the REPLI-g Midi Kit (Qiagen, Inc.). When we were unable to acquire fresh material, total genomic DNA (gDNA) was extracted from silica-dried material using the DNeasy Plant Mini kit (Qiagen, Inc.), and treated with RNase A at 60°C for 1.5 hours to remove any residual RNA contamination. For each species, an Illumina library with the insert size of 350±50 bp was prepared from five micrograms of DNA following the protocols of Bentley et al. [52]. All libraries were sequenced on the Genome Analyzer II (Illumina, Inc.) with 100 bp paired-end runs at the FAS Center for Systems Biology at Harvard University (Table S1).
Sequence assembly and alignment
Illumina reads were assembled de novo in ABySS v1.2.1 [53] using default parameters (Table S1). The assembled contigs were annotated against published mitochondrial and plastid genomes from 27 seed plants (Table S3) with BLASTN v2.2.23 [54] using an e-value ≤10−5. Gene sequences from all species were then queried against themselves using BLASTN v2.2.23. BLASTN hits with an e-value ≤10−10 were passed to MCL v08-312 [55] for Markov clustering. Only those gene clusters that included at least Cycas/Spirodela (for outgroup rooting), Rafflesia/Sapria (sequences under investigation), Ricinus (close relative of Rafflesiaceae), and Vitis (close relative of Tetrastigma) were retained. The nucleotide sequences of each gene were first aligned using MAFFT v6.624 [56], and then manually inspected and realigned if necessary.
Gene copy number estimation
To assess gene copy number and corresponding genomic compartment localization of our assembled gene sequences, we mapped the Illumina gDNA reads from R. cantleyi [28], R. tuan-mudae, and S. himalayana to gene sequences identified here and to the 1305 nuclear genes identified from R. cantleyi [28] and R. tuan-mudae [18] using Bowtie v0.12.7 [57] (Table S2). To avoid complications with intron regions, we first divided each Illumina read into multiple 25 bp fragments following Kim and Salzberg [58], and then mapped each 25-mer with zero mismatches and unique mapping.
Phylogenetic analyses and alternative topology tests
Our ML analyses were conducted for all genes using RAxML v7.2.8 [59] with the GTR+Γ nucleotide substitution model. The best-scoring ML tree and BP for each gene were obtained using the rapid bootstrap algorithm [60] with 500 replicates (Figures S1 and S2). For nad5, we also performed ML analyses on three gene regions separately (i.e., nad5 exon A, intron A/B, and exon B; Figure 2A, see also Figure S1C), which allowed us to determine the location of homologous recombination more accurately (see above).
Alternative topology tests were performed in an ML framework using the approximately unbiased (AU) test [61] as implemented in scaleboot v0.3-3 [62] (Table 1). To generate constrained ML trees for genes that show evidence of HGT, we enforced all transgenic and native (when present) sequences from Rafflesiaceae to be monophyletic with Ricinus, and then conducted ML searches using these constraints.
Gene expression level analyses
To estimate the gene expression level in R. cantleyi, the Illumina cDNA reads from Xi et al. [28] were mapped onto the assembled R. cantleyi mitochondrial gene sequences using Bowtie v0.12.7 [57] as described above. cDNA reads that mapped onto each gene sequence were then summed and further normalized to reads per kilobase per million reads (RPKM [63]; Table 1, see also Figure S4).
Contamination and the determination of HGT in Rafflesiaceae
Tremendous care was taken to avoid and/or detect host or lab contamination during our sample preparation and data analyses. First, our DNA sample preparation and genome library sequencing of Rafflesiaceae were performed separate from any work involving Tetrastigma; thus, laboratory contamination of our Rafflesiaceae DNAs with Tetrastigma is unlikely. Second, the plastid genome has apparently been lost in Rafflesiaceae [31]. If there were any host contamination, the host's plastid gene sequences should be easily detected in our sequence data. This was not the case. Third, the mitochondrial genome sequences of R. cantleyi and R. tuan-mudae were generated from two different sources, i.e., a fresh flower bud using sucrose gradient centrifugation and silica-dried perigone lobes using total gDNA extraction, respectively (Table S1). If one of these samples, or genome libraries, were contaminated, we would not expect to have identified the identical set of transgenes from these samples. Similarly, for S. himalayana, all transgenes identified from the genome library prepared using sucrose gradient centrifugation were verified in our second library of this species that was prepared from total gDNA (Table S2). Fourth, most transgenic sequences identified here possess some amount of sequence divergence when directly compared with homologs from their current host. For example, all 15 transgenic sequences from Rafflesia show some degree of sequence divergence when directly compared with homologs from their host species, T. rafflesiae (mean DNA sequence distance = 0.042189). Similarly, 26 of 30 transgenic sequences from Sapria show some degree of sequence divergence when directly compared with homologs from their host species, T. cruciatum (mean DNA sequence distance = 0.020265) (Figure S1A and S1D). These sequence distances are significantly greater (p-value<0.01, Welch's t test) than those between the two included Tetrastigma species (mean DNA sequence distance = 0.001984). This is despite the fact that these two Tetrastigma species have diverged from each other at least 10 Ma [49]. Furthermore, three transgenic sequences from Rafflesia and 13 transgenic sequences from Sapria contain nonsense mutations (Table 1). These results strongly indicate that some period of evolution has elapsed since the time of HGT. Fifth, all seven transgenes from R. cantleyi show evidence of gene expression based on its transcriptome (Table 1), and levels of expression are not significantly different between transgenes and native genes (p-value = 0.19, Welch's t test; Figure S4). Lastly, and perhaps most importantly, seven of our assembled contigs contain both transgenes and native genes (Figure 2) indicating that these transgenes are clearly integrated into the mitochondrial genome of Rafflesiaceae.
Supporting Information
Phylograms of all horizontally transferred genes in the mitochondrial genomes of Rafflesia cantleyi, Rafflesia tuan-mudae, and Sapria himalayana. Maximum likelihood bootstrap percentages (BP) were summarized from 500 bootstrap replicates, and only BP values greater than 50% are shown. Gene sequences from Rafflesiaceae and the host Tetrastigma are highlighted in red and blue, respectively. H and V indicate sequences of horizontal and vertical transmission, respectively. Number of aligned characters (chars) and scale bar (substitutions per site) are shown for each gene. (A) Phylograms for the 16 mitochondrial genes where HGT was detected. (B) Phylograms of the four mitochondrial genes with RNA editing sites excluded from our alignments. (C) Phylograms for the three gene regions of nad5: exons A and B and intron A/B. (D) Phylograms for the 14 genes of plastid origin where HGT was detected.
(PDF)
Phylograms of all vertically transferred and unassigned genes in the mitochondrial genomes of Rafflesia cantleyi, Rafflesia tuan-mudae, and Sapria himalayana. Maximum likelihood bootstrap percentages (BP) were summarized from 500 bootstrap replicates, and only BP values greater than 50% are shown. Gene sequences from Rafflesiaceae and the host Tetrastigma are highlighted in red and blue, respectively. V indicates sequences of vertical transmission. Number of aligned characters (chars) and scale bar (substitutions per site) are shown for each gene.
(PDF)
Gene organization of three assembled contigs (A–C) for Sapria himalayana (Rafflesiaceae) and Vitis vinifera (Vitaceae). The red boxes indicate Vitis-like genes. Pseudogenes are represented by striped boxes, and the sequence length (in kilobases [kb]) is indicated to the right of each assembled contig.
(PDF)
Boxplot of gene expression levels of horizontally (HGT) and vertically (VGT) transferred mitochondrial gene sequences in Rafflesia cantleyi. The number of gene sequences for each category is shown in parentheses; RPKM = reads per kilobase per million reads.
(PDF)
Newly sequenced species in this study with associated assembly statistics.
(PDF)
Estimated gene copy number for Rafflesia cantleyi, Rafflesia tuan-mudae, and Sapria himalayana. The Illumina genomic DNA reads from R. cantleyi, R. tuan-mudae, and S. himalayana were mapped onto the 38 mitochondrial genes, 14 genes of plastid origin, and 1,305 nuclear genes from R. cantleyi and R. tuan-mudae. Illumina reads that mapped onto each gene were then summed and further normalized to reads per base pair.
(XLS)
Data sources of gene sequences included in our phylogenetic analyses.
(PDF)
Acknowledgments
We thank Renate Hellmiss, Matthew Klooster, Kenneth Wurdack, and members of the Davis laboratory for advice and discussion. We would also like to thank the anonymous reviewers for their valuable comments and suggestions to improve the quality of the paper.
Funding Statement
This work was supported by the National Science Foundation (NSF) Assembling the Tree of Life grant DEB-0622764 and NSF DEB-1120243 (to CCD) and by the China Scholarship Council Program for Visiting Scholars (to YW). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Richardson AO, Palmer JD (2007) Horizontal gene transfer in plants. J Exp Bot 58: 1–9. [DOI] [PubMed] [Google Scholar]
- 2. Bock R (2010) The give-and-take of DNA: horizontal gene transfer in plants. Trends Plant Sci 15: 11–22. [DOI] [PubMed] [Google Scholar]
- 3. Won H, Renner SS (2003) Horizontal gene transfer from flowering plants to Gnetum . Proc Natl Acad Sci USA 100: 10824–10829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bergthorsson U, Adams KL, Thomason B, Palmer JD (2003) Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature 424: 197–201. [DOI] [PubMed] [Google Scholar]
- 5. Bergthorsson U, Richardson AO, Young GJ, Goertzen LR, Palmer JD (2004) Massive horizontal transfer of mitochondrial genes from diverse land plant donors to the basal angiosperm Amborella . Proc Natl Acad Sci USA 101: 17747–17752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sanchez-Puerta MV, Cho Y, Mower JP, Alverson AJ, Palmer JD (2008) Frequent, phylogenetically local horizontal transfer of the cox1 group I intron in flowering plant mitochondria. Mol Biol Evol 25: 1762–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mower JP, Stefanovic S, Hao W, Gummow JS, Jain K, et al. (2010) Horizontal acquisition of multiple mitochondrial genes from a parasitic plant followed by gene conversion with host mitochondrial genes. BMC Biol 8: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Davis CC, Wurdack KJ (2004) Host-to-parasite gene transfer in flowering plants: Phylogenetic evidence from Malpighiales. Science 305: 676–678. [DOI] [PubMed] [Google Scholar]
- 9. Mower JP, Stefanovic S, Young GJ, Palmer JD (2004) Gene transfer from parasitic to host plants. Nature 432: 165–166. [DOI] [PubMed] [Google Scholar]
- 10. Nickrent DL, Blarer A, Qiu YL, Vidal-Russell R, Anderson FE (2004) Phylogenetic inference in Rafflesiales: the influence of rate heterogeneity and horizontal gene transfer. BMC Evol Biol 4: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Park JM, Manen JF, Schneeweiss GM (2007) Horizontal gene transfer of a plastid gene in the non-photosynthetic flowering plants Orobanche and Phelipanche (Orobanchaceae). Mol Phylogenet Evol 43: 974–985. [DOI] [PubMed] [Google Scholar]
- 12. Yoshida S, Maruyama S, Nozaki H, Shirasu K (2010) Horizontal gene transfer by the parasitic plant Striga hermonthica . Science 328: 1128. [DOI] [PubMed] [Google Scholar]
- 13. Barkman TJ, McNeal JR, Lim SH, Coat G, Croom HB, et al. (2007) Mitochondrial DNA suggests at least 11 origins of parasitism in angiosperms and reveals genomic chimerism in parasitic plants. BMC Evol Biol 7: 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stegemann S, Bock R (2009) Exchange of genetic material between cells in plant tissue grafts. Science 324: 649–651. [DOI] [PubMed] [Google Scholar]
- 15. Stegemann S, Keuthe M, Greiner S, Bock R (2012) Horizontal transfer of chloroplast genomes between plant species. Proc Natl Acad Sci USA 109: 2434–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Barkman TJ, Lim SH, Salleh KM, Nais J (2004) Mitochondrial DNA sequences reveal the photosynthetic relatives of Rafflesia, the world's largest flower. Proc Natl Acad Sci USA 101: 787–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Davis CC, Latvis M, Nickrent DL, Wurdack KJ, Baum DA (2007) Floral gigantism in Rafflesiaceae. Science 315: 1812. [DOI] [PubMed] [Google Scholar]
- 18. Wurdack KJ, Davis CC (2009) Malpighiales phylogenetics: gaining ground on one of the most recalcitrant clades in the angiosperm tree of life. Am J Bot 96: 1551–1570. [DOI] [PubMed] [Google Scholar]
- 19. Chan AP, Crabtree J, Zhao Q, Lorenzi H, Orvis J, et al. (2010) Draft genome sequence of the oilseed species Ricinus communis . Nat Biotechnol 28: 951–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rivarola M, Foster JT, Chan AP, Williams AL, Rice DW, et al. (2011) Castor bean organelle genome sequencing and worldwide genetic diversity analysis. PLoS ONE 6: e21743 doi:10.1371/journal.pone.0021743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jansen RK, Kaittanis C, Saski C, Lee SB, Tomkins J, et al. (2006) Phylogenetic analyses of Vitis (Vitaceae) based on complete chloroplast genome sequences: effects of taxon sampling and phylogenetic methods on resolving relationships among rosids. BMC Evol Biol 6: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, et al. (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463–467. [DOI] [PubMed] [Google Scholar]
- 23. Goremykin VV, Salamini F, Velasco R, Viola R (2009) Mitochondrial DNA of Vitis vinifera and the issue of rampant horizontal gene transfer. Mol Biol Evol 26: 99–110. [DOI] [PubMed] [Google Scholar]
- 24. Wikström N, Savolainen V, Chase MW (2001) Evolution of the angiosperms: calibrating the family tree. Proc R Soc B 268: 2211–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Magallón S, Castillo A (2009) Angiosperm diversification through time. Am J Bot 96: 349–365. [DOI] [PubMed] [Google Scholar]
- 26. Wang H, Moore MJ, Soltis PS, Bell CD, Brockington SF, et al. (2009) Rosid radiation and the rapid rise of angiosperm-dominated forests. Proc Natl Acad Sci USA 106: 3853–3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bell CD, Soltis DE, Soltis PS (2010) The age and diversification of the angiosperms re-revisited. Am J Bot 97: 1296–1303. [DOI] [PubMed] [Google Scholar]
- 28. Xi Z, Bradley RK, Wurdack KJ, Wong KM, Sugumaran M, et al. (2012) Horizontal transfer of expressed genes in a parasitic flowering plant. BMC Genomics 13: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Adams KL, Qiu YL, Stoutemyer M, Palmer JD (2002) Punctuated evolution of mitochondrial gene content: High and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc Natl Acad Sci USA 99: 9905–9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jansen RK, Raubeson LA, Boore JL, DePamphilis CW, Chumley TW, et al. (2005) Methods for obtaining and analyzing whole chloroplast genome sequences. Method Enzymol 395: 348–384. [DOI] [PubMed] [Google Scholar]
- 31. Nickrent DL, Yan OY, Duff RJ, dePamphilis CW (1997) Do nonasterid holoparasitic flowering plants have plastid genomes? Plant Mol Biol 34: 717–729. [DOI] [PubMed] [Google Scholar]
- 32. Draper CK, Hays JB (2000) Replication of chloroplast, mitochondrial and nuclear DNA during growth of unirradiated and UVB-irradiated Arabidopsis leaves. Plant J 23: 255–265. [DOI] [PubMed] [Google Scholar]
- 33. Hiesel R, Wissinger B, Schuster W, Brennicke A (1989) RNA editing in plant mitochondria. Science 246: 1632–1634. [DOI] [PubMed] [Google Scholar]
- 34.Nais J (2001) Rafflesia of the world: Kota Kinabalu: Sabah Parks.
- 35. Veldkamp JF (2008) The correct name for the Tetrastigma (Vitaceae) host of Rafflesia (Rafflesiaceae) in Malesia and a (not so) new species. Reinwardtia 12: 261–265. [Google Scholar]
- 36. Soejima A, Wen J (2006) Phylogenetic analysis of the grape family (Vitaceae) based on three chloroplast markers. Am J Bot 93: 278–287. [DOI] [PubMed] [Google Scholar]
- 37. Ren H, Lu LM, Soejima A, Luke Q, Zhang DX, et al. (2011) Phylogenetic analysis of the grape family (Vitaceae) based on the noncoding plastid trnC-petN, trnH-psbA, and trnL-F sequences. Taxon 60: 629–637. [Google Scholar]
- 38. Bremer B, Bremer K, Chase MW, Fay MF, Reveal JL, et al. (2009) An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot J Linn Soc 161: 105–121. [Google Scholar]
- 39. Hillis DM, Bull JJ (1993) An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol 42: 182–192. [Google Scholar]
- 40. Qiu Y-L, Li L, Wang B, Xue J-Y, Hendry TA, et al. (2010) Angiosperm phylogeny inferred from sequences of four mitochondrial genes. J Syst Evol 48: 391–425. [Google Scholar]
- 41. Thomas CM, Nielsen KM (2005) Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nature Rev Microbiol 3: 711–721. [DOI] [PubMed] [Google Scholar]
- 42. Hao W, Palmer JD (2009) Fine-scale mergers of chloroplast and mitochondrial genes create functional, transcompartmentally chimeric mitochondrial genes. Proc Natl Acad Sci USA 106: 16728–16733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Arimura S, Yamamoto J, Aida GP, Nakazono M, Tsutsumi N (2004) Frequent fusion and fission of plant mitochondria with unequal nucleoid distribution. Proc Natl Acad Sci USA 101: 7805–7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sheahan MB, McCurdy DW, Rose RJ (2005) Mitochondria as a connected population: ensuring continuity of the mitochondrial genome during plant cell dedifferentiation through massive mitochondrial fusion. Plant J 44: 744–755. [DOI] [PubMed] [Google Scholar]
- 45. Logan DC (2010) Mitochondrial fusion, division and positioning in plants. Biochem Soc Trans 38: 789–795. [DOI] [PubMed] [Google Scholar]
- 46. Woloszynska M, Bocer T, Mackiewicz P, Janska H (2004) A fragment of chloroplast DNA was transferred horizontally, probably from non-eudicots, to mitochondrial genome of Phaseolus . Plant Mol Biol 56: 811–820. [DOI] [PubMed] [Google Scholar]
- 47. Sanchez-Puerta MV, Abbona CC, Zhuo S, Tepe EJ, Bohs L, et al. (2011) Multiple recent horizontal transfers of the cox1 intron in Solanaceae and extended co-conversion of flanking exons. BMC Evol Biol 11: 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bendiksby M, Schumacher T, Gussarova G, Nais J, Mat-Salleh K, et al. (2010) Elucidating the evolutionary history of the Southeast Asian, holoparasitic, giant-flowered Rafflesiaceae: Pliocene vicariance, morphological convergence and character displacement. Mol Phylogenet Evol 57: 620–633. [DOI] [PubMed] [Google Scholar]
- 49. Chen PT, Wen J, Chen LQ (2011) Spatial and temporal diversification of Tetrastigma Planch. (Vitaceae). Gard Bull Singapore 63: 313–333. [Google Scholar]
- 50. Davis CC, Anderson WR, Wurdack KJ (2005) Gene transfer from a parasitic flowering plant to a fern. Proc R Soc B 272: 2237–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hao W (2010) OrgConv: detection of gene conversion using consensus sequences and its application in plant mitochondrial and chloroplast homologs. BMC Bioinformatics 11: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, et al. (2008) Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456: 53–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJM, et al. (2009) ABySS: A parallel assembler for short read sequence data. Genome Res 19: 1117–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Enright AJ, van Dongen S, Ouzounis CA (2002) An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 30: 1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30: 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kim D, Salzberg SL (2011) TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol 12: R72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Stamatakis A (2006) RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- 60. Stamatakis A, Hoover P, Rougemont J (2008) A rapid bootstrap algorithm for the RAxML web servers. Syst Biol 57: 758–771. [DOI] [PubMed] [Google Scholar]
- 61. Shimodaira H (2002) An approximately unbiased test of phylogenetic tree selection. Syst Biol 51: 492–508. [DOI] [PubMed] [Google Scholar]
- 62. Shimodaira H (2008) Testing regions with nonsmooth boundaries via multiscale bootstrap. J Stat Plan Infer 138: 1227–1241. [Google Scholar]
- 63. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylograms of all horizontally transferred genes in the mitochondrial genomes of Rafflesia cantleyi, Rafflesia tuan-mudae, and Sapria himalayana. Maximum likelihood bootstrap percentages (BP) were summarized from 500 bootstrap replicates, and only BP values greater than 50% are shown. Gene sequences from Rafflesiaceae and the host Tetrastigma are highlighted in red and blue, respectively. H and V indicate sequences of horizontal and vertical transmission, respectively. Number of aligned characters (chars) and scale bar (substitutions per site) are shown for each gene. (A) Phylograms for the 16 mitochondrial genes where HGT was detected. (B) Phylograms of the four mitochondrial genes with RNA editing sites excluded from our alignments. (C) Phylograms for the three gene regions of nad5: exons A and B and intron A/B. (D) Phylograms for the 14 genes of plastid origin where HGT was detected.
(PDF)
Phylograms of all vertically transferred and unassigned genes in the mitochondrial genomes of Rafflesia cantleyi, Rafflesia tuan-mudae, and Sapria himalayana. Maximum likelihood bootstrap percentages (BP) were summarized from 500 bootstrap replicates, and only BP values greater than 50% are shown. Gene sequences from Rafflesiaceae and the host Tetrastigma are highlighted in red and blue, respectively. V indicates sequences of vertical transmission. Number of aligned characters (chars) and scale bar (substitutions per site) are shown for each gene.
(PDF)
Gene organization of three assembled contigs (A–C) for Sapria himalayana (Rafflesiaceae) and Vitis vinifera (Vitaceae). The red boxes indicate Vitis-like genes. Pseudogenes are represented by striped boxes, and the sequence length (in kilobases [kb]) is indicated to the right of each assembled contig.
(PDF)
Boxplot of gene expression levels of horizontally (HGT) and vertically (VGT) transferred mitochondrial gene sequences in Rafflesia cantleyi. The number of gene sequences for each category is shown in parentheses; RPKM = reads per kilobase per million reads.
(PDF)
Newly sequenced species in this study with associated assembly statistics.
(PDF)
Estimated gene copy number for Rafflesia cantleyi, Rafflesia tuan-mudae, and Sapria himalayana. The Illumina genomic DNA reads from R. cantleyi, R. tuan-mudae, and S. himalayana were mapped onto the 38 mitochondrial genes, 14 genes of plastid origin, and 1,305 nuclear genes from R. cantleyi and R. tuan-mudae. Illumina reads that mapped onto each gene were then summed and further normalized to reads per base pair.
(XLS)
Data sources of gene sequences included in our phylogenetic analyses.
(PDF)