Abstract
The unconventional prefoldin URI/RMP, in humans, and its orthologue in yeast, Bud27, have been proposed to participate in the biogenesis of the RNA polymerases. However, this role of Bud27 has not been confirmed and is poorly elucidated. Our data help clarify the mechanisms governing biogenesis of the three eukaryotic RNA pols. We show evidence that Bud27 is the first example of a protein that participates in the biogenesis of the three eukaryotic RNA polymerases and the first example of a protein modulating their assembly instead of their nuclear transport. In addition we demonstrate that the role of Bud27 in RNA pols biogenesis depends on Rpb5. In fact, lack of BUD27 affects growth and leads to a substantial accumulation of the three RNA polymerases in the cytoplasm, defects offset by the overexpression of RPB5. Supporting this, our data demonstrate that the lack of Bud27 affects the correct assembly of Rpb5 and Rpb6 to the three RNA polymerases, suggesting that this process occurs in the cytoplasm and is a required step prior to nuclear import. Also, our data support the view that Rpb5 and Rpb6 assemble somewhat later than the rest of the complexes. Furthermore, Bud27 Rpb5-binding but not PFD-binding domain is necessary for RNA polymerases biogenesis. In agreement, we also demonstrate genetic interactions between BUD27, RPB5, and RPB6. Bud27 shuttles between the nucleus and the cytoplasm in an Xpo1-independent manner, and also independently of microtubule polarization and possibly independently of its association with the RNA pols. Our data also suggest that the role of Bud27 in RNA pols biogenesis is independent of the chaperone prefoldin (PFD) complex and of Iwr1. Finally, the role of URI seems to be conserved in humans, suggesting conserved mechanisms in RNA pols biogenesis.
Author Summary
The mechanisms governing the assembly and the transport of the three eukaryotic RNA polymerases to the nucleus are in discussion. Interesting papers have demonstrated the participation of some proteins in the assembly of the nuclear RNA polymerases and in their transport to the nucleus, but the mechanisms involved are poorly understood. Our data help clarify the mechanisms governing biogenesis of the three eukaryotic RNA pols and demonstrate that the prefoldin Bud27 of Saccharomyces cerevisiae mediates the correct assembly of the three complexes prior to their translocation to the nucleus, in a process which is dependent on Rpb5. In addition, our data support the view that, during the assembly of the RNA pols, Rpb5 and Rpb6 assemble rather late compared to the rest of the complexes. Furthermore, this role of Bud27 seems to be specific, as it is not extended to other prefoldin members. Finally, the role of Bud27 seems to be conserved in humans, suggesting conserved mechanisms in RNA pols biogenesis.
Introduction
Eukaryotic RNA polymerases are a family of multimeric enzymes, RNA pol I, II, and III, responsible for the specific synthesis of different RNAs. RNA pol I is specialized in the synthesis of the pre-rRNA precursor of the three largest rRNA and typically account for about 75% of the entire transcription output in fast-growing yeast cells. RNA pol III transcribes mostly tRNAs and 5S rRNA, together with several short non-translated RNAs, while transcription corresponds to about 15% of the total RNA. RNA pol II, the enzyme that produces all mRNAs and many non-coding ones, transcribes most of the nuclear genome but nevertheless contributes to less than 10% of total RNA in growing cells. RNA pol I, II, and III are composed of 14, 12, and 17 subunits respectively, with a catalytic core formed by the two largest subunits highly conserved through evolution and five common subunits to the three enzymes [1]–[3]. Despite intensive studies concerning the structure and the transcriptional regulation of the three RNA polymerases [4], [5], little is known about the mechanisms governing their assembly and their nuclear import.
Noteworthy findings in both human and yeast demonstrate the participation of different proteins in the transport of the RNA pol II to the nucleus, Iwr1 and Npa3 in yeast, and GPN1 (RPAP4) and GPN3 in humans [6]–[10]. It has also been suggested that RPAP2 plays a role in import on the basis that it is cytoplasmic, binds fully assembled enzyme and shuttles in a CRM1-dependent manner [11]. However, no data concerning proteins involved in the nuclear transport of the RNA pol I or III are available. In addition, proteomic analysis in humans cells seek to decipher the mechanisms of RNA pol II biogenesis and assembly identifying a number of polymerase-associated factors. Among these, HSP90 and its R2TP/Prefoldin-like chaperone, including hSpagh (RPAP3), are clearly involved in these processes [8], [12].
In humans, R2TP/Prefoldin-like complex contains Rpb5, a common subunit to the three eukaryotic RNA polymerases [2], as well as the unconventional prefoldin Rpb5 interactor (URI/RMP), a member of the prefoldin (PFD) family of ATP-independent molecular chaperones [8],[13]. URI physically binds Rpb5, other nuclear proteins involved in transcription, including the general transcription factor TFIIF [14]–[16] and components of the Paf-1 complex that promotes RNA pol II CTD phosphorylation and histone modification during transcription elongation [17]. Notably, its yeast homologue Bud27 also binds Rpb5 [18]. URI was originally characterized in human and yeast cells as regulator of gene expression controlled by TOR (for target of Rapamycin) pathway [18]. Furthermore, URI has been linked to translation initiation [19], transcription regulation, chromatin stability or DNA damage response [13], [20]. URI is located mainly in the cytoplasm, although nuclear and perinuclear localization has also been observed in different organisms [20]–[22]. However, in Saccharomyces cerevisiae, only a cytoplasmic localization has been detected [19]. In addition, URI is believed to function as a scaffold protein able to assemble additional members of PFD family (through its PFD- and Rpb5-binding domains) in both human and yeast [13], [23] and different authors have proposed a role in the cytoplasmic assembly of the human RNA pol II [8], [12], [13].
Despite that some experiments, using immunoprecipitation and mass-spectrometry, have shown that URI interacts with RNA polymerases and other intermediary subcomplexes involved in RNA pol II cytoplasmic assembly, very little is known about the role of URI in RNA polymerases biogenesis. In this report, we investigate the role of the URI yeast homologue Bud27 in RNA polymerases assembly and its relationship with Rpb5. Bud27 interacts with different subunits of the three RNA polymerases, with the common subunit Rpb5 and Rpb10, as well as with a component of the prefoldin-like complex, Yke2 (Pfd6). Deletion of BUD27 leads to a substantial accumulation of the three RNA polymerases in the cytoplasm, a defect offset by the overexpression of RPB5. Supporting this, our data demonstrate that the lack of Bud27 affects the correct assembly of Rpb5 and Rpb6 to the three RNA polymerases, suggesting that this process occurs in the cytoplasm and is a required step prior to nuclear import. As previously proposed in human [12], Rpb6 appears to assemble rather late after Rpb5 assembly. In agreement, we observed a genetic interaction between BUD27, RPB5 and RPB6. In addition, Bud27 PFD- or Rpb5-binding domains are not necessary for RNA polymerases biogenesis. Finally, our data demonstrate that Bud27 shuttles between the cytoplasm and nucleus, as shown by the deletion of the NES domain, contrary to what has been previously described, a mechanism that is independent of the Xpo1-mediated pathway. URI silencing suggests that similar role for this protein accounts in humans.
Results
Bud27 interacts physically with the three eukaryotic RNA polymerases
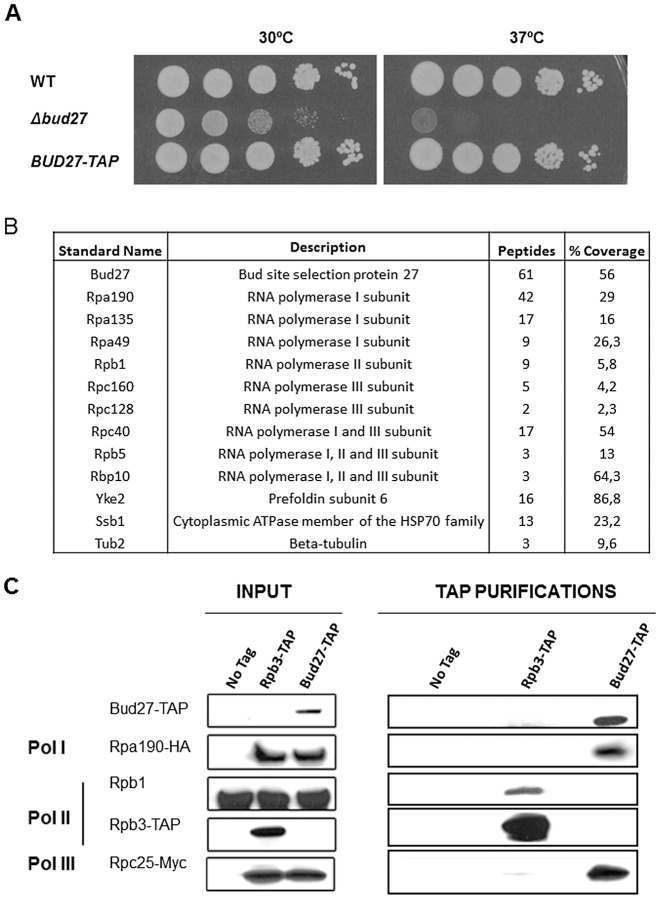
Immunoprecipitation studies and protein identification by mass spectrometry have shown that URI interacts with components of the RNA pol II in human cells [8], [12]–[14]. In addition, work in yeast [24], [25], using systematic characterization of complexes by TAP and mass spectrometry predicted the association of the URI orthologue, Bud27, with multiple components of the RNA pol II machinery. However, only the physical interaction between Bud27 and Rpb5 in yeast has been further demonstrated and no clear data concerning the association of Bud27 with the other two RNA polymerases have been reported. To gain insights into the association of Bud27 with the three RNA pols and to identify proteins that associate with Bud27 in yeast, we used the BUD27 gene TAP-tagged at its 3′ end in TAP purifications. As shown, the Bud27-TAP cells grow normally (Figure 1A). Bud27-TAP was affinity-purified from a whole-cell lysate by two consecutive affinity columns (IgG-Sepharose and Calmodulin-Sepharose). After the second purification, 10 proteins were specifically enriched. It bears noting that most of these proteins are members of the three RNA polymerases: Rpa190, Rpa135 and Rpa49 (RNA pol I); Rpc160 and Rpc128 (pol III); Rpc40 (pol I and III); Rpb1 (pol II); Rpb10 and Rpb5 (pol I, II and III) (Figure 1B). In addition, we also identified Yke2 (Pfd6), a member of the Gim/prefoldin protein complex involved in the folding of alfa-tubulin, beta-tubulin, and actin and also a member of the RPAP3/R2TP/prefoldin-like complex participating as intermediary of the RNA pol II assembly in humans [13], [26]. Curiously beta-tubulin and Ssb1, a chaperone member of the HSP70 family [27], were present in the affinity-purified Bud27 preparation.
Figure 1. Bud27 physically interacts with the three RNA polymerases.
A) Growth of BUD27-TAP, Wild-type and a Δbud27 cells inYPD at the indicated temperatures. B) Summary of the proteins interacting with Bud27 in a TAP purification analysis. C) Western blot of protein co-purified with Bud27-TAP (3), Rpb3-TAP (2), and non- tagged wild-type strains (1). anti-HA, anti-Myc, anti-Rpb1 (8WG16), and anti-PAP antibodies were used. No tag: wild-type strain BY4741; Rpb3-TAP: Rpb3-TAP, Rpa190-HA (RNA pol I) and Rpc25-Myc (RNA pol III) strain; Bud27-TAP: Bud27-TAP, Rpa190-HA (RNA pol I) and Rpc25-Myc (RNA pol III) strain.
To confirm the interaction between Bud27 and the three RNA polymerases, we purified Bud27-TAP from a strain containing also Rpa190-HA tagged (RNA pol I) and Rpc25-Myc tagged (RNA pol III) proteins. As shown in Figure 1C, an Rpa190-HA reacting band was revealed. No such band was detected when the TAP purification was performed in control strain BY4741 or in control strain YFN229 containing Rpb3 TAP but also tagged forms of the other two RNA polymerases (Rpa190-HA, and Rpc25-Myc), indicating that Rpa190-HA does not interact with the TAP module and that no anti-Rpa190-HA-reacting material was adsorbed nonspecifically to the beads. Similarly, an Rpc25-Myc reacting band (by using anti-Myc antibodies) was noted when Bud27-TAP was purified, but not in the control strain BY4741 or in a control strain containing Rpb3-TAP. Finally, no reacting band was revealed for Rpb1, although it clearly co-purified in a control strain containing Rpb3-TAP. Thus, we cannot rule out that Bud27 and RNA pol II could associate only transiently in vivo or less efficiently. These observations indicate that interactions between Bud27 and the RNA pol I and III are specific, and also suggest that this could similarly account for RNA pol II.
Bud27 shuttles between the nucleus and cytoplasm in an Xpo1-independent manner
Our data demonstrating a physical interaction between Bud27 and the three RNA polymerases, as well as the reported localization of Bud27 in the cytoplasm [19], suggest a role for this prefoldin in the cytoplasmic biogenesis of the RNA pols. To clarify the localization of Bud27 in the cell and to explore if this protein shuttles between cytoplasm and nucleus, as is the case in humans and Drosophila [20], [21], we used a functional BUD27-GFP gene fusion, cloned in a centromeric plasmid, expressed from a Tet-repressible promoter [25]. The functionality of this Bud27-GFP fusion protein was confirmed by its ability to complement the temperature sensitivity of a Δbud27 mutant strain (Figure S1). As shown in Figure 2A, Bud27-GFP was preferentially localized at the cytoplasm.
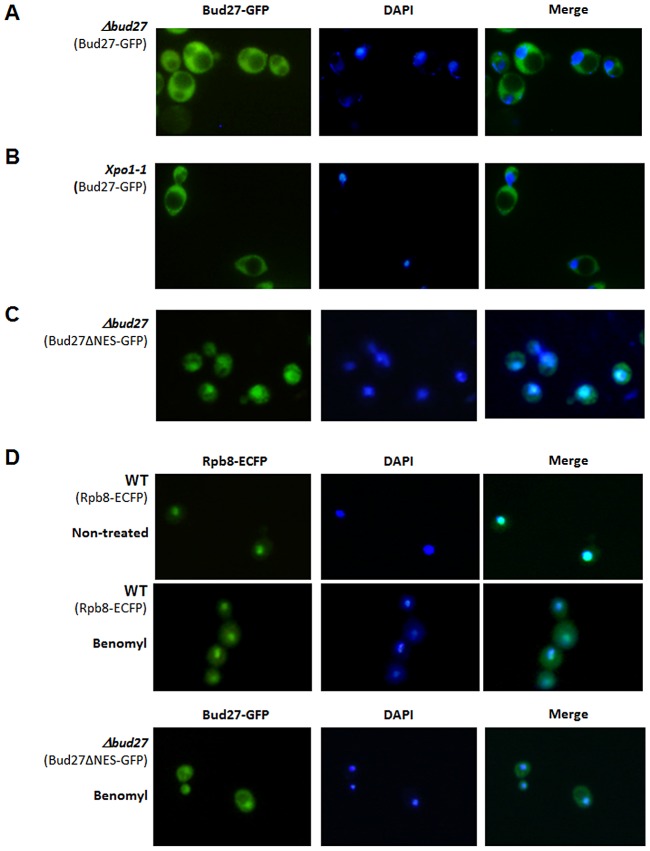
Figure 2. Bud27 shuttles between nucleus and cytoplasm.
Live cell imaging of Bud27-Gfp (centromeric plasmid pCM189) in Δbud27 mutant cells at 30°C (A) and Xpo-1 mutant cells for 1 h at 37°C (B). C) Live cell imaging of Bud27ΔNES-GFP in Δbud27 mutant cells at 30°C, showing nuclear localization. D) Rpb8-ECFP (C-terminal ECFP tagged Rpb8) and Bud27ΔNES-GFP in wild-type or Δbud27 mutant cells treated with benomyl (60 µg/ml) at 30°C.
A more detailed analysis of Bud27 amino acid sequence using the NetNES 1.1 and cNLS mapper servers [28], [29] predicted a possible leucine-rich nuclear export signal (NES) between positions 686 and 695 (LRDEIRDFQL) and a nuclear localization signal (NLS; amino acids 562 to 595). Proteins with a NES signal are actively translocated to the cytoplasm via the action of nuclear export pathway mediated by an evolutionarily conserved CRM1/exportin protein Xpo1, suggesting that Bud27 could shuttle between the nucleus and cytoplasm in an Xpo1-dependent manner. To verify this possibility, we examined the localization of Bud27 in xpo1-1 cells transformed with the above-mentioned plasmid after shifting from 30°C to 37°C for up to 5 h, a condition under which the Xpo1-dependent protein export was blocked [30]. However, no nuclear Bud27-GFP accumulation was found following a shift to 37°C for up to 5 h (Figure 2B).
On the other hand, and as the existence of NES and NLS signals in Bud27 suggest that this is a shuttling protein, we deleted the NES sequence from Bud27-GFP. Our results demonstrated that this led to a nuclear accumulation of Bud27 (Figure 2C). In addition, the deletion of this sequence did not affect the ability of the fusion protein to complement the growth defect caused by the bud27 null mutation (Figure S1). To gain insight into the mechanism by which Bud27 translocates to the nucleus, we analysed Bud27ΔNES-GFP localization after adding benomyl, a drug demonstrated to promote depolarization of microtubules and accumulation or the largest subunit of the RNA pol II, Rpb1, in the cytoplasm [8]. As expected, benomyl led to the accumulation of RNA pols in the cytoplasm, as shown by monitoring Rpb8-ECFP in vivo (Figure 2D upper panel). Contrary, nuclear localization of Bud27ΔNES-GFP is not significantly altered by the addition of benomyl (Figure 2D lower panel).
These data together demonstrate that Bud27 shuttles between nucleus and cytoplasm and suggest that the NES sequence is required for the nuclear export of Bud27 in an Xpo1-independent manner. In addition, the data recorded using benomyl suggest that the translocation of Bud27 to the nucleus is independent of microtubule polarization.
Lack of Bud27 results in RNA polymerases cytoplasmic accumulation
To help elucidate the function of Bud27 and of its association with the RNA pols, and to investigate the effect of Bud27 in the assembly and/or transport of these enzymes to the nucleus, we tested the hypothesis that Bud27 is needed for correct localization of the three RNA pols. The reason to suspect a role for this prefoldin in the cytoplasmic biogenesis of the three RNA pols came from the observation that Bud27 is localized mainly in the cytoplasm of yeast cells while the three RNA pols in the nucleus.
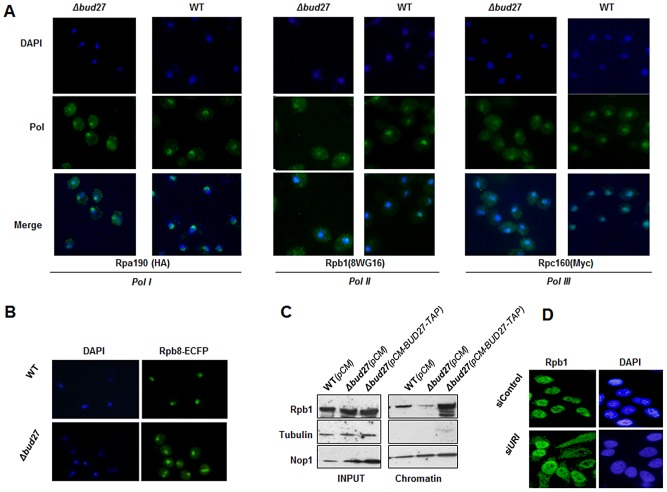
Then we performed immunocytochemistry experiments in a wild-type and a Δbud27 mutant strain containing an Rpa190-HA tagged (RNA pol I) and Rpc160-Myc tagged (RNA pol III) proteins, using anti-HA, anti-Rpb1 (8WG16) and anti-Myc antibodies, to analyse the intracellular localization of the largest subunits of the RNA pol I, II, and III, respectively. In a wild-type strain (Figure 3A), fluorescence for Rpb1 and Rpc160 was restricted to the nucleus, while for Rpa190 it was mainly nucleolar, indicating that, as expected, RNA pol II and III were localized in the nucleus and RNA pol I, mainly in the nucleolus. However, deletion of BUD27 resulted in the accumulation of the three RNA pols in the cytoplasm, although nuclear and nucleolar, but more diffuse, staining was also observed. To corroborate these results and to monitor the localization of another subunit shared by the three RNA polymerases, we genomically tagged Rpb8 with a C-terminal ECFP tag and we monitored its localization by live cell imaging (Figure 3B). As in the case for Rpa190, Rpb1, and Rpc160, fluorescence for Rpb8 was detected mainly in the nucleus of a wild-type strain, while a clear cytoplasmic accumulation was found in a Δbud27 mutant strain. These results indicate that the three RNA enzymes are partially mislocalized, according to the differences in the amount of Rpb1 associated with chromatin fractions (Figure 3C). In addition, nuclear localization of the three RNA pols seems not to depend on Bud27 localization, since Bud27 variant that lacks NES sequence and that is accumulated in the nucleus did not impair the nuclear localization of the RNA pol I, II or III (Figure S2A).
Figure 3. Lack of Bud27 led to RNA pol I, II, and III cytoplasmic accumulation.
A) Immunocytochemistry experiments using antibodies against Rpa190-HA (anti-HA), Rpb1 (8WG16), and Rpc160-Myc (anti-Myc) in wild-type and Δbud27 mutant cells with tagged Rpa190-HA (RNA pol I) and Rpc160-Myc (RNA pol III), at 30°C. B) Live cell imaging of Rpb8-ECFP in wild-type and Δbud27 mutant cells, at 30°C. C) Western blot of chromatin fractions from wild-type and Δbud27 mutant cells harbouring an empty vector (pCM) or a vector overexpressing BUD27 (pCM-BUD27-TAP). Tubulin and Nop1 were used as controls of non-chromatin and chromatin fractions, respectively. D) Rpb1 immunolocalisation analysis (8WG16) in human pulmonary fibroblast under silencing of URI. As a control, cells without siRNA heteroduplex.
Also, we investigated whether cells resumed growth and RNA pols nuclear localization when a Δbud27 mutant strain was complemented with a BUD27-TAP gene fusion into a centromeric plasmid expressed from a Tet-repressible promoter [25]. As expected, BUD27-TAP fully restored growth of the Δbud27 mutant strain at the restrictive temperature of 37°C (Figure S1). In addition, Rpb8-ECFP signal was again restricted to the nucleus, suggesting that the three RNA pols again became nuclear (Figure S2B). Furthermore, these data correlate with an increase in the amount of Rpb1 associated with chromatin fractions in the Δbud27 mutant strain overexpressing the BUD27-TAP gene fusion (Figure 3C).
All together, these data indicate that Bud27 is necessary for correct nuclear localization of RNA pols in the S. cerevisiae nucleus, and suggest that it may play a role in assembly and/or nuclear transport of the three enzymes.
Silencing of URI results in cytoplasmic accumulation of RPB1 in human cells
URI is an evolutionarily conserved member of the prefoldin family among eukaryotes [18], [22]. Then, to start elucidating if in humans URI IS also involved in the biogenesis of the RNA pols, we performed siRNA silencing experiments in human pulmonary fibroblast and monitored the effect of URI depletion on Rpb1 intracellular localization. As shown by q-RT PCR, URI mRNA expression decreased to 40% at 100 nM of siURI (Figure S3). Furthermore, silencing of URI resulted in the accumulation of Rpb1 in the cytoplasm of treated cells (Figure 3D), as revealed by immunocytochemistry experiments using 8WG16 antibodies. Contrary, control experiment did not affect nuclear localization of Rpb1. These data suggest that, as it is the case for Bud27, URI modulates the translocation of RNA pol II to the nucleus, pointing to a common role for these conserved proteins in the biogenesis of, at least, the RNA pol II.
Bud27 is required for correct assembly of the three RNA polymerases
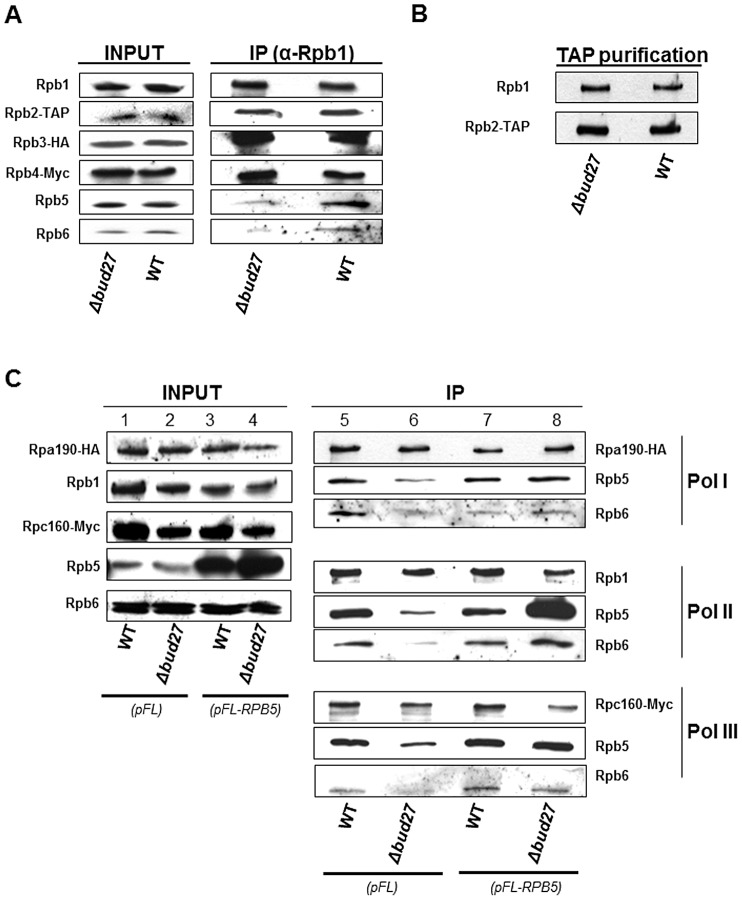
To investigate the effect of Bud27 in RNA pols complex assembly, we immunoprecipitated RNA pol II from a wild-type and a Δbud27 mutant strain containing functional tagged versions of different RNA pol II subunits (Rpb2-TAP, Rpb3-HA and Rpb4-Myc). We performed immunoprecipitation experiments using anti-Rpb1 antibodies (8WG16) and analysed different RNA pol II subunits corresponding to the different assembly intermediate previously described, Rpb1 together with Rpb4/5/7/8/9 and Rpb2 together with Rpb3/10/11/12 [10], [12], [31]. Our results revealed no significant differences in the yield or subunit composition between the two polymerases for Rpb1, Rpb2, Rpb3, or Rpb4 (Figure 4A). It is worth noting that Rpb2 purification gave also similar amount of Rpb1/Rpb2 (Figure 4B).
Figure 4. Lack of Bud27 affects assembly of RNA pol I, II, and III.
A) RNA pol II was immunoprecipitated with anti-Rpb1 antibodies (8WG16) from a strain containing Rpb2-TAP, Rpb3-HA and Rpb4-Myc. RNA pol II subunits were analysed with Anti-HA, anti-Rpb1 (8WG16), anti-Myc, anti-PAP, anti-Rpb5 and anti-Rpb6 antibodies. B) Western blot of Rpb1 co-purified with RPB2-TAP. Anti-Rpb1 (8WG16) and anti-PAP antibodies were used. C) RNA pol I, II and III were immunoprecipitated with anti-HA, anti-Rpb1 (8WG16) and anti-Myc antibodies from a wild-type and a Δbud27 mutant strains containing Rpa190-HA (RNA pol I) and Rpc160-Myc (RNA pol III), transformed with a plasmid overexpressing RPB5 (pFL-RPB5), or with an empty vector (pFL). Rpa190-HA, Rpb1, Rpc160-Myc, Rpb5, and Rpb6 were analysed by Western blot with the antibodies indicated above.
Also, we analysed Rpb5, a common subunit shared by the three RNA polymerases and demonstrated by us and others [18] to physically interact with Bud27. Surprisingly, the amount of Rpb5 clearly decreased in RNA pol II immunoprecipitated from a Δbud27 mutant strain (Figure 4A), indicating that lack of Bud27 affects RNA pol II assembly. To corroborate these results and to investigate whether Bud27 also participates in the assembly of the other two RNA pols (I and III), we immunoprecipitated the three RNA pols from a wild-type and a Δbud27 mutant strain containing functional tagged versions of different RNA pols subunits (Rpa190-HA and Rpc160-Myc). The immunoprecipitation of the three largest RNA pol subunits, using anti-HA, anti-Rpb1 (8WG16), and anti-Myc antibodies, again revealed significant differences in yield between Rpb5 and the largest subunits of the three RNA pols between wild-type and mutant polymerase complexes (Figure 4C, lines 5 and 6 ).
Rpb6, another common subunit to the three RNA pols seems to assemble rather late in humans although this mechanism is unclear [12]. Thus, we also analysed the amount of Rpb6 in immunoprecipitated RNA pol II between the wild-type and Δbud27 mutant strains. Again, surprisingly, the amount of Rpb6 clearly decreased in RNA pol II mutant complex (Figure 4A and 4C, lines 5 and 6), but also in RNA pol I and III (Figure 4C, lines 5 and 6). These data indicate that Bud27 has a role in the assembly of the three RNA pols, by interfering with the correct assembly of Rpb5 and Rpb6 in the complexes. Furthermore, the fact that no significant differences were detected between the wild-type and the mutant strain in whole-cell extracts for any of the RNA pols subunits analysed suggests that when Bud27 lacks, the non-assembled subunits are not rapidly degraded.
To extend our analysis, we performed Rpb3-TAP purification from wild type and Δbud27 mutant containing functional tagged version of Rpb3 (Rpb3-TAP), as Rpb3-TAP purification has been largely and successfully used to purify RNA pol II [32]–[34]. The protein mixture obtained in each case was subjected to multidimensional protein identification technology (MudPIT) [35] and to separation by gel electrophoresis. From our analysis we can conclude that all RNAPII subunits are associated to Rpb3 in absence of Bud27 (Table S1), but however, the yield of Rpb3 recovery drops abruptly in Δbud27 cells. It is also significant the reduction of Rpb3 co-purifying proteins. Taking together, our results suggest that lack of Bud27 led to an instable RNA pol II complex and then, that Bud27 is also necessary to maintain stability of the enzyme.
RPB5 overexpression corrects temperature sensitivity, rapamycin sensitivity, nuclear RNA pols localization, and RNA pols assembly in cells lacking Bud27
The data above strongly indicate that Bud27 is required for assembly of the three RNA pols and that this mechanism depends on correct assembly or stabilization of Rpb5. Because Rpb5 was identified as an interactor with Bud27, we considered the possibility that these proteins are functionally linked.
In an attempt to clarify the relationship between Rpb5 and Bud27, we explored whether RPB5 overexpression corrects the temperature sensitivity of the Δbud27 mutant strain. Notably, increasing dosage of Rpb5 suffices to rescue the growth defect of the strain lacking Bud27 at a restrictive temperature (Figure 5A), as is the case for BUD27 overexpression (Figure S1). Bud27 in yeast and URI in human cells were originally characterized as regulators of gene expression controlled by TOR (for target of Rapamycin) pathway [18]. As opposed to a S. cerevisiae wild-type strain, the Δbud27 mutant was slightly sensitive to Rapamycin (Figure 5A). Again, RPB5 overexpression corrected the sensitivity of the Δbud27 mutant cells to this drug (Figure 5A).
Figure 5. Δbud27 mutant phenotypes are corrected by overexpression of different BUD27 constructions and RPB5.
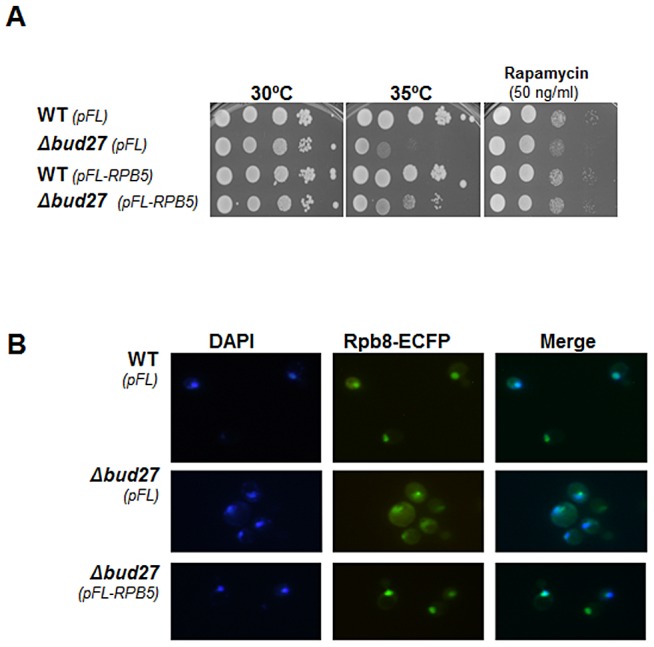
A) Growth of wild-type and Δbud27 mutant strains transformed with vectors overexpressing RPB5, at different temperatures or in the presence of rapamycin. pCM and pFL correspond to the control empty vectors. B) Live cell imaging of Rpb8-ECFP in wild-type and Δbud27 mutant cells at 30°C, containing empty vector (pFL) or overexpressing RPB5 (pFL-RPB5).
Considering the functional connection between BUD27 and RPB5, we investigated whether increasing Rpb5 dosage could also correct Bud27-dependent RNA pols localisation. Then we monitored the localization of the three RNA pols by analysing Rpb8-ECFP by live cell imaging. Rpb8 was observed mainly in the nucleus when RPB5 was overexpressed, as compared to the same strain containing a void plasmid as a control (Figure 5B), as in the case of BUD27 overexpression (Figure S1). Similar results were found by performing immunolocalization experiments using anti-HA, anti-Rpb1 (8WG16) and anti-Myc antibodies in a Δbud27 mutant strain containing functional tagged versions of different RNA pols subunits (Rpa190-HA and Rpc160-Myc; Figure S4). Taken together, our data showing that lack of Bud27 does not impair the nuclear localization of RNA pols under RPB5 overexpression, suggest that Bud27 is not the main requisite for their nuclear import.
Also, we investigated whether RPB5 overexpression was sufficient to rescue the defects in RNA pols assembly observed in the Δbud27 mutant strain that could also explain the correction in RNA pols mislocalization. We immunoprecipitated the three RNA pols from a wild-type and a Δbud27 mutant strain containing functional tagged versions of different RNA pols subunits (Rpa190-HA and Rpc160-Myc) under RPB5 overexpression. Immunoprecipitation of the three largest RNA pols subunits, using anti-HA, anti-Rpb1 (8WG16) and anti-Myc antibodies, revealed no significant differences in the yield between Rpb5 and the largest subunits of the three RNA pols between wild-type and mutant polymerase complexes (Figure 5B) with respect to the same strains harbouring a void plasmid as a control. In addition, similar results were found for Rpb6. These results demonstrate that increasing dosage of Rpb5 is sufficient to correct the RNA pols assembly defects caused by the lack of Bud27, suggesting that it accounts for the rescue of nuclear RNA pols localization.
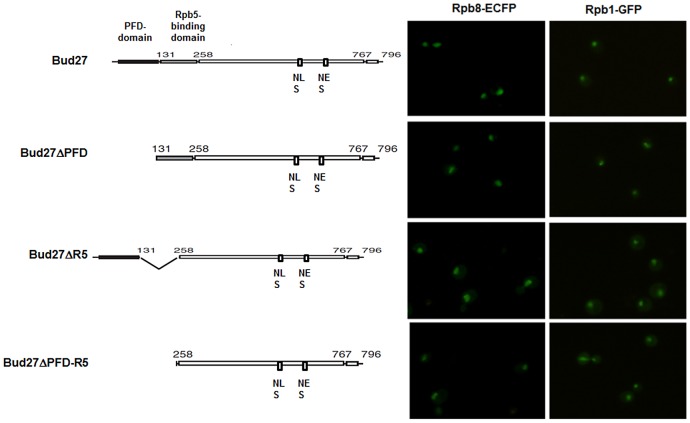
Bud27 Rpb5-binding domain is required for nuclear RNA pols localization, although both Prefoldin and Rpb5-binding domains are dispensable for growth
To test which conserved domains of Bud27 are important for growth, we transformed Δbud27 cells with plasmids expressing full-length Bud27 or proteins deleted for the indicated domains, and tested their ability to complement the temperature-sensitive phenotype (Figure S1). Surprisingly, neither the PFD or the Rpb5-binding domain nor both were functionally important in vivo, as previously indicated [19].
To assess whether the above-mentioned conserved domains participated in the nuclear localization of the three RNA pols, we monitored Rpb8-ECFP and Rpb1-GFP by live cell imaging in a Δbud27cells with plasmids containing full-length BUD27 or BUD27 deleted for the PFD, Rpb5-binding domains, or both. As shown in Figure 6, deletion of Rpb5-binding domain led to a clear Rpb1-GFP cytoplasmic accumulation. Similarly, a more diffuse cytoplasmic Rpb8-ECFP signal was observed. However, PFD-binding domain was not crucial for the correct nuclear localization of the three RNA pols. Thus, we conclude that nuclear localization of the three RNA pols mediated by Bud27 is dependent on Rpb5-binding domain.
Figure 6. PFD, Rpb5-binding domains, or both are dispensable for RNA pol I, II, and III nuclear localization.
Live cell imaging of Rpb8-ECFP and Rpb1-GFP in Δbud27 mutant cells at 30°C, containing vectors overexpressing whole BUD27 or BUD27 deleted for PFD, Rpb5-binding domains or both.
The role of Bud27 in RNA pols biogenesis is independent of the chaperone prefoldin (PFD) complex
URI (Bud27) is believed to function as a scaffold protein able to assemble additional members of chaperone prefoldin (PFD) family through its PFD and Rpb5-binding domains in both human and yeast. Human and yeast PFD is a complex composed of six different subunits, PFD1-PFD6, referred to as the prefoldin/GimC complex, which functions as a molecular chaperone and delivers newly synthesised unfolded proteins to cytosolic chaperonin TRiC/CCT to facilitate the folding of proteins [13], [23]. In humans, PFDs and URI are constituents of an 11-subunit complex associated to the RNA pol II, namely the RPAP3/R2TP/prefoldin-like complex, suggested to participate in RNA pol II assembly [12], [13]. In addition, data from yeast and mammalian cells have shown that the functions of the prefoldin and CCT chaperone complexes are eliminated by removal of individual subunits [36].
To evaluate whether the deletion of other components of the prefoldin complex in yeast also participate in the RNA pols nuclear localization, we analysed Rpb8-ECFP and Rpb1-GFP localization by live cell imaging in strains deleted for different prefoldins: YKE2 (PFD6), shown by us and others [23] to physically interact with Bud27, with Rpb5 [24] and genetically with other components of the RNA pol I and III machinery [37], [38]; GIM6 (PFD1), which genetically interacts with BUD27 [19] and with subunits of RNA pol I and III machinery [37], [39], [40]; GIM4 (PFD2), which interacts neither with BUD27 nor with the RNA pols. Surprisingly, in contrast to Bud27, these three components of the prefoldin complex are dispensable for the nuclear localization of the three RNA pols at 30°C or even at 37°C (Figure 7, for 30°C), indicating that the role of Bud27 in the biogenesis of the RNA pols seems to be independent from the rest of the prefoldin complex.
Figure 7. RNA pol I, II, and III nuclear localization is independent of the chaperone prefoldin (PFD) complex.
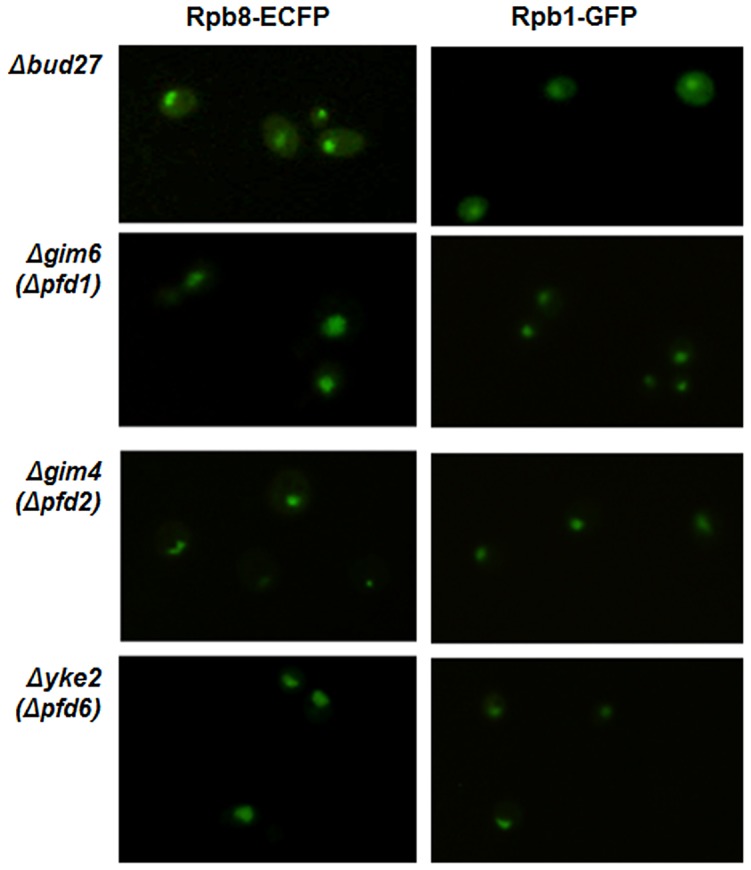
Live cell imaging of Rpb8-ECFP and Rpb1-GFP in Δbud27, Δgim6 (Δpfd1), Δgim4 (Δpfd2), and Δyke2 (Δpfd6) mutant cells containing C-terminal ECFP tagged Rpb8 and C-terminal GFP tagged Rpb1, at 30°C.
The role of Bud27 in RNA pols biogenesis is independent of Iwr1
The conserved protein Iwr1 was originally identified as a protein that co-purified with almost every subunit of RNA pol II and that interacts with the basal transcription machinery and regulates the transcription of specific genes [30]. Recently, it has been shown that Iwr1 specifically binds RNA pol II between Rpb1 and Rpb2 and directs its nuclear import, although it is not involved in RNA pol I or III transport [6], [10]. Curiously, IWR1 genetically interacts with BUD27 [41] and RPB5 [42] and physically with Bud27 [24] although we could not reproduce this interaction by our TAP purification. Based on these data, and to rule out the possibility that accumulation of RNA pol II in the cytoplasm in cells lacking Bud27 is not an indirect effect due to the mislocation of Iwr1, we analysed localization of Iwr1 in Δbud27 cells. For this experiment, cells were transformed with a plasmid expressing an Iwr1 protein lacking its NES domain and shown to accumulate in the nucleus, instead of a plasmid with entire IWR1, since it leads to a diffuse signal and no clear nuclear localization [30]. As shown (Figure 8), deletion of BUD27 does not affect the nuclear localization of Iwr1-ΔNES. These data suggest that nuclear localization of Iwr1 does not require interaction with Bud27.
Figure 8. Iwr1 nuclear localization is independent of Bud27.
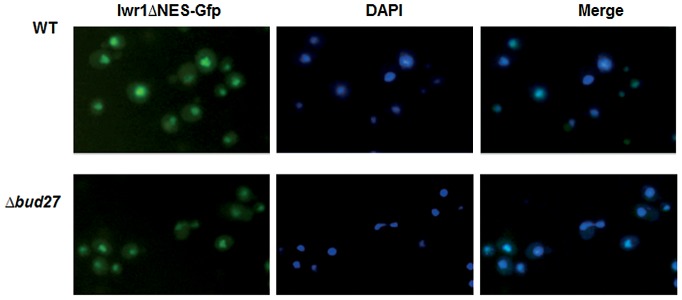
Live cell imaging of Iwr1-Gfp deleted for its NES domain in wild-type and Δbud27 cells at 30°C.
Discussion
Human URI/RMP and its orthologue in yeast, Bud27, is the most studied member of the prefoldin-like (PFD) family of ATP-independent molecular chaperones, also called unconventional prefoldin Rpb5 interactor [18]. Based on recent data of large-scale proteomic screen of RNA polymerases in human cells, URI has been shown to be a component of the HSP90/R2TP complex and has been proposed to participate in the biogenesis of RNA polymerases [8], [12]. However, this role has not been confirmed. In this work, we show evidence that Bud27 in S. cerevisiae is the first example of a protein that participates in the biogenesis of all RNA polymerases, and mediates the assembly of the three transcriptional complexes. Furthermore, Bud27 seems not to play a role in the nuclear transport of the RNA pols. Our data also suggest similar role for human URI.
Consistent with a role for Bud27 in RNA pols biogenesis, we demonstrated by TAP purification and immunoprecipitation experiments that Bud27 interacts with different subunits of the three RNA pols. Our data agrees with those of Krogan et al. showing physical interactions between Bud27 and RNA pol II [24] or between URI or Bud27 and Rpb5, a common subunit shared by the three RNA pols [14], [18]. Moreover other authors identified, in human cells, URI as a component of the R2TP/prefoldin-like complex which binds the largest subunits of the RNA pol II [12], [43]. Finally, in accordance with these physical associations, genetic interactions between BUD27 and different components of the three transcriptional machineries have been found in yeast by us (Mirón-García, unpublished data) and others [44].
Biochemical and structural studies of RNA pols have proposed a detailed model of these enzymes, but however, little is known on how they assemble into the complexes or how they are transported from the cytoplasm to the nucleus. Our work provides new data to elucidate the mechanisms governing RNA pols biogenesis and localization and complement those in human and yeast [6]–[8], [12]. Bud27 is the first protein so far demonstrated to participate in the biogenesis of the three RNA pols, modulating their assembly. Accumulation of the three RNA pols in the cytoplasm, as a consequence of BUD27 deletion accounts for a defect in nuclear transport. However, lack of Bud27 does not impair RNA pols nuclear localization under RPB5 overexpression, suggesting that Bud27 is dispensable for their nuclear import. Only two proteins in S. cerevisiae had been shown to participate in RNA pol II biogenesis so far, Iwr1 and Npa3, and only the Npa3 homologue RPAP4/GPN1 in humans. Notably, these proteins are necessary for nuclear transport but none of them are involved in pol assembly [6]–[8].
Our immunoprecipitation experiments in Δbud27 mutant strain and the analysis of different subunits corresponding to the different assembly intermediate previously described [12], [31], demonstrate that Bud27 mediates the assembly of the three RNA pols. In fact, the lack of Bud27 alters the correct assembly of Rpb5 and Rpb6 in the three RNA complexes and led to a more instable enzyme. Results are also consistent with RNA pols assembly in the cytoplasm as a prerequisite for their nuclear import and agree with recent observations in yeast and humans [6], [8], [12]. In addition, co-purification and gel filtration analysis (unpublished data) suggest that RNA pols intermediaries do not appear in yeast in the absence of Bud27. These data, together with the fact that only differences in the yield of Rpb5 and Rpb6 were observed and that RPB5 overexpression corrects not only assembly but also nuclear RNA pols transport, point to the fact that Bud27 mediates RNA pols assembly in an Rpb5-dependent manner, in agreement with data confirming its role in protein folding [23]. Our data point to a role of Bud27 in the correct folding of Rpb5 to the rest of the complex, since lack of Bud27 leads to transcription defects not related to a decrease in nuclear RNA pols amount but to RPB5-dependent processes (Mirón-García, in preparation). Furthermore, we cannot rule out that Bud27 could act to stabilize the interaction between Rpb5 and the rest of the RNA pols complexes, and hence their integrity. Interestingly, the Rpb5-binding domain but not the conserved PFD-binding domain of Bud27 is essential for this role, since only Δbud27 cells expressing a version of Bud27 lacking the Rpb5-binding domain show cytoplasmic RNA pols localization. In addition, the defect in Rpb6 assembly is also consistent with the fact that RPB6 overexpression partially corrects the temperature sensitivity of the Δbud27 mutant (our unpublished data) and with the physical contact between Rpb5 and Rpb6 on the RNA pol II structure [45]. Furthermore, as BUD27 is not essential and its deletion seems only to affect part of the RNA pols complexes, it appears that Bud27 participates in coordination with other proteins to address its role in RNA pols biogenesis. These results also provide information concerning the mechanisms governing the assembly of Rpb5 and Rpb6 into the rest of the complexes. Our data suggest that Rpb6 could assemble rather late once Rpb5 is assembled, as previously proposed in human [12]. However, we cannot disregard the possibility that it assembles before and Rpb5 would be necessary to stabilize its contact with the other components of the RNA pols. Alternatively, we cannot rule out that Rpb5 could be assembled in early steps of the RNA pols biogenesis participating in maintaining the integrity of the RNA pols, although this possibility seems less unlikely.
As is the case for Bud27 in yeast, HSP90 and its cochaperone RPAP3 in human cells have been shown to coordinate the assembly of RNA pol II and its involvement in RNA pol I and III assembly has been suggested [43]. Curiously, HSP90 and RPAP3 are members of the RTP2/prefoldin-like complex, a molecular machine dedicated to the assembly of multi-molecular protein complexes, such as the RNA pol II. It contains 11 components, among them URI and Rpb5. On the contrary, in yeast the R2TP complex contains five proteins and Rpb5 and Bud27 have not been found as bona-fide constituents [43]. Interestingly, prefoldin 6 (Yke2), which is part of the human RTP2 complex also appears as a Bud27 interactor when purified via TAP. Thus is temptating to speculate that in yeast, also Bud27 and Rpb5 could associate with the RT2P complex. Moreover, physical interactions between Bud27 and a member of the Hsp70 family chaperones (Ssb1), as well as with the beta-tubulin chain of the microtubules (Tub2) have been identified (Figure 1B). This is consistent with previous data from two-hybrid analysis [19], [23]. Notably, polymerization of tubulins into microtubules requires prefoldins and chaperonin CCT complex, which has been shown to interact with RNA pol II subunits [8], [36]. Impairing microtubule assembly, both in humans and yeast, leads to a RNA pol II mislocalization in the cytoplasm [8]. Thus it is possible that these interactions are functional, althought further work will be necessary to address this issue.
As shown here RNA pols nuclear localization is dependent on Rpb5-binding domain. However, Bud27 PFD and Rpb5-binding conserved domains are not required for growth, neither for its role in translation [19]. Then, these domains could be also important for other roles of Bud27 in the nucleus, such as for its interaction with other transcriptional regulators [14], [16], [20]. Consistent with this possibility Bud27 shuttles between the cytoplasm and nucleus via an Xpo1-independent pathway. Discrepant results have been reported concerning Bud27 localization. According to Desplaces et al., Bud27 is excluded from the nucleus in yeast [19]. However, in humans and Drosophila [20], [21] nuclear localization for Bud27 has also been reported. Moreover, physical association between Rpb5, Bud27, and transcription factors TFIIF, as well as between Rpb5 and TFIIB [14], [16], let us to propose that Bud27 could compete with these transcription factors to bind Rpb5.
Finally, silencing experiments in human cells, account for a conserved role of URI in RNA pols biogenesis, suggesting similar mechanisms that must be deciphered.
Materials and Methods
Yeast strains, plasmids, genetic manipulations, media, and genetic analysis
Common yeast media, growth conditions, and genetic techniques were used as described elsewhere [46]. Rapamycin (LCLAbs, USA) and Benomyl (Sigma-Aldrich) was used at the indicated concentrations.
Strains, plasmids and primers are listed in Table 1, Table 2, and Table 3.
Table 1. S. cerevisiae strains.
| Strain | Genotype | Origin |
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Euroscarf |
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Euroscarf |
| Y01246 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 YJL179W(GIM6, PFD1)::kanMX4 | Euroscarf |
| Y00243 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 YEL003W(GIM4,PFD2)::kanMX4 | Euroscarf |
| Y04149 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YLR200W(YKE2, PFD6)::kanMX4 | Euroscarf |
| Y05642 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YFL023W(BUD27)::kanMX4 | Euroscarf |
| ATCC201388 (BUD27-GFP) | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YFL023W(BUD27)::GFP::HIS3MX6 | Invitrogen |
| YSC1178 (BUD27-TAP) | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YFL023W(BUD27)::TAP::HIS3MX6 | Open Biosystems |
| YSC1178 (RPB2-TAP) | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YOR151C(RPB2)::TAP::HIS3Mx6 | Open Biosystems |
| W303-1A | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 | [52] |
| W303-1B | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 | [52] |
| YPH500 | MATα ade2-101 his3-Δ200 leu2Δ1 lys2-801 trp1-Δ63 ura3-52 | [53] |
| MW3522 | MATa ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 YOR341W(RPA190)::3HA::HIS3 | [49] |
| XPO1 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 YGR218W(XPO1)::LEU2 [pKW440 (XPO1 in pRS313)] | [54] |
| xpo1-1 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 YGR218W(XPO1)::LEU [pKW457 (xpo1-1 in pRS313)] | [54] |
| YVV50-4c | MATa ade2-101 his3-Δ200 leu2Δ1 lys2-801 trp1-Δ63 ura3-52 YIL021W(RPB3)::HA::KanMX4 | [55] |
| D473-4A | MATα ade2-1 his3-11,15 leu2Δ1 lys2-801 trp1-Δ63 ura3-1 YBR154C(RPB5)::URA3::LEU2 [ pFL44L-RPB5] | Gift from P. Thuriaux |
| LMY3.1 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YDR007W(TRP1)::kanMX4; YJL140W(RPB4)::Myc18::TRP1 | Gift from S. Chávez |
| MW3608 | MATα ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 YOR116C(RPC160)::13Myc::TRP | Gift from M. Werner |
| SL876a | MATa his3-Δ200 leu2-3,112 trp1-Δ63 ura3-52 YIL021W(RPB3)::TAP::KIURA | Gift from P. Thuriaux |
| YCZ106 | MATα ade2-101 his3-Δ200 leu2Δ1 lys2-801 trp1-Δ63 ura3-52 YKL144C(RPC25)::Myc::KanMX6 | Gift from P. Thuriaux |
| D495-1c | MATa his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 YKL144C(RPC25)::Myc::KanMX6 YIL021W(RPB3)::TAP::URA3 YOR341W(RPA190)::3HA::HIS3 | This workYFN56×YCZ106 |
| D610-11A | MATα his3Δ1 leu2-3,112 lys2-801 met15Δ0 trp1-Δ63 ura3-1 YFL023W(BUD27)::kanMX4 YBR154C(RPB5)::URA3::LEU2 [ pFL44L-RPB5] | This workY05642×D473-4A |
| YFN56 | MATa his3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52 YIL021W(RPB3)::TAP::KIURA YOR341W(RPA190)::3HA::HIS3 | This workSL876a×MW3522 |
| YFN105 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 trp1-Δ63 YFL023W(BUD27)::kanMX4 | This workBY4741×D610-11A |
| YFN106 | MATα his3Δ1 leu2-3,112 lys801 met15Δ0 ura3Δ0 trp1-Δ63 YFL023W(BUD27)kanMX4 | This workBY4741×D610-11A |
| YFN223 | MATα ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 YOR341W(RPA190)::3HA::HIS3 YOR116C(RPC160)-13Myc::TRP | This workD495-1c×MW3608 |
| YFN229 | MATα ade2-1 his3-11,15 leu2-3,112 lys2-801 trp1-1 ura3-1 YKL144C(RPC25)::Myc::KanMX6 YOR341W(RPA190)::3HA::HIS3 YIL021W(RPB3)::TAP::URA3 | This workD495-1c×W303-1A |
| YFN233 | MATa ade2-101 his3-Δ200 leu2Δ0 trp1-Δ63 ura3Δ0 YOR341W(RPA190)::3HA::HIS3 YOR116W(RPC160)-13Myc::TRP YFL023W(BUD27)::kanMX4 | This workYFN223×YFN105 |
| YFN255 | MATa ade2-1 his3-11,15 leu2Δ0 lys2-801 trp1-1 ura3-1 YKL144C(RPC25)::Myc::KanMX6 YOR341W(RPA190)::3HA::HIS3 YFL023W(BUD27)::TAP::HIS3MX6 | This workYFN229×YSC1178 (BUD27-TAP) |
| YFN289 | MATα his3-11,15 leu2-3,112 met15Δ0 trp1-1 ura3-1 YOR151C(RPB2)::TAP::HIS3Mx6 | This workYSC1178(RPB2-TAP)×W303-1B |
| YFN308 | MATa his3Δ1 leu2Δ0 met15D0 trp1-1 ura3Δ0 YOR151C(RPB2)::TAP::HIS3Mx6 YJL140W(RPB4)::Myc18::TRP1 | This workYFN289×LMY3.1 |
| YFN323 | MATα ade2-101 his3Δ1 leu2-3,112 lys2-801 ura3-52 YFL023W(BUD27)::kanMX4 YIL021W(RPB3)::HA::KanMX4 | This workYFN106×YVV50-4c |
| YFN329 | MATα his3Δ1 leu2-3,112 lys2-801 trp1-Δ63 ura3-52 YFL023W(BUD27)::kanMX4 YIL021W(RPB3)::HA::KanMX4 YOR151C(RPB2)::TAP::HIS3Mx6 YJL140W(RPB4)::Myc18-TRP1 | This workYFN323×YFN308 |
| YFN331 | MATa his3Δ1 leu2-3,112 lys2-801 trp1-Δ63 ura3Δ0 YIL021W(RPB3)::HA::KanMX4 YOR151C(RPB2)::TAP::HIS3Mx6 YJL140W(RPB4)::Myc18-TRP1 | This workYFN323×YFN308 |
| YFN334 | MATa his3Δ1 leu2Δ0 met15Δ0 trp1-Δ63 ura3Δ0 YFL023W(BUD27)::kanMX4 YOR224C(RPB8)::ECFP::SpHIS5 | This work |
| YFN335 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 lys2Δ YOR224C(RPB8)::ECFP::SpHIS5 | This work |
| YFN348 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 YJL179W(GIM6, PFD1)::kanMX4 YOR224C(RPB8)::ECFP::SpHIS5 | This work |
| YFN349 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 YEL003W(GIM4, PFD2)::kanMX4 YOR224C(RPB8)::ECFP::SpHIS5 | This work |
| YFN350 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YLR200W(YKE2, PFD6)::kanMX4 YOR224C(RPB8)::ECFP::SpHIS5 | This work |
| FY86 | MATα leu2Δ1 ura3-52 his3Δ200 RPB1-GFP::his5 | Gift from F. Estruch |
| YFN416 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 RPB1-GFP::his5 | This work |
| YFN417 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 trp1-Δ63 YFL023W(BUD27)::kanMX4 RPB1-GFP::his5 | This work |
| YFN418 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 YJL179W(GIM6, PFD1)::kanMX4 RPB1-GFP::his5 | This work |
| YFN419 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 YEL003W(GIM4,PFD2)::kanMX4 RPB1-GFP::his5 | This work |
| YFN420 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YLR200W(YKE2, PFD6)::kanMX4 RPB1-GFP::his5 | This work |
Table 2. Plasmids.
| Name | Yeast markers | Origin |
| pCM189 | ORI (CEN) URA3 | [25] |
| pCM189-BUD27 | ORI (CEN) URA3 | This work |
| pCM189-BUD27-GFP | ORI (CEN) URA3 | This work |
| pCM189-BUD27ΔNES -GFP | ORI (CEN) URA3 | This work |
| pCM189-BUD27-TAP | ORI (CEN) URA3 | This work |
| pCM189-BUD27ΔPFD-TAP | ORI (CEN) URA3 | This work |
| pCM189-BUD27ΔR5-TAP | ORI (CEN) URA3 | This work |
| pCM189-BUD27ΔPFDR5-TAP | ORI (CEN) URA3 | This work |
| pFL44L | ORI (2 µm) URA3 | [56] |
| pFL44L-RPB5 | ORI (2 µm) URA3 | [57] |
| pCM189-IWR1ΔNES | ORI (CEN) LEU2 | Gómez-Navarro et al, submitted |
| pKT210 | [48] |
Table 3. Primers.
| Name | Sequence |
| Rpb8ECFP-501 | GAAATTTGAATAACTTGAAGCAAGAGAACGCTTATCTTTTGATTCGTCGTggtgacggtgctggttta |
| Rpb8ECFP-301 | CACTTTTATAAAGTATTATTTATATTACTAGTAGCAGTAAGTGATCGCCC tcgatgaattcgagctcg |
| Rpb1-508 | CCCAACATCTCCAGGCTACAGC |
| Rpb1-310 | CGCAAGCCATGATTACTGG |
| hsURI1_02 | GCCUGAUAAAUUGUCUUAUUU |
| hsURI1_02_as | AUAAGACAAUUUAUCAGGCUU |
| hsURI1E9-f01 | ATTGACGACGATGATGGTGA |
| hsURI1E10-r01 | GCCAGTGCTGTTCTTTCGTT |
| PPIA 501 | TTCATCTGCACTGCCAAGAC |
| PPIA 301 | TCGAGTTGTCCACAGTCAGC |
Protein tagging
Rpb8-ECFP and Rpb1-GFP tagging was performed by yeast recombination of a PCR fragment as described in Longtine et al. [47] amplified from plasmid pKT210 [48] or from chromosomal DNA from strain FY86, using Rpb8ECFP-501 and Rpb8ECFP-301 or Rpb1-308 and Rpb1-310 primers, respectively (see Table 3).
Protein immunoprecipitation and TAP purification
400 ml cells growing exponentially (A600∼0.6–0.8) in yeast extract-peptone-dextrose (YPD) medium or synthetic minimal (SD) were washed twice with ultrapure water and lysis buffer (50 mM HEPES [pH 7.5], 120 mM NaCl, 1 mM EDTA, 0.3% Chaps 50%). Cells were resuspended in 1 ml lysis buffer supplemented with 1x protease inhibitor cocktail (Complete, Roche), 0.5 mM PMSF, 2 mM sodium orthovanadate and 1 mM sodium fluoride and whole-cell extracts were prepared using a MixerMill MM400 RETSCH (3 min 30 Hz). Immunoprecipitations were carried out as described elsewhere [49] with some modifications: 150 µl of whole-cell extract (2000 µg) and lysis buffer for all washes were used. 35 µl of Dynabeads M-280 Sheep anti-Mouse IgG (Invitrogen) were used with 9E10 anti C-Myc antibody (1 µg, Santa Cruz Biotechnology), 8WG16 anti-Rpb1 antibody (1.5 µg, Covance) and 12CA5 anti-HA antibodies (0.4 µg, ROCHE). For TAP purification, the same protocol was used with Dynabeads Pan Mouse IgG (Invitrogen). The affinity-purified proteins were released from the beads by boiling for 10 min. Eluted proteins were analysed by Western blot with different antibodies: 9E10 anti-C-Myc, 12CA5 anti-HA, 8WG16 anti-Rpb1, PAP, anti-POLR2C (1Y26, Abcam), and anti-Rpb6 or anti-Rpb5 (a gift from M. Werner).
TAP purification for protein identification by mass spectrometry was performed as described previously [50]. Bud27-TAP fusion protein and associated proteins were recovered from cell extracts by affinity selection on an IgG matrix. After washing, the TEV protease is added to release the bound material. The eluate is incubated with calmodulin-coated beads in the presence of calcium. After washing, the bound material is released with EGTA. This enriched final fraction was analyzed by mass spectrometry using the MudPIT approach as described in [35].
Immunolocalization and fluorescence microscopy
Cells were grown at 30°C in YPD or SD medium (A600∼0.5–0.7), fixed with 37% w/v formaldehyde at room temperature for 2 h with slow shaking, and then centrifuged and washed twice with PBS. Cells were resuspended in spheroplasting buffer (1.2 M sorbitol, 0.1 M K-phosphate buffer pH 6.5) and cell wall digested with 125 µg/ml zymolyase 20T (USBiological) and 22.7 mM 2-mercaptoethanol (SIGMA) by incubation for 1 h at 37°C without shaking. The spheroplasts were washed twice with PBST (PBS with 0.05% Tween 20) and then resuspended in the same solution. Cell suspension was added to an AAS (3-aminopropyltriethoxysilane, Sigma) slide, incubated at room temperature until slide was dry and washed twice with PBST. Then, 50 µl of PBS-BSA (1 mg/ml BSA) were added, and the slides. After incubation for 30 min in a humid chamber were washed three times with PBS. Next, 50 µl of 1∶100 dilution of the primary antibodies (8WG16, anti C-Myc or anti-HA) in PBS-BSA (1 mg/ml BSA) were added and incubated 2 h at room temperature in a humid chamber. Slides were then washed three times with PBS, and incubated for 1 h, in the dark, at room temperature in a humid chamber with 50 µl of 1∶100 dilution of secondary antibody (Cy2 antimouse; Jackson Labs). The slides were washed three times with PBS and incubated for 5 min with 50 µl of 1 µg/ml DAPI (in PBS). After washing three times with PBS, slides were finally covered with a Vectashield (Vector Laboratories) mounting solution.
Human cells were fixed for 15 min at room temperature with 4% (v/v) paraformaldehyde in PBS. Following fixation, cells were washed three times for 15 min with PBS, treated with 50 mM ammonium chloride for 30 min and permeabilised for 20 min with PBS containing 0.1% (v/v) Triton X-100 (wash solution). Blocking solution (wash solution with 5% (w/v) BSA) was then added for 30 min and then, cells incubated for 1 h with 1∶100 dilution of the primary antibodies (8WG16) in blocking solution. Cells were washed three times for 15 min with wash solution and then incubated with 1∶100 dilution of secondary antibody (Cy2 antimouse; Jackson Labs) for 1 h in blocking solution. After washing three times with wash solution, 15 min each, slides were finally covered with a Vectashield (Vector Laboratories) mounting solution containing DAPI.
The fluorescence intensity was scored with a fluorescence microscope (Olympus BX51).
Chromatin isolation
Chromatin isolation was performed as previously described [51] with some modifications. Briefly, about 5×108 cells growing exponentially (A600∼0.6–0.8) were resuspended in 3 ml of 100 mM PIPES/KOH (pH 9.4) containing 10 mM DTT and 0.1% sodium azide and then incubated at room temperature for 10 min. Cells were spun down, resuspended in 2 ml of 50 mM phosphate buffer (pH 7.5), containing 0.6 M Sorbitol, 10 mM DTT, and 4 µl of 20 mg/ml zymoliase and incubated 10 min at 37°C in a water bath to spheroplast formation. Spheroplasts were then pelleted at 4°C, washed with 50 mM HEPES-HOK buffer (pH 7.5) containing 100 mM KCl, 2.5 mM MgCl2 and 0.4 M Sorbitol, resuspended in equal volume (∼80 µl) of EBX buffer (50 mM HEPES/KOH (pH 7.5), 100 mM KCl, 2.5 mM MgCl2, 0.25% Tritón-X100, 0.5 mM PMSF, 0.5 mM DTT, cocktail protease inhibitors Complete Roche 1x) and incubated for 3 min on ice. This whole cell extract was laid onto 400 µl of EBX-S buffer (EBX with sucrose 30%) and centrifuged at 12000 rpm for 10 min. After the sucrose gradient a chromatin pellet became visible and was washed with 400 µl of EBX buffer and finally resuspended in 100 µl of the same solution. A 1/10 dilution of chromatin pellet was used for SDS-PAGE and Western blotted with antibodies against Rpb1 (8WG16), alfa-tubulin (T5168; Sigma-Aldrich) and Nop1 (28F2; Abcam).
RNA silencing (siRNA)
URI, prefoldin-like chaperone (Gene ID: 8725) gene was silenced by transfection with the siRNA heteroduplex hsURI1_02 and hsURI1_02_as (Table S1). Human pulmonary fibroblast (HPF) were cultured in growth medium (GM), consisting of DMEM supplemented with 10% fetal bovine serum, 2 mM l-glutamine, and 50 U/ml penicillin–streptomycin. Cells were seeded in twenty four-well plates (30,000 cells per well) and transfected in triplicate with 40–200 nM heteroduplex oligonucleotides using the Lipofectamine 2000 Transfection Reagent (Invitrogen), following the manufacturer's protocol, and incubated for 24 h at 37°C. Control cells were treated in the same conditions without siRNA heteroduplex. The experiments were performed three times (three replicates). After indicated time, cells were either harvested for RNA extraction or fixed with PFA 4% in PBS for inmunolocalisation analysis.
RNA isolation and quantitative real-time PCR (q–RT–PCR)
Total RNA was isolated from URI silenced HPF cells using the SV Total RNA Isolation System (Promega), according to the manufacturers. Retrotranscription was performed from 200 ng of total RNA using the Maxima First Strand cDNA Synthesis Kit (Fermentas) in a final volume of 20 µl, according to the manufacturer's protocol. As a control, each sample was subjected to the same process without reverse transcriptase.
URI mRNA accumulation was analyzed by q-RT-PCR with oligonucleotides hsURI1E9-f01 and hsURI1E10-r01 using cDNA corresponding to 10 ng. Human cyclophilin A (PPIA) was used as an internal control. Each PCR reaction was performed at least three times, with three independent samples. All oligonucleotides used are indicated in Table 3.
Supporting Information
Δbud27 mutant phenotypes are corrected by overexpression of different BUD27 constructions. Growth of wild-type and Δbud27 mutant strains transformed with different constructions containing whole BUD27 or deleted forms of BUD27, at different temperatures; pCM and pFL correspond to the control empty vectors.
(TIF)
RNA pols localisation. A) Deletion of Bud27 NES domain does not impair RNA pols nuclear localisation (Rpb8-ECFP). Δbud27 mutant cells overexpressing Bud27ΔNES-GFP were grown at 30°C and localisation of Bud27ΔNES-GFP and Rpb8-ECFP was analysed in vivo. B) BUD27 overexpression (pCM-BUD27-TAP) corrects nuclear localization of Rpb8-ECFP in Δbud27 mutant cells at 30°C when compared to the same strain containing an empty plasmid (pCM).
(TIF)
siRNA silencing of URI. URI mRNA accumulation was determined in human pulmonary fibroblast grown in GM medium at 37°C transfected with 40, 100 and 200 nM of siRNA(URI) heteroduplex. CONTROL cells were treated in the same conditions without siRNA heteroduplex.
(TIF)
RPB5 overexpression rescues nuclear RNA pols localisation. Immunocytochemistry experiments using antibodies against Rpa190-HA (anti-HA), Rpb1 (8WG16), and Rpc160-Myc (anti-Myc) in Δbud27 mutant cells with tagged Rpa190-HA (RNA pol I) and Rpc160-Myc (RNA pol III), at 30°C, transformed with a plasmid overexpressing RPB5 (pFL-RPB5), or with an empty vector (pFL).
(TIF)
Rpb3-TAP purification from wild type and Δbud27 mutant containing functional tagged version of Rpb3 (Rpb3-TAP). The protein mixture obtained in each case was subjected to multidimensional protein identification technology (MudPIT) [35] and their ratios versus Rpb3 calculated.
(DOC)
Acknowledgments
This work is dedicated to the memory of Pierre Thuriaux. We thank Dr. M. Werner for providing antibodies, Dr. F. Estruch for IWR1 constructions, and the “Centro de Intrumentación Científico-Técnico, CICT; Universidad de Jaén.”
Funding Statement
This work was supported by grants from the Spanish Ministry of Education and Science, Ministry of Science and Innovation, and FEDER (BFU2010-21975-C03-02 Spain; http://www.idi.mineco.gob.es/portal/site/MICINN/) to FN and SR-N (BFU2011-23418; http://www.idi.mineco.gob.es/portal/site/MICINN/) and from Junta de Andalucía (BIO258, P10-CVI6521, P08-CVI-03508; http://www.juntadeandalucia.es/organismos/economiainnovacioncienciayempleo.html) to FN. AIG-G and VG-M are recipients of predoctoral fellowships from MEC, and FH-T of postdoctoral fellowships from Junta de Andalucía. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Werner F, Grohmann D (2011) Evolution of multisubunit RNA polymerases in the three domains of life. Nat Rev Microbiol 9: 85–98. [DOI] [PubMed] [Google Scholar]
- 2. Zaros C, Briand JF, Boulard Y, Labarre-Mariotte S, Garcia-Lopez MC, et al. (2007) Functional organization of the Rpb5 subunit shared by the three yeast RNA polymerases. Nucleic Acids Res 35: 634–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Werner M, Thuriaux P, Soutourina J (2009) Structure-function analysis of RNA polymerases I and III. Curr Opin Struct Biol 19: 740–745. [DOI] [PubMed] [Google Scholar]
- 4. Cramer P, Armache KJ, Baumli S, Benkert S, Brueckner F, et al. (2008) Structure of eukaryotic RNA polymerases. Annu Rev Biophys 37: 337–352. [DOI] [PubMed] [Google Scholar]
- 5. Fernandez-Tornero C, Bottcher B, Riva M, Carles C, Steuerwald U, et al. (2007) Insights into transcription initiation and termination from the electron microscopy structure of yeast RNA polymerase III. Mol Cell 25: 813–823. [DOI] [PubMed] [Google Scholar]
- 6. Czeko E, Seizl M, Augsberger C, Mielke T, Cramer P (2011) Iwr1 Directs RNA Polymerase II Nuclear Import. Mol Cell 42: 261–266. [DOI] [PubMed] [Google Scholar]
- 7. Staresincic L, Walker J, Dirac-Svejstrup AB, Mitter R, Svejstrup JQ (2011) GTP-dependent binding and nuclear transport of RNA polymerase II by NPA3. J Biol Chem [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Forget D, Lacombe AA, Cloutier P, Al-Khoury R, Bouchard A, et al. (2010) The protein interaction network of the human transcription machinery reveals a role for the conserved GTPase RPAP4/GPN1 and microtubule assembly in nuclear import and biogenesis of RNA polymerase II. Mol Cell Proteomics 9: 2827–2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carre C, Shiekhattar R (2011) Human GTPases Associate with RNA Polymerase II To Mediate Its Nuclear Import. Mol Cell Biol 31: 3953–3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wild T, Cramer P (2012) Biogenesis of multisubunit RNA polymerases. Trends Biochem Sci 37: 99–105. [DOI] [PubMed] [Google Scholar]
- 11. Egloff S, Zaborowska J, Laitem C, Kiss T, Murphy S (2012) Ser7 phosphorylation of the CTD recruits the RPAP2 Ser5 phosphatase to snRNA genes. Mol Cell 45: 111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boulon S, Pradet-Balade B, Verheggen C, Molle D, Boireau S, et al. (2010) HSP90 and its R2TP/Prefoldin-like cochaperone are involved in the cytoplasmic assembly of RNA polymerase II. Mol Cell 39: 912–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cloutier P, Coulombe B (2010) New insights into the biogenesis of nuclear RNA polymerases? Biochem Cell Biol 88: 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dorjsuren D, Lin Y, Wei W, Yamashita T, Nomura T, et al. (1998) RMP, a novel RNA polymerase II subunit 5-interacting protein, counteracts transactivation by hepatitis B virus X protein. Mol Cell Biol 18: 7546–7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Le TT, Zhang S, Hayashi N, Yasukawa M, Delgermaa L, et al. (2005) Mutational analysis of human RNA polymerase II subunit 5 (RPB5): the residues critical for interactions with TFIIF subunit RAP30 and hepatitis B virus X protein. J Biochem (Tokyo) 138: 215–224. [DOI] [PubMed] [Google Scholar]
- 16. Wei W, Gu JX, Zhu CQ, Sun FY, Dorjsuren D, et al. (2003) Interaction with general transcription factor IIF (TFIIF) is required for the suppression of activated transcription by RPB5-mediating protein (RMP). Cell Res 13: 111–120. [DOI] [PubMed] [Google Scholar]
- 17. Yart A, Gstaiger M, Wirbelauer C, Pecnik M, Anastasiou D, et al. (2005) The HRPT2 tumor suppressor gene product parafibromin associates with human PAF1 and RNA polymerase II. Mol Cell Biol 25: 5052–5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gstaiger M, Luke B, Hess D, Oakeley EJ, Wirbelauer C, et al. (2003) Control of nutrient-sensitive transcription programs by the unconventional prefoldin URI. Science 302: 1208–1212. [DOI] [PubMed] [Google Scholar]
- 19. Deplazes A, Mockli N, Luke B, Auerbach D, Peter M (2009) Yeast Uri1p promotes translation initiation and may provide a link to cotranslational quality control. EMBO J 28: 1429–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mita P, Savas JN, Djouder N, Yates JR 3rd, Ha S, et al. (2011) REGULATION OF ANDROGEN RECEPTOR MEDIATED TRANSCRIPTION BY RPB5 BINDING PROTEIN URI/RMP. Mol Cell Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kirchner J, Vissi E, Gross S, Szoor B, Rudenko A, et al. (2008) Drosophila Uri, a PP1alpha binding protein, is essential for viability, maintenance of DNA integrity and normal transcriptional activity. BMC Mol Biol 9: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Delgermaa L, Hayashi N, Dorjsuren D, Nomura T, Thuy le TT, et al. (2004) Subcellular localization of RPB5-mediating protein and its putative functional partner. Mol Cell Biol 24: 8556–8566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mockli N, Deplazes A, Hassa PO, Zhang Z, Peter M, et al. (2007) Yeast split-ubiquitin-based cytosolic screening system to detect interactions between transcriptionally active proteins. Biotechniques 42: 725–730. [DOI] [PubMed] [Google Scholar]
- 24. Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, et al. (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440: 637–643. [DOI] [PubMed] [Google Scholar]
- 25. Gari E, Piedrafita L, Aldea M, Herrero E (1997) A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast 13: 837–848. [DOI] [PubMed] [Google Scholar]
- 26. Geissler S, Siegers K, Schiebel E (1998) A novel protein complex promoting formation of functional alpha- and gamma-tubulin. EMBO J 17: 952–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lopez N, Halladay J, Walter W, Craig EA (1999) SSB, encoding a ribosome-associated chaperone, is coordinately regulated with ribosomal protein genes. J Bacteriol 181: 3136–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kosugi S, Hasebe M, Tomita M, Yanagawa H (2009) Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci U S A 106: 10171–10176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. la Cour T, Kiemer L, Molgaard A, Gupta R, Skriver K, et al. (2004) Analysis and prediction of leucine-rich nuclear export signals. Protein Eng Des Sel 17: 527–536. [DOI] [PubMed] [Google Scholar]
- 30. Peiro-Chova L, Estruch F (2009) The yeast RNA polymerase II-associated factor Iwr1p is involved in the basal and regulated transcription of specific genes. J Biol Chem 284: 28958–28967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Corden J (2011) Going nuclear: transcribers in transit. Mol Cell 42: 143–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mosley AL, Sardiu ME, Pattenden SG, Workman JL, Florens L, et al. (2011) Highly reproducible label free quantitative proteomic analysis of RNA polymerase complexes. Mol Cell Proteomics 10: M110 000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lotan R, Bar-On VG, Harel-Sharvit L, Duek L, Melamed D, et al. (2005) The RNA polymerase II subunit Rpb4p mediates decay of a specific class of mRNAs. Genes Dev 19: 3004–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sole C, Nadal-Ribelles M, Kraft C, Peter M, Posas F, et al. (2011) Control of Ubp3 ubiquitin protease activity by the Hog1 SAPK modulates transcription upon osmostress. EMBO J 30: 3274–3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Graumann J, Dunipace LA, Seol JH, McDonald WH, Yates JR 3rd, et al. (2004) Applicability of tandem affinity purification MudPIT to pathway proteomics in yeast. Mol Cell Proteomics 3: 226–237. [DOI] [PubMed] [Google Scholar]
- 36. Miyazawa M, Tashiro E, Kitaura H, Maita H, Suto H, et al. (2011) Prefoldin subunits are protected from ubiquitin-proteasome system-mediated degradation by forming complex with other constituent subunits. J Biol Chem 286: 19191–19203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, et al. (2007) Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446: 806–810. [DOI] [PubMed] [Google Scholar]
- 38. Wilmes GM, Bergkessel M, Bandyopadhyay S, Shales M, Braberg H, et al. (2008) A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol Cell 32: 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ye P, Peyser BD, Pan X, Boeke JD, Spencer FA, et al. (2005) Gene function prediction from congruent synthetic lethal interactions in yeast. Mol Syst Biol 1: 2005 0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zheng J, Benschop JJ, Shales M, Kemmeren P, Greenblatt J, et al. (2010) Epistatic relationships reveal the functional organization of yeast transcription factors. Mol Syst Biol 6: 420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, et al. (2010) The genetic landscape of a cell. Science 327: 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Esberg A, Moqtaderi Z, Fan X, Lu J, Struhl K, et al. (2011) Iwr1 protein is important for preinitiation complex formation by all three nuclear RNA polymerases in Saccharomyces cerevisiae. PLoS ONE 6: e20829 doi:10.1371/journal.pone.0020829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Boulon S, Bertrand E, Pradet-Balade B (2012) HSP90 and the R2TP co-chaperone complex: Building multi-protein machineries essential for cell growth and gene expression. RNA Biol 9. [DOI] [PubMed] [Google Scholar]
- 44. Koh JL, Ding H, Costanzo M, Baryshnikova A, Toufighi K, et al. (2010) DRYGIN: a database of quantitative genetic interaction networks in yeast. Nucleic Acids Res 38: D502–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cramer P, Bushnell DA, Kornberg RD (2001) Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292: 1863–1876. [DOI] [PubMed] [Google Scholar]
- 46. Garcia-Lopez MC, Pelechano V, Miron-Garcia MC, Garrido-Godino AI, Garcia A, et al. (2011) The conserved foot domain of RNA pol II associates with proteins involved in transcriptional initiation and/or early elongation. Genetics 189: 1235–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, et al. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- 48. Sheff MA, Thorn KS (2004) Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 21: 661–670. [DOI] [PubMed] [Google Scholar]
- 49. Soutourina J, Bordas-Le Floch V, Gendrel G, Flores A, Ducrot C, et al. (2006) Rsc4 connects the chromatin remodeler RSC to RNA polymerases. Mol Cell Biol 26: 4920–4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liang C, Stillman B (1997) Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev 11: 3375–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Thomas BJ, Rothstein R (1989) Elevated recombination rates in transcriptionally active DNA. Cell 56: 619–630. [DOI] [PubMed] [Google Scholar]
- 53. Sikorski RS, Hieter P (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Stade K, Ford CS, Guthrie C, Weis K (1997) Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90: 1041–1050. [DOI] [PubMed] [Google Scholar]
- 55. Van Mullem V, Wery M, Werner M, Vandenhaute J, Thuriaux P (2002) The Rpb9 subunit of RNA polymerase II binds transcription factor TFIIE and interferes with the SAGA and elongator histone acetyltransferases. J Biol Chem 277: 10220–10225. [DOI] [PubMed] [Google Scholar]
- 56. Bonneaud N, Ozier-Kalogeropoulos O, Li GY, Labouesse M, Minvielle-Sebastia L, et al. (1991) A family of low and high copy replicative, integrative and single- stranded S. cerevisiae/E. coli shuttle vectors. Yeast 7: 609–615. [DOI] [PubMed] [Google Scholar]
- 57. Rubbi L, Labarre-Mariotte S, Chedin S, Thuriaux P (1999) Functional characterization of ABC10alpha, an essential polypeptide shared by all three forms of eukaryotic DNA-dependent RNA polymerases. J Biol Chem 274: 31485–31492. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Δbud27 mutant phenotypes are corrected by overexpression of different BUD27 constructions. Growth of wild-type and Δbud27 mutant strains transformed with different constructions containing whole BUD27 or deleted forms of BUD27, at different temperatures; pCM and pFL correspond to the control empty vectors.
(TIF)
RNA pols localisation. A) Deletion of Bud27 NES domain does not impair RNA pols nuclear localisation (Rpb8-ECFP). Δbud27 mutant cells overexpressing Bud27ΔNES-GFP were grown at 30°C and localisation of Bud27ΔNES-GFP and Rpb8-ECFP was analysed in vivo. B) BUD27 overexpression (pCM-BUD27-TAP) corrects nuclear localization of Rpb8-ECFP in Δbud27 mutant cells at 30°C when compared to the same strain containing an empty plasmid (pCM).
(TIF)
siRNA silencing of URI. URI mRNA accumulation was determined in human pulmonary fibroblast grown in GM medium at 37°C transfected with 40, 100 and 200 nM of siRNA(URI) heteroduplex. CONTROL cells were treated in the same conditions without siRNA heteroduplex.
(TIF)
RPB5 overexpression rescues nuclear RNA pols localisation. Immunocytochemistry experiments using antibodies against Rpa190-HA (anti-HA), Rpb1 (8WG16), and Rpc160-Myc (anti-Myc) in Δbud27 mutant cells with tagged Rpa190-HA (RNA pol I) and Rpc160-Myc (RNA pol III), at 30°C, transformed with a plasmid overexpressing RPB5 (pFL-RPB5), or with an empty vector (pFL).
(TIF)
Rpb3-TAP purification from wild type and Δbud27 mutant containing functional tagged version of Rpb3 (Rpb3-TAP). The protein mixture obtained in each case was subjected to multidimensional protein identification technology (MudPIT) [35] and their ratios versus Rpb3 calculated.
(DOC)