Abstract
Small genomic rearrangements and copy-number variations (CNVs) involving a single gene have been associated recently with many neurocognitive phenotypes, including intellectual disability (ID), behavioral abnormalities, and autistic spectrum disorders (ASDs). Such small CNVs in the Autism susceptibility candidate 2 (AUTS2) gene have been shown to be associated with seizures, ID, and ASDs. We report four patients with small CNVs ranging in size between 133–319 kb that disrupt AUTS2. Two patients have duplications involving single exons, whereas two have deletions that removed multiple exons. All patients had developmental delay, whereas two patients had a diagnosis of ASDs. The CNVs were detected by an exon-targeted array CGH with dense oligonucleotide coverage in exons of genes known or hypothesized to be causative of multiple human phenotypes. Our report further shows that disruption of AUTS2 results in a variety of neurobehavioral phenotypes. More importantly, it demonstrates the utility of targeted exon array as a highly sensitive clinical diagnostic tool for the detection of small genomic rearrangements in the clinically relevant regions of the human genome.
Keywords: intellectual disability, targeted exon array, AUTS2
Introduction
The availability of high-resolution genome analysis by array comparative genomic hybridization (aCGH) has lead to the discovery of a wide range of benign and pathogenic copy-number variations (CNVs) in humans.1, 2, 3, 4 Although some of the relatively larger CNVs have been associated with recognizable genomic disorders,5, 6, 7, 8, 9, 10, 11, 12 others have been implicated in causation of clinically overlapping neuro-developmental phenotypes, including autism spectrum disorders (ASDs), developmental delay (DD), and intellectual disability (ID).13, 14, 15, 16, 17, 18, 19 More recently, smaller deletions or intragenic deletions disrupting a single gene have been associated with neurodevelopmental phenotypes.19, 20, 21, 22 Increasing the number of array probes provides higher resolution and can hence detect such smaller CNVs. However, such an approach can be associated with increase in the costs of clinical tests and the number of clinically irrelevant CNVs detected. One way to overcome this difficulty would be with the use of a targeted exon array with a dense coverage of protein-coding genes that are known or suspected to be causative of human phenotypes.20 Use of such targeted testing can detect CNVs in the genes that are clinically relevant.
In this report, as a proof of concept, we describe the detection of CNVs in exon(s) of the Autism susceptibility candidate 2 (AUTS2, MIM 607270) gene in patients evaluated for developmental delay. Detection of such small CNVs in specific genes can help identify the genetic causes for various human phenotypes and may also help delineating the pivotal roles of specific protein domains in disease causation.
Patients and Methods
The clinical information was obtained from the health-care provider by the use of a standardized checklist. The research was reviewed and approved by the Institutional Review Board of Baylor College of Medicine (BCM). DNA was extracted from the whole blood using the Puregene DNA Blood Kit (Gentra, Minneapolis, MN, USA), according to the manufacturer's instructions. We performed aCGH analysis on the clinical microarray platform routinely used in our institution. The microarrays were designed in the Medical Genetics Laboratory (MGL) at BCM. The patient samples were interrogated using a custom-designed array with ∼180 000 interrogating oligonucleotides (60 mer), manufactured by Agilent Technologies, Inc. (Santa Clara, CA, USA). This array contains the ‘best-performing' oligos selected from Agilent's online library (eArray; https://earray.chem.agilent.com/earray/) and has been further optimized using empirical data. This array is designed to provide interrogation of all known microdeletion and microduplication syndrome regions, pericentromeric and subtelomeric regions, and computationally predicted NAHR-mediated genomic instability regions flanked by low-copy repeats, as previously described.23 In addition, ∼1800 known or candidate disease genes have exonic coverage (101 644 probes in 24 319 exons; average of 4.2 probes/exon). The entire genome is covered with an average resolution of 30 kb, excluding low-copy repeats and other repetitive sequences. Further details are available at (https://www.bcm.edu/geneticlabs/). The procedures for DNA digestion, labeling, and hybridization, and data analysis, were performed as previously described.24
Results
Among 6054 DNA samples from subjects with ID, DD, or ASDs, we identified six patients with CNVs involving the AUTS2 gene in 7q11.22. This CNV was not identified in 6214 patient samples analyzed during the same period, where the requisition for testing did not mention ID, DD, or ASD as an indication. From six patients with CNVs in AUTS2, we were able to obtain detailed clinical information on four patients. The clinical phenotypes of the patients are summarized in Table 1. Patient 1 was referred for evaluation of developmental delay. The prenatal and birth history were unremarkable. The developmental milestones were significantly delayed as evidenced by walking at 21 months of age and developing first words at 4 years. Physical examination was remarkable for camptodactyly and mild scoliosis of thoracic spine. The developmental evaluation by the individualized education program from the school revealed that her performance was at the level of a 5-year-old individual at the chronological age of 10 years. Patient 2 was referred for evaluation of global developmental delay. This subject had moderate to severe delay as evidenced by crawling at 14 months, standing at 17 months, and walking at two years. Her first words were at 2 years of age and at the age of 2.5 years, her vocabulary was limited to three words. She had no craniofacial dysmorphisms or abnormalities on physical examination. Neuro-imaging and a metabolic work up, including plasma amino acids, urine organic acids and acyl-carnitine profile were normal. Patients 3 and 4 are siblings, who presented with developmental delay. The sister in the sibship had radio-ulnar synostosis, microcephaly, and dysmorphic features such as triangular facies, and mid-face hypoplasia. Her ID was graded as mild to moderate. Based on qualitative impairments in social interaction and communication that included avoidance of eye contact, inconsistent responses to questions, inability to maintain conversation, repetitive language, and lack of social reciprocity, she was diagnosed with pervasive developmental disorder (PDD). The brother aged 16 years had developmental delay, generalized tonic-clonic seizures, and behavioral abnormalities. He had significant language delay, significant impairment in social interaction, lack of emotional reciprocity, and stereotypic motor movements, and was diagnosed with PDD. The seizures in this subject began at age of 6 years, and have been well controlled by treatment with carbamazepine. In all these patients, exon-targeted aCGH identified small CNVs, ranging in size between 133–319 kb (Table 1).
Table 1. Clinical Phenotype of patients with CNVs in AUTS2.
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | |
|---|---|---|---|---|
| Age | 10 | 3 | 10 | 3 |
| Gender | F | F | F | M |
| Height (centile) | 25 | 17 | 90 | NA |
| Weight (centile) | 50 | 3 | 40 | NA |
| Head circumference (centile) | 90–97 | 11 | <3rd | 50 |
| DD | + | + | + | + |
| Autistic spectrum disorders | − | − | + | + |
| Seizures | − | − | − | + |
| Brain MRI/CT | Nl | Nl | NA | NA |
| Dysmorphic features | + | − | + | + |
| Other features | FTT, scoliosis, atrial septal defect | None | Radioulnar synostosis | None |
| CNV | Deletion | Deletion | Duplication | Duplication |
| Inheritance | De novo | CNV not present in the mother | Maternal | Maternal |
| Size (kb) | 133 | 319 | 179 | 179 |
| Genomic cordinates | 69 750 889–69 884 307 | 69 569 118–69 887 732 | 69 450 913–69 630 331 | 69 450 913–69 630 331 |
Abbreviations: AUTS2, Autism susceptibility candidate 2; CNV, copy-number variations; CT, computed tomography; DD, developmental delay; F, female; M, male; MRI, magnetic resonance imaging; NA, datum not available; Nl, normal; DD, developmental delay.
+ denotes the presence of a characteristic whereas – denotes absence. The genomic coordinates are based on hg18 genome build.
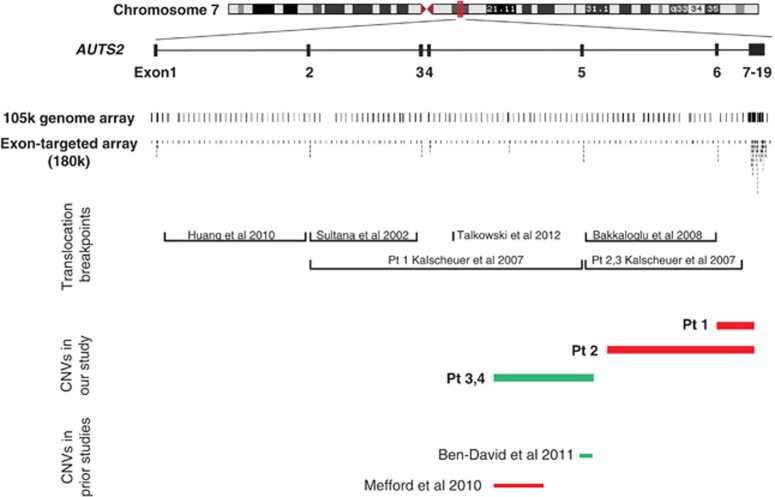
The deletions in patients 1 and 2 were intragenic, involving exons 6–14 (Figure 1). Patients 3 and 4 had a CNV inherited from the mother that duplicated exon 5. The mother of patients 3 and 4 has microcephaly and mild ID. Patient 4 had an additional duplication at 15q11.2 between BP1/BP2, which was inherited from the phenotypically normal father. The CNV in patients 1 was a de novo rearrangement. The deletion observed in patient 2 was not present in the phenotypically normal mother and the father was unavailable for testing. The CNVs in each patient were confirmed either with FISH or a repeat analysis using the exon-targeted platform.
Figure 1.
Results of targeted exon-specific aCGH showing CNVs in four patients with developmental delay and/or ASDs. The oligonucleotide coverage in the 180k exon-targeted array clearly shows more oligos per exon as compared to the 105k genome array. The previously published reports on either translocation breakpoints (Huang et al,33 Sultana et al,30 Kalscheuer et al32) or inversion breakpoints (Talkowski et al,34 Bakkaloglu et al 31) disrupting Autism susceptibility candidate 2 (AUTS2) have been depicted. Red bars depict loss and green bars depict gain of copy number. Patients 3 and 4 with ASDs, as well as three of the previously reported patients with ASDs, have alterations in the 5′ region of the gene. Patients 1 and 2 with multiple-exon deletions have developmental delay but not ASDs.
Discussion
The AUTS2 gene spans 1.2 Mb of genomic DNA on chromosome 7q11.22, and consists of 19 exons encoding for a protein of yet undetermined function. By use of in situ hybridization, immunohistochemistry, and immunoblotting, Bedogni et al25 showed high levels of AUTS2 mRNA and protein in the developing murine cerebral cortex and cerebellum. Early embryonic expression of AUTS2 was predominant in the cortical plate, and postnatal expression was observed in frontal cortex, hippocampus, cerebellum, and the basal ganglia nuclei.25
Haploinsufficiency of AUTS2 (three reports of apparently balanced translocation and two reports of inversion) has been associated with ASDs, ID, seizures, and attention deficit hyperactivity disorder.26, 27, 28, 29, 30, 31, 34 Sultana et al30 reported monozygotic twins with ID and autism with an apparently balanced t(7;20) (q11.2; p11.2) that disrupted AUTS2. Kalscheuer et al32 described translocations involving AUTS2 in three unrelated individuals with mild to moderate ID. Huang et al33 reported autism in a patient with an apparently balanced t(6;7)(q14;q11.2). An inversion disrupting both the AUTS2 and contactin-associated protein-like 2 (CNTNAP2) genes was reported in a child with ASD and ID.31 More recently, by sequencing patients with balanced chromosomal abnormalities, Talkowski et al34 showed the disruption of AUTS2 in a subject with ASD, mild learning disability, poor oral motor coordination, tongue protrusion, and flexion contractures. The disruption of AUTS2 in this patient because of an inversion was in intron 4 with the other breakpoint disrupting PTPRN2. Whereas these reports point that haploinsufficiency in AUTS2 may lead to neurobehavioral phenotypes, the interpretation of data in cases with translocation and inversion is complicated by the disruption of other genes.
As of yet, there have been only two reports of intragenic CNVs disrupting AUTS2.26, 29 Ben-David et al26 reported a duplication of 140 kb in size, involving exon 5 that resulted in monoallelic expression of AUTS2 in a patient with ASDs. Mefford et al29 used whole-genome oligonucleotide aCGH in a cohort of 517 individuals with idiopathic epilepsy and found two patients with small intragenic deletions. Recently, Girirajan et al35 used a custom aCGH platform with high probe density for 107 genomic hotspots and found CNVs in AUTS2 gene in two unrelated families with dyslexia and a subject with ID. Interestingly, these authors also found a statistically significant enrichment AUTS2 CNVs in subjects with neurodevelopmental phenotypes as compared with control population. However all these studies were done on a research basis on a selected patient population. Our data constitute the first report on two intragenic deletions and two small duplications, involving AUTS2 in a clinical setting by exon-targeted array CGH. The ID in patients 3 and 4 and their mother segregated with CNV that duplicated exon 5 of AUTS2. Our results support the existing literature regarding the pathogenicity of CNVs involving AUTS2.
CNVs involving one or more exons in a gene can affect gene expression dosage in multiple ways. Alteration in the reading frame may result in a premature stop codon followed by nonsense-mediated decay. Alternatively, an in-frame loss or gain may lead to an altered protein product that may be associated with either loss or gain of function.36, 37 Thus, CNVs involving exons may lead to a null allele or a hypomorphic allele, leading to haploinsufficiency or alternatively, to antimorphic or neomorphic mutant alleles with toxic gain of function. Hence CNVs in the same gene may expectedly confer various phenotypes, depending on its effects on particular protein domains and protein function. Detection of such small CNVs in the genes known or hypothesized to convey human phenotypes is best accomplished with the use of aCGH with targeted exon coverage as these typically escape detection with standard whole genomic untargeted arrays of similar probe number.20 It is to be noted that very small rearrangements like those involving single exons may need confirmation by additional methods before reporting.
In the case of CNVs involving AUTS2, previous reports have shown that patients with ASDs have disruption of the gene proximal to exon 6.30, 32, 34, 38 Our results are consistent with this observation as patients 3 and 4 with duplication of exon 5 have ASDs. Both these duplications are not expected to alter the reading frame, but can result in addition of amino-acid residues to the amino terminus of the protein. These results, although preliminary, may imply that a particular motif at the amino-terminus of AUTS2 may be critical for the neurodevelopmental processes critical for behavior and social interaction.
In conclusion, our study adds to the existing literature supporting the role of AUTS2 in normal neurological development and that its altered expression may result in ASDs and ID. In addition, our results highlight the utility of targeted exon aCGH in detecting small CNVs in genes associated with human phenotypes.
Acknowledgments
This work was supported in part by fellowship grants by the National Urea Cycle Disorders foundation and LCRC from Osteogenesis Imperfecta Foundation (SNSC), DK081735–01A1, NIH/NIGMS T32 contract grant number GM07526 (AE).
Drs Nagamani, Erez, Patel, Stankiewicz and Cheung are faculty members of the Department of Molecular and Human Genetics at Baylor College of Medicine, which derives revenue from the chromosomal microarray analysis offered in the Medical Genetics Laboratory. The remaining authors declare no conflict of interest.
References
- Iafrate AJ, Feuk L, Rivera MN, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Troge J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Lupski JR. Genomic disorders ten years on. Genome Med. 2009;1:42. doi: 10.1186/gm42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunetti-Pierri N, Berg JS, Scaglia F, et al. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet. 2008;40:1466–1471. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford HC, Sharp AJ, Baker C, et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med. 2008;359:1685–1699. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamani SC, Erez A, Shen J, et al. Clinical spectrum associated with recurrent genomic rearrangements in chromosome 17q12. Eur J Hum Genet. 18:278–284. doi: 10.1038/ejhg.2009.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamani SC, Zhang F, Shchelochkov OA, et al. Microdeletions including YWHAE in the Miller-Dieker syndrome region on chromosome 17p13.3 result in facial dysmorphisms, growth restriction, and cognitive impairment. J Med Genet. 2009;46:825–833. doi: 10.1136/jmg.2009.067637. [DOI] [PubMed] [Google Scholar]
- Sharp AJ, Hansen S, Selzer RR, et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet. 2006;38:1038–1042. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- Sharp AJ, Selzer RR, Veltman JA, et al. Characterization of a recurrent 15q24 microdeletion syndrome. Hum Mol Genet. 2007;16:567–572. doi: 10.1093/hmg/ddm016. [DOI] [PubMed] [Google Scholar]
- Shinawi M, Liu P, Kang SH, et al. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J Med Genet. 2010;47:332–341. doi: 10.1136/jmg.2009.073015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballif BC, Hornor SA, Jenkins E, et al. Discovery of a previously unrecognized microdeletion syndrome of 16p11.2-p12.2. Nat Genet. 2007;39:1071–1073. doi: 10.1038/ng2107. [DOI] [PubMed] [Google Scholar]
- Friedman JM, Baross A, Delaney AD, et al. Oligonucleotide microarray analysis of genomic imbalance in children with mental retardation. Am J Hum Genet. 2006;79:500–513. doi: 10.1086/507471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froyen G, Van Esch H, Bauters M, et al. Detection of genomic copy number changes in patients with idiopathic mental retardation by high-resolution X-array-CGH: important role for increased gene dosage of XLMR genes. Hum Mutat. 2007;28:1034–1042. doi: 10.1002/humu.20564. [DOI] [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menten B, Maas N, Thienpont B, et al. Emerging patterns of cryptic chromosomal imbalance in patients with idiopathic mental retardation and multiple congenital anomalies: a new series of 140 patients and review of published reports. J Med Genet. 2006;43:625–633. doi: 10.1136/jmg.2005.039453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatmari P, Paterson AD, Zwaigenbaum L, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniowiecka-Kowalnik B, Nesteruk M, Peters SU, et al. Intragenic rearrangements in NRXN1 in three families with autism spectrum disorder, developmental delay, and speech delay. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:983–993. doi: 10.1002/ajmg.b.31064. [DOI] [PubMed] [Google Scholar]
- Boone PM, Bacino CA, Shaw CA, et al. Detection of clinically relevant exonic copy-number changes by array CGH. Hum Mutat. 2010;31:1326–1342. doi: 10.1002/humu.21360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celestino-Soper PB, Shaw CA, Sanders SJ, et al. Use of array CGH to detect exonic copy number variants throughout the genome in autism families detects a novel deletion in TMLHE. Hum Mol Genet. 2011;20:4360–4370. doi: 10.1093/hmg/ddr363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhail FM, Lose EJ, Robin NH, et al. Clinically relevant single gene or intragenic deletions encompassing critical neurodevelopmental genes in patients with developmental delay, mental retardation, and/or autism spectrum disorders. Am J Med Genet A. 2011;155A:2386–2396. doi: 10.1002/ajmg.a.34177. [DOI] [PubMed] [Google Scholar]
- El-Hattab AW, Smolarek TA, Walker ME, et al. Redefined genomic architecture in 15q24 directed by patient deletion/duplication breakpoint mapping. Hum Genet. 2009;126:589–602. doi: 10.1007/s00439-009-0706-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Z, Kang SH, Shaw CA, et al. Bacterial artificial chromosome-emulation oligonucleotide arrays for targeted clinical array-comparative genomic hybridization analyses. Genet Med. 2008;10:278–289. doi: 10.1097/GIM.0b013e31816b4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedogni F, Hodge RD, Nelson BR, et al. Autism susceptibility candidate 2 (Auts2) encodes a nuclear protein expressed in developing brain regions implicated in autism neuropathology. Gene Expr Patterns. 2010;10:9–15. doi: 10.1016/j.gep.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David E, Granot-Hershkovitz E, Monderer-Rothkoff G, et al. Identification of a functional rare variant in autism using genome-wide screen for monoallelic expression. Hum Mol Genet. 2011;20:3632–3641. doi: 10.1093/hmg/ddr283. [DOI] [PubMed] [Google Scholar]
- de la Barra F, Skoknic V, Alliende A, Raimann E, Cortes F, Lacassie Y. [Twins with autism and mental retardation associated with balanced (7;20) chromosomal translocation] Rev Chil Pediatr. 1986;57:549–554. [PubMed] [Google Scholar]
- Elia J, Gai X, Xie HM, et al. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol Psychiatry. 2010;15:637–646. doi: 10.1038/mp.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford HC, Muhle H, Ostertag P, et al. Genome-wide copy number variation in epilepsy: novel susceptibility loci in idiopathic generalized and focal epilepsies. PLoS Genet. 2010;6:e1000962. doi: 10.1371/journal.pgen.1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana R, Yu CE, Yu J, et al. Identification of a novel gene on chromosome 7q11.2 interrupted by a translocation breakpoint in a pair of autistic twins. Genomics. 2002;80:129–134. doi: 10.1006/geno.2002.6810. [DOI] [PubMed] [Google Scholar]
- Bakkaloglu B, O'Roak BJ, Louvi A, et al. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am J Hum Genet. 2008;82:165–173. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalscheuer VM, FitzPatrick D, Tommerup N, et al. Mutations in autism susceptibility candidate 2 (AUTS2) in patients with mental retardation. Hum Genet. 2007;121:501–509. doi: 10.1007/s00439-006-0284-0. [DOI] [PubMed] [Google Scholar]
- Huang XL, Zou YS, Maher TA, Newton S, Milunsky JM. A de novo balanced translocation breakpoint truncating the autism susceptibility candidate 2 (AUTS2) gene in a patient with autism. Am J Med Genet A. 2010;152A:2112–2114. doi: 10.1002/ajmg.a.33497. [DOI] [PubMed] [Google Scholar]
- Talkowski ME, Rosenfeld JA, Blumenthal I, et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149:525–537. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Brkanac Z, Coe BP, et al. Relative burden of large CNVs on a range of neurodevelopmental phenotypes. PLoS Genet. 2011;7:e1002334. doi: 10.1371/journal.pgen.1002334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifton RP, Dluhy RG, Powers M, et al. A chimaeric 11 beta-hydroxylase/aldosterone synthase gene causes glucocorticoid-remediable aldosteronism and human hypertension. Nature. 1992;355:262–265. doi: 10.1038/355262a0. [DOI] [PubMed] [Google Scholar]
- Yatsenko AN, Shroyer NF, Lewis RA, Lupski JR. An ABCA4 genomic deletion in patients with Stargardt disease. Hum Mutat. 2003;21:636–644. doi: 10.1002/humu.10219. [DOI] [PubMed] [Google Scholar]
- Poot M, van der Smagt JJ, Brilstra EH, Bourgeron T. Disentangling the myriad genomics of complex disorders, specifically focusing on autism, epilepsy, and schizophrenia. Cytogenet Genome Res. 2011;135:228–240. doi: 10.1159/000334064. [DOI] [PubMed] [Google Scholar]