Abstract
Human Y chromosomes belonging to the haplogroup R1b1-P25, although very common in Europe, are usually rare in Africa. However, recently published studies have reported high frequencies of this haplogroup in the central-western region of the African continent and proposed that this represents a ‘back-to-Africa' migration during prehistoric times. To obtain a deeper insight into the history of these lineages, we characterised the paternal genetic background of a population in Equatorial Guinea, a Central-West African country located near the region in which the highest frequencies of the R1b1 haplogroup in Africa have been found to date. In our sample, the large majority (78.6%) of the sequences belong to subclades in haplogroup E, which are the most frequent in Bantu groups. However, the frequency of the R1b1 haplogroup in our sample (17.0%) was higher than that previously observed for the majority of the African continent. Of these R1b1 samples, nine are defined by the V88 marker, which was recently discovered in Africa. As high microsatellite variance was found inside this haplogroup in Central-West Africa and a decrease in this variance was observed towards Northeast Africa, our findings do not support the previously hypothesised movement of Chadic-speaking people from the North across the Sahara as the explanation for these R1b1 lineages in Central-West Africa. The present findings are also compatible with an origin of the V88-derived allele in the Central-West Africa, and its presence in North Africa may be better explained as the result of a migration from the south during the mid-Holocene.
Keywords: Central-West Africa, Equatorial Guinea, human male lineages, Y chromosome, haplogroup R-V88, back to Africa hypothesis
Introduction
Genetic data, particularly from non-recombining regions of the genome, have always had a major role in the understanding of the past and present of the human species. In this context, the male-specific region of the Y chromosome, which is the largest non-recombining region in the human genome, has become a valuable tool for the study of historical and pre-historical human movements, among other anthropological questions.1
The fact that the Y-SNP haplogroups are so stable and are distributed in such a specific geographic location allows the understanding of human migrations according to their distribution. However, in Africa, the study of the major Y chromosome haplogroups has not been very detailed and has been essentially restricted to the principal clades observed in this continent, namely the non-monophyletic lineages of clade A and haplogroups B and E. Although haplogroup E is widespread across sub-Saharan Africa, lineages from A and B, the most basal branches in the Y chromosome tree, are usually restricted to specific populations, especially pastoralists and hunter-gatherers.2, 3, 4
Apart from the aforementioned haplogroups, some studies have reported unexpectedly high frequencies of haplogroup R1b1-P25 in some African populations.5, 6, 7 This haplogroup is thought to have originated in Europe, and its high frequencies in Central-West African countries have been explained through a migration back to Africa in prehistoric times, mediated in Africa by speakers of the Chadic family of Afro-Asiatic languages.6, 7 The arrival of this ethnic group to Lake Chad from the Proto-Afro-Asiatic homeland in Eastern Africa has been explained by two different hypotheses. Blench's theory (the ‘inter-Saharan' hypothesis)8 suggests that Chadic speakers arrived at the Chad Basin through an east to west migration through the Sahel, whereas Ehret's theory (the ‘Trans-Saharan' hypothesis)9 suggests that they arrived from the north through a migration across the Sahara desert. The latter hypothesis has been used as the explanation for the high frequencies of the R1b1-P25 haplogroup in Central-West Africa, mainly due to its presence in speakers of other Afro-Asiatic languages in North Africa.7, 10 Nevertheless, there is still an on-going debate about the most suitable explanation for these observations.10, 11
To obtain a better and deeper understanding about this migration, a more detailed study of populations in all of the regions across the proposed paths is necessary. Equatorial Guinea is a Central-West African country located below the Chad Basin that could also have been influenced by this migration. The Y-SNP haplogroups in this country are still uncharacterised, although it borders Gabon and Cameroon, where the highest frequencies of the R1b1 haplogroup in Africa have been found.7
The main objective of the present work is to better characterise the male lineages from Equatorial Guinea, particularly lineages of the Y chromosome haplogroup R1b1, to investigate the Chadic migration route hypothesis. The comparison of our results with previously published data will hopefully contribute not only to a better characterisation of the still understudied African genetic diversity but also to a better understanding of the unexpectedly high frequencies of the R1b1 haplogroup in this region of Africa.
Materials and methods
Population sample and DNA extraction
A total of 112 unrelated males from Equatorial Guinea (living in Madrid, Spain, at the time of sample collection) were analysed for 17 STRs and 49 biallelic polymorphisms (single-nucleotide polymorphisms (SNPs)) in the male-specific region of the Y chromosome.
Sample collection was performed in 2001, from adult males that were born and had lived in Equatorial Guinea before moving to Spain for work, keeping their Guinean nationality. The great majority of the population in Equatorial Guinea is of Bantu origin (the largest tribe, Fang, represents 85.7% of the population), according to the 1994 census.12 This strong Bantu influence is also noted in the language; apart from the more recent official European languages (Spanish and French) and two creole languages (admixture with Indo-European), all of the others belong to the Narrow Bantu linguistic group.13 Although no information was available concerning the ethnic background of each male, it is expected that they mostly represent a mixture of Fang and Bubi groups from different regions in Equatorial Guinea.
DNA was extracted using a standard Chelex-100 (Bio-Rad, Hercules, CA, USA) method.14
Y-STR typing
The AmpFℓSTR Yfiler PCR Amplification Kit (Applied Biosystems, Foster City, CA, USA) was used to analyse 17 STR markers located in the male-specific region of the Y chromosome. An ABI 3130 Genetic Analyser (Applied Biosystems) was used for Y-STR typing, and the results were analysed using GeneMapper ID software v3.2 (Applied Biosystems). The Y-STR alleles were designated according to the ISFG recommendations.15 Several additional Y-STRs had already been studied in a subset of these samples using the PowerPlex Y System (Promega Corporation, Madison, WI, USA), following the protocol described by Arroyo-Pardo and collaborators.16
Y-SNP typing
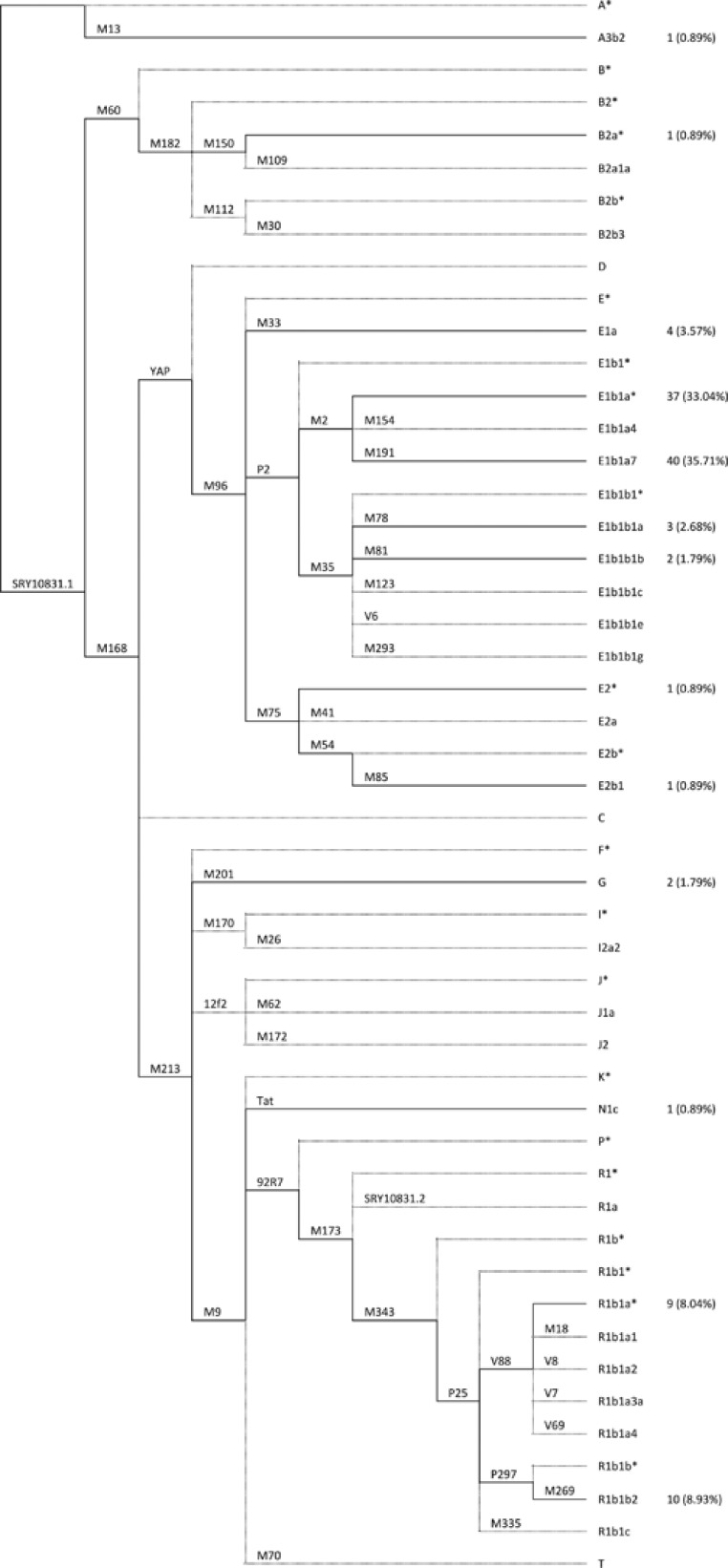
The analysis of 49 Y-SNPs allowed the identification of 49 different Y chromosome haplogroups (Figure 1). The SNP markers used in this study were selected based on the Y chromosome parsimony tree17 to characterise the haplogroups most frequently found in sub-Saharan Africa as well as to identify those that are thought to have been brought by the Europeans to Africa during the last five centuries.3, 5, 18, 19, 20, 21, 22 Moreover, eight additional SNPs inside haplogroup R1b were also included in our set to increase the resolution of this haplogroup.7
Figure 1.
Phylogenetic tree of the Y chromosome haplogroups studied. Biallelic markers are displayed in each branch. Haplogroups are named according to Karafet et al.17
A hierarchical approach (Supplementary Figure S1) based on the phylogeny reported by Karafet et al17 was used, eliminating the need to type all SNPs to define each haplogroup. This method was performed using single and multiplex PCR, RFLP, SNaPshot and direct DNA sequencing analysis. The Y Alu polymorphic element (YAP), Y-SNPs M41, M54 and M75, and three multiplexes (Multiplex 1 plus M13, Multiplex E and Multiplex B) were analysed as described by Gomes et al3 Based on the results from Multiplex 1 plus M13, a new set of Y-SNPs was typed: (i) if only the markers SRY10831.1 and M213 carried the derived allele, Multiplex 2 (described by Brión et al23) was performed under the same conditions used for the other three multiplexes; (ii) in chromosomes carrying the derived allele at marker P25, M269 was typed using the RFLP technique. The amplification PCR for this marker was conducted using the primers described by Beleza et al,18 and the conditions were optimised from Gomes et al3 for the multiplexes used. Touchdown PCR was performed with a first set of 5 cycles with a higher annealing temperature of 65 °C for 90 s and a second set of 30 cycles in which the temperature was decreased to 62 °C, also for 90 s. After confirming the amplification in a polyacrylamide gel using the silver staining method, 2 μl of the amplified product were incubated overnight at 37°C with 0.05 μl of the restriction enzyme MvaI at a concentration of 10 U/μl, 0.5 μl of 10 × buffer R+ and 2.45 μl of water. As the restriction site for MvaI is present only when the SNP is in the derived state, the two alleles can be discriminated in a silver-stained polyacrylamide gel.
The eight remaining SNPs in the set—M18, M335, M343, P297, V7, V8, V69 and V88—were typed to improve the resolution of the R1b haplogroup. This was performed through direct DNA sequencing in samples carrying the derived allele at marker P25 and the ancestral allele at M269. For all but one of the markers, new primers were designed for this analysis (Supplementary Table S1). The PCR amplification conditions were the same as those used for the aforementioned multiplexes, except in the case of the V88 marker. This marker is located in a region of the Y chromosome that has significant homology to the X chromosome. Therefore, to increase the specificity for the Y chromosome, the annealing temperature used during the amplification PCR was increased to 63°C. The PCR product purification, sequencing analysis and extended product purification were performed according to Gomes et al.3 The products of the sequencing reaction were run on an ABI PRISM 3130xl Genetic Analyser (Applied Biosystems) and were analysed using DNA Sequencing Analysis Software v5.2 (Applied Biosystems).
Data analysis
For the analysis of both Y-STRs and Y-SNPs, diversity values and pairwise genetic distances (FST) were calculated with the software Arlequin 3.5.1.224 Y-SNP haplogroup frequencies were determined by direct counting.
Pairwise genetic distances were visualised in a two-dimensional graphic through the multidimensional scaling (MDS) method implemented in the software STATISTICA 7.0.25
Phylogenetic networks were constructed to investigate the genetic relationships within haplogroup R1b1-P25 using the software Network 4.6.0.0 (http://www.fluxus-engineering.com/sharenet.htm) with sequential application of the reduced median26 and the median-joining27 methods to resolve extensive reticulation. Differential microsatellite weighting (inversely proportional to variance) was applied to obtain the most parsimonious network in accordance with Qamar et al.28
Levene's test was used to assess the homogeneity of variances in IBM SPSS Statistics, version 19.
Results and Discussion
Characterisation of the male lineages of Equatorial Guinea
In the 112 samples analysed, we were able to identify 104 different haplotypes and 13 different haplogroups (Figure 1 and Supplementary Table S2). The majority of the Y-SNP lineages found in this study (almost 80%) belong to haplogroup E, namely bearing the M2-derived allele, the most common haplogroup in sub-Saharan Africa Bantu populations. Apart from this haplogroup and five other chromosomes, all the remaining samples belong to haplogroup R, namely R1b1-P25, a lineage that is rare in Africa and is found mainly in Europe and Asia. Nevertheless, high frequencies of these lineages in some African populations have been previously reported by several authors.5, 6, 7, 29, 30
Lineages in clade A, although almost entirely restricted to Africa, have been described in Bantu populations at low frequencies. These lineages are mostly present in Nilo-Saharan speakers (for example, reference2, 5, 6, 21) and therefore, a high frequency was not expected in our sample. This expectation was confirmed; only one of the chromosomes in our sample belongs to this haplogroup, more specifically to the haplogroup A3b2-M13, which is more frequently observed among Nilotes than other African groups.3 Similarly, only one chromosome in our sample was found to belong to haplogroup B, namely to the branch that is most common in Bantu individuals (defined by the derived allele at M150).2, 5, 18, 21
The major haplogroup E, the most diverse clade in the Y chromosome tree, is widespread across the African continent, where its highest frequencies are found and is also present in the Middle East, southern Europe, and Central and South Asia.17 This clade was the most frequent in our sample, representing almost 80% of all the chromosomes, similar to results for other sub-Saharan populations (for example, reference5, 30). The most frequent sub-lineage found in our sample inside this clade was the haplogroup E1b1a-M2, proposed as a marker of Bantu expansion.22, 30 Nevertheless, some other typically non-Bantu lineages were also found in our sample. One is the E1a-M33 haplogroup, which is usually not found further South in the Bantu expansion route (for example, Angola18) but is detected at higher frequencies above the starting point of these migrations (for example, Guinea-Bissau31). Therefore, the presence of this lineage could be an indication of a genetic pool that existed before the beginning of the Bantu expansion and was not spread by this movement. Other non-Bantu lineages found within this haplogroup are E1b1b1a-M78 and E1b1b1-M81, which have been frequently found in North Africa and also in Europe.32
Both haplogroups G and N (accounting for two and one chromosomes, respectively) are rare in Africa, and their low frequency in this sample can most likely be explained by a recent Eurasian influx. Altogether, the proportion of recent Eurasian admixture found in our sample is approximately 15% (haplogroups E1b1b1b-M81, G-M201, N1c-Tat, R1b1b2-M269 and two chromosomes belonging to E1b1b1a-M78—see criteria below), which is easily explained by the well-reported European arrivals to this territory within the last five centuries.
For the Y-SNP data, the haplogroup diversity of our sample (0.7526±0.0261), although higher than that found in other West African populations,18 is still lower than for other regions, a fact that has been explained by a loss of diversity during the Bantu migrations.3, 5, 18 For the Y-STRs, high levels of haplotype diversity (0.9987±0.0014) and of the mean number of pairwise differences (9.5048±4.3932) were found.
The results obtained from Equatorial Guinea were compared with available data for other African populations5, 33, 34, 35 (see Supplementary Figure S2 and Table 1). However, Africa is still a poorly studied continent, and data for comparative analysis were not always accessible. Furthermore, the data for all populations need to be reduced to the same resolution level, requiring a minimal common set of Y-STRs and further constraining the amount of data available for comparisons. To overcome potential bias due to recent historical factors (for example, colonisation by Europeans), the levels of genetic diversity were calculated in different African samples and within each lineage (with n>5) after excluding chromosomes that had most likely been recently introduced by Europeans (including haplogroups E1b1b1, F, G, N1c, R1a, R1b1b2 and T). Because chromosomes belonging to haplogroup E1b1b1a-M78 can be considered of either African or European ancestry, a search for identical haplotypes was performed on the YHRD,36 and only those that did not present a match in European populations were maintained in the analysis.
Table 1. Diversity indices estimated with 10 Y-STRs and Y-SNP haplogroup diversities, excluding those chromosomes that most likely have been recently introduced by Europeans (see text for excluded haplogroups).
| Y-STRs | Y-SNPs | ||||
|---|---|---|---|---|---|
| |
|
Gene diversity |
MNPD a |
WIMP |
Gene diversity |
| Equatorial guinea | 95 | 0.9915±0.0036 | 4.643449±2.297308 | 3.0191407 | 0.3351±0.0605 |
| E1b1a*-M2 | 37 | 0.9805±0.0115 | 3.321321±1.745204 | — | — |
| E1b1a7-M191 | 40 | 0.9692±0.0165 | 2.846154±1.531191 | — | — |
| R1b1a*-V88 | 9 | 1.0000±0.0524 | 2.972222±1.712433 | — | — |
| Uganda | 117 | 0.9841±0.0040 | 5.802829±2.795409 | 3.1275240 | 0.7974±0.0183 |
| A3b2-M13 | 39 | 0.9379±0.0275 | 3.260459±1.716166 | — | — |
| B2a1*-M218 | 20 | 0.8842±0.0416 | 3.905263±2.043998 | — | — |
| B2b*-M112 | 13 | 0.7692±0.1031 | 3.102564±1.721209 | — | — |
| E1b1a*-M2 | 8 | 0.8571±0.1083 | 2.857143±1.678080 | — | — |
| E1b1a7-M191 | 6 | 0.9333±0.1217 | 2.800000±1.714643 | — | — |
| E1b1b1*-M35 | 7 | 0.7143±0.1267 | 2.285714±1.420252 | — | — |
| E2a-M41 | 13 | 0.8974±0.0537 | 3.666667±1.983080 | — | — |
| Guinea-bissau | 29 | 0.9975±0.0099 | 5.246305±2.611534 | 3.8374383 | 0.4064±0.1012 |
| E1b1a*-M2 | 22 | 0.9957±0.0153 | 4.194805±2.165882 | — | — |
| Gabon (Bantu) | 794 | 0.9917±0.0009 | 4.026361±2.045803 | 3.7290481 | 0.3564±0.0211 |
| B2a-M150 | 49 | 0.9694±0.0127 | 4.088163±2.072229 | — | — |
| E*-M96 | 10 | 1.0000±0.0447 | 3.377778±1.888082 | — | — |
| E1b1a-M2 | 631 | 0.9880±0.0013 | 3.725565±1.883845 | — | — |
| E2-M75 | 49 | 0.9651±0.0124 | 3.379252±1.761024 | — | — |
| R1b1*-P25 | 44 | 0.9873±0.0089 | 3.955603±2.018674 | — | — |
| Gabon (Pygmies) | 33 | 0.9356±0.0259 | 6.507576±3.157549 | 4.3515151 | 0.5739±0.0902 |
| B2b-M112 | 21 | 0.8619±0.0551 | 5.695238±2.842610 | — | — |
| Cameroon (Bantu) | 28 | 0.9841±0.0131 | 5.187831±2.588411 | 3.1802722 | 0.4735±0.0790 |
| B2a-M150 | 8 | 0.9643±0.0772 | 3.714286±2.095457 | — | — |
| E1b1a-M2 | 19 | 0.9708±0.0239 | 3.122807±1.693930 | — | — |
| Cameroon (Pygmies) | 27 | 0.9630±0.0204 | 5.150997±2.575040 | 3.6851854 | 0.6980±0.0650 |
| B2b-M112 | 7 | 0.9524±0.0955 | 5.047619±2.787667 | — | — |
| E1b1a-M2 | 13 | 0.9359±0.0507 | 3.192308±1.762954 | — | — |
| Cabinda | 68 | 0.9838±0.0061 | 4.110623±2.072840 | 2.8441449 | 0.2647±0.0667 |
| B2a1a-M109 | 6 | 0.9333±0.1217 | 3.000000±1.816590 | — | — |
| E1b1a*-M2 | 34 | 0.9626±0.0170 | 2.811052±1.521228 | — | — |
| E1b1a7-M191 | 24 | 0.9529±0.0309 | 2.826087±1.544030 | — | — |
Abbreviations: MNPD, mean number of pairwise differences; WIMP, weighted mean intralineage mean pairwise difference; Y-SNP, single-nucleotide polymorphism on chromosome Y; Y-STR, short tandem repeats on chromosome Y.
MNPD within haplogroups was calculated for haplogroups with n>5.
The diversity values observed for Y-SNPs are lower for Bantu populations (Table 1), which may be an indication of the homogeneity that characterises sub-Saharan African populations after the replacement of the previously existent lineages as a consequence of the Bantu expansion.3, 5, 18 In contrast, the levels of Y-STR diversity observed are high and generally similar across populations (Table 1), with the highest values observed mainly in Bantu samples. Moreover, the mean number of pairwise differences (MNPD) values are more heterogeneous, and the Bantu samples generally presented the lowest values. Additionally, a population further south along the route of the Bantu migrations (that is, Cabinda) presents a lower MNPD value that those near the migration origin (that is, Equatorial Guinea and Bantu sample from Cameroon), which could be an indication of genetic pool drift along the Bantu expansion path, responsible for the loss of variation in populations on the edge of the route. Nevertheless, an analysis of the MNPD within each lineage shows that no significant differences are found within each population, indicating that no haplogroup suffered a strong founding effect in those populations. This result was also confirmed through the analysis of WIMP (weighted mean intralineage mean pairwise difference),37 which shows a pattern similar to that of MNPD.
Population comparisons were performed using a classical genetic distance method (FST). A stepwise-based model (RST), which accounts for the number of differences observed at each locus assuming the single stepwise model for the formation of new alleles, has been developed as a statistic for microsatellite data.38 However, the weight given to mutations by this method does not reflect the demographic events involved in Y-STR evolution in sub-Saharan populations because it underestimates the importance of genetic drift.
For most of both Y-STR and Y-SNP data comparisons performed, a statistically significant value was obtained for pairwise differences (P<0.05). In our sample, the only non-significant values obtained were with Cabinda (for both markers), the Bantu sample from Cameroon (for Y-STRs), and with Guinea-Bissau and the Bantu sample from Gabon (for Y-SNPs) (Supplementary Tables S3 and S4).
An MDS analysis was performed to allow an easier interpretation of the pairwise distance values matrix (Supplementary Figures S3 and S4). Through an analysis of both MDS plots obtained, a cluster composed by the Bantu samples was observed, including our sample from Equatorial Guinea. In addition, the non-Bantu populations are spread across the plot. This is another indication of the importance of ethnographic information when studying sub-Saharan populations because high distance values are obtained for different samples (labelled according to their linguistic affiliation) within a country (for example, Pygmy and Bantu samples from Gabon).
Haplogroup R and the ‘back to Africa' hypothesis
Haplogroup R is the most common haplogroup in European populations, and although it is usually rare in Africa, chromosomes bearing the P25-derived allele (lineage R1b1) have been reported at frequencies as high as 95% in some Central African populations.5, 6, 7, 29, 30 This haplogroup is thought to have originated in Central Asia approximately 40 000 years BP and then migrated westward into Europe, achieving its highest frequencies in the western region of this continent.39, 40 Although Balaresque et al41 proposed the hypothesis of a European spread of haplogroup R1b1b2-M269 during the Neolithic, the distribution of the M269 sub-haplogroups and their Y-STR diversities proved to be compatible with a pre-Neolithic diffusion of M269 in Europe.42, 43 The reported high frequencies of this haplogroup in Central-West Africa led to the proposal of a ‘back to Africa' migration as the justification for the otherwise unexpected presence of this haplogroup in the region.
In our sample, R1b1 was the second most frequently observed haplogroup, which was present in 17% of the sample. Ten out of the nineteen chromosomes that belong to this haplogroup present the M269-derived allele, a typical European marker and may therefore indicate recent European influx. Although European arrivals within the last five centuries have been well reported in this country, European influx was not significant in the neighbouring regions, such as Cameroon and Gabon, where this Eurasian haplogroup is rarely observed.5, 7 The remaining chromosomes presented the V88-derived allele, which was recently reported to be present in all of the typed R1b1*-P25 African chromosome.7 Furthermore, in our sample, most of the V88 chromosomes presented non-consensus alleles at the DYS385 marker, suggesting a different additional sub-lineage within this haplogroup.
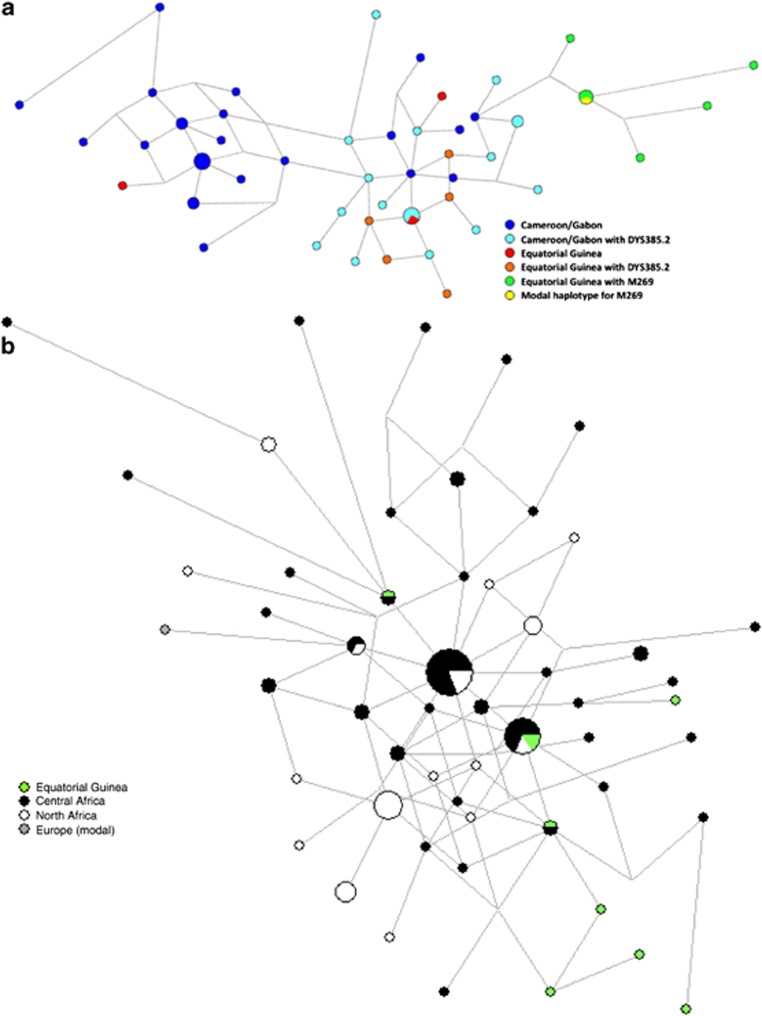
A phylogenetic network of R1b1 lineages based on 10 Y-STR haplotypes was constructed with samples from Cameroon and Gabon5 and from the present study (Figure 2a). A clear separation of the R1b1b2-M269 samples was observed; they clustered with the European modal haplotype, supporting their European ancestry. The non-consensus alleles observed in both studies do not show a well-defined separation from the samples with consensus alleles. However, a cluster containing haplotypes with both consensus and non-consensus alleles (representing two different lineages) and another exclusively with consensus alleles could be indicative of at least three different lineages within the R1b1-P25( × M269) haplogroup. Nonetheless, it is important to note that the data from Berniell-Lee et al5 were published before the recently discovered mutations within P25.7 Cruciani et al7 did not find any intermediate variant alleles at the locus DYS385, which were all found within haplogroup R1b1a-V88 in the present work (with only two chromosomes presenting consensus alleles). This result may indicate the presence of different sub-lineages within this haplogroup that are yet to be discovered.
Figure 2.
Phylogenetic network constructed with information from (a) 10 Y-STR haplotypes (DYS19, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS437, DYS438 and DYS439) within the R1b1-P25 haplogroup; and (b) 7 Y-STR haplotypes (DYS19, DYS391, DYS393, DYS439, DYS460, DYS461 and Y GATA A10) for the R1b1a-V88 haplogroup. Microsatellite haplotypes are represented by circles, with size proportional to their frequency in the samples and colours corresponding to their geographic regions.
Phylogenetic comparisons regarding the recent found lineages by Cruciani et al7 were also performed based on information from seven Y-STRs (four typed in the present work and three additional from a previous study of the same Equatorial Guinea samples by Arroyo-Pardo et al16), with R1b1a-V88 samples from North and Central Africa, Europe and from the present work (Figure 2b). The network results show that the Equatorial Guinea samples appear to be related to the remaining African samples. Furthermore, the R1b1a samples from the present work seem to characterise an old lineage due to the highly dispersed pattern presented and do not show the typical signs of a recent origin or founder effects.
The variance of these microsatellite data in the African continent (present study and Cruciani et al7) was also analysed for the V88 marker (Table 2) because the diversity of each lineage reflects its age.41 As observed in Table 2, a higher value of average variance can be observed in the sample from Equatorial Guinea, indicating an ancient origin for these lineages. Furthermore, the highest variance values were found in the Equatorial Guinea population for four out of the seven markers analysed. Nonetheless, Levene's test only revealed significant heterogeneity between the variance values for DYS439, DYS461 and Y GATA A10 (Table 2). To specifically assess the significance of the values observed in Equatorial Guinea, a homogeneity test was also performed between our sample and data from Central and North Africa (Table 2). In both cases, significant differences were only observed in two of the seven comparisons, which demonstrate that the small size of the samples hinder the extrapolation of our conclusions from the samples to the populations. Moreover, it would be valuable to calculate these levels in less broad areas, but this is not possible when relying on the published data. It is also worth noting that a recent discussion highlighted the influence of the microsatellite mutation rate on age estimates,42 and thus, the set of markers used must be taken into consideration when estimating the age of the lineages.
Table 2. Variance values observed for V88 mutation for each STR locus in Equatorial Guinea and in Central and North Africa7, and Levene's test values for the three populations together and comparing Equatorial Guinea with each of the other populations.
| Levene test | ||||||
|---|---|---|---|---|---|---|
| EG (n=9) | CA (n=66) | NA (n=32) | All | EG vs CA | EG vs NA | |
| DYS19 | 0.00000 | 0.13054 | 0.21673 | 0.054 | 0.003* | 0.093 |
| DYS391 | 0.25000 | 0.22284 | 0.22480 | 0.320 | 0.264 | 0.154 |
| DYS393 | 0.25000 | 0.05781 | 0.06048 | 0.435 | 0.221 | 0.321 |
| DYS439 | 1.25000 | 0.54662 | 0.28931 | 0.027* | 0.619 | 0.090 |
| DYS460 | 0.25000 | 0.38252 | 0.11290 | 0.546 | 0.599 | 0.976 |
| DYS461 | 0.00000 | 0.18089 | 0.32157 | 0.000* | 0.117 | 0.000* |
| Y GATA A10 | 1.00000 | 0.38438 | 0.47984 | 0.001* | 0.003* | 0.025* |
| Average | 0.42857 | 0.27223 | 0.24366 | — | — | — |
Abbreviations: CA, Central Africa; EG, Equatorial Guinea; NA, North Africa; STR, short tandem repeat.
The variance values that are italicized and bold are the highest observed for the corresponding STR locus.
*P<0.05.
The origin of the V88 lineages
Although the recently advanced hypothesis that the V88 lineages migrated with Proto-Chadic speakers from the North Africa through the Central Sahara into the Lake Chad Basin,7 given that a high variance was found in lineages from haplogroup R1b1a in the sample from Equatorial Guinea, our results are also compatible with an origin of the V88 lineages in Central-West Africa.
Assuming that Central-West Africa is in fact the place of origin for V88, the arrival of Chadic groups in the Lake Chad Basin, coming from the North, is equally likely as the alternative hypothesis of a migration mediated by the Proto-Chadic speaking people coming from East to West Africa (‘Inter-Saharan' hypothesis), which was previously defended by Lancaster.11
According to Blench's ‘inter-Saharan' hypothesis, Chadic speakers originated during the eastward migration of a pastoralist Cushitic group, from the Nile towards the Lake Chad, with subsequent dispersion in different directions around the lake. The pastoralist nature of the groups involved in the dispersion of haplogroup R1b1a in Central Africa is also supported by the V88 distribution as well as by the distribution of its subclade V69 (the only one found in the African continent).7 In fact, although the origin and dispersion of these lineages seem to be much older than the beginning of the Bantu expansion, which began in this same region in Central Africa,44, 45 its frequency decreases drastically towards the South.
Furthermore, in opposition to the previously suggested direction of migration,7 the presence of these lineages in North Africa could also be explained as the result of a migration of V88 carriers from South to North, possibly during the mid-Holocene.
In summary, altogether, our data are compatible with an origin of the V88-derived allele in Central-West Africa and a later migration (or migrations) across the Sahara to North Africa. Assuming this origin for V88, both the ‘Trans-Saharan' and ‘Inter-Saharan' hypotheses for the arrival of Chadic groups in the Lake Chad Basin are equally likely, as already described in other studies,8, 46 but this model is also compatible with the dispersion of V88 from Central-West to North Africa, contrary to what was proposed by Cruciani et al.7 Nevertheless, further evidence will be required to support this hypothesis, and additional data are crucial to obtain a deeper understanding of the origin and history of this haplogroup, namely from the populations in the proposed paths of migration.
Acknowledgments
This work was supported by the Fundação para a Ciência e Tecnologia through a PhD grant SFRH/BD/36045/2007. IPATIMUP is an Associate Laboratory of the Portuguese Ministry of Science, Technology and Higher Education and is partially supported by FCT, the Portuguese Foundation for Science and Technology.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Jobling MA, Tyler-Smith C. The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet. 2003;4:598–612. doi: 10.1038/nrg1124. [DOI] [PubMed] [Google Scholar]
- Batini C, Ferri G, Destro-Bisol G, et al. Signatures of the preagricultural peopling processes in sub-saharan africa as revealed by the phylogeography of early Y chromosome lineages. Mol Biol Evol. 2011;28:2603–2613. doi: 10.1093/molbev/msr089. [DOI] [PubMed] [Google Scholar]
- Gomes V, Sánchez-Diz P, Amorim A, et al. Digging deeper into East African human Y chromosome lineages. Hum Genet. 2010;127:603–613. doi: 10.1007/s00439-010-0808-5. [DOI] [PubMed] [Google Scholar]
- Cruciani F, Trombetta B, Massaia A, et al. A revised root for the human Y chromosomal phylogenetic tree: the origin of patrilineal diversity in Africa. Am J Hum Genet. 2011;88:814–818. doi: 10.1016/j.ajhg.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berniell-Lee G, Calafell F, Bosch E, et al. Genetic and demographic implications of the Bantu expansion: insights from human paternal lineages. Mol Biol Evol. 2009;26:1581–1589. doi: 10.1093/molbev/msp069. [DOI] [PubMed] [Google Scholar]
- Cruciani F, Santolamazza P, Shen P, et al. A back migration from Asia to sub-Saharan Africa is supported by high-resolution analysis of human Y-chromosome haplotypes. Am J Hum Genet. 2002;70:1197–1214. doi: 10.1086/340257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruciani F, Trombetta B, Sellitto D, et al. Human Y chromosome haplogroup R-V88: a paternal genetic record of early mid Holocene trans-Saharan connections and the spread of Chadic languages. Eur J Hum Genet. 2010;18:800–807. doi: 10.1038/ejhg.2009.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blench R. Archaeology, Language, and the African Past. Oxford, UK: Altamira Press; 2006. [Google Scholar]
- Ehret C. The Civilizations of Africa: A History to 1800. Oxford, UK: University of Virginia Press; 2002. [Google Scholar]
- Cruciani F, Trombetta B, Sellitto D, et al. Chadic languages and Y haplogroups Reply. Eur J Hum Genet. 2010;18:1186–1187. doi: 10.1038/ejhg.2009.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster A. Chadic languages and Y haplogroups. Eur J Hum Genet. 2010;18:1185–1185. doi: 10.1038/ejhg.2010.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The World Factbook . Central Intelligence Agency. Washington, DC; 2009. [Google Scholar]
- Lewis MP.Ethnologue: Languages of the World6th ednOnline version http://www.ethnologue.com/SIL . International: Dallas, TX; 2009 [Google Scholar]
- Lareu MV, Phillips CP, Carracedo A, et al. Investigation of the STR locus HUMTH01 using PCR and two electrophoresis formats: UK and Galician Caucasian population surveys and usefulness in paternity investigations. Forensic Sci Int. 1994;66:41–52. doi: 10.1016/0379-0738(94)90318-2. [DOI] [PubMed] [Google Scholar]
- Gusmão L, Butler JM, Carracedo A, et al. DNA Commission of the International Society of Forensic Genetics (ISFG): an update of the recommendations on the use of Y-STRs in forensic analysis. Int J Legal Med. 2006;120:191–200. doi: 10.1007/s00414-005-0026-1. [DOI] [PubMed] [Google Scholar]
- Arroyo-Pardo E, Gusmão L, López-Parra AM, et al. Genetic variability of 16 Y-chromosome STRs in a sample from Equatorial Guinea (Central Africa) Forensic Sci Int. 2005;149:109–113. doi: 10.1016/j.forsciint.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Karafet TM, Mendez FL, Meilerman MB, et al. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 2008;18:830–838. doi: 10.1101/gr.7172008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beleza S, Gusmão L, Amorim A, et al. The genetic legacy of western Bantu migrations. Hum Genet. 2005;117:366–375. doi: 10.1007/s00439-005-1290-3. [DOI] [PubMed] [Google Scholar]
- Hassan HY, Underhill PA, Cavalli-Sforza LL, et al. Y-chromosome variation among sudanese: restricted gene flow, concordance with language, geography, and history. Am J Phys Anthropol. 2008;137:316–323. doi: 10.1002/ajpa.20876. [DOI] [PubMed] [Google Scholar]
- Semino O, Santachiara-Benerecetti AS, Falaschi F, et al. Ethiopians and Khoisan share the deepest clades of the human Y-chromosome phylogeny. Am J Hum Genet. 2002;70:265–268. doi: 10.1086/338306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Gonder MK, Henn BM, et al. History of click-speaking Populations of Africa inferred from mtDNA and Y chromosome genetic variation. Mol Biol Evol. 2007;24:2180–2195. doi: 10.1093/molbev/msm155. [DOI] [PubMed] [Google Scholar]
- Underhill PA, Passarino G, Lin AA, et al. The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann Hum Genet. 2001;65:43–62. doi: 10.1046/j.1469-1809.2001.6510043.x. [DOI] [PubMed] [Google Scholar]
- Brión M, Sobrino B, Blanco-Verea A, et al. Hierarchical analysis of 30 y-chromosome SNPs in European populations. Int J Legal Med. 2005;119:10–15. doi: 10.1007/s00414-004-0439-2. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resources. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- StatSoft Inc.: STATISTICA (data analysis software system), version 7, 2004
- Bandelt HJ, Forster P, Sykes BC, et al. Mitochondrial portraits of human populations using median networks. Genetics. 1995;141:743–753. doi: 10.1093/genetics/141.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Qamar R, Ayub Q, Mohyuddin A, et al. Y-chromosomal DNA variation in Pakistan. Am J Hum Genet. 2002;70:1107–1124. doi: 10.1086/339929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis JR, Rowold DJ, Regueiro M, et al. The Levant versus the Horn of Africa: Evidence for bidirectional corridors of human migrations. Am J Hum Genet. 2004;74:532–544. doi: 10.1086/382286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood ET, Stover DA, Ehret C, et al. Contrasting patterns of Y chromosome and mtDNA variation in Africa: evidence for sex-biased demographic processes. Eur J Hum Genet. 2005;13:867–876. doi: 10.1038/sj.ejhg.5201408. [DOI] [PubMed] [Google Scholar]
- Rosa A, Ornelas C, Jobling MA, et al. Y-chromosomal diversity in the population of Guinea-Bissau: a multiethnic perspective. BMC Evol Biol. 2007;7:124. doi: 10.1186/1471-2148-7-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruciani F, La Fratta R, Santolamazza P, et al. Phylogeographic analysis of haplogroup E3b (E-M215) y chromosomes reveals multiple migratory events within and out of Africa. Am J Hum Genet. 2004;74:1014–1022. doi: 10.1086/386294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho M, Brito P, Bento AM, et al. Paternal and maternal lineages in Guinea-Bissau population. Forensic Sci Int Genet. 2011;5:114–116. doi: 10.1016/j.fsigen.2010.10.007. [DOI] [PubMed] [Google Scholar]
- Coelho M, Sequeira F, Luiselli D, et al. On the edge of Bantu expansions: mtDNA, Y chromosome and lactase persistence genetic variation in southwestern Angola. BMC Evol Biol. 2009;9:80. doi: 10.1186/1471-2148-9-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes V, Alves C, Amorim A, et al. Nilotes from Karamoja, Uganda: Haplotype data defined by 17 Y-chromosome STRs. Forensic Sci Int Genet. 2010;4:E83–E86. doi: 10.1016/j.fsigen.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Willuweit S, Roewer L. Y chromosome haplotype reference database (YHRD): update. Forensic Sci Int Genet. 2007;1:83–87. doi: 10.1016/j.fsigen.2007.01.017. [DOI] [PubMed] [Google Scholar]
- Hurles ME, Nicholson J, Bosch E, et al. Y chromosomal evidence for the origins of oceanic-speaking peoples. Genetics. 2002;160:289–303. doi: 10.1093/genetics/160.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M. A measure of population subdivision based on microsatellite allele frequencies. Genetics. 1995;139:457–462. doi: 10.1093/genetics/139.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semino O, Passarino G, Oefner PJ, et al. The genetic legacy of paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective. Science. 2000;290:1155–1159. doi: 10.1126/science.290.5494.1155. [DOI] [PubMed] [Google Scholar]
- Wells RS, Yuldasheva N, Ruzibakiev R, et al. The Eurasian Heartland: A continental perspective on Y-chromosome diversity. Proc Natl Acad Sci USA. 2001;98:10244–10249. doi: 10.1073/pnas.171305098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaresque P, Bowden GR, Adams SM, et al. A predominantly neolithic origin for European paternal lineages. PLoS Biol. 2010;8:e1000285. doi: 10.1371/journal.pbio.1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby GB, Brisighelli F, Sanchez-Diz P, et al. The peopling of Europe and the cautionary tale of Y chromosome lineage R-M269. Proc Biol Sci Royal Soc. 2012;279:884–892. doi: 10.1098/rspb.2011.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myres NM, Rootsi S, Lin AA, et al. A major Y-chromosome haplogroup R1b Holocene era founder effect in Central and Western Europe. Eur J Hum Genet. 2011;19:95–101. doi: 10.1038/ejhg.2010.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaza S, Salas A, Calafell F, et al. Insights into the western Bantu dispersal: mtDNA lineage analysis in Angola. Hum Genet. 2004;115:439–447. doi: 10.1007/s00439-004-1164-0. [DOI] [PubMed] [Google Scholar]
- Vansina J. New linguistic evidence and the Bantu Expansion. J Afr Hist. 1995;36:173–195. [Google Scholar]
- Cerny V, Pereira L, Musilova E, et al. Genetic Structure of Pastoral and Farmer Populations in the African Sahel. Mol Biol Evol. 2011;28:2491–2500. doi: 10.1093/molbev/msr067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.