Abstract
Human Cripto-1 (CR-1) plays an important role in regulating embryonic development while also regulating various stages of tumor progression. However, mechanisms that regulate CR-1 expression during embryogenesis and tumorigenesis are still not well defined. In the present study, we investigated the effects of two nuclear receptors, liver receptor homolog (LRH)-1 and germ cell nuclear factor receptor (GCNF) and epigenetic modifications on CR-1 gene expression in NTERA-2 human embryonal carcinoma cells and in breast cancer cells. CR-1 expression in NTERA-2 cells was positively regulated by LRH-1 through direct binding to a DR0 element within the CR-1 promoter, while GCNF strongly suppressed CR-1 expression in these cells. In addition, the CR-1 promoter was unmethylated in NTERA-2 cells, while T47D, ZR75-1 and MCF7 breast cancer cells showed high levels of CR-1 promoter methylation and low CR-1 mRNA and protein expression. Treatment of breast cancer cells with a demethylating agent and histone deacetylase inhibitors reduced methylation of the CR-1 promoter and reactivated CR-1 mRNA and protein expression in these cells, promoting migration and invasion of breast cancer cells. Analysis of a breast cancer tissue array revealed that CR-1 was highly expressed in the majority of human breast tumors, suggesting that CR-1 expression in breast cancer cell lines might not be representative of in vivo expression. Collectively, these findings offer some insight into the transcriptional regulation of CR-1 gene expression and its critical role in the pathogenesis of human cancer.
Keywords: Cripto-1, GCNF, LRH-1, DNA methylation
Introduction
Human CR-1 is an embryonic gene that is a member of the Epidermal Growth Factor (EGF)/CR-1-FRL-1-Cryptic (CFC) family and that performs pivotal functions during embryonic development and oncogenic transformation (Bianco et al., 2010; de Castro et al., 2010). During embryogenesis, CR-1 functions as an obligatory co-receptor for the transforming growth factor-β (TGF-β) family members, Nodal and Growth and Differentiation factor 1 and 3 (GDF1/3) (Bianco et al., 2010; de Castro et al., 2010). CR-1 is essential for Nodal to induce activation of serine-threonine kinase activin type I receptors Alk4 and Alk7 and activin type II receptor complex, which in turn triggers phosphorylation of cytoplasmic receptor-activated Smad proteins, Smad-2 and Smad-3, that in a complex with Smad-4 translocate into the nucleus and interact with other co-activators such as FoxH1 to activate transcription of specific target genes (Bianco et al., 2002; Yeo and Whitman, 2001). CR-1 together with Nodal performs key regulatory functions during embryonic development, such as regulating anterior/posterior axis formation and left/right body asymmetry, mesoendoderm specification during gastrulation and promoting cardiomyocyte differentiation (Bianco et al., 2010). In this context, deletion of the mouse CR-1 gene by homologous recombination during embryonic development is lethal and mouse embryos die around day 6.5 of embyogenesis as a result of defects in mesoderm formation, axial organization and cardiac development (Ding et al., 1998; Xu et al., 1998). CR-1 has also been shown to be expressed in human and mouse undifferentiated embryonic stem (ES) cells, regulating pluripotentiality and self-renewal (Assou et al., 2007; Bianco et al., 2010; Wei et al., 2005). Furthermore, CR-1 is a direct downstream target gene of the pluripotentiality transciption factors Oct-4 and Nanog and is re-expressed in induced puripotent stem cells (iPSCs) derived from adult differentiated cells engineered to express Oct-4, Nanog, Kfl4 and c-myc (Aasen et al., 2008; Aoi et al., 2008; Chang et al., 2009; Loh et al., 2006). In the adult, CR-1 is expressed at very low levels, probably in the stem cell compartment of adult tissues (Bianco et al., 2010). For instance, CR-1 can regulate self-renewal of hematopoietic stem cells in the hypoxic bone marrow niche together with the heath shock 78 kDa glucose-regulated protein (GRP78) (Miharada et al., 2011). In contrast, high levels of CR-1 expression are found in different types of human carcinomas, including breast, colon, pancreas, stomach, lung, testis, prostate and ovary (de Castro et al., 2010). Experiments in vitro have indeed confirmed that CR-1 can function as an oncogene by increasing migration, invasion and epithelial to mesenchymal transition of several human and mouse mammary epithelial cells and by promoting tumor angiogenesis in vitro and in vivo (Bianco et al., 2005; Nagaoka et al., 2012; Rangel et al., 2012). However, regulatory mechanisms that might drive CR-1 re-expression in cancer cells are not well defined. We have previously shown, using a CR-1 promoter luciferase report assay, that the promoter region of the CR-1 gene contains Smad binding elements, hypoxia responsive elements and T-cell factor/lymphoid enhancer factor (Tcf/Lef) binding elements (Bianco et al., 2009; Hamada et al., 2007; Mancino et al., 2008a). In fact, CR-1 gene expression is modulated by TGF-β family members, the transcription factor hypoxia inducible factor-1 α (HIF-1α) and the canonical Wnt/β-catenin signaling pathway. CR-1 is also directly repressed by the orphan nuclear receptor germ cell nuclear factor (GCNF) during retinoic acid induced differentiation of human embryonal carcinoma cells following binding of GCNF to a DR0 motif in the human CR-1 promoter region (Hentschke et al., 2006). GCNF is also required to repress expression of Oct-4, Nanog and Sox-2 upon differentation of ES cells with retinoic acid (Gu et al., 2005b). Furthermore, GCNF binding to the Oct-4 promoter triggers initiation of promoter DNA methylation by recruitment of methyl-CpG binding domain and DNA methylatransferases to the Oct-4 promoter thereby initiating epigenetic gen silencing of the Oct4 locus during ES cell differentiation (Gu et al., 2011). DR0 elements can also bind to other orphan nuclear receptors, such as liver receptor homolog-1 (LRH-1), which is essential to maintain Oct-4 expression in undifferentiated ES cells (Gu et al., 2005a). Therefore, GCNF and LRH-1 regulate ES cells pluripotency and differentiation by competing for the same regulatory element within the Oct-4 promoter. In the present study, we investigated the effects of GNCF and LRH-1 orphan nuclear receptors on CR-1 gene expression in human embryonal and breast carcinoma cell lines as this relates to the methylation status of the CR-1 gene. We also analyzed expression of CR-1, GCNF and LRH-1 in human breast ductal invasive carcinomas using a tissue microarray.
Materials and Methods
Cell Culture
Human NTERA-2 embryonal carcinoma cells were grown in McCoy’s 5A medium containing 15% fetal bovine serum (FBS). NCCIT human embryonal carcinoma cells and MCF7 human breast cancer cells were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% FBS. ZR75-1 and T47D human breast cancer cells were grown in RPMI-1640 medium containing 10% FBS. For the sphere forming assay, MCF7 Neo and MCF7 CR-1 cells (Normanno et al., 2004) were seeded in 24-well ultra-low attachment plates at 1000 cells/well in 500 mictroliters of MammoCult® Human Medium Kit (Catalog # 05620, Stem Cell Technologies, Vancouver, Canada). Spheres were counted between days 7 and 10 after plating, using Gel Count TM - Oxford OPTRONIX version 1.03.
Treatment of NTERA-2 embryonal carcinoma cells with retinoic acid (RA)
NTERA-2 cells were seeded in 60 mm plates at a density of 8×105 cells/dish. The following day, cells were treated with RA (Sigma, St. Lois, MO) (5 or 10 μM) for 48 and 72 hours. Cells were then lysed for RNA and protein analysis as described below.
Treatment of breast cancer cells with 5-aza-2′-Deoxycytidine (5-aza-dC), Thricostatin A (TSA) and Valproic acid (VA)
MCF7, T47D, and ZR75-1 breast cancer cells were seeded in 100 mm plates (5×105 cells/plate). The following day, culture medium was changed and cells were treated with 2.5 μM 5-aza-dC (Sigma) for 96 hours or 100 ng/ml TSA (Sigma) or 3 μM VA (Sigma) for 12 hours. For the combination treatments, 5-aza-dC was present in the medium for 96 hours and TSA or VA were added for the last 12 hours. Cells were then lysed for RNA and protein analysis, as described below.
Migration and invasion assays
Migration and invasion assays were performed in fibronectin-coated or matrigel-coated Boyden chambers (Chemicon, Temecula, CA), as previously described (Bianco et al., 2008). MCF7 cells were pretreated with 2.5 μM 5-aza-dC (Sigma) for 72 hr. Cells were then seeded in 12-well plates (2×105 per well) and incubated in the upper Boyden chambers with 100 ng/ml TSA (Sigma) or 3 μM VA (Sigma) for 12 hours. In all the experiments 5% FBS was used as chemoattractant in the lower chamber. These experiments were repeated three times with duplicate samples. For the migration and invasion assays performed with siRNA transfected cells, MCF7 cells after 48 hr treatment with 2.5 μM 5-aza-dC were transfected with 100 nM of ON-TARGETplus smartpool siRNA anti-human CR-1 (Dharmacon, Chicago, IL) or with 100 nM of ON-TARGETplus non-targeting pool siRNA (Dharmacon) using the DharmaFECT 1 (Dharmacon) transfection reagent, as suggested by the supplier. The following day MCF7 cells were plated in Boyden chambers for migration and invasion assays in the presence of TSA or VA, as described above.
Transfection of NTERA-2 cells with siRNAs
NTERA-2 cells were seeded in complete medium in 60 mm plates (5×105 cells/plate). The following day the cells were transfected with 100 nM of ON-TARGETplus smartpool siRNA anti-human LRH-1 or anti-human GCNF (Dharmacon, Chicago, IL) or with 100 nM of ON-TARGETplus non-targeting pool siRNA (Dharmacon) using the DharmaFECT 1 (Dharmacon) transfection reagent, as suggested by the supplier. Forty-eight hours after transfection, cells were lysed for RNA or protein extraction and analyzed by real-time PCR or Western blot analysis.
Dual-luciferase assay
Dual-luciferase reporter assays were performed as previously described (Mancino et al., 2008a). Briefly, NTERA-2 cells (2.5×105 cells/well in 12-well plates) were co-transfected with a 2.5 kb human CR-1 promoter luciferase vector (0.5 μg/well) and increasing concentrations of either GCNF or LRH-1 expression vectors (kindly provided by Dr. Cooney AJ, Baylor College of Medicine, Houston, TX) using Fugene 6 (Roche, Nutley, NJ). A renilla luciferase control reporter vector (Promega, Madison, WI) was co-transfected into the cells to normalize for transfection efficiency. Twenty-four hours after transfection, the cells were lysed and luciferase activity was measured using the Dual Luciferase Reporter Assay Kit (Promega), according to the manufacturer’s instructions.
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed as previously described, using the ChIP kit (Upstate Biotechnology, Lake Placid, NY, USA) (Bianco et al., 2009). Briefly, NTERA-2 cells (1×106 cells in 100 mm-diameter plates) were incubated under normoxia or hypoxia (0.5% O2) (BioSpherix, Redfield, NY, USA) for 24 hours. Following crosslinking using 1% formaldeyde at 37°C for 10 minutes, NTERA-2 cells were resuspended in SDS lysis buffer (Upstate Biotechnology) and DNA was sheared to small fragments by sonication. After preclearing the recovered supernatant with salmon sperm DNA/protein A agorose slurry, samples were incubated with either anti-LRH-1α rabbit polyclonal antibody (ab18293, Abcam, Cambridge, MA) or an isotype control IgG overnight at 4°C. After incubation with salmon sperm DNA/protein A agorose beads, the immunoprecipitated DNA/protein complexes were washed and eluted from the beads with 1% SDS and 0.1 M NaHCO3 solution. Protein/DNA crosslinks were reversed by adding 5 M NaCl at 65°C for 4 hours and DNA was recovered by phenol/chloroform extraction and ethanol precipitation. PCR was done on the extracted DNA using human CR-1 promoter specific primers (DR0): forward (5′-TTT GTT GTT GAA GAA GGA GAA TCC-3′) and reverse (5′-GGT CGT AGC AGA AGC AGG AGC AAG G-3′) with Platinum PCR Super mix (Invitrogen) for 30 cycles for 30 sec at 94° C, 30 sec at 60° C, and 30 sec at 72° C. The input control and hypoxia samples prior to immunoprecipitation were used as loading control for the PCR.
Quantitative real-time PCR
NTERA-2, NCCIT, MCF7, T47D and ZR75-1 cells were seeded in 60 mm plates (5×105 cells/plate) and 48 hours later total RNA was isolated using RNeasy mini kit (Qiagen, Valencia, CA, USA) according to manufacturer’s instruction. RNA was also isolated from NTERA-2 cells treated with RA (5 or 10 μM) for 48 and 72 hours. One microgram of total RNA was used for cDNA synthesis using the RETROscript kit (Ambion Applied Biosystems, Foster City, CA, USA) following the manufacturer’s protocol. Quantitative real-time PCR was performed on Startagene MX300P using Brilliant II SYBR Green PCR master mix (Stratagene, Cedar Creek, TX). Quantification of mRNA expression was performed by using the DDct method (Dct sample-Dct calibrator). The thermal cycling conditions and the primers for GAPDH and CR-1 have been previously described (Bianco et al., 2006)). The primers used for GCNF and LRH-1 are: GCNF forward 5′-GTG TGT GGA GAT AAA GTG TCT GGG-3′; GCNF reverse 5′-CTT AAG TCC ATT GGC TCG GAT GAG-3′; LRH-1 forward 5′-TGC AGG CTG AAG AAT ACC TCT-3′; LRH-1 reverse 5′-GCA TGC AAC ATT TCA ATG AG-3′. For the real time PCR on MCF7 Neo and MCF7 CR-1 sphere RNA, the following primers were used: NANOG forward 5′-CTGTGATTTGTGGGCCTGAA 3′ and reverse 5′-TGTTTGCCTTTGGGACTGGT3′; OCT-4 forward 5 ′-CTTGCTGCAGAAGTGGGTGGAGGAA-3′ and 5′-CTGCAGTGTGGGTTTCGGGCA-3 ′; CD133 forward 5 ′-AGATTGTCTACTATGAAGCAGGGA-3′ and 5′-TCTGTCGCTGGTGCATTTCT.
Western blot analysis
Protein extraction and Western blot analysis were performed as previously described (Bianco et al., 2002). The following primary antibodies were used: anti CR-1 rabbit monoclonal antibody (1:1000, Epitomics, Burlingame, CA); anti-LRH-1 mouse monoclonal antibody (1:1000, R&D systems, Minneapolis, MN); anti-GCNF antibody (1:1000, Abcam) and anti-β-actin mouse monoclonal antibody (1:20000, Sigma).
DNA methylation of the human CR-1 promoter
Genomic DNA was isolated using Purelink Genomic DNA isolation kit (Invitrogen), according to manufacturer’s instruction. Methylation of genomic DNA was analyzed using the Methyl-profiler DNA methylation Enzyme kit (SABioscience, Frederick, MD). Genomic DNA (0.5 μg for each digestion) was digested with methylation-sensitive restriction enzyme (Enzyme A), with methylation-dependent restriction enzyme (Enzyme B), with both Enzymes A and B or with no enzymes (mock digestion) for 4 hours at 37° C. After inactivation of the enzymes at 65° C for 20 minutes, DNA digestions were amplified with specific primers for CpG islands in the CR-1 promoter (SABioscience) using the Methyl-Profiler DNA Methylation qPCR assay (SABioscience), according to the manufacturer’s instruction. The product of mock digestion contains the input genomic DNA, the product of methylation-sensitive restriction enzyme digestion contains hypermethylated DNA, while the product of methylation-dependent restriction enzyme contains unmethylated DNA. The double digestion was used to assess the success of both enzymatic digestions.
Immunofluorescence analysis
MCF7 cells were seeded in 4 well Chamber slides (LAB-TEK II, Nulge Nunc International, Naperville, IL) at 10×104 cells/well. After twenty-four hours, cells were treated with 5-aza-dC and TSA or VA, as described above. Immunofluorescence analysis was then performed as previously described using an anti-CR-1 rabbit polyclonal antibody (Rockland, Gilbertsville, PA, USA) diluted at 1:100 (Bianco et al., 2008). Slides for immunofluorescence analysis were stained with 4′, 6′-diamino-2-phenylindole (DAPI).
Tissue microarray analysis
Breast tumor tissue arrays were purchased from Biomax US (Rockville, MD) and contain 70 duplicated cases of invasive breast ductal carcinomas. Expression of CR-1, LRH-1 and GCNF was assessed using immunohistochemistry (IHC), as previously described (Bianco et al., 2009). The antibodies used were: anti CR-1 rabbit polyclonal antibody (1:500 dilution) (Rockland), anti-LRH-1 rabbit polyclonal (1:200 dilution) (ab18293, Abcam, Cambridge MA) and anti-GCNF rabbit polyclonal (1:200 dilution) (GeneTex, Irvine, CA). Analysis of CR-1, LRH-1 and GCNF immunostaining was performed by two independent observers. Each of the two samples from every tumor core was examined and scored separately. Immunohistochemistal staining was scored as negative (0), weak (1), moderate (2) and strong (3).
Statistical analysis
The statistical significance of the various groups in the different experiments was calculated with the nonparametric Mann-Whitney U-test using the Statistical Package for Social Sciences software package, version 11.0 (SPSS, Chicago, IL, USA). Statistical tests were two-sided and data were considered statistically significant at P <0.05. Chi Square analysis and Student T test were used to assess P values in the tissue microarray analysis. The Pearson’s linear correlation coefficient was used to calculate the statistical relationships between CR-1, GCNF and LRH-1 expression in breast carcinomas.
Results
LRH-1 binds to the CR-1 promoter and positively regulates CR-1 promoter luciferase activity in NTERA-2 human embryonal carcinoma cells
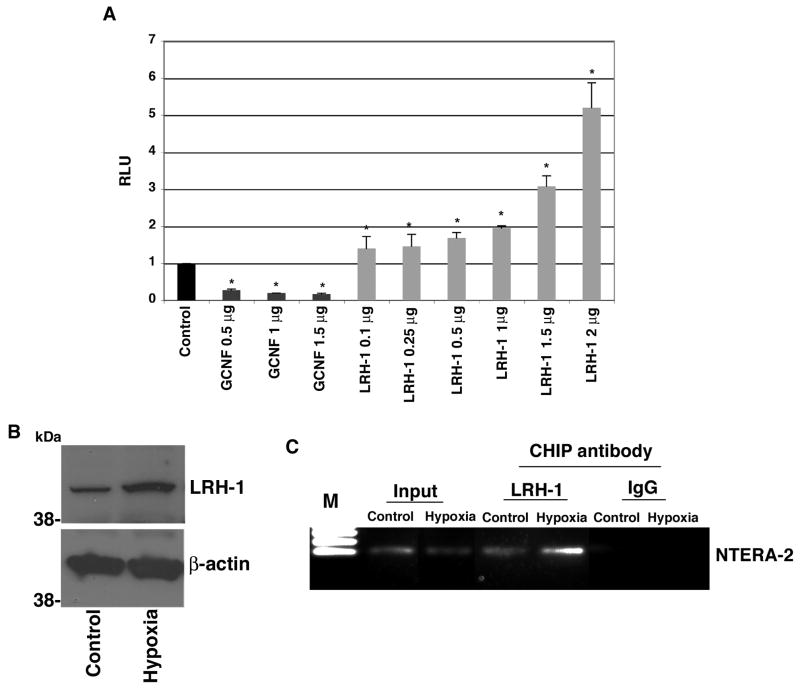
Previous studies have shown that the nuclear receptor GCNF is induced in response to RA treatment in NTERA-2 cells and that it binds to a DR0 element in the CR-1 promoter, thereby repressing CR-1 transcription during RA-induced differentiation of NTERA-2 cells (Hentschke et al., 2006). Since DR0 elements can also bind other nuclear receptors (Mullen et al., 2007), we investigated whether another nuclear receptor, LRH-1, can also bind and modulate CR-1 promoter activity in NTERA-2 cells. NTERA-2 cells were transfected with a full-length CR-1 promoter luciferase reporter vector together with LRH-1 or GCNF expression vectors (Mancino et al., 2008b). In agreement with previous results (Hentschke et al., 2006), GCNF strongly suppressed CR-1 promoter luciferase activity in NTERA-2 cells (Fig. 1A). In contrast, LRH-1 induced a dose-dependent increase in CR-1 promoter luciferase activity in NTERA-2 cells, as compared to control cells expressing the CR-1 promoter luciferase vector only (Fig. 1A). Therefore, these results suggest that GCNF and LRH-1 nuclear receptors have opposite regulatory effects on CR-1 promoter activity. We then investigated if LRH-1, like GCNF, can directly interact with the CR-1 promoter using a ChIP assay (Hentschke et al., 2006). We have previously shown that HIF-1α, induced by hypoxic conditions, enhances CR-1 gene expression in embryonal carcinoma cells (Bianco et al., 2009). Since analysis of the LRH-1 promoter region identified a hypoxia responsive element (HRE) (Bianco and Salomon, unpublished data), we assessed whether hypoxic conditions would enhance LRH-1 expression and recruit LRH-1 to the CR-1 promoter. Therefore, NTERA-2 cells were grown under normoxic (20% oxygen) or hypoxic (0.5% oxygen) conditions for 24 hours. As shown in Fig. 1B, hypoxia induced an increase in LRH-1 protein expression, supporting the presence of a functional HRE element in the LRH-1 promoter. ChIP assay was then performed to measure recruitment of LRH-1 to the DR0 region of the human CR-1 promoter. As shown in Fig. 1C, the anti-LRH-1 antibody, but not the control IgG antibody, precipitated the human CR-1 promoter spanning the specific DR0 region in NTERA-2 cells and a significant increase in LRH-1 binding to DNA was detected in NTERA-2 cells following hypoxia treatment as compared to control unstimulated cells. Thus, LRH-1 enhances CR-1 promoter luciferase activity through binding to a specific DR0 motif within the CR-1 promoter in NTERA-2 cells. A positive modulation of CR-1 luciferase reporter acitivity by LRH-1 was also detected in NCCIT human embryonal carcinoma cells (data not shown).
Figure 1. LRH-1 binds to the CR-1 promoter and enhances CR-1 promoter luciferase activity in NTERA-2 cells.
A: Dual luciferase assay of transiently co-transfected NTERA-2 cells with a full-length CR-1 promoter luciferase reporter vector and GCNF or LRH-1 expression plasmids. These results are the mean ± SD of triplicates of three separate experiments (*P<0.01). B: Western blot analysis for LRH-1 and β-actin in NTERA-2 cells grown in normoxic (20% oxygen) or hypoxic (0.5% oxygen) conditions for 24 hours. C: ChIP assay in NTETRA-2 cells following 24 hours of incubation in a hypoxic chamber. The cross-linked protein-DNA complexes were immunoprecipitated with anti-LRH-1 antibody or with an isotype control IgG as negative control, and the purified DNA was amplified by PCR using specific primers spanning the DR0 region within the CR-1 promoter.
LRH-1 is required to maintain high levels of CR-1 expression in NTERA-2 cells
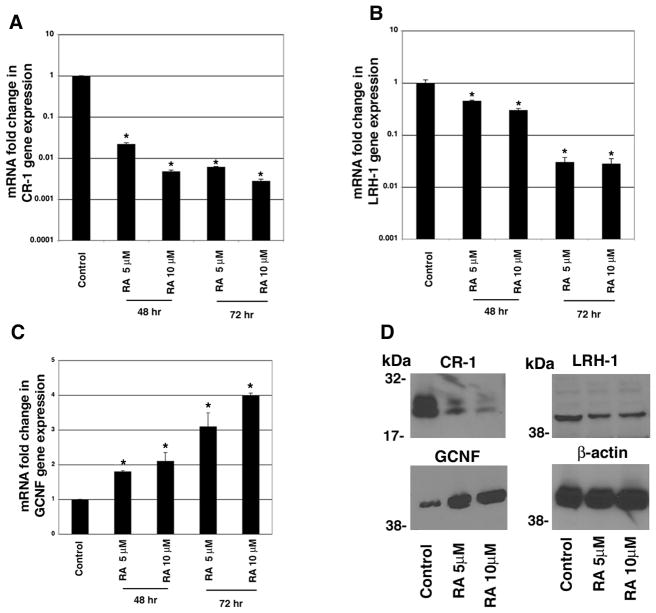
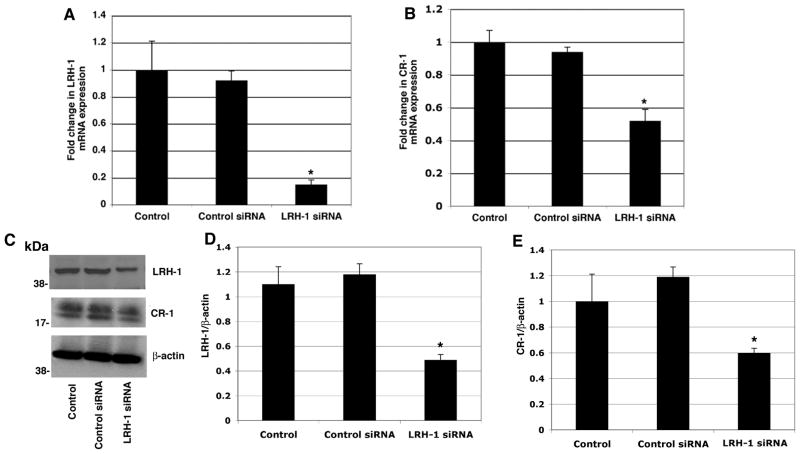
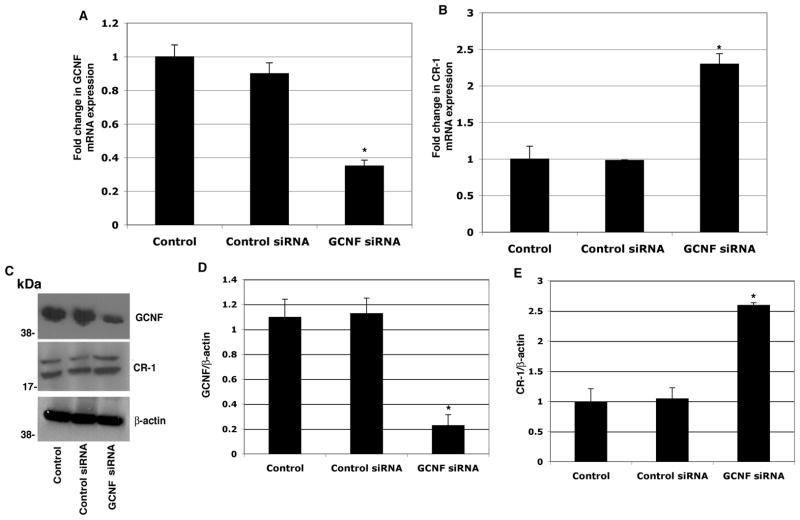
GCNF upon RA treatment is recruited to the CR-1 promoter and mediates repression of CR-1 expression in NTERA-2 cells (Hentschke et al., 2006). We therefore investigated the effects of RA treatment on GCNF, LRH-1 and CR-1 expression in NTERA-2 cells. A significant dose-dependent downregulation of CR-1 mRNA and protein expression was observed following treatment with RA for 48 and 72 hours, as assessed by real-time PCR and Western blot analysis (Fig. 2A and D). In contrast, GCNF mRNA and protein were found to be upregulated by RA treatment in NTERA-2 cells (Fig. 2C and D). LRH-1, similarly to CR-1, was also decreased in a dose-dependent manner by RA treatment in NTERA-2 cells (Fig. 2B and D). To further understand the involvement of LRH-1 in regulating CR-1 gene expression, LRH-1 expression was knocked down with small interfering RNA (siRNA) in NTERA-2 cells. Transfection of a pool of specific LRH-1 siRNAs in NTERA-2 cells strongly decreased LRH-1 mRNA and protein expression, as compared to NTERA-2 cells transfected with a control non-silencing siRNA (Fig. 3A, C, D). Interestingly, downregulation of LRH-1 in NTERA-2 cells was accompanied by an approximately 40–60% reduction in CR-1 mRNA and protein expression, as compared to cells transfected with control non-silencing siRNA (Fig. 3B, C, E). Conversely, CR-1 mRNA and protein expression were significantly enhanced following GCNF siRNA transfection in NTERA-2 cells, as shown in Fig. 4. Therefore, these results support a role of the nuclear receptor LRH-1 in positively regulating CR-1 mRNA and protein expression in NTERA-2 cells, while confirming the repressive activity of GCNF on CR-1 expression in NTERA-2 cells.
Figure 2. RA treatment downregulates CR-1 and LRH-1 and upregulates GCNF expression in NTERA-2 cells.
Quantitative real time PCR for CR-1 (A), LRH-1 (B) and GCNF (C) in NTERA-2 cells treated with RA for 48 and 72 hours. Control cells were treated with vehicle DMSO. mRNA levels were normalized to GAPDH expression. The values shown are averages ± SD for duplicate samples from one of three experiments. *P<0.05, as compared to control DMSO treated NTERA-2 cells. D: Western blot analysis for CR-1, LRH-1, GCNF and βactin in NTERA-2 cells treated for 72 hours with 5 or 10 μM RA. Control cells were treated with vehicle DMSO.
Figure 3. Knockdown of LRH-1 expression by siRNA downregulates CR-1 mRNA and protein expression in NTERA-2 cells.
Real time PCR for LRH-1 (A) and CR-1 (B) in NTERA-2 cells transfected with control or LRH-1 siRNAs. Data are representative of three experiments with duplicate samples. *P<0.05, as compared to control non-transfected samples. C: Western blot analysis for LRH-1, CR-1 and β-actin in siRNA transfected cells. D, E: Fold difference in LRH-1 and CR-1 expression by densitometric analysis of LRH-1 or CR-1 in NTERA-2 cell lysates normalized to β-actin content. *P<0.05, as compared to control non-transfected samples.
Figure 4. Knockdown of GCNF expression by siRNA upregulates CR-1 mRNA and protein expression in NTERA-2 cells.
Real time PCR for GCNF (A) and CR-1 (B) in NTERA-2 cells transfected with control or GCNF siRNAs. Data are representative of three experiments with duplicate samples. *P<0.05, as compared to control non-transfected samples. C Western blot analysis for GCNF, CR-1 and β-actin in siRNA transfected cells. D, E: Fold difference in GCNF and CR-1 expression by densitometric analysis of GCNF or CR-1 in NTERA-2 cell lysates normalized to β-actin content. *P<0.05, as compared to control non-transfected samples.
Methylation of the CR-1 promoter in human breast carcinoma cell lines
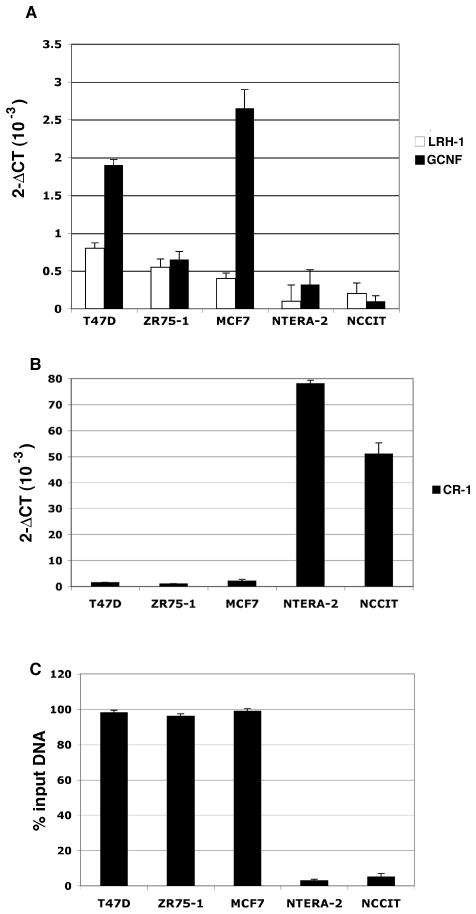
To further delineate the role performed by the nuclear receptors GCNF and LRH-1 in regulating CR-1 gene expression in different cell lines, we examined GCNF and LRH-1 expression in several human breast cancer cell lines (MCF7, T47D and ZR75-1). We therefore assessed by real-time PCR the expression levels of GCNF and LRH-1 in human embryonal and breast carcinoma cells. Human breast cancer cells showed high levels of GCNF and LRH-1 mRNA expression as compared to NTERA-2 and NCCIT cells (Fig. 5A). In contrast, CR-1 was highly expressed in embryonal carcinoma cells while all the breast cancer cell lines showed low levels of CR-1 mRNA (Fig. 5B). Transfection of T47D, ZR75-1 and MCF-7 with a GCNF siRNA was not able to restore CR-1 expression in these cells (data not shown). Since GCNF has been shown to promote repression and silencing of genes through epigenetic modifications (Gu et al., 2006; Sato et al., 2006), especially DNA methylation, we investigated if methylation of the CR-1 promoter might be responsible for CR-1 silencing in breast cancer cells. We have previously identified a cluster of CpG nucleotides (CpG island) in a region surrounding the transcriptional start site (TSS) of the human CR-1 gene (Watanabe et al., 2010). We therefore assessed methylation of the CR-1 promoter in breast and embryonal carcinoma cells using a genomic DNA methylation PCR assay. Following digestion of genomic DNA isolated from breast and embryonal carcinoma cells with methylation-sensitive and methylation-dependent restriction enzymes, we amplified CpG islands within the CR-1 promoter by real-time PCR. As shown in Fig. 5C, more than 95% of the CR-1 promoter was highly methylated in all the breast cancer cell lines. In contrast, the CR-1 promoter was unmethylated in the NCCIT and NTERA-2 embryonal carcinoma cells. Thus, hypermethylation of the CR-1 promoter in breast cancer cell lines might result in inhibition of CR-1 expression in these cells.
Fig. 5. GCNF, LRH-1 and CR-1 mRNA expression and methylation status of the CR-1 promoter in human breast and embryonal carcinoma cells.
A, B: Real time PCR for GCNF, LRH-1 and CR-1 in breast T47D, MCF7 and ZR75-1 and embryonal NCCIT and NTERA-2 carcinoma cells. Data were normalized to GAPDH expression and are representative of three experiments with duplicate samples. C: Methylation of the CR-1 promoter in breast and embryonal carcinoma cells after enzymatic digestion of genomic DNA with methylation sensitive or methylation dependent restriction enzymes (see Materials and Methods). DNA was subsequently amplified using specific primers spanning CpG islands in the CR-1 promoter. Experiment was repeated three times on duplicate samples.
DNA methyltransferase (DNMT) and/or histone deacetylase (HDAC) inhibitors restore CR-1 mRNA and protein expression in breast cancer cells
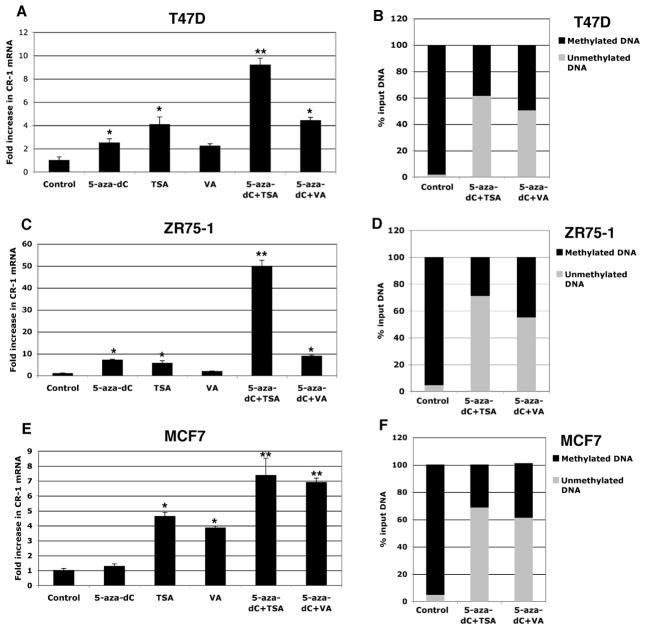
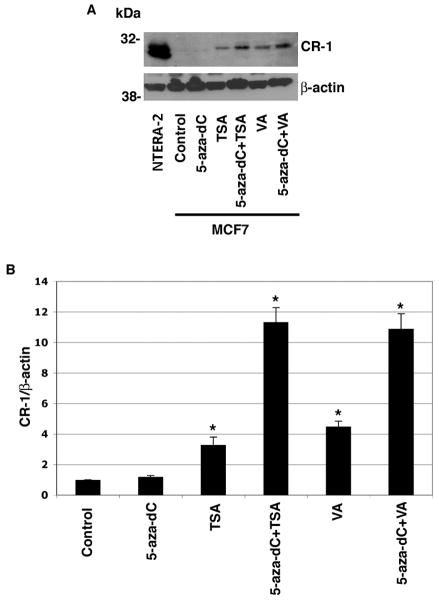
To ascertain if hypermethylation of the CR-1 promoter results in transcriptional silencing of the CR-1 gene in breast cancer cells, MCF7, T47D and ZR75-1 cells were treated with inhibitors of DNMT (5-aza-dC) and/or HDAC (Thricostatin A [TSA] or Valproic acid [VA]). Treatment of breast cancer cell lines with 5-aza-dC, TSA or VA alone induced a modest but statistically significant increase in CR-1 mRNA expression, as assessed by real-time PCR (Fig. 6A, C and E). Combination of 5-aza-dC with TSA or VA strongly enhanced CR-1 mRNA expression in all of the breast cancer cell lines. Methylation specific PCR analysis also confirmed that treatment with 5-aza-dC in combination with TSA or VA partially, but significantly, demethylated the CpG islands within the CR-1 promoter in MCF7, ZR75-1 and T47D cells (Fig. 6B, D and F). We finally evaluated if demethylation of the CR-1 promoter by 5-aza-dC, TSA and VA treatments can facilitate re-expression of CR-1 protein in breast cancer cells. We therefore performed Western blot analysis and immunofluorescence staining for CR-1 in MCF7 cells treated with 5-aza-dC, TSA and VA. CR-1 protein could not be detected by Western blot analysis in MCF7 cells, while NTERA-2 cells showed high levels of CR-1 protein expression (Fig 7A). Re-expression of CR-1 protein was induced by TSA or VA treatments alone or by a combination of 5-aza-dC with TSA or VA in MCF7 cells (Fig. 7A and B). In addition, MCF7 treated with 5-aza-dC in combination with TSA or VA showed a strong punctate membrane immunofluorescent staining for CR-1 as compared to control untreated MCF7 cells (Fig. 8). Therefore, demethylation of CpG islands within the CR-1 promoter induces re-expression of CR-1 mRNA and protein in breast cancer cell lines.
Fig. 6. DNMT and HDAC inhibitors reactivate CR-1 mRNA expression in breast cancer cell lines.
A, C, E: Real time PCR for CR-1 in T47D, ZR75-1 and MCF7 cells treated with 5-aza-dC, TSA or VA alone or in combination. Real time data were normalized to GAPDH expression. Data are representative of three experiments with duplicate samples. *P<0.05. **P<0.01. B, D, F: Methylation of the CR-1 promoter in drug-treated and vehicle-treated T47D, ZR-75-1 and MCF7 cells. A representative example of two independent experiments with duplicate samples is presented.
Fig. 7. DNMT and HDAC inhibitors reactivate CR-1 protein expression in MCF7 cells.
A: Western blot analysis for CR-1 in drug-treated and vehicle-treated MCF-7 cells. NTERA-2 cells were used as positive control. β-actin was used as a protein loading control. B: Fold difference in CR-1 protein expression by densitometric analysis in MCF7 cell lysates normalized to β-actin content. *P<0.01, as compared to control vehicle-treated cells.
Fig. 8. Immunofluorescence staining for CR-1 in vehicle-treated or DNMT and HDAC inhibitors-treated MCF7 cells.
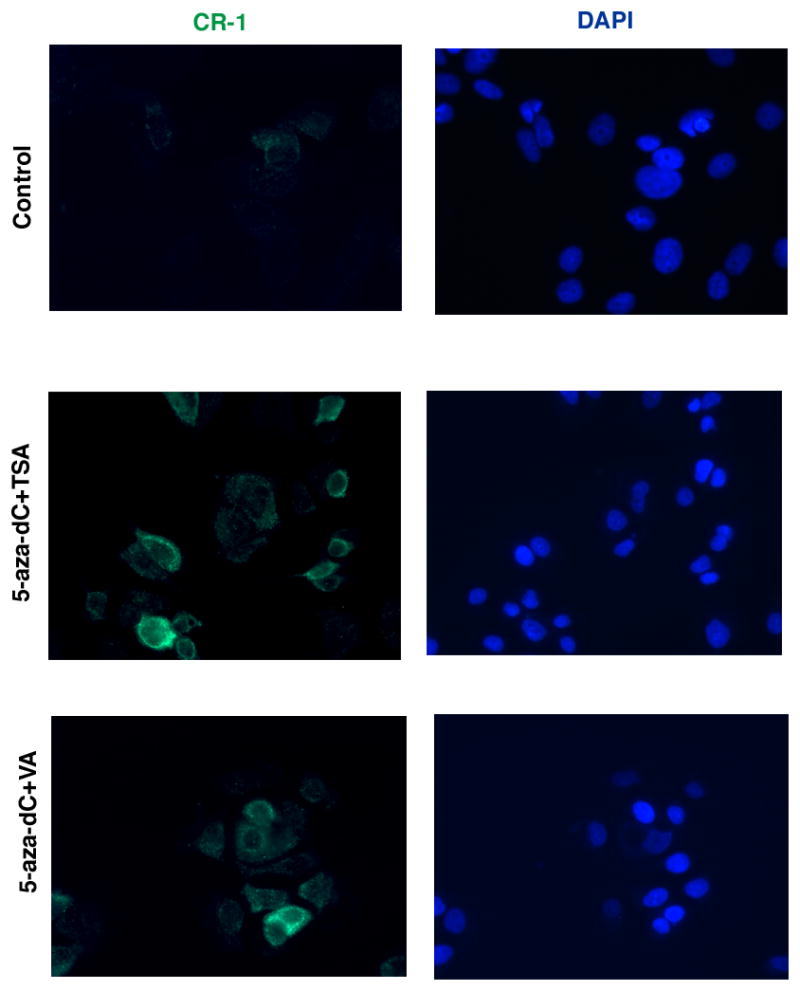
DAPI staining of nuclei is shown in blue.
Demethylation of the CR-1 promoter in MCF7 cells enhances their migratory and invasive behavior
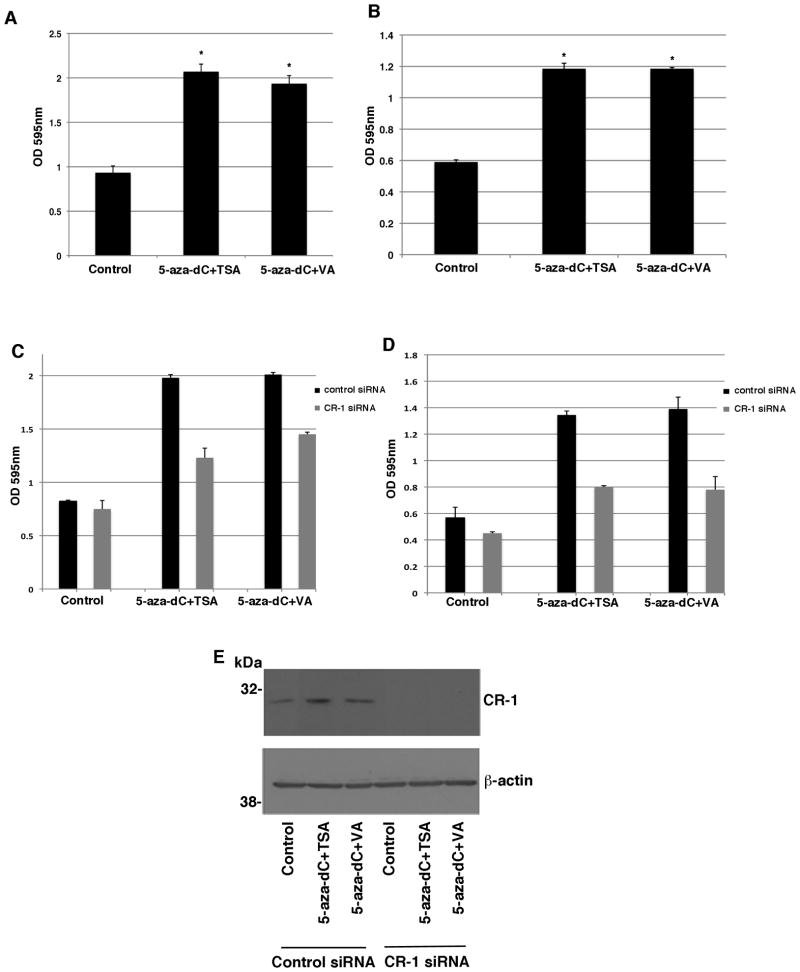
To evaluate if re-expression of CR-1 mRNA and protein in breast cancer cells upon treatment with demethylating and HDAC agents is biologically significant, we performed migration and invasion assays in MCF7 cells treated with 5-aza-dC, TSA and VA. A significant increase in cell migration and invasion was observed in MCF7 cells treated with 5-aza-dC in combination with TSA or VA as compared to untreated MCF7 cells, suggesting that re-expression of CR-1 in MCF7 cells might be associated with a more migratory and invasive behavior (Fig. 9A and B). Transfection of a pool of anti-CR-1 siRNAs in MCF7 treated with 5-aza-dC in combination with TSA or VA significantly reduced migration and invasion of these cells as compared to MCF7 cells transfected with a control siRNA (Fig. 9C and D). CR-1 protein expression was completely abrogated in control and 5-aza-dC, TSA or VA treated MCF7 cells following transfection with a CR-1 siRNA pool, as shown in figure 9E. Thus, these results suggest that the enhanced motility of MCF7 cells treated with demethylating and HDCA agents is in part dependent upon CR-1 re-expression in these cells.
Fig. 9. DNMT and HDAC inhibitors enhance migration and invasion of MCF7 cells in a CR-1-dependent manner.
Migration (A) and invasion (B) assays of 5-aza-dC, TSA and VA treated MCF7 cells. MCF7 cells were also transfected with control or CR-1 siRNAs and their migratory (C) and invasive (D) behavior was assessed. E: Western blot analysis for CR-1 and β-actin in MCF7 control and treated cells that have been transfected with control or CR-1 siRNA pools.
CR-1 enhances embryonic stem cell markers gene expression in MCF7 breast cancer cell tumor spheres
Cripto-1, together with Oct4 and Nanog, has been shown to regulate pluripotentiality, self-renewal and differentiation of mouse and human ES cells (Rangel et al., 2012). We therefore assessed whether Cripto-1 overexpression in MCF7 breast cancer cells was associated with an increase in the expression of other embryonic stem cell markers. To enrich for subpopulations of cells with stem-like properties, we grew MCF7 Neo control cells and MCF7 stably expressing CR-1 (MCF7 CR-1) cells (Normanno et al., 2004) in low attachment plates, which allow cells to form non-adherent spheres. Both MCF7 Neo and MCF7 CR-1 cells showed sphere forming capacity (Fig. 10A and B). Interestingly, MCF7 CR-1 cells grew at a much slower rate than MCF7 Neo cells (Fig. 10C). However, spheres derived from MCF7 CR-1 cells were significantly larger than spheres derived from the parental cell line (Fig. 10D), suggesting that MCF7 CR-1 spheres have higher self-renewal capacity but lower proliferation rate than MCF7 Neo spheres. Similar results have been described by Strizzi and colleagues in melanoma cells (Strizzi et al., 2008). Real time PCR analysis of tumor spheres derived from MCF7 Neo and MCF7 CR-1 cells showed a significant increase of embryonic stem cell markers, including Nanog, Oct4 and CD133, in MCF7 CR-1 tumor spheres (Fig. 10E–H). As expected, CR-1 was highly expressed in MCF7 CR-1 tumor spheres (Fig. 10E). These results, therefore, indicate that MCF7 CR-1 tumor spheres express higher levels of embryonic stem-cell markers as compared to the control parental cell line.
Fig. 10. CR-1 overexpression in MCF7 tumor spheres is associated with increased expression of embryonic stem cell markers.
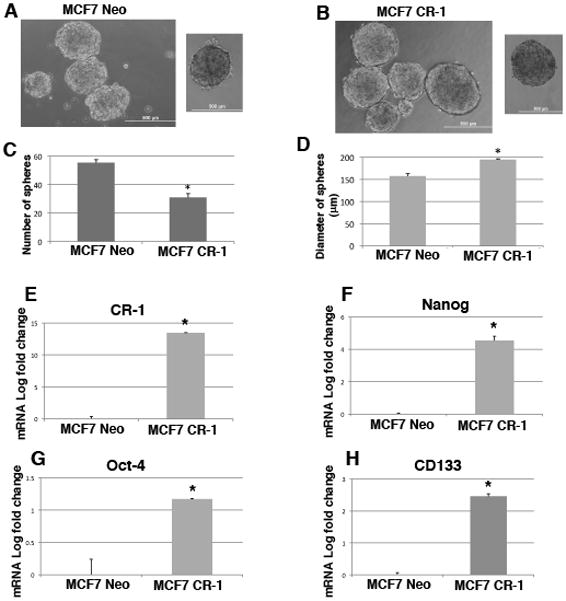
Representative images of MCF7 Neo (A) and MCF7 CR-1 (B) spheres after 7 days in culture in ultra-low attachment plates. The number (C) and the size (D) of spheres derived from MCF7 Neo and MCF7 CR-1 cells were assessed. Quantitative real time PCR for CR-1 (E), Nanog (F), Oct-4 (G) and CD133 (H) in MCF7 Neo and MCF7 CR-1 spheres. Data are representative of two independent experiments performed in a 24 well plate for each cell line. *P<0.0001.
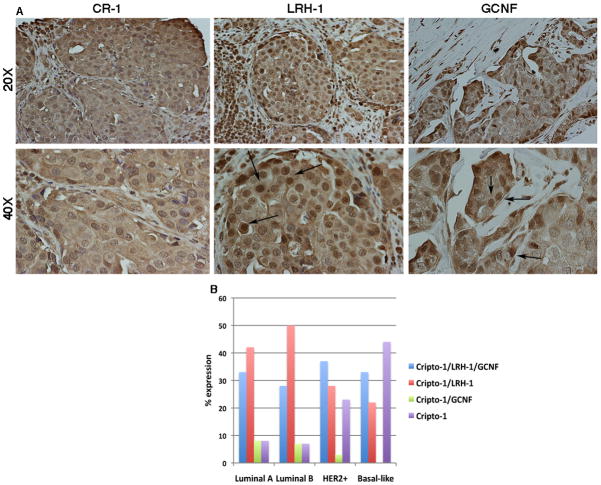
Expression of CR-1, LRH-1 and GCNF in human invasive ductal breast carcinomas
To evaluate the expression of CR-1, LRH-1 and GCNF in human breast tumors, we performed immunohistochemical analysis on a tissue microarray of 70 duplicated specimens of human invasive ductal breast carcinomas. CR-1 expression was detected in the majority of breast tumors (92%, 65/70) and showed a strong cytoplasmic staining (Fig. 11A and Table 1). LRH-1 was also highly expressed in breast cancer specimens (70%, 49/70) and as expected was detected mostly in the nucleus of the cells (Fig. 11A and Table 1). In contrast, GCNF was expressed in only 40% (28/70) of human breast tumors and, similarly to LRH-1, showed a nuclear staining (Fig. 11A and Table 1). Analysis of CR-1, GCNF and LRH-1 expression with clinical pathological parameters showed a statistical significant association of CR-1 expression and small tumor size (<5 cm) (Table 1). Furthermore a positive linear correlation was found between CR-1 and GCNF expression and LRH-1 and GCNF expression in breast carcinomas (Table 1). We also analyzed expression of CR-1, GCNF and LRH-1 in breast carcinomas that were divided according to their gene expression profile, including Luminal A, Luminal B, HER2 positive and basal-like molecular subtypes. CR-1 together with LRH-1 and/or GCNF was equally expressed among the four molecular subtypes (Figure 11B and Table 2). In contrast, CR-1 in the absence of LRH-1 and GCNF expression was significantly enriched in HER2 positive and basal-like tumors (27.3%) as compared to Luminal A and Luminal B tumors (7.7%).
Fig. 11. Expression of CR-1, LRH-1 and GCNF in human invasive ductal breast carcinomas.
A: Immunohistochemical analysis shows positive staining for CR-1, LRH-1 and GCNF. LRH-1 and GCNF show cytoplasmic and nuclear staining. Arrows are pointing to some cells with LRH-1 and GCNF nuclear staining. Original magnifications are 20X and 40X. B: Expression of CR-1, LRH-1 and GCNF in breast cancer molecular subtypes as assessed by immunohistochemical staining.
Table 1.
| % CR-1 (n) | % GCNF (n) | % LRH-1 (n) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Staining Score | ≤1 | 2 | 3 | ≤1 | 2 | 3 | ≤1 | 2 | 3 |
| Overall Distribution (N=70) | 21 (15) | 44 (31) | 34 (24) | 73 (51) | 14 (10) | 13 (9) | 47 (33) | 20 (14) | 33 (23) |
Cripto-1 versus GCNF (R2=0.0856; 95% Cl: 0.062, 0.494; P = 0.0141)
Cripto-1 versus LRH1 (R2=0.0288; 95% CI: −0.067, 0.389; P = 0.160)
GCNF versus LRH1 (R2=0.0742; 95% CI: 0.04, 0.476; P = 0.0228)
Table 2.
| % CR-1 (n) | % GCNF (n) | % LRH-1 (n) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Staining Score | ≤1 | 2 | 3 | ≤1 | 2 | 3 | ≤1 | 2 | 3 |
| Tumor Size | |||||||||
| < 5 cm (n=54) | 20 (11) | 41 (22) | 39 (21) | 72 (39) | 15 (8) | 13 (7) | 46 (25) | 19 (10) | 35 (19) |
| > 5 cm (n=16) | 25 (4) | 56 (9) | 19 (3) | 69 (11) | 13 (2) | 19 (3) | 50 (8) | 25 (4) | 25 (4) |
| *P value | P = 0.008 | P = 0.515 | P = 0.266 | ||||||
| Hormone Receptor | |||||||||
| Positive (n=30) | 27 (8) | 40 (12) | 33 (10) | 74 (22) | 13 (4) | 13 (4) | 37 (11) | 27 (8) | 36 (11) |
| Negative (n=40) | 17 (7) | 45 (18) | 38 (15) | 72.5 (29) | 15 (6) | 12.5 (5) | 52.5 (21) | 17.5 (7) | 30 (12) |
| *P value | P = 0.254 | P = 0.919 | P = 0.072 | ||||||
| HER2 | |||||||||
| Positive (n=52) | 21 (11) | 42 (22) | 37 (19) | 73 (38) | 13.5 (7) | 13.5 (7) | 46 (24) | 17 (9) | 37 (19) |
| Negative (n=18) | 16 (3) | 48 (9) | 33 (6) | 72 (13) | 21 (4) | 5 (1) | 55 (10) | 21 (4) | 21 (4) |
| *P value | P = 0.605 | P = 0.085 | P = 0.066 | ||||||
Chi-square; n= number of breast cancer tissue samples; CI= confidence interval. Staining score: 1 (weak staining), 2 (moderate staining), 3 (strong staining).
Discussion
Embryonic stem cells and cancer cells share common regulatory mechanisms and signaling pathways. CR-1 is a typical example of an oncofetal protein that performs key regulatory functions during embryonic development and oncogenic transformation in vitro and in vivo (Bianco et al., 2010). However, factors that might modulate CR-1 gene expression during embryogenesis and tumorigenesis are not well defined. We have previously demonstrated that CR-1 is a downstream target gene of TGF-β family members, such as TGF-β1 and BMP4, the transcription factor HIF-1α and the canonical Wnt/β-catenin signaling pathway (Bianco et al., 2009; Hamada et al., 2007; Mancino et al., 2008a). TGF-β1, HIF-1α and the canonical Wnt/β-catenin signaling pathway significantly enhance CR-1 mRNA and protein expression in ES cells and several cancer cell lines, while BMP-4 strongly down-regulates CR-1 mRNA and protein expression in embryonal and colon cancer cells. Regulation of CR-1 gene expression by these factors modulates CR-1 biological activity in ES cells and cancer cell lines. For example, HIF-1α, upon binding to hypoxia responsive elements within the promoter of mouse and human Cripto-1 genes, accelerates and enhances differentiation of mouse ES cells toward beating cardiomyocytes (Bianco et al., 2009). Furthermore, hypoxic conditions and HIF-1α might also be involved in regulating CR-1 expression in vivo, since a dramatic increase in CR-1 expression is detected in cardiac tissue samples derived from patients who had suffered ischemic heart disease (Bianco et al., 2009). In contrast, BMP-4 is a strong repressor of CR-1 expression and inhibits migration, invasion and colony formation in soft-agar of LS174-T colon cancer cells (Mancino et al., 2008a). Addition of a CR-1 recombinant protein together with BMP-4 is able to override BMP-4 inhibitory effect on cell migration in LS174-T cells, indicating that CR-1 is a key modulator of cell migration in colon cancer cells. Another repressor of CR-1 expression is GCNF. Hentschke and colleagues have identified a DR0 motif at position −219 to −207 upstream of the ATG codon start site of the human CR-1 gene (Hentschke et al., 2006). GCNF directly binds to the DR0 motif of the CR-1 promoter and represses its transcription during RA-induced differentiation of NTERA-2 cells. Furthermore, GCNF also down-regulates expression of CR-3, a CR-1 pseudogene located on chromosome X that retains an open reading frame with six amino acid differences from CR-1, during RA differentiation of NTERA-2 cells (Hentschke et al., 2006). Interestingly, CR-3 has been found to be expressed in several tumors and to be biologically functional in signaling assays in vitro (Sun et al., 2008). In the present study, we demonstrate that another orphan nuclear receptor, LRH-1, directly regulates CR-1 expression by interacting with the DR0 motif within the CR-1 promoter region. Interestingly, while GCNF is a strong repressor of CR-1 expression in NTERA-2 cells upon RA treatment, LRH-1 positively modulates CR-1 expression in embryonal carcinoma cells. In fact, transfection of LRH-1 siRNAs into NTERA-2 cells downregulates CR-1 expression, indicating that LRH-1 is required to maintain CR-1 expression in these cells. In contrast, transfection of GCNF siRNAs into NTERA-2 cells increases CR-1 expression, confirming the inhibitory effect of GCNF on CR-1 expression in embryonal carcinoma cells. The mechanisms by which GCNF and LRH-1 modulate gene expression through binding to the same DR0 regulatory element is still unclear. It has been proposed that GCNF and LRH-1 compete for the same regulatory motift and during RA differentiation GCNF is induced and displaces LRH-1 from the DR0 motif, initiating transcriptional repression of target genes (Mullen et al., 2007). Interestingly, we see a strong downregulation of LRH-1 expression upon RA treatment of NTERA-2 cells that mirrors CR-1 downregulation, suggesting that a decrease in LRH-1 expression might favor the repressor activity of GCNF on the CR-1 promoter in NTERA-2 cells. Regulation of CR-1 expression by GCNF and LRH-1 might be also operative in ES cells. CR-1, together with Oct-4, Nanog and Sox-2, is important in maintenance of the pluripotent phenotype of ES cells and is downregulated upon differentiation of ES cells (Bianco et al., 2010). CR-1 is also a direct downstream target gene of the transcription factors Oct-4 and Nanog, while CR-1 together with Nodal and Activin, is essential for initiating and maintaining the expression of Nanog (Loh et al., 2006; Vallier et al., 2009). Therefore, CR-1 is part of a transcriptional regulatory loop that maintains pluripotency gene expression in ES cells. GCNF and LRH-1 are also involved in regulating pluripotency and differentiation of ES cells. For instance, GCNF binds to DR0 elements within the Oct-4 and Nanog promoters, repressing pluripotency gene expression during RA induced differentiation of ES cells (Gu et al., 2005b). In contrast, LRH-1 is required to maintain Oct-4 expression in ES cells. In fact, disruption of LRH-1 gene results in early embryonic death (E 6.5) due to a loss of Oct-4 gene expression at the epiblast stage of embryonic development (Gu et al., 2005a). CR-1 regulation by GCNF and LRH-1 in embryonal carcinoma cells closely resembles Oct-4 regulation by these orphan nuclear receptors in ES cells. CR-1 gene modulation by GCNF and LRH-1 might also occur indirectly in ES cells through induction or repression of Oct-4 that can then directly regulate CR-1 expression. However, our findings of direct binding of LRH-1 to the CR-1 promoter in NTERA-2 cells suggest that a direct modulation of CR-1 gene expression by LRH-1 and GCNF might also occur in ES cells. We also demonstrate that the CR-1 promoter is highly methylated in breast cancer cell lines where we found high levels of GCNF mRNA expression. Analysis of the repression mechanism of GCNF has shown that epigenetic modifications play an important role in the gene repression and silencing function of GCNF (Mullen et al., 2007). Indeed, GCNF binding to the Oct-4 promoter triggers initiation of DNA promoter methylation by recruitment of Methyl CpG-binding domain factors (MBD3 and MBD2) and DNMT 3A, inducing silencing of the Oct-4 gene during ES cell differentiation (Gu et al., 2011). We have previously identified two CpG-rich regions in the human CR-1 promoter, one located approximately 0.5–0.8 kb upstream of the transcriptional start site and another region surrounding the transcriptional start site (Watanabe et al., 2010). RA treatment of NTERA-2 cells strongly induces methylation of the CR-1 promoter in a time-dependent fashion. Therefore, GCNF might be directly involved in methylation of the CR-1 promoter in NTERA-2 cells, mediating repression of the CR-1 gene in early stages of differentiation while inducing CR-1 promoter methylation in the later stages. Treatment of breast cancer cell lines with DNMT and/or HDAC inhibitors leads to re-activation of CR-1 mRNA and functional protein, supporting a model in which DNMTs and HDACs might be involved in the transcriptional control of CR-1 in breast cancer cell lines. More importantly, MCF7 cells, treated with DNMT and HDAC inhibitors re-express CR-1 and recapitulate the profile of MCF7 cells overexpressing CR-1, showing an enhanced migratory and invasive capacity in Boyden chamber assays as compared to control untreated MCF7 cells (Normanno et al., 2004). In addition, MCF7 stably expressing CR-1 show an increased expression of embryonic stem cell markers, such as Nanog, Oct4 and CD133, indicating that CR-1 might be expressed in a subpopulation of breast cancer cells with stem-like characteristics, also known as cancer stem cells. Cripto-1 has, indeed, been identified in a small subpopulation of stem-like cancer cells in human prostate and pancreatic tumor cells and in human melanomas (Rangel et al., 2012). Although DNMT and HDAC inhibitors might re-activate expression of other genes involved in cell motility, CR-1 is indeed a key player in MCF7 cells enhanced migration and invasion in response to DNMT and HDAC inhibitors treatment. In fact, CR-1 siRNAs not only interfere with CR-1 re-expression in MCF7 cells treated with DNMT and HDAC inhibitors (Fig. 9A), but also significantly reduce the enhanced migratory behavior of MCF7 cells in response to these drugs (Fig. 9C and D). While in vitro breast cancer cell lines express low levels of CR-1 probably due to a hypermethylation of the CR-1 promoter, a completely different scenario is found in vivo in breast cancer tumor specimens. Our immunohistochemichal study on human invasive ductal breast carcinoma tissue samples demonstrate that CR-1 and LRH-1 are expressed in a high percentage of human breast tumors (92% and 70%, respectively) while GCNF is expressed in only 40% of human breast carcinomas. However, a positive correlation was found in breast carcinomas between CR-1 and GCNF expression. High levels of CR-1 and LRH-1 expression in breast tumors are in agreement with other studies reported in the literature (Miki et al., 2006; Rangel et al., 2012). CR-1 overexpression in human breast tumors is associated with poor prognosis and decreased survival, indicating that it might play a role in the progression of this disease (Gong et al., 2007). LRH-1 is a downstream target gene of estrogen receptor α in MCF7 cells and in a feed forward manner regulates estrogen receptor expression in breast cancer cells and aromatase expression in mammary adipose tissue (Annicotte et al., 2005; Clyne et al., 2004; Thiruchelvam et al., 2011). Similarly to CR-1, LRH-1 promotes migration and invasion of breast cancer cell lines, suggesting that LRH-1 might enhance breast cancer cell motility indirectly through CR-1 (Chand et al., 2010). While immunohistochemical studies in human breast carcinomas have found a positive association of LRH-1 immunoreactivity with hormone receptor status (Miki et al., 2006), we were unable to find a significant positive correlation between LRH-1 expression and ER positive and/or PR positive breast tumors (Table 1), probably due to the small number of tumor samples analyzed. GCNF expression in human tumors is still unexplored and here we demonstrate for the first time that GNCF is expressed in 40% of human breast carcinomas. High levels of GCNF expression have been reported in MDA-MD-231 breast cancer cells and are in agreement with our findings in MCF7, T47D and ZR75-1 breast cancer cells lines (Koohi et al., 2005). Analysis of CR-1, LRH-1 and GCNF expression in Luminal A, Luminal B, HER2 positive and basal-like molecular subtypes of mammary tumors revealed an enrichment of CR-1 positive tumors, in the absence of LRH-1 and GCNF expression, in HER-2 positive/basal-like (27.3%) mammary tumors versus Luminal A/Luminal B (7.7%) mammary tumors (Table 2). In contrast, breast tumors co-expressing CR-1 with LRH-1 and/or GCNF showed little preferential distribution among the four molecular subtypes of breast cancer. Gene expression profile studies have revealed similarities between mammary epithelial progenitors and specific breast cancer subtypes. For example, luminal progenitors share similarities with HER2 positive and basal-like mammary tumors, whereas Luminal A and Luminal B tumors resemble more differentiated mature luminal mammary cells (Lim et al., 2009). The enrichment of CR-1 expression in HER2 positive/basal-like tumors, which derive from a common luminal progenitor, suggests that CR-1 might play a role in these subpopulations of breast tumors independently of GCNF and LRH-1 regulation.
In conclusion, we have further characterized CR-1 gene expression regulation by GCNF and LRH-1 and by DNA promoter methylation in embryonal and breast carcinoma cells. Co-expression of CR-1 with LRH-1 and GCNF is also found in human invasive ductal breast carcinomas indicating that these two orphan nuclear receptors may regulate CR-1 expression in vivo. Lack of CR-1 expression in breast cancer cell lines is in disagreement with high levels of CR-1 expression found in human breast tumors. We have previously demonstrated that CR-1 expression in tumor cell lines is heterogeneous with subpopulations of cells expressing high and low levels of CR-1 protein (Watanabe et al., 2010). Cripto-1 high and Cripto-1 low subpopulations in NTERA 2 cells exist in a dynamic equilibrium and microenviromental factors, such as cell confluence or serum deprivation, can influence CR-1 expression in this cell line. Culture of breast cancer cells in vitro might be associated with a loss of a CR-1 high subpopulation, probably due to high levels of GCNF that trigger methylation of the CR-1 promoter gene. A similar scenario is found in colon cancer cells, such as GEO and COLO205 cell lines, which overtime in tissue culture lose CR-1 expression. Remarkably, injection of these cells into immunocompromized mice give rise to tumors that are highly positive for CR-1 expression (Bianco C and Salomon DS unpublished data), indicating that a high grade of phenotypic plasticity characterize human cancer cells.
Table 3.
| Marker | Luminal A/B % of n=26 | HER2+/basal-like % of n=44 | P (T-Test) |
|---|---|---|---|
| CR-1 | 7.7 (n=2) | 27.3 (n=12) | 0.007 |
| LRH-1 | 3.8 (n=1) | 0 (n=0) | N/C |
| GCNF | 0 (n=0) | 2.3 (n=1) | N/C |
| CR-1/LRH-1 | 46.2 (n=12) | 27.3 (n=12) | NS |
| CR-1/GCNF | 7.7 (n=2) | 2.3 (n=1) | N/C |
| CR-1/LRH-1/GCNF | 30.8 (n=8) | 36.4 (n=16) | NS |
n=number of breast cancer tissue samples
N/C= non calculated (must have at least n=2 in both groups)
NS= non significant (P>0.05)
Acknowledgments
Contract grant sponsor: Intramural Research Program, NIH, National Cancer Institute, Center for Cancer Research; Contract grant number; N/A. Contract grant sponsor: Eisenberg Research Scholar Fund; Contract grant number; N/A.
References
- Aasen T, Raya A, Barrero MJ, Garreta E, Consiglio A, Gonzalez F, Vassena R, Bilic J, Pekarik V, Tiscornia G, Edel M, Boue S, Izpisua Belmonte JC. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat Biotechnol. 2008;26(11):1276–1284. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- Annicotte JS, Chavey C, Servant N, Teyssier J, Bardin A, Licznar A, Badia E, Pujol P, Vignon F, Maudelonde T, Lazennec G, Cavailles V, Fajas L. The nuclear receptor liver receptor homolog-1 is an estrogen receptor target gene. Oncogene. 2005;24(55):8167–8175. doi: 10.1038/sj.onc.1208950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoi T, Yae K, Nakagawa M, Ichisaka T, Okita K, Takahashi K, Chiba T, Yamanaka S. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science. 2008;321(5889):699–702. doi: 10.1126/science.1154884. [DOI] [PubMed] [Google Scholar]
- Assou S, Lecarrour T, Tondeur S, Strom S, Gabelle A, Marty S, Nadal L, Pantesco V, Reme T, Hugnot JP, Gasca S, Hovatta O, Hamamah S, Klein B, De Vos J. A meta-analysis of human embryonic stem cells transcriptome integrated into a web-based expression atlas. Stem Cells. 2007;25:961–973. doi: 10.1634/stemcells.2006-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco C, Adkins HB, Wechselberger C, Seno M, Normanno N, De Luca A, Sun Y, Khan N, Kenney N, Ebert A, Williams KP, Sanicola M, Salomon DS. Cripto-1 activates nodal- and ALK4-dependent and -independent signaling pathways in mammary epithelial Cells. Mol Cell Biol. 2002;22(8):2586–2597. doi: 10.1128/MCB.22.8.2586-2597.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco C, Cotten C, Lonardo E, Strizzi L, Baraty C, Mancino M, Gonzales M, Watanabe K, Nagaoka T, Berry C, Arai AE, Minchiotti G, Salomon DS. Cripto-1 is required for hypoxia to induce cardiac differentiation of mouse embryonic stem cells. Am J Pathol. 2009;175(5):2146–2158. doi: 10.2353/ajpath.2009.090218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco C, Rangel MC, Castro NP, Nagaoka T, Rollman K, Gonzales M, Salomon DS. Role of Cripto-1 in stem cell maintenance and malignant progression. Am J Pathol. 2010;177(2):532–540. doi: 10.2353/ajpath.2010.100102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco C, Strizzi L, Ebert A, Chang C, Rehman A, Normanno N, Guedez L, Salloum R, Ginsburg E, Sun Y, Khan N, Hirota M, Wallace-Jones B, Wechselberger C, Vonderhaar BK, Tosato G, Stetler-Stevenson WG, Sanicola M, Salomon DS. Role of human cripto-1 in tumor angiogenesis. J Natl Cancer Inst. 2005;97(2):132–141. doi: 10.1093/jnci/dji011. [DOI] [PubMed] [Google Scholar]
- Bianco C, Strizzi L, Mancino M, Rehman A, Hamada S, Watanabe K, De Luca A, Jones B, Balogh G, Russo J, Mailo D, Palaia R, D’Aiuto G, Botti G, Perrone F, Salomon DS, Normanno N. Identification of cripto-1 as a novel serologic marker for breast and colon cancer. Clin Cancer Res. 2006;12(17):5158–5164. doi: 10.1158/1078-0432.CCR-06-0274. [DOI] [PubMed] [Google Scholar]
- Bianco C, Strizzi L, Mancino M, Watanabe K, Gonzales M, Hamada S, Raafat A, Sahlah L, Chang C, Sotgia F, Normanno N, Lisanti M, Salomon DS. Regulation of Cripto-1 signaling and biological activity by caveolin-1 in mammary epithelial cells. Am J Pathol. 2008;172(2):345–357. doi: 10.2353/ajpath.2008.070696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chand AL, Herridge KA, Thompson EW, Clyne CD. The orphan nuclear receptor LRH-1 promotes breast cancer motility and invasion. Endocr Relat Cancer. 2010;17(4):965–975. doi: 10.1677/ERC-10-0179. [DOI] [PubMed] [Google Scholar]
- Chang CW, Lai YS, Pawlik KM, Liu K, Sun CW, Li C, Schoeb TR, Townes TM. Polycistronic lentiviral vector for “hit and run” reprogramming of adult skin fibroblasts to induced pluripotent stem cells. Stem Cells. 2009;27(5):1042–1049. doi: 10.1002/stem.39. [DOI] [PubMed] [Google Scholar]
- Clyne CD, Kovacic A, Speed CJ, Zhou J, Pezzi V, Simpson ER. Regulation of aromatase expression by the nuclear receptor LRH-1 in adipose tissue. Mol Cell Endocrinol. 2004;215(1–2):39–44. doi: 10.1016/j.mce.2003.11.001. [DOI] [PubMed] [Google Scholar]
- de Castro NP, Rangel MC, Nagaoka T, Salomon DS, Bianco C. Cripto-1: an embryonic gene that promotes tumorigenesis. Future Oncol. 2010;6(7):1127–1142. doi: 10.2217/fon.10.68. [DOI] [PubMed] [Google Scholar]
- Ding J, Yang L, Yan YT, Chen A, Desai N, Wynshaw-Boris A, Shen MM. Cripto is required for correct orientation of the anterior-posterior axis in the mouse embryo. Nature. 1998;395(6703):702–707. doi: 10.1038/27215. [DOI] [PubMed] [Google Scholar]
- Gong YP, Yarrow PM, Carmalt HL, Kwun SY, Kennedy CW, Lin BP, Xing PX, Gillett DJ. Overexpression of Cripto and its prognostic significance in breast cancer: a study with long-term survival. Eur J Surg Oncol. 2007;33(4):438–443. doi: 10.1016/j.ejso.2006.10.014. [DOI] [PubMed] [Google Scholar]
- Gu P, Goodwin B, Chung AC, Xu X, Wheeler DA, Price RR, Galardi C, Peng L, Latour AM, Koller BH, Gossen J, Kliewer SA, Cooney AJ. Orphan nuclear receptor LRH-1 is required to maintain Oct4 expression at the epiblast stage of embryonic development. Molecular and cellular biology. 2005a;25(9):3492–3505. doi: 10.1128/MCB.25.9.3492-3505.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu P, Le Menuet D, Chung AC, Cooney AJ. Differential recruitment of methylated CpG binding domains by the orphan receptor GCNF initiates the repression and silencing of Oct4 expression. Molecular and cellular biology. 2006;26(24):9471–9483. doi: 10.1128/MCB.00898-06. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gu P, LeMenuet D, Chung AC, Mancini M, Wheeler DA, Cooney AJ. Orphan nuclear receptor GCNF is required for the repression of pluripotency genes during retinoic acid-induced embryonic stem cell differentiation. Molecular and cellular biology. 2005b;25(19):8507–8519. doi: 10.1128/MCB.25.19.8507-8519.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Gu P, Xu X, Le Menuet D, Chung AC, Cooney AJ. Differential recruitment of methyl CpG-binding domain factors and DNA methyltransferases by the orphan receptor germ cell nuclear factor initiates the repression and silencing of Oct4. Stem Cells. 2011;29(7):1041–1051. doi: 10.1002/stem.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada S, Watanabe K, Hirota M, Bianco C, Strizzi L, Mancino M, Gonzales M, Salomon DS. beta-Catenin/TCF/LEF regulate expression of the short form human Cripto-1. Biochem Biophys Res Commun. 2007;355(1):240–244. doi: 10.1016/j.bbrc.2007.01.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschke M, Kurth I, Borgmeyer U, Hubner CA. Germ cell nuclear factor is a repressor of CRIPTO-1 and CRIPTO-3. The Journal of biological chemistry. 2006;281(44):33497–33504. doi: 10.1074/jbc.M606975200. [DOI] [PubMed] [Google Scholar]
- Koohi MK, Ivell R, Walther N. Transcriptional activation of the oxytocin promoter by oestrogens uses a novel non-classical mechanism of oestrogen receptor action. J Neuroendocrinol. 2005;17(4):197–207. doi: 10.1111/j.1365-2826.2005.01298.x. [DOI] [PubMed] [Google Scholar]
- Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, Asselin-Labat ML, Gyorki DE, Ward T, Partanen A, Feleppa F, Huschtscha LI, Thorne HJ, Fox SB, Yan M, French JD, Brown MA, Smyth GK, Visvader JE, Lindeman GJ. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nature medicine. 2009;15(8):907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, Wong KY, Sung KW, Lee CW, Zhao XD, Chiu KP, Lipovich L, Kuznetsov VA, Robson P, Stanton LW, Wei CL, Ruan Y, Lim B, Ng HH. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38(4):431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Mancino M, Strizzi L, Wechselberger C, Watanabe K, Gonzales M, Hamada S, Normanno N, Salomon DS, Bianco C. Regulation of human Cripto-1 gene expression by TGF-beta1 and BMP-4 in embryonal and colon cancer cells. Journal of cellular physiology. 2008a;215(1):192–203. doi: 10.1002/jcp.21301. [DOI] [PubMed] [Google Scholar]
- Mancino M, Strizzi L, Wechselberger C, Watanabe K, Gonzales M, Hamada S, Normanno N, Salomon DS, Bianco C. Regulation of human Cripto-1 gene expression by TGF-beta1 and BMP-4 in embryonal and colon cancer cells. J Cell Physiol. 2008b;215(1):192–203. doi: 10.1002/jcp.21301. [DOI] [PubMed] [Google Scholar]
- Miharada K, Karlsson G, Rehn M, Rorby E, Siva K, Cammenga J, Karlsson S. Cripto regulates hematopoietic stem cells as a hypoxic-niche-related factor through cell surface receptor GRP78. Cell Stem Cell. 2011;9(4):330–344. doi: 10.1016/j.stem.2011.07.016. [DOI] [PubMed] [Google Scholar]
- Miki Y, Clyne CD, Suzuki T, Moriya T, Shibuya R, Nakamura Y, Ishida T, Yabuki N, Kitada K, Hayashi S, Sasano H. Immunolocalization of liver receptor homologue-1 (LRH-1) in human breast carcinoma: possible regulator of insitu steroidogenesis. Cancer Lett. 2006;244(1):24–33. doi: 10.1016/j.canlet.2005.11.038. [DOI] [PubMed] [Google Scholar]
- Mullen EM, Gu P, Cooney AJ. Nuclear Receptors in Regulation of Mouse ES Cell Pluripotency and Differentiation. PPAR Res. 2007;2007:61563. doi: 10.1155/2007/61563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka T, Karasawa H, Castro NP, Rangel MC, Salomon DS, Bianco C. An evolving web of signaling networks regulated by Cripto-1. Growth Factors. 2012;30(1):13–21. doi: 10.3109/08977194.2011.641962. [DOI] [PubMed] [Google Scholar]
- Normanno N, De Luca A, Bianco C, Maiello MR, Carriero MV, Rehman A, Wechselberger C, Arra C, Strizzi L, Sanicola M, Salomon DS. Cripto-1 overexpression leads to enhanced invasiveness and resistance to anoikis in human MCF-7 breast cancer cells. J Cell Physiol. 2004;198(1):31–39. doi: 10.1002/jcp.10375. [DOI] [PubMed] [Google Scholar]
- Rangel MC, Karasawa H, Castro NP, Nagaoka T, Salomon DS, Bianco C. Role of Cripto-1 during Epithelial-To-Mesenchymal Transition in Development and Cancer. Am J Pathol. 2012 doi: 10.1016/j.ajpath.2012.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Kondo M, Arai K. The orphan nuclear receptor GCNF recruits DNA methyltransferase for Oct-3/4 silencing. Biochem Biophys Res Commun. 2006;344(3):845–851. doi: 10.1016/j.bbrc.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Strizzi L, Abbott DE, Salomon DS, Hendrix MJ. Potential of Cripto-1 in defining stem cell-like characteristics in human malignat melanoma. Cell Cycle. 2008;7 (13):1931–1935. doi: 10.4161/cc.7.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Orozco O, Olson DL, Choi E, Garber E, Tizard R, Szak S, Sanicola M, Carulli JP. CRIPTO3, a presumed pseudogene, is expressed in cancer. Biochem Biophys Res Commun. 2008;377(1):215–220. doi: 10.1016/j.bbrc.2008.09.113. [DOI] [PubMed] [Google Scholar]
- Thiruchelvam PT, Lai CF, Hua H, Thomas RS, Hurtado A, Hudson W, Bayly AR, Kyle FJ, Periyasamy M, Photiou A, Spivey AC, Ortlund EA, Whitby RJ, Carroll JS, Coombes RC, Buluwela L, Ali S. The liver receptor homolog-1 regulates estrogen receptor expression in breast cancer cells. Breast Cancer Res Treat. 2011;127(2):385–396. doi: 10.1007/s10549-010-0994-9. [DOI] [PubMed] [Google Scholar]
- Vallier L, Mendjan S, Brown S, Chng Z, Teo A, Smithers LE, Trotter MW, Cho CH, Martinez A, Rugg-Gunn P, Brons G, Pedersen RA. Activin/Nodal signalling maintains pluripotency by controlling Nanog expression. Development. 2009;136(8):1339–1349. doi: 10.1242/dev.033951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Meyer MJ, Strizzi L, Lee JM, Gonzales M, Bianco C, Nagaoka T, Farid SS, Margaryan N, Hendrix MJ, Vonderhaar BK, Salomon DS. Cripto-1 is a cell surface marker for a tumorigenic, undifferentiated subpopulation in human embryonal carcinoma cells. Stem Cells. 2010;28(8):1303–1314. doi: 10.1002/stem.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CL, Miura T, Robson P, Lim SK, Xu XQ, Lee MY, Gupta S, Stanton L, Luo Y, Schmitt J, Thies S, Wang W, Khrebtukova I, Zhou D, Liu ET, Ruan YJ, Rao M, Lim B. Transcriptome profiling of human and murine ESCs identifies divergent paths required to maintain the stem cell state. Stem Cells. 2005;23(2):166–185. doi: 10.1634/stemcells.2004-0162. [DOI] [PubMed] [Google Scholar]
- Xu C, Liguori G, Adamson ED, Persico MG. Specific arrest of cardiogenesis in cultured embryonic stem cells lacking Cripto-1. Dev Biol. 1998;196(2):237–247. doi: 10.1006/dbio.1998.8862. [DOI] [PubMed] [Google Scholar]
- Yeo C, Whitman M. Nodal signals to Smads through Cripto-dependent and Cripto-independent mechanisms. Mol Cell. 2001;7(5):949–957. doi: 10.1016/s1097-2765(01)00249-0. [DOI] [PubMed] [Google Scholar]