Abstract
Mapping protein interactions and their dynamics is crucial to defining physiologic states, building effective models for understanding cell function, and to allow more effective targeting of new drugs. Crosslinking studies can estimate the proximity of proteins, determine sites of protein–protein interactions, and have the potential to provide a snapshot of dynamic interactions by covalently locking them in place for analysis. Several major challenges are associated with the use of crosslinkers in mass spectrometry, particularly in complex mixtures. We describe the synthesis and characterization of a MS-cleavable crosslinker containing cyclic amines, which address some of these challenges. The DC4 crosslinker contains two intrinsic positive charges, which allow crosslinked peptides to fragment into their component peptides by collision-induced dissociation (CID) or in-source decay. Initial fragmentation events result in cleavage on either side of the positive charges so crosslinked peptides are identified as pairs of ions separated by defined masses. The structures of the component peptides can then be robustly determined by MS3 because their fragmentation products rearrange to generate a mobile proton. The DC4 crosslinking reagent is stable to storage, highly reactive, highly soluble (1 M solutions), quite labile to CID, and MS3 results in productive backbone fragmentation.
Keywords: Mass spectrometry, Crosslinking, Aldolase, Subunit interactions, Protein crosslinker
Introduction
Mass spectrometry-based methods have been extensively used to characterize protein structures and their interactions due to their sensitivity, speed, and accuracy [1–7]. Purification techniques such as tandem affinity purification (TAP) tagging and affinity purification have been used to first purify complexes and then identify their components using mass spectrometry. Hydrogen–deuterium exchange, timed oxidation, and limited proteolysis, in combination with mass spectrometry, as well as mass spectrometry of intact complexes are methods used for evaluation of protein secondary, tertiary, and quaternary structures [1–7]. Each of these methods has limitations when applied to global analysis of protein interactions, whether it is in the relatively lower throughput of hydrogen–deuterium exchange, the loss of weakly associated proteins during TAP tagging, and other methods that require isolation of protein complexes or lower sensitivities for intact protein analyses.
Chemical crosslinking combined with mass spectrometry can provide complementary information to these methods and has several potential advantages [8]. The weaker binding partners that might be lost during affinity isolation are not lost during chemical crosslinking because crosslinking locks protein interactions in place [9]. Interacting partners can be determined, spatial information can be obtained, relatively small quantities of protein are required, and data can be generated from crosslinking experiments more rapidly than some other methods.
A number of challenges have been traditionally associated with mass spectrometric analysis of crosslinked peptides [9–12]. Crosslinked peptides (type 2 crosslinks – see Table 1) in a proteolytic digest are generally very low in abundance relative to noncrosslinked peptides. Sample complexity is increased by the presence of side reactions, intracrosslinked peptides (type 1) and dead-end reactions [type 0 in which one end of the crosslinker reacts with protein and the other end with a nucleophile (e.g., water)] [13,14]. Tandem mass spectra of crosslinked peptides often undergo diminished fragmentation relative to linear peptides by collision-induced dissociation or are inherently complex due to overlapping b- and y-ion series from both peptides [12, 15].
Table 1.
During a Crosslinking Reaction, Three Types of Modifications can Occur, Resulting in Three Types of Peptide Products. Dead-End Products Result when the Crosslinking Reagent Reacts on One End with Protein and Does Not Couple on the Other End or Reacts with a Small Molecule (e.g., Water) on the Other End. Loop Links Occur When Both Ends of the Crosslinking Reagent React within the Protein but No Proteolytic Cleavage Occurs between the Two Sites. When Proteolytic Cleavage Occurs between the Two Sites or the Sites are on Different Proteins, a Crosslinked Peptide is the Result
| Structure | Common Name | Crosslink Type |
|---|---|---|
| Dead-Ends | Type 0 | |
| Loop Links | Type 1 | |
| Crosslinks | Type 2 |
The nature of the crosslinking chemistry can also introduce challenges. For example, N-hydroxysuccinimide ester is a common reactive group used to crosslink proteins and reacts primarily with free amines such as the epsilon nitrogen in lysyl residues. There is evidence that seryl, threonyl, and tyrosyl residues react as well [16, 17], but appropriate data analysis algorithms can take advantage of these extended specificities to potentially identify additional sites of crosslinking. In addition, proteins crosslinked at lysyl residues have fewer sites available for tryptic digestion, resulting in larger peptides. These larger peptides may not fragment as effectively by collision-induced dissociation (CID) around the crosslinked site and may have complex fragmentation patterns making the spectra difficult to identify and interpret. To overcome these obstacles, several crosslinking reagents are commercially available and new crosslinkers are being actively developed by various research groups. Properties of these new crosslinkers include a broader range of chemical reactivities, improved fragmentation properties in MS/MS, and the addition of affinity ligands [18–30]. It is worth noting that most of these affinity methods also target the dead-end reaction products, which are in large excess over the type 2 crosslinked peptides and thus the informative peptides still constitute a minor population of the enriched peptides. In addition, many crosslinking reagents are not water soluble or have limited solubility. The organic solvents required to solubilize these crosslinkers can directly affect protein interactions and structures, leading to experimental artifacts.
Identification of the rare crosslinked peptides has also been facilitated by use of a mixture of heavy and light isotopically labeled crosslinking reagents and chemical tags [31]. Mass analysis of the peptides identifies ions separated by the distinctive mass corresponding to the number of hydrogen atoms substituted by deuterium [9, 32–34]. Dead-end peptides also produce pairs of ions and computational schemes have been developed which filter these spectra out as part of the identification process [35–38]. Crosslinked peptides (type 2) that produce marker ions upon fragmentation facilitate the identification of the spectra and to our knowledge few crosslinkers have been identified to do so, specifically with type 2 peptides [14, 39].
To overcome the poor fragmentation properties of crosslinked peptides, both chemically cleavable and mass spectrometry cleavable crosslinking reagents have been developed [40–43]. Chemical cleavage of the crosslinked peptides prior to mass spectrometry leads to ambiguity regarding which two cleaved peptides were crosslinked, particularly in complex mixtures. More desirable are crosslinkers that fragment efficiently in MS/MS mode to yield two major fragment ions corresponding to the component peptides which can subsequently be subjected to MS3 for identification. Thus, MS/MS provides a clear precursor/product relationship indicating unambiguously that the two product peptides were covalently crosslinked. Several recent efforts have addressed the design of these MS-cleavable crosslinking reagents, leading to new crosslinkers with improved functionality [15, 22, 44–51], including electron transfer dissociation (ETD) cleavable reagents [30, 52] but further improvements are still needed.
Balancing the properties of the reagents to obtain good reactivity in solution and stability in MS mode while maximizing fragmentation of the crosslinker for a variety of peptides in MS/MS mode is challenging. Many of the crosslinking studies use model proteins but publications analyzing crosslinks of more complex systems are only recently starting to appear, in part because of computational limitations in data analysis and also because reagents that work for model peptides do not necessarily work well for the diverse peptides derived from intact proteins [26, 53].
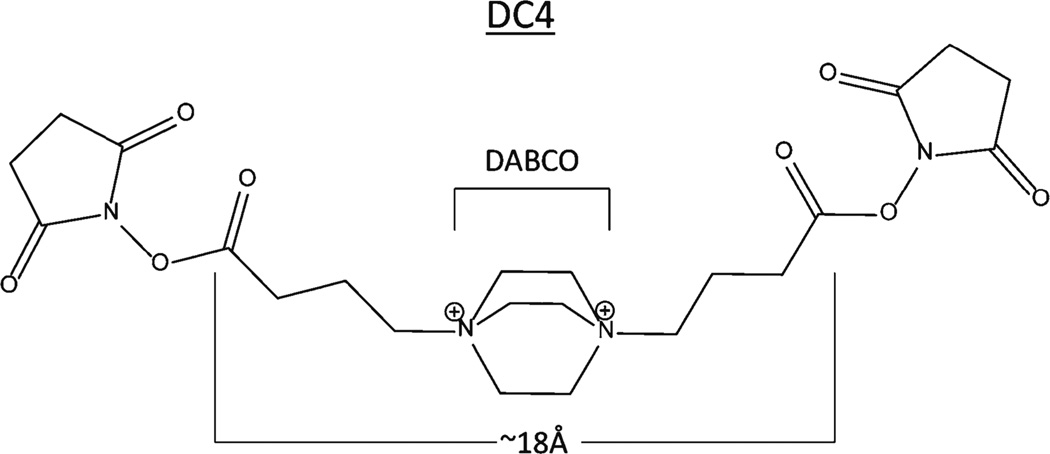
We have completed the synthesis and initial characterization of a new crosslinking reagent DC4, (Figure 1) based on cyclic diamine structures highly susceptible to fragmentation and having a number of other desirable properties. The DC4 crosslinker has been characterized with model peptides to determine its fragmentation behavior in both electrospray ionization (ESI) and matrix-assisted laser desorption ionization (MALDI) and then validated using the multimeric model protein aldolase. Good efficiencies of fragmentation were observed with both ionization methods. Fragmentation occurs within the crosslinking moiety adjacent to both nitrogen atoms, producing two intact peptide fragments that can be further fragmented, yielding a robust series of b- and y-ions for peptide identification.
Figure 1.
The crosslinker synthesized was named DC4 and spans approximately 18Å. The 1,4-diazabicyclo[2.2.2]octane moiety within DC4 is abbreviated as DABCO and will be referred to as such throughout this paper as such
Experimental
Materials
N-terminally acetylated peptides Ac-QIGKGVAR, Ac-QHAKLGR, were synthesized by the Biomedical Research Core Facility at the University of Michigan Medical School using standard Fmoc-based methods, a PTI Symphony synthesizer and purification by reversed-phase HPLC. All peptides were at least 90% pure and had the expected intact masses and fragmentation patterns. Trypsin was TPCK treated from Worthington Biochemical Corporation. Deuterated solvents were purchased from Cambridge Isotope (Andover, MA, USA). Rabbit aldolase and all other reagents were purchased from Sigma Aldrich or Fisher Scientific and used without further purification.
Instrumentation for Crosslinker Synthesis
Melting points were determined in open capillary tubes on a Laboratory Devices Mel-Temp apparatus and are uncorrected. The NMR spectra were recorded on a Bruker instrument at 500 MHz for 1H and 125 MHz for 13C spectra. Chemical shift values are recorded in δ units (ppm). Mass spectra of the synthesized compounds were recorded on a Micromass LCT time-of-flight mass spectrometer with an electrospray ionization source.
Synthesis
2,5-Dioxopyrrolidin-1-yl 4-bromobutanoate (1): Compound 1 was prepared as previously described by Almirante et al. with minor modifications [54]. Briefly, a mixture of 4-bromobutyric acid (24 g, 144 mmol), N-hydroxysuccinimide (19.8 g, 172 mmol), N,N-dicyclohexylcarbodiimide (35.5 g, 172 mmol) 4-(dimethylamino)pyridine (3.5 g, 28.6 mmol), and dichloromethane (300 mL) was stirred overnight at room temperature. Precipitated N,N-dicyclohexylurea was filtered off, washed with dichloromethane, and the filtrate was concentrated to an oil that was purified by flash silica gel chromatography eluting with 30:70 ethyl acetate:hexanes. Approximately 200 mL fractions were collected and each was concentrated and assayed by 1H NMR. Fractions containing pure product were combined to give 3 (23.3 g, 61.4 %) as a white solid: mp 58–64 °C; 1H NMR (d6DMSO) δ 3.6 (t, 2H), 2.8 (m, 6H), 2.2 (m, 2H); 13C NMR (d6DMSO) δ 170.64 (C=O), 168.64 (C=O), 33.38 (CH2Br), 29.47 (CH2), 27.97 (CH2), 25.92 (CH2).
1,4-Bis(4-((2,5-dioxopyrrolidin-1-yl)oxy)-4-oxobutyl)-1,4-diazabicyclo[2.2.2]octane-1,4-diium dibromide (2): A mixture of NHS ester (1) (2 g, 7.5 mmol), 1,4-diazabicyclo[2.2.2]octane (DABCO; 403 mg, 3.6 mmol), and DMSO (6 mL) was stirred at room temperature for 1 wk. Acetonitrile was added to precipitate the product, which was collected by filtration, washed with acetonitrile, and dried to yield (2) (1.7 g, 73.6 %) as a white solid: mp 230 °C (dec); 1H NMR (d6DMSO) δ 3.9 (s, 6H), 3.6 (t, 4H), 2.8 (m, 12H), 2.1 (m, 4H); 13C NMR (d6DMSO) δ 170.64 (C=O), 168.64 (C=O), 62.4 (CH2), 51.08 (CH2), 27.42 (CH2), 25.94 (CH2), 17.55 (CH2); MS (ES+) m/z 559.2, 561.2 (MBr)+.
Crosslinking Reaction
In all experiments, 100 mM crosslinker was prepared immediately before use in 250 mM HEPES, pH 7.0. The acetylated standard peptides (10 ug of a 1 ug/uL solution) were reacted overnight with a 5 molar ratio of crosslinker. Fifty micrograms of a freshly prepared 5 µg/µL solution of aldolase was crosslinked using a 25-fold molar ratio of crosslinker to lysine residues for 30 min.
Mass Spectrometry
Prior to mass spectrometry, the crosslinked synthetic peptides were dried using a vacuum centrifuge. The dry peptides were reconstituted in 0.1% trifluoroacetic acid and Millipore C18 ZipTips were used to desalt the fractions according to the manufacturer’s instructions. The pH of the crosslinked aldolase was adjusted to 8.0 with 100 mM triethylammonium bicarbonate and then the solution was digested with trypsin in a 1:20 substrate to enzyme ratio overnight at 37 °C. The tryptic digest was further digested with endoprotease GluC (50:1) overnight at 37 °C. An HPLC equipped with a Gilson 811 C dynamic mixer, Gilson 307 and 305 pumps, Scientific Systems model CP-21 Lo Pulse; Amersham Biosystems 759A absorbance detector set at 214 nm, and Isco Foxy 200 fraction collector was used with a C18 PLRP-S 5 µ 100Å column to separate the peptides. Solvent A was 5% acetonitrile, 0.1% trifluoroacetic acid (TFA) while Solvent B was 90% acetonitrile 0.1% TFA. The flow rate was set to 1 mL/min with the following binary gradient: 0 min, 6.5% B, 14 min, 6.5% B, 17 min, 18% B, 62.4 min, 40%B, 77.8 min, 50% B, 83.4 min, 70% B, 84.8 min, 100% B, 86.2 min, 100% B, 87.6 min, 6.5% B, 96 min, 6.5% B. One minute fractions were collected and then dried using a Jouan speedvap. Dried fractions were reconstituted in 20 uL of 0.1% TFA, sonicated 30 min or more and spotted onto a MALDI target using 5 mg/mL dihydroxybenzoic acid, 5 mg/mL α-cyano-4-hydroxycinnamic acid in 50% acetonitrile 0.1% TFA as matrix.
ESI-LTQ-Orbitrap-MS
The crosslinked standard peptides were analyzed with a hybrid linear quadrupole ion trap-Orbitrap mass spectrometer (ThermoFisher Scientific, Inc. model LTQ-Orbitrap XL; San Jose, CA, USA). They were introduced by infusion using a TriVersa Nanomate nanospray ionization source from Advion BioSciences (Ithaca, NY, USA). Peptides were delivered with a pneumatic displacement pressure of 0.45 psi and an applied spray voltage of 1.63 kV. The heated capillary temperature was set at 200 °C. Monoisotopic precursor ions were selected for multistage tandem mass spectrometric acquisition using an isolation window of 3.0 (m/z units) and excitation energy settings of 35 for both MS2 and 35 for MS3 spectra. All CID spectra were collected in FT (Orbitrap) mode for high mass accuracy (>10 ppm) and peak resolution (30,000 at m/z 400).
MALDI-TOF/TOF-MS
The crosslinked aldolase digest spectra were acquired in positive reflector mode in the 4800 TOF/TOF mass spectrometer (Applied Biosystems/MSD SCIEX). MS spectra were acquired from 600 to 3500m/z in each fraction. CID was achieved with a collision energy of 2 kV and atmosphere was used as the collision gas in the medium gas setting (~10−6 Torr). The crosslinked standard peptides were manually spotted onto a 4800 MALDI sample plate and then MALDI matrix spotted over the samples (5 mg/mL of ↦-cyano-4-hydroxycinnamic acid, 5 mg/mL 2,5-dihydroxybenzoic acid in 50% acetonitrile, 0.1% trifluoroacetic acid). The eight most abundant peaks in each well were automatically selected for MS/MS fragmentation.
Identification of Crosslinked Peptides with a Custom Algorithm and Database Searching Software
A custom database containing all standard proteins used in our lab was searched using the Mascot search engine. The searches were completed using a peptide mass tolerance of 0.5 Da, fragment mass tolerance of 0.6 Da, and no enzyme. Variable modifications included the deamidation of glutamine and asparagine residues, oxidation of methionine, pyro-Glu, and the modification of lysine residues or the N-terminus that results from in source decay (in-source decay) of the crosslinker (+68 Da).
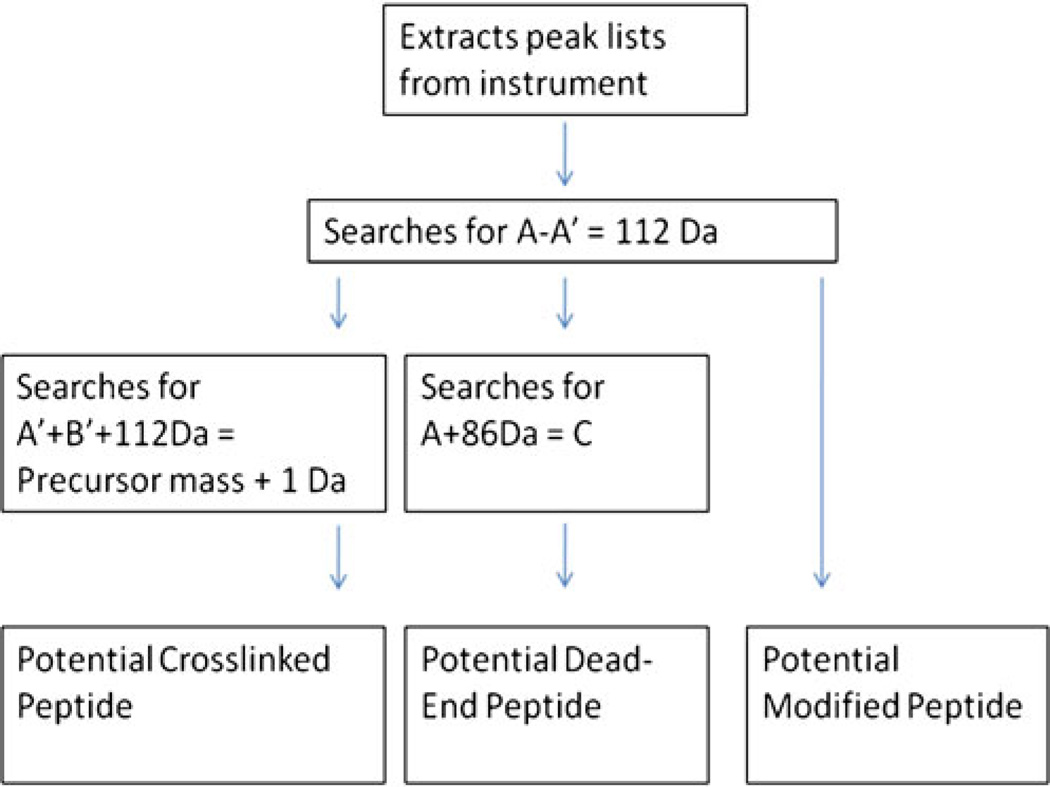
Algorithms written in Javawere used to perform three filters of the observed masses to identify potentially modified peptides according to Scheme 1. Peak lists from the MS mode were extracted from the ABI 4800 TOF/TOF. A 0.5 Da mass tolerance was allowed for the searches. The software first searched for differences of 112 Da between two peaks. In the second filter, candidate dead-end peptides were identified by searching for a set of three peaks: a precursor mass, a loss of 86 Da from the precursor, and then a subsequent loss of 112 Da or a total 198 Da loss from the precursor. In the third filter, potential crosslinked peptides were identified by searching the pairs of peaks separated by 112 Da for a set in which the lower of the two masses plus 112 Da added up to a precursor mass observed in the spectrum. Additional MS/MS spectra were obtained based on these results. The source code can be obtained by contacting the authors.
Scheme 1.
Filtering scheme to identify crosslinked peptides
Results and Discussion
Crosslinker Design
In the past decade, there has been an increase in research involving chemical crosslinking combined with mass spectrometry, and several crosslinkers have been developed to address one or more of the inherent difficulties associated with this technique. This project attempted to address several of those challenges. Crosslinked peptides have been reported to fragment poorly via CID (31, 44, 46), consistent with our observations. For this reason and to overcome some other limitations of current crosslinking reagents, we designed the crosslinker to be readily cleavable by CID and in-source decay (ISD), allowing the individual peptides released in MS2 or MS1 (via in-source decay) to be identified via MS3. Additional desired properties included generation of distinctive marker ions for the various reaction products, excellent solubility, ease of synthesis, good reactivity, self-quenching, and good stability in dry form. Substituted amines are known to undergo facile fragmentation via CID and several crosslinking and tagging reagents have been designed to include these structures [12, 52], including commercial reagents like the isobaric tag iTRAQ [55]. The energy required to induce fragmentation of these structures is similar to the energy required for peptide backbone fragmentation, allowing cleavage to still occur along the peptide backbone. DC4 contains quaternary amine moieties to further increase crosslinker lability over the peptide backbone and to generate fragmentation products containing mobilized protons.
Current crosslinkers that have been used to map tertiary or quaternary structure of proteins and complexes span a range from the “zero length” crosslinker formaldehyde to much longer crosslinkers of 40 or more Å [22, 24, 26]. The arm span of DC4 was estimated to be 18Å from one leaving group to the other, situating this crosslinker in the middle range of reported crosslinker lengths. As discussed below for the aldolase study, the results summarized in Table 2 clearly demonstrate that this crosslinker is effective for lysyl residue amino groups from approximately 18Å separation down to very short separations as deduced from the crystal structure. For studies of very large complexes we expect that longer crosslinkers will be needed to complement DC4 as well as shorter crosslinks to further constrain distances in certain cases.
Table 2.
Type Two Crosslinked Peptides Identified from Proteolytic Digest of Crosslinked Aldolase.
| Crosslink type | Peptide A | Peptide B | Distance (Å) | Precursor mass |
Fragment masses |
|---|---|---|---|---|---|
| 2 (Inter) | ILPDGDHDLKR (190–200) | QKKE (11–14) | 7 | 2221.2 | 651.4, 763.3, 875.4, 1278.8, 1346.8, 1458.8 |
| 2 (Inter) | KVLAAVYK (207–214) | QKKE (11–14) | 10 | 1920.1 | 959.6, 1071.7, 651.2, 763.4, 875.6 |
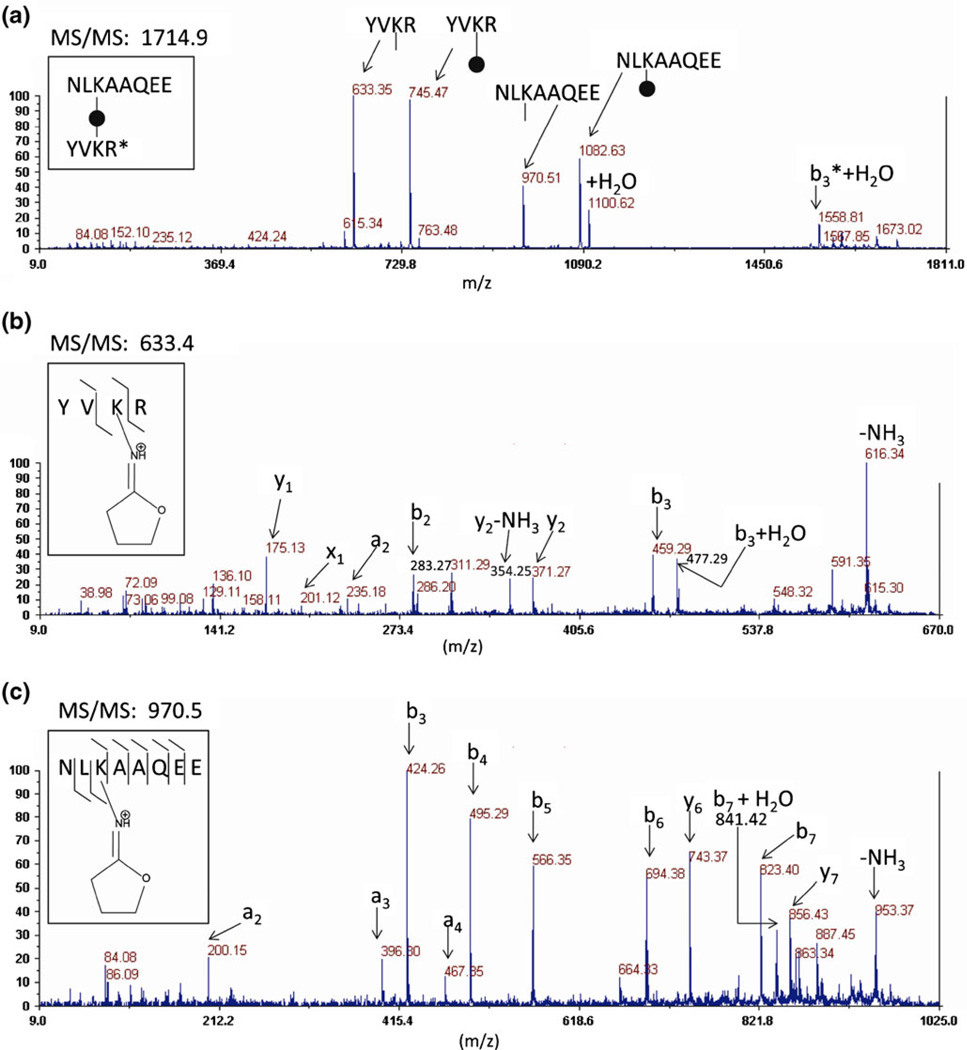
| 2 (Intra) | YVKR (327–330) | NLKAAQEE (319–326) | 14 | 1714.9 | 633.4, 745.5, 970.5, 1082.6 |
| 2 (Inter) | ILPDGDHDLKR (190–200) | *PHSHPALTPE (1–10) | 15 | 2611.3 | 1153.6, 1265.7, 1346.7, 1458.8 |
| 2 (Intra) | CVLKIGE (149–155) | STGSIAKR (35–42) | 17 | 1827.9 | 887.5, 999.6, 941.5, 829.5 |
| 2 (Intra) | YVKR (327–330) | STGSIAKR (35–42) | 23 | 1631.9 | 999.6, 887.5, 745.5, 633.4 |
| 2 (Intra) | STGSIAKR (35–42) | NLKAAQEE (319–326) | 30 | 1969.0 | 999.6, 887.4, 1082.5, 970.5 |
| 2 (Intra) | STGSIAKR (35–42) | *PHSHPALTPE (1–10) | 30 | 2152.1 | 887.5, 999.6, 1153.5, 1265.7 |
| 2 (Intra) | *PHSHPALTPE (1–10) | KDGADFAK (139–146) | 32 | 2184.1 | 1153.5, 1265.6, 1031.5 |
| 2 (Inter) | YVKR (327–330) | QKKE (11–14) | 39 | 1507.9 | 875.5, 763.4, 745.5, 633.3 |
Bold Type Represents Modified Residues.
The * Represents the N-Terminus of the Protein
MS Fragmentation of Model DC4-Crosslinked Peptides
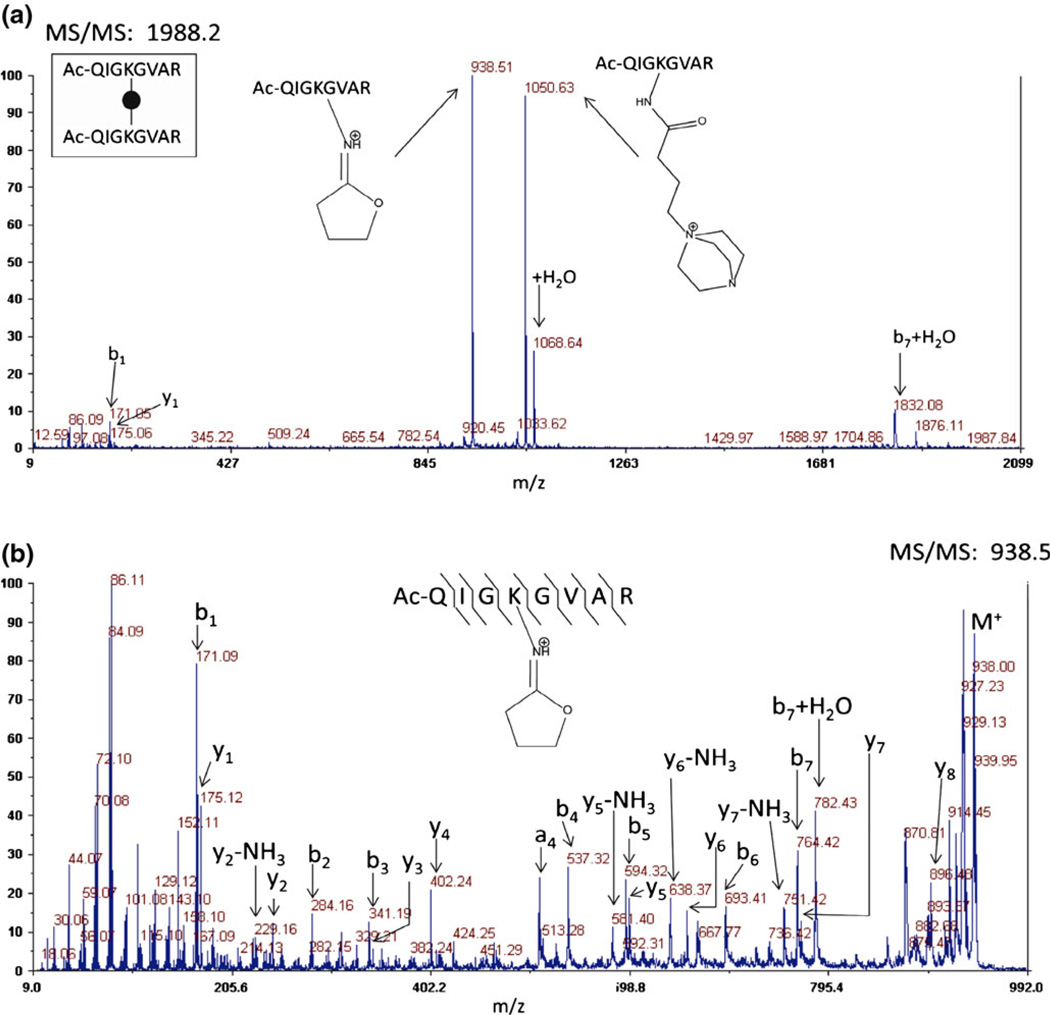
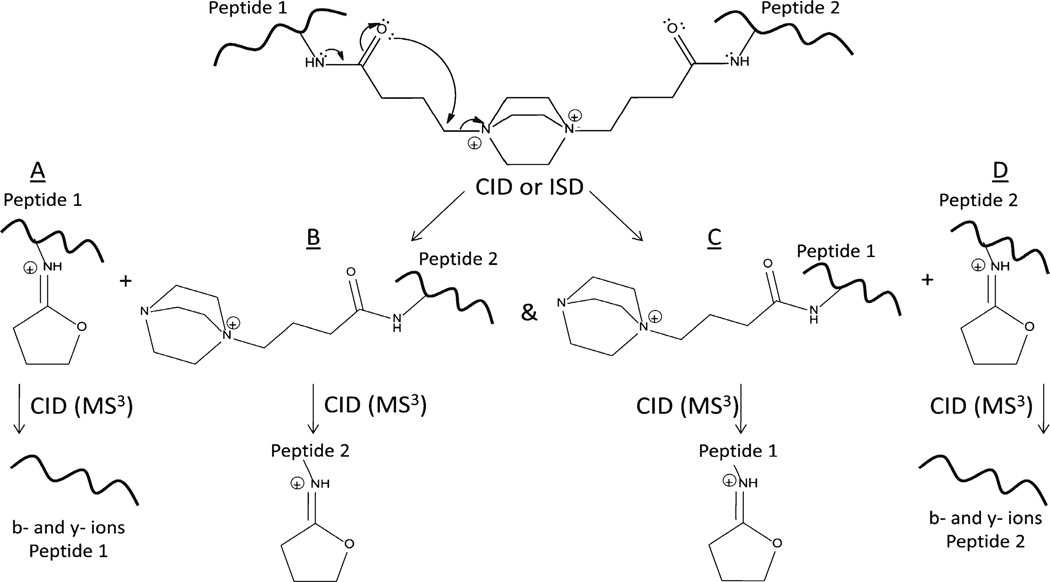
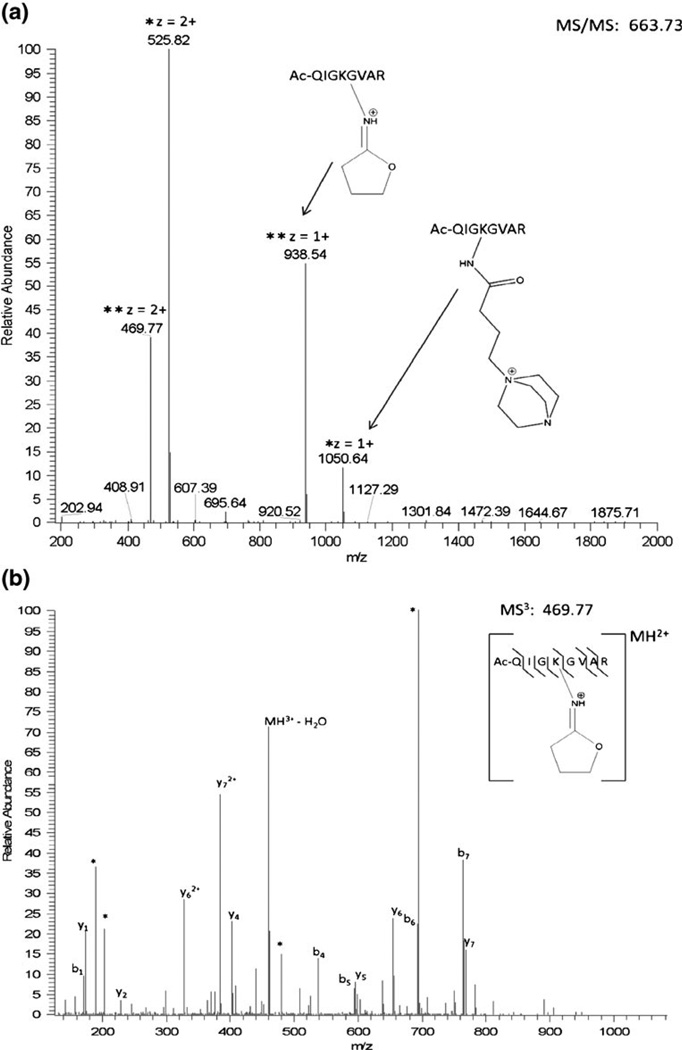
The N-terminally acetylated peptides Ac-QIGKGVAR and Ac- QHAKLGR were prepared in solution concentrated enough to force the ternary reaction of two peptides with DC4 through mass action. As expected, crosslinked peptides, dead-end reaction products, and in-source decay cleavage products from both the dead-end reaction and the crosslinked peptide products were observed in the initial mass spectrum via MALDI-TOF-TOF-MS. No peptides containing unmodified lysyl residues were observed. Although there are two intrinsic positive charges in the crosslinking moiety, the mass of the crosslinked peptide represents a loss of a proton to yield the singly charged ion (M –H)+ in MALDI-TOF-MS. No evidence of a doubly charged crosslinked peptide exists in the MALDI spectra. This is likely due to formation of an internal ion pair between the carboxyl group of the C-terminus and one of the quaternary amines [56, 57]. CID of the crosslinked peptide results in two fragments, 1050.6 and 938.5, both from cleavage at the sites of intrinsic positive charge (Figure 2a). The fragment at 1050.6 Da is due to cleavage at the intrinsic positive charge on the nitrogen containing ring (DABCO), retaining the DABCO moiety. The second fragment results from a rearrangement reaction which eliminates the DABCO ring. The same two cleavage products are visible in the original mass spectrum resulting from in-source decay. Pseudo MS3 (MS2 of the in-source decay product) of the elimination product (938.5) produces a spectrum with a robust series of b- and y-ions from which the component peptide may be identified (Figure 2b). Rearrangement of quaternary amines has been previously described by He and Reilly [58], a similar mechanism involving a sulfonium crosslinker was proposed by Lu et al. [47], and the proposed mechanism for DC4 is shown here in Figure 3. This mechanism is consistent with the data presented here and, importantly, it mobilizes an existing proton on the rearranged product when the original precursor was not protonated. This mobile proton can then facilitate backbone fragmentation yielding the b- and y-ion series.
Figure 2.
(a) CID fragmentation on the MALDI-TOF-TOF of the crosslinked synthetic peptide m/z 1988.2 (homodimer of AcQIGKGVAR) results in two major cleavage events at the intrinsic positive charges which are also observed in the first mass spectrum via in-source decay. (b) Pseudo MS3 of the fragment without an intrinsic positive charge produces a robust series of b- and y-ions which allows for peptide identifications
Figure 3.
Although the crosslinker contains two intrinsic positive charges, peptides modified with DC4 are observed as singly charged ions in MALDI, most likely due to the loss of a proton from a carboxylic acid moiety and ion pair formation (not shown). Upon fragmentation of the singly charged precursor by either in-source decay or CID, the rearrangement produces two singly charged species by mobilizing an existing proton as shown. The difference between species A and C as well as B and D is 112 Da resulting from the loss of the DABCO moiety and is used as a selection filter for crosslinked peptides
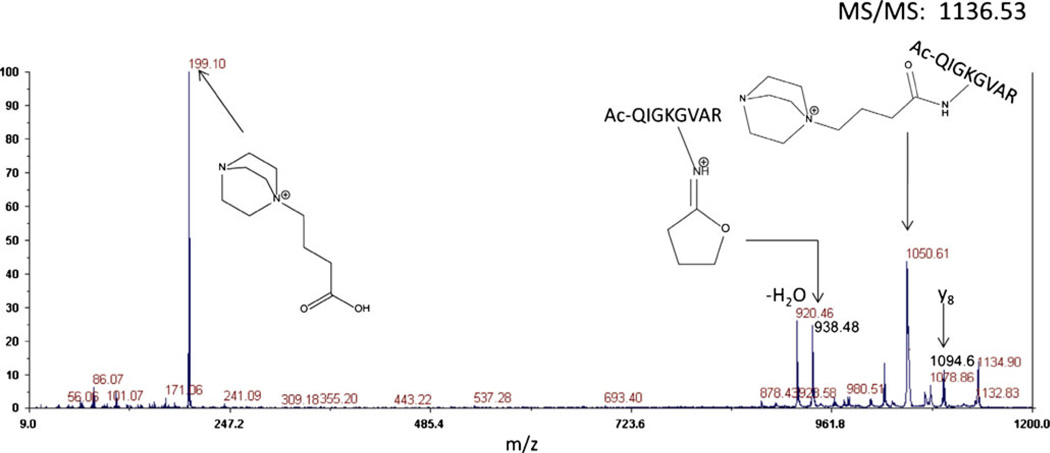
Tandem mass spectrometry of the dead-end product results in a diagnostic ion clearly visible at m/z 199.1. (Figure 4) The presence of this ion unambiguously identifies this spectrum as being from a modified peptide, although it has been observed occasionally in crosslinked peptides and valid crosslinked peptides may also contain dead-end reaction products. This cannot be used as an absolute exclusion criterion although it is useful. The other major peaks in the MS/MS spectrum result from fragmentation at the intrinsic positive charges. Low level b- and y-ions are also observed, however, the same fragments due to cleavage at the intrinsic positive charges are present in the mass spectrum from in-source decay. CID of the in-source decay product which has lost the DABCO moiety yields a robust sequence of b- and y-ions. The fragmentation of crosslinked peptides was also evaluated in the ESI-LTQ-Orbitrap-MS via CID and similar fragmentation events were observed as shown in Figures 5a and b.
Figure 4.
CID fragmentation via MALDI-TOF-TOF of a dead-end reaction product (m/z 1136.3) from a proteolytic digest of crosslinked aldolase. Similar fragmentation is observed as for the crosslinked peptide (Figure 2), but a diagnostic ion at 199.1 is observed, confirming this spectrum as being from a dead-end (type 0) product
Figure 5.
(a) CID on the LTQ-Orbitrap of the crosslinked homodimer synthetic peptide (Ac-QIGKGVAR) results in the same two major cleavage events observed in MALDI at the intrinsic positive charges. (b) MS3 of the fragment without an intrinsic positive charge produces a robust series of b- and y-ions for peptide identification
Algorithm Development
The in source decay fragmentation of DC4 crosslinked peptides provides a unique series of ions that can be taken advantage of in developing a computational approach to identifying crosslinked peptides. An algorithm was developed to identify modified peptides using a simple filtering schema (Scheme 1). The software application based on this algorithm initially searches MS2 spectra (or in-source decay in MS spectra from MALDI) for differences of 112 Da between two peaks. Output from this first filter may contain dead-end peptides, crosslinked peptides, or multiply modified peptides. Next, the dead-end peptides are culled out by searching for precursor ions that have an 86 Da loss, followed by a 112 Da loss for a total loss of 198 Da from the precursors. These results are tagged as potential dead-end peptides. The crosslinked peptides are identified by first searching for the pairs of peaks separated by 112 Da, then the software determines if the observed peptide pairs sum (while correcting for the DABCO mass of 112 Da) to a precursor ion in the spectrum. Those ions that satisfy these criteria are returned to the user as potential crosslinked peptides and are then subjected to MS3 and detailed de novo analysis.
Application of DC4 to Aldolase
Aldolase crosslinking by DC4 was optimized in solution by reaction at a range of crosslinker to protein ratios and the products evaluated by SDS gel electrophoresis (Supplemental Materials Figure 1) with the objective of maximizing crosslinks within tetramers while minimizing crosslinks between tetramers (nonspecific interactions). Aldolase tetramers were crosslinked at the optimum ratio of DC4 (minimizing nonspecific interactions while maximizing crosslinking efficiency) for mass spectrometry analysis. CID of the in-source decay fragments from DC4 crosslinked peptides generated sufficient backbone fragmentation in the aldolase digest to identify the amino acid sequence of the crosslinked peptides as shown in Figure 6 and in Supplemental Materials Figures 2–19. The majority of the spectra from modified residues displayed preferential fragmentation, consisting predominantly of b-ions from the N-terminal side of the modified residues and y-ions from the C-terminal side. Not surprisingly, this fragmentation property is somewhat sequence dependent.
Figure 6.
(a) CID of DC4-crosslinked peptides from a proteolytic digest of aldolase produces two pairs of peaks where the lower mass is separated from the higher mass by 112 Da. The black circle represents the central DABCO moiety and the lines on either side represent the carbon spacer arm in the crosslinker. Those same four peaks are observed in the mass spectrum from in-source decay and CID of the lower mass of each peptide pair [(b), (c)] yields the b- and y-ions for peptide identification
Both crosslinked peptides and dead-end peptides were identified from proteolytic digests of DC4-crosslinked aldolase and are shown in Tables 2 and 3, respectively. These modified peptides were evaluated for their consistency with the crystal structure for aldolase [59], and it was found that both the dead-end and crosslinked peptide residues observed were solvent exposed. Six of the crosslinked peptides identified had inter-Lys distances between 7 and 23Å. Although the DABCO moiety is rigid, the reactive side arms are quite flexible and capable of coupling to Lys residues, spaced closely together, consistent with the shortest distance observed between two crosslinked Lys residues being 7Å. The side chain residue distance measurements were based on the rigid crystal structure and some changes in solution phase are likely since aldolase exhibits multiple crystal forms and portions of the molecule exhibit considerable flexibility between crystal structures and in solution [60–63].
Table 3.
The Type Zero Crosslinked Peptides (Dead-Ends) Identified from a Proteolytic Digest of Crosslinked Aldolase.
| Peptide sequence | Precursor mass |
Fragment masses |
|---|---|---|
| VDKGVVPLAGTNGE (108–121) | 1621.8 | 1535.8, 1423.8 |
| IVAPGKGILAADE (22–34) | 1519.8 | 1433.8, 1321.7 |
| NLKAQEE (319–325) | 1168.6 | 1082.6, 970.5 |
| KDGADFAK (139–146) | 1117.5 | 1031.5, 919.5 |
| KVLAAVYK (207–214) | 1157.6 | 1071.6, 959.5 |
| CVLKIGE (149–155) | 1027.5 | 941.5, 829.4 |
| QKKE (11–14) | 961.5 | 875.5, 763.4, 651.3 |
| STGIASKR (35–42) | 1085.6 | 999.5, 887.5 |
| *PHSHPALTPE (1–10) | 1351.6 | 1265.6, 1153.5 |
| YVKR (327–330) | 831.5 | 745.4, 633.4 |
| ILPDGDHDLKR (191–201) | 1544.8 | 1458.7, 1346.7 |
Bold Type Represents Modified Residues.
The * Represents the N-Terminus of the Protein
The other four crosslinked peptides had inter-lysyl residue distances from 30 to 39Å. Although these distances are longer than we would initially expect based on the length of DC4 alone and a rigid protein structure, they are consistent with flexibility of protein backbones and lysyl side chains in solution. Here, several of the lysyl residues were observed to exhibit crosslinking with more than one other lysyl residue, consistent with earlier claims of flexibility in those regions. This flexibility allows the crosslinker to sample a larger region on the surface of the protein. As an example, Lys 41 was observed crosslinked to both Lys-152, which was 17Å away, and Lys-321, 23Å away, but also to Lys 329, 30Å away and to the N-terminus, also 30Å away. Deuterium exchange studies [60] in solution indicate very high exchange rates for the short helical region containing Lys-41 and high B-values observed for this region in the crystal structure [61, 63] provide further evidence for the flexibility of this region. This high level of flexibility is consistent with the observation for Lys-41 crosslinked to Lys-321 and to the N-terminus, both at 30Å. Likewise, the four N-terminal residues show considerable variation between crystal structures, indicating this region is flexible and consistent with our identification of crosslinks between the N-terminus and Lys-199, Lys-139, and Lys-41 with distances of 15, 32, and 30Å from the N-terminus, respectively. The crosslink exhibiting the longest span between Lys residues is between Lys 13 and Lys 329. The latter lysyl residue lies within the highly flexible C-terminal region involved in binding to F-actin [62]. It has been previously suggested that crosslinker length is not necessarily a precise indicator of inter-residue spacing when flexibility of the peptide backbone and lysyl side chains are factored in [64,65], and our results are certainly consistent with this assertion.
All the peptides constituting type 2 crosslinked peptides were also observed as type 0 reaction products as shown in Table 3. Two peptides (VDKGVVPLAGTNGE, IVAPGKGILAADE) were only observed as the type 0 products. The closest lysyl residues for Lys-110 and Lys-27 are only 10 and 13Å away, respectively, suggesting that the crosslinked products may not have been suitable for analysis (low ionization efficiency or large fragment size). Two lysyl residues, Lys-214 and Lys-146, were observed only as unmodified forms, suggesting that they were not as accessible to reagent as those that exhibited modification. Inspection of these residues in the crystal structure revealed that they are much less accessible than other lysyl residues observed to be modified in this study, as illustrated in Supplementary Material Figure 20, where we show a ribbon model with the pertinent lysyl residues highlighted. The two lysyl residues are completely obscured from all angles in the space-filling model.
Conclusions
We have designed and tested a new crosslinking reagent that overcomes some of the difficulties associated with the use of crosslinking reagents with mass spectrometry. The DC4 crosslinking reagent is stable as a solid over long-term storage, highly reactive, highly soluble (1 M solutions), and quite labile to CID. When fragmented by either CID or ISD, DC4 crosslinked peptides fragment characteristically into four rearranged products. Even though the precursor ions are not protonated, the fragmentation products rearrange to generate a mobile proton, and MS3 results in efficient backbone fragmentation for peptide identification. Database searches can then identify which peptides were crosslinked, and based on this characteristic fragmentation behavior, an algorithm was developed to identify which peptides were crosslinked. This crosslinking strategy was used on the model tetrameric protein, aldolase, and the crosslinked peptides identified were consistent with the structure.
Supplementary Material
Acknowledgments
The authors acknowledge funding in part for this work by NIH #1P41RR018627 and NIH #R01GM09583201. They thank the members of the Michigan Proteome Consortium for their assistance in this effort. Peptides were synthesized by the Biomedical Research Core Facility. The authors acknowledge that Bryan E. Smith wrote the filtering algorithm for identifying candidate crosslinked peptides.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s13361-011-0288-4) contains supplementary material, which is available to authorized users.
References
- 1.Berggard T, Linse S, James P. Methods for the detection and analysis of protein–protein interactions. Proteomics. 2007;7:2833–2842. doi: 10.1002/pmic.200700131. [DOI] [PubMed] [Google Scholar]
- 2.Gingras A, Gstaiger M, Raught B, Aebersold R. Analysis of protein complexes using mass spectrometry. Nat. Rev. Mol. Cell Biol. 2007;8:645–654. doi: 10.1038/nrm2208. [DOI] [PubMed] [Google Scholar]
- 3.Benesch J, Ruotolo B, Simmons D, Robinson C. Protein complexes in the gas phase: Technology for structural genomics and proteomics. Chem. Rev. 2007;107:3544–3567. doi: 10.1021/cr068289b. [DOI] [PubMed] [Google Scholar]
- 4.Konermann L, Tong X, Pan Y. Hydrogen exchange mass spectrometry for studying protein structure and dynamics. J. Mass. Spectrom. 2008;43:1021–1036. doi: 10.1002/jms.1435. [DOI] [PubMed] [Google Scholar]
- 5.Heck A. Native mass spectrometry: A bridge between interactomics and structural. Biol. nat. Methods. 2008;5:92–933. doi: 10.1038/nmeth.1265. [DOI] [PubMed] [Google Scholar]
- 6.Loo J. Studying noncovalent protein complexes by electrospray ionization mass spectrometry. Mass Spectrom. Rev. 1997;16:1–23. doi: 10.1002/(SICI)1098-2787(1997)16:1<1::AID-MAS1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 7.Yang W, Steen H, Freeman M. Proteomic approaches to the analysis of multiprotein signaling complexes. Proteomics. 2008;8:832–851. doi: 10.1002/pmic.200700650. [DOI] [PubMed] [Google Scholar]
- 8.Rappsilber J, Siniossoglou S, Hurt E, Mann M. A generic strategy to analyze the spatial organization of Multi-protein complexes by crosslinking and mass spectrometry. Anal. Chem. 2000;72:267–275. doi: 10.1021/ac991081o. [DOI] [PubMed] [Google Scholar]
- 9.Seebacher J, Mallick P, Zhang N, Eddes JS, Aebersold R, Gelb MH. Protein Cross-linking analysis using mass spectrometry, Isotopecoded Cross-linkers, and integrated computational data processing. J. Proteome Res. 2006;5:2270–2282. doi: 10.1021/pr060154z. [DOI] [PubMed] [Google Scholar]
- 10.Lee JY, Lachner L, Nunnari J, Phinney B. Shotgun Cross-linking analysis for studying quaternary and tertiary protein structures. J. Proteome Res. 2007;6:3908–3917. doi: 10.1021/pr070234i. [DOI] [PubMed] [Google Scholar]
- 11.Sinz A. Chemical Cross-linking and mass spectrometry to map threedimensional protein structures and Protein–protein interactions. Mass Spectrom. Rev. 2006;25:663–682. doi: 10.1002/mas.20082. [DOI] [PubMed] [Google Scholar]
- 12.Back JW, Hartog AF, Dekker HL, Muijser AO, de Koning LJ, de Jong L. A new crosslinker for mass spectrometry analysis of the quaternary structure of protein complexes. J. Am. Soc. Mass Spectrom. 2001;12:222–227. doi: 10.1016/S1044-0305(00)00212-9. [DOI] [PubMed] [Google Scholar]
- 13.Swaim CL, Smith JB, Smith DL. Unexpected products from the reaction of the synthetic Cross-linker 3,3'-dithiobis(sulfosuccinimidyl propionate), DTSSP with peptides. J. Am. Soc. Mass Spectrom. 2004;15:736–749. doi: 10.1016/j.jasms.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 14.Iglesias AH, Santos LFA, Gozzo FC. Collision-induced dissociation of Lys-lys intramolecular Cross-linked peptides. J. Am. Soc. Mass. Spectrom. 2009;20:557–566. doi: 10.1016/j.jasms.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 15.Soderblom E, Bobay B, Cavanagh J, Goshe M. Tandem mass spectrometry acquisition approaches to enhance Identification of Protein–protein interactions using Low-energy Collision-induced dissociative chemical crosslinking reagents. Rapid Commun. Mass Spectrom. 2007;21:3395–3408. doi: 10.1002/rcm.3213. [DOI] [PubMed] [Google Scholar]
- 16.Kalkhof S, Sinz A. Chances and pitfalls of chemical Cross-linking with Amine-reactive N-hydroxysuccinimide Esters. Anal. Bioanal. Chem. 2008;392:305–312. doi: 10.1007/s00216-008-2231-5. [DOI] [PubMed] [Google Scholar]
- 17.Madler S, Bich C, Touboul D, Zenobi R. Chemical Cross-linking with NHS Esters: A systematic study on Amino acid reactivities. J. Mass Spectrom. 2009;44:694–706. doi: 10.1002/jms.1544. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Q, Crosland E, Fabris D. Nested Arg-specific bifunctional crosslinkers for MS-based structural analysis of proteins and protein assemblies. Anal. Chim. Acta. 2008;627:117–128. doi: 10.1016/j.aca.2008.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Novak P, Kruppa GH. Intra-molecular Cross-linking of acidic residues for protein structure studies. Eur. J. Mass Spectrom. 2008;14:355–365. doi: 10.1255/ejms.963. [DOI] [PubMed] [Google Scholar]
- 20.Krauth F, Ihling CH, Ruttinger HH, Sinz A. Heterobifunctional Isotope-labeled Amine-reactive Photo-cross-linker for structural investigation of proteins by MALDI-TOF/TOF and ESI-LTQ-orbitrap mass spectrometry. Rapid Commun. Mass Spectrom. 2009;23:2811–2818. doi: 10.1002/rcm.4188. [DOI] [PubMed] [Google Scholar]
- 21.Kang S, Mou L, Lanman J, Velu S, Brouillette WJ, Prevelige PE., Jr Synthesis of Biotin-tagged chemical Cross-linkers and their applications for mass spectrometry. Rapid Commun. Mass Spectrom. 2009;23:1719–1726. doi: 10.1002/rcm.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang H, Tang X, Munske GR, Tolic N, Anderson GA, Bruce JE. Identification of Protein–protein interactions and topologies in living cells with chemical Cross-linking and mass spectrometry. Mol. Cell. Proteom. 2009;8:409–420. doi: 10.1074/mcp.M800232-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujii N, Jacobsen RB, Wood NL, Schoeniger JS, Guy RK. A novel protein crosslinking reagent for the determination of moderate resolution protein structures by mass spectrometry (MS3D) Bioorg Med. Chem. Lett. 2004;14:427–429. doi: 10.1016/j.bmcl.2003.10.043. [DOI] [PubMed] [Google Scholar]
- 24.Sinz A, Kalkhof S, Ihling C. Mapping protein interfaces by a trifunctional Cross-linker combined with MALDI-TOF and ESI-FTICR mass spectrometry. J. Am. Soc. Mass Spectrom. 2005;16:1921–1931. doi: 10.1016/j.jasms.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 25.Trester-Zedlitz M, Kamada K, Burley SK, Fenyo D, Chait BT, Muir TW. A modular Cross-linking approach for exploring protein interactions. J. Am. Chem. Soc. 2003;125:2416–2425. doi: 10.1021/ja026917a. [DOI] [PubMed] [Google Scholar]
- 26.Zhang H, Tang X, Munske GR, Zakharova N, Yang L, Zheng C, Wolff MA, Tolic N, Anderson GA, Shi L, Marshall MJ, Frederickson JK, Bruce JE. In vivo identification of the outer membrane protein OmcA-MtrC interaction network in Shewanella oneidensis MR-1 cells using novel hydrophobic chemical Cross-linkers. J. Proteome Res. 2008;7:1712–1720. doi: 10.1021/pr7007658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hur GH, Meier JL, Baskin J, Codelli JA, Bertozzi RC, Marahiel MA, Burkart MD. Crosslinking studies of Protein–protein interactions in nonribosomal peptide biosynthesis. Chem. Biol. 2009;16:372–381. doi: 10.1016/j.chembiol.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nessen MA, Kramer G, Back J, Baskin JM, Smeenk LEJ, de Koning LJ, van Maarseveev JH, de Jong L, Bertozzi CR, Hiemstra H, de Koster CG. Selective enrichment of Azide-containing peptides from complex mixtures. Proteome Res. 2009;8:3702–3711. doi: 10.1021/pr900257z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chowdhury SM, Du X, Tolic N, Wu S, Moore RJ, Mayer MU, Smith RD, Adkins JN. Identification of Cross-linked peptides containing a CLICK-based enrichment Group using sequential CID and ETD tandem mass spectrometry. Anal. Chem. 2009;81:5524–5532. doi: 10.1021/ac900853k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ihling Ch, Schmidt A, Kalkhof S, Schulz D, Stingl C, Mechtler K, Haack M, Beck-Sickinger A, Cooper D, Sinz A. Isotopelabeled Cross-linkers and fourier transform ion cyclotron resonance mass spectrometry for structural analysis of a protein/peptide complex. J. Am. Soc. Mass Spectrom. 2006;17:1100–1113. doi: 10.1016/j.jasms.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 31.Petrotchenko EV, Serpa JJ, Borchers CH. Use of a combination of isotopically coded Cross-linkers and isotopically Coded N-terminal modification reagents for selective identification of Inter-peptide crosslinks. Anal. Chem. 2010;82:817–823. doi: 10.1021/ac901637v. [DOI] [PubMed] [Google Scholar]
- 32.Chu F, Mahrus S, Craik CS, Burlingame AL. Isotope-Coded and Affinity-Tagged Cross-Linking (ICATXL): An efficient strategy to probe protein interaction surfaces. J. Am. Chem. Soc. 2006;128:10362–10363. doi: 10.1021/ja0614159. [DOI] [PubMed] [Google Scholar]
- 33.Muller DR, Schindler P, Towbin H, Wirth U, Voshol HH, Hoving S, Steinmetz MO. Isotope-Tagged Cross-Linking Reagents: A new tool in mass spectrometry protein interaction analysis. Anal. Chem. 2001;73:1927–1934. doi: 10.1021/ac001379a. [DOI] [PubMed] [Google Scholar]
- 34.Collins CJ, Schilling B, Young M, Dollinger G, Guy RK. Isotopically labeled crosslinking reagents: Resolution of mass degeneracy in the identification of crosslinked peptides. Bioorg. Med. Chem. Lett. 2003;13:4023–4026. doi: 10.1016/j.bmcl.2003.08.053. [DOI] [PubMed] [Google Scholar]
- 35.Rinner O, Seebacher J, Walzthoeni T, Mueller LN, Beck M, Schmidt A, Mueller M, Aebersold R. Identification of Cross-linked peptides from large sequence databases. Nat. Methods. 2008;5:315–318. doi: 10.1038/nmeth.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schilling B, Row RH, Gibson BW, Guo X, Young MM. MS2Assign, automated assignment and nomenclature of tandem mass spectra of chemically crosslinked peptides. J. Am. Soc. Mass Spectrom. 2003;14:834–850. doi: 10.1016/S1044-0305(03)00327-1. [DOI] [PubMed] [Google Scholar]
- 37.Lee YJ. Mass spectrometric analysis of Cross-linking sites for the structure of proteins and protein complexes. Mol. Biosyst. 2008;4:816–823. doi: 10.1039/b801810c. [DOI] [PubMed] [Google Scholar]
- 38.Maiolica A, Cittaro D, Borsotti D, Sennels L, Ciferri C, Tarricone C, Musacchio A, Rappsilber J. Structural analysis of Multi-protein complexes by Cross-linking, mass spectrometry and database searching. Mol. Cell. Proteom. 2007;6:2200–2211. doi: 10.1074/mcp.M700274-MCP200. [DOI] [PubMed] [Google Scholar]
- 39.Santos LFA, Iglesias AH, Gozzo FC. Fragmentation features of intermolecular Cross-linked peptides using N-hydroxy-succinimide esters by MALDI- and ESI-MS/MS for use in structural proteomics. J. Mass Spectrom. 2011;46:742–750. doi: 10.1002/jms.1951. [DOI] [PubMed] [Google Scholar]
- 40.Vellucci D, Kao A, Kaake RM, Rychnovsky SD, Huang L. Selective enrichment and identification of Azide-Tagged Cross-Linked peptides using chemical ligation and mass spectrometry. J. Am. Soc. Mass Spectrom. 2010;21:1432–1445. doi: 10.1016/j.jasms.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.King GJ, Jones A, Kobe B, Huber T, Mouradov D, Hume DA, Ross IL. Identification of Disulfide-containing chemical Cross-links in proteins using MALDI-TOF/TOF mass spectrometry. Anal. Chem. 2008;80:5036–5043. doi: 10.1021/ac702277q. [DOI] [PubMed] [Google Scholar]
- 42.Petrotchenko EV, Olkhovik VK, Borchers CH. Isotopically coded cleavable Cross-linker for studying Protein–Protein interaction and protein complexes. Mol. Cell. Proteom. 2005;4:1167–1179. doi: 10.1074/mcp.T400016-MCP200. [DOI] [PubMed] [Google Scholar]
- 43.Muller MQ, Dreiocker F, Ihling CH, Schafer M, Sinz A. Cleavable Cross-linker for protein structure analysis: Reliable identification of Cross-linking products by tandem MS. Anal. Chem. 2010;82:6958–6968. doi: 10.1021/ac101241t. [DOI] [PubMed] [Google Scholar]
- 44.Petrotchenko EV, Xiao K, Cable J, Chen Y, Dokholyan NV, Borchers CH. BiPS, a photocleavable, isotopically coded, fluorescent crosslinker for structural proteomics. Mol. Cell. Proteom. 2009;8:273–296. doi: 10.1074/mcp.M800265-MCP200. [DOI] [PubMed] [Google Scholar]
- 45.Gardner MW, Vasicek LA, Shabbir S, Anslyn EV, Brodbelt JS. Chromogenic Cross-linker for the characterization of protein structure by infrared multiphoton dissociation mass spectrometry. Anal. Chem. 2008;80:4807–4819. doi: 10.1021/ac800625x. [DOI] [PubMed] [Google Scholar]
- 46.Tang X, Munske G, Siems W, Bruce J. Mass spectrometry identifiable Cross-linking strategy for studying Protein–Protein interactions. Anal. Chem. 2005;77:311–318. doi: 10.1021/ac0488762. [DOI] [PubMed] [Google Scholar]
- 47.Lu Y, Tansova M, Borhan B, Reid G. Ionic reagent for controlling the Gas-phase fragmentation reactions of Cross-linked peptides. Anal. Chem. 2008;80:9279–9287. doi: 10.1021/ac801625e. [DOI] [PubMed] [Google Scholar]
- 48.Kasper TT, Back JW, Vitale M, Hartog AF, Roseboom W, de Koning LJ, van Maarseveen JH, Muijsers AO, de Koster GD, de Jong L. An aptly positioned azido group in the Spacer of a Protein Cross-linker for facile mapping of lysines in close proximity. Chem. BioChem. 2007;8:1281–1292. doi: 10.1002/cbic.200700150. [DOI] [PubMed] [Google Scholar]
- 49.Yang L, Tank X, Weisbrod CR, Munske GR, Eng JK, von Haller PD, Kaiser NK, Bruce JE. A photocleavable and mass spectrometry identifiable Cross-linker for protein interaction studies. Anal. Chem. 2010;82:3556–3566. doi: 10.1021/ac902615g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muller MQ, Dreiocker F, Ihling CH, Schafer M, Sinz A. Fragmentation behavior of a Thiourea-based reagent for protein structure analysis by Collision-induced dissociative chemical Crosslinking. J Mass Spectrom. 2010;45:880–891. doi: 10.1002/jms.1775. [DOI] [PubMed] [Google Scholar]
- 51.Dreoicker F, Muller MQ, Sinz A, Schafer M. Collision-induced dissociative chemical Cross-linking reagent for protein structure characterization: Applied Edman chemistry in the gas phase. J. Mass Spectrom. 2010;45:178–189. doi: 10.1002/jms.1702. [DOI] [PubMed] [Google Scholar]
- 52.Trnka MJ, Burlingame AL. Topographic studies of the GroELGroES chaperonin complex by chemical Cross-linking using diformyl ethynylbenzene. Mol. Cell. Proteom. 2010;9:2306–2317. doi: 10.1074/mcp.M110.003764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen ZA, Jawhar A, Fischer L, Buchen C, Tahir S, Kamenski T, Rasmussen M, Larivier L, Bukowski-Wills J, Nilges M, Cramer P, Rappsilber J. Architecture of the RNA polymerase II-TFIF complex revealed by Cross-linking and mass spectrometry. EMBO. J. 2010;29:717–726. doi: 10.1038/emboj.2009.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Almirante N, Ferrario M, Ongini E. Process for preparing Nitrooxy Esters, Nitrooxy Thioesters Nitrooxy Carbonates, and Nitrooxy Thiocarbonates, intermediates useful in said process and preparation thereof. WO Patent WO 2006/008196 A1. 2006 Jan 26; [Google Scholar]
- 55.Ross P, Huang Y, Marchese J, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin D. Multiplexed protein quantitation in Saccharomyces cerevisiae using Amine-reactive isobaric tagging reagents. Mol. Cell. Proteom. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 56.Nordhoff E, Kirpekar R, Roepstorff P. Mass spectrometry of nucleic acids. Mass Spectrom. Rev. 1996;15:67–138. doi: 10.1002/(SICI)1098-2787(1996)15:2<67::AID-MAS1>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 57.Freitas MA, Marshal AG. Rate and extent of Gas-phase hydrogen/ deuterium exchange of Bradykinins: Evidence for Peptide Zwitterions in the gas phase. Int. J. Mass Spectrom. 1999;182:221–231. [Google Scholar]
- 58.He Y, Reilly JP. Does a charge tag really provide a fixed charge? Angew. Chem. Int. Ed. 2008;47:2463–2465. doi: 10.1002/anie.200705048. [DOI] [PubMed] [Google Scholar]
- 59.Maurady A, Zdanov A, De Moissac D, Beaudry D, Sygusch J. A conserved glutamate residue exhibits multifunctional catalytic Roles in D-Fructose-1,6-Bisphosphate Aldolases. J. Biol. Chem. 2002;277:9474–9483. doi: 10.1074/jbc.M107600200. [DOI] [PubMed] [Google Scholar]
- 60.Dalby A, Dauter Z, Littlechild JA. Crystal structure of human muscle aldolase complexed with fructose 1,6-Bisphosphate: mechanistic implications. Protein Sci. 1999;8:291–297. doi: 10.1110/ps.8.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Z, Post CB, Smith DL. Amide hydrogen exchange determined by mass spectrometry: Application to rabbit muscle aldolase. Biochemistry. 1996;35:779–791. doi: 10.1021/bi952227q. [DOI] [PubMed] [Google Scholar]
- 62.Wang J, Morris AJ, Tolan DR, Pagliaro J. The Molecular nature of the F-Actin binding activity of aldolase revealed with Site-directed mutants. J. Biol. Chem. 1996;271:6861–6865. [PubMed] [Google Scholar]
- 63.Pan H, Smith DL. Quaternary structure of aldolase leads to differences in its folding and unfolding intermediates. Biochemistry. 2003;42:5713–5721. doi: 10.1021/bi027388q. [DOI] [PubMed] [Google Scholar]
- 64.Leitner A, Walzthoeni T, Kahraman A, Herzog F, Rinner O, Beck M, Aebersold R. Probing native protein structures by chemical Cross-linking, mass spectrometry, and bioinformatics. Mol. Cell. Proteom. 2010;9:1634–1649. doi: 10.1074/mcp.R000001-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Green NS, Reisler E, Houk KN. Quantitative evaluation of the lengths of homobifunctional protein Cross-linking reagents used as molecular rulers. Protein Sci. 2001;10:1293–1304. doi: 10.1110/ps.51201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.