Abstract
Background
Thousands of common single nucleotide polymorphisms (SNPs) are weakly associated with schizophrenia. It is likely that subsets of disease-associated SNPs are associated with distinct heritable disease-associated phenotypes. Therefore, we examined the shared genetic susceptibility modulating schizophrenia and brain volume.
Methods
Odds ratios for genome-wide SNP data were calculated in the sample collected by the Psychiatric GWAS Consortium (8,690 schizophrenia patients and 11,831 controls, excluding subjects from the present study). These were used to calculate individual polygenic schizophrenia (“risk”) scores (PSSs) in an independent sample of 152 schizophrenia patients and 142 healthy controls with available structural MRI scans.
Results
In the entire group, the PSS was significantly associated with total brain volume (R2=0.048, p=1.6×10−4) and white matter volume (R2=0.051, p=8.6×10−5) equally in patients and controls. The number of (independent) SNPs that substantially influenced both disease risk and white matter (n=2,020) was much smaller than the entire set of SNPs that modulated disease status (n=14,751). From the set of 2,020 SNPs, a group of 186 SNPs showed most evidence for association with white matter volume and an explorative functional analysis showed that these SNPs were located in genes with neuronal functions.
Conclusions
These results indicate that a relatively small subset of schizophrenia genetic risk variants is related to the (normal) development of white matter. This in turn suggests that disruptions in white matter growth increase the susceptibility to develop schizophrenia.
Keywords: structural MRI, imaging, endophenotype, SNPs, psychiatric, genome-wide
Introduction
Schizophrenia is a disabling mental disorder with a heritability of around 80% (1). The Psychiatric Genomics Consortium (PGC) recently published a large genome-wide association study (GWAS) on schizophrenia (2); this analysis of 17,836 cases and 33,859 controls yielded seven loci with common alleles that subtly increase schizophrenia risk. However, there are most likely many more single nucleotide polymorphisms (SNPs) involved in schizophrenia susceptibility: Purcell and colleagues described the additive effects of thousands of disease-associated SNPs combined into a single polygenic schizophrenia score (PSS) (3). This PSS based on >30,000 (mostly independent) SNPs explained around 3% of the variance in schizophrenia in an independent sample. Another recent study estimated that 23% of the variation in liability to schizophrenia is captured by the combined effect of >900,000 SNPs (4). These data support a complex mode of inheritance, with thousands of genetic variants of small effect contributing to disease. This large genetic heterogeneity is further complicated by substantial variation in clinical presentation, disease course and associated phenotypes. It is likely that subsets of disease-associated SNPs are associated with distinct heritable disease-associated phenotypes (also called endophenotypes (5)). One such phenotype is brain volume and it is well suited to be linked to disease associated SNPs. Brain volume is robustly associated with schizophrenia, with average reductions in total brain volume of about 3% in schizophrenia patients compared to healthy individuals (6;7). It is highly heritable in healthy subjects as well as in schizophrenia patients (8–11) and reduced brain volumes are inherited together with illness in families (12). In fact, the largest twin study (n=684) to date recently reported that 77% of the phenotypic overlap between schizophrenia and total brain volume was of genetic origin (11), with white matter loss in schizophrenia patients largely (94%) attributable to genetic factors (although gray matter volume was determined by unique and common environmental factors (11)). Thus, white matter volume is an excellent candidate endophenotype to be linked to schizophrenia-associated SNPs.
The aim of the current study was to investigate the combined effect of schizophrenia-associated loci on brain volume in order to answer several questions. First, is brain volume in schizophrenia patients indeed determined by disease-associated SNPs, and if so, by which proportion of these SNPs? Second, do disease-associated SNPs affect brain volume in patients only, or do they modulate brain volume in general? Finally, is the involvement of genetic factors on white matter volume in particular, as previously suggested by heritability calculations from twin studies, supported by genotype data?
Methods and materials
Discovery sample
Data from the Schizophrenia Psychiatric GWAS Consortium (PGC) was used as a discovery sample to identify the schizophrenia risk variants, their p-values and odds ratios. Analysis and quality control was performed according to the consortium’s standards (2). All subjects from the PGC sample were included, except for the 1,342 cases and controls from our own schizophrenia genome-wide association study (UCLA/UMC Utrecht).
Target sample
The target sample consisted of 152 schizophrenia patients and 142 controls with available magnetic resonance imaging (MRI) data. Subjects were included for the Genetic Risk and Outcome of Psychosis (GROUP) study (n=162) (13) and a study described previously (14) (n=132), performed in the University Medical Centre Utrecht. For both patients and controls psychopathology was assessed using the Comprehensive Assessment of Symptoms and History (15). Of the target sample 138 subjects were diagnosed with schizophrenia and 14 with schizo-affective disorder. Unaffected subjects had no history of psychiatric illness except for four subjects who had a history of depressive disorder, anxiety disorder, obsessive-compulsive disorder and adjustment disorder respectively. None of the control subjects had first-degree family members with psychotic illness.
MRI analysis
Brain images were acquired on either a Philips NT or a Philips Achieva scanner at 1.5 Tesla. Scanner type showed no main effect or interaction effect with disease status on total brain and white matter volume (after correction for age, gender and intracranial volume). Gray matter volumes were slightly lower with the Philips Achieva scanner (mean 615.0 ml (sd 25.0) versus 624.3 ml (sd 26.7), p=0.002). Correcting for scanner type did not influence the results. MRI acquisition and processing methods have been previously described (6;16). Post-processing was done on the neuro-imaging computer network of the Department of Psychiatry at the University Medical Centre Utrecht. All images were coded to ensure blindness for subject identification. Scans were put into Talairach frame (no scaling), and corrected for inhomogeneities in the magnetic field (17). Volume measures of the intracranium, total brain, cerebral gray and white matter were determined. Quantitative assessment of the intracranial volume was performed with use of a full-automated computer program based on histogram analyses followed by mathematical morphological operators in either the DE-TSE image (NT) or a single-shot echo planar imaging scan (as part of a diffusion tensor imaging series) together with a magnetization transfer imaging scan (Achieva). All intracranial segmentations were visually checked and corrected where necessary. Quantitative assessment of the total brain, gray and white matter volumes were performed based on histogram analyses followed by mathematical morphological operators in the 3D-FFE image, using the intracranial volume as mask (18).
Genetic analysis
Subjects in the target sample were genotyped at UCLA Neurosciences Genomics Core (UNGC) using the Illumina HumanHap550 beadchip. Initial quality control was performed by the PGC, removing individuals with more than 5% missing SNPs or with evidence of more than random genetic similarity (c.q. distant relatedness) and SNPs on chromosomes X, Y and mitochondrial DNA. Only SNPs genotyped in the target sample were included (in the discovery sample part of the SNPs were imputed due to the use of different genotyping platforms). These SNPs were filtered based on minor allele frequencies of less than 0.02 (removing 4,528 SNPs) and >1% missing genotypes per SNP (7,552 SNPs). There was no evidence of deviation from Hardy-Weinberg equilibrium with p< 1×10−6, non-random genotyping errors with p< 1×10−6, such as systematic batch effects. There was a marginally increased call rate in patients compared to controls (e.g. for SNPs with a p-value for association with schizophrenia <0.01 : mean genotyping rate in patients: 4,038/4,040 versus 4,036/4,040 in controls, p=0.02). To remove all SNPs in linkage disequilibrium, SNPs were pruned based on a pair wise r2 threshold of 0.25 and a sliding window of 50 SNPs wide, shifting 5 SNPs at each step, using PLINK (19). In this way another 341,261 SNPs (74.3%) were removed, leaving 117,924 SNPs for analysis.
Statistical analysis
Measures of total brain, cerebral gray and white matter volume were corrected for the covariates age, sex and intracranial volume by taking the unstandardized residuals of the volumes using linear regression in the total group. For each subject the unstandardized residual was added to the intercept + betai*meani, where i represents the different covariates. Intracranial volume explained a large part of the variation in brain volume, resulting in a correlation between the uncorrected and corrected brain volumes of 0.36. Intracranial volumes were corrected for age and sex in a similar way. All corrected brain volume measures were normally distributed in the total group and in the patient and control groups separately.
For each individual in the target sample a polygenic schizophrenia score (PSS) was calculated using PLINK (19). For each SNP, the number of ‘risk variants’ an individual carried (0, 1 or 2) was multiplied by the logarithm of the odds ratio for that particular variant. ‘Risk variants’ are the alleles (nominally) associated with disease, including both true risk alleles and stochastic variation. Sets of SNPs with p-values below different cutoffs for effect on schizophrenia (p-value cutoffs for effect on schizophrenia or Pcutoff-SZ) were defined. First, the following Pcutoff-SZ were used: 0.01; 0.06; 0.1; 0.2; 0.3; 0.4 and 0.5. When the largest R2 values were found at relatively low cutoffs, we added smaller Pcutoff-SZ (0.002; 0.004; 0.006; 0.008; 0.02; 0.04 and 0.08). The score was summed over all SNPs in the Pcutoff-SZ-SNP sets for each individual in the target sample to obtain the individual polygenic scores.
We first performed a logistic regression with disease status as dependent (outcome) variable and subsequently performed linear regressions using total brain, gray matter and white matter volumes as dependent (outcome) variables. Sex and intracranial volume were analyzed as negative controls. Total brain volume and intracranial volume develop at a similar rate until the age of 14, after which intracranial volume is practically stable (20). Total brain volume corrected for intracranial volume thus represents the brain volume changes in late adolescent and adult life. Because intracranial volume is not substantially different between schizophrenia patients and healthy controls, it is not likely to be influenced by the PSS. A full model with the PSS and the covariates was compared with a model only containing the covariates, to investigate whether PSS significantly improved phenotype prediction. We compared the difference in R2 adjusted for the number of predictors in the model, a measure of the variance explained. For disease status Nagelkerke’s pseudo R2 was used. Ten population stratification dimensions were used as covariates in the analyses as measures of hidden genetic population substructures. These dimensions were calculated in PLINK based on identity-by-state pair wise distances between individuals using the multidimensional scaling plot option. Other covariates were the number of non-missing SNPs used for scoring, the inbreeding coefficient (the ratio of the observed versus expected number of homozygous alleles, calculated in PLINK) and the number of copy number variants (CNVs). For a description of CNV calling see (21). For the brain volume analyses we subsequently included disease status and the interaction between disease status and PSS in the analyses. P-values < 0.008 were considered significant (α 0.05/6 phenotypes (disease status, total brain, white matter, gray matter and intracranial volume and sex), not corrected for the different Pcutoff-SZ, since these are highly correlated).
Functional analyses
Refseq genes within 100kb from the SNPs associated with both schizophrenia and white matter (n=186) were identified. These genes were functionally clustered using the database for annotation, visualization and integrated discovery (DAVID)(22). This database uses functional annotations, including gene ontology (GO) terms, KEGG pathways and Biocarta pathways to cluster genes into biological meaningful groups. These settings were used: high classification stringency and minimal 4 genes per group.
Results
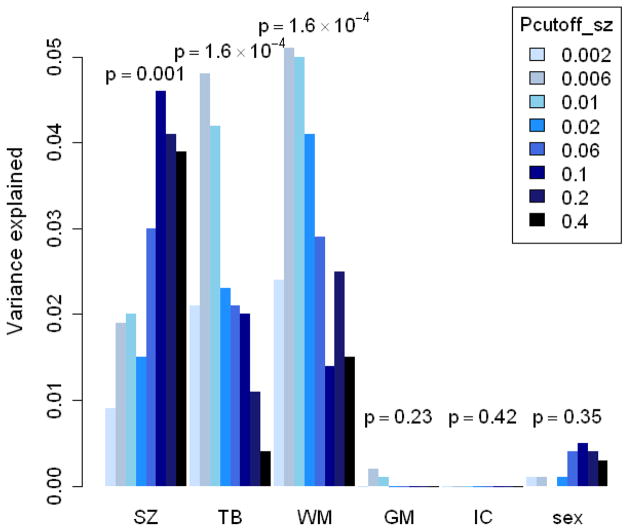
For demographic information see table I. While there were significantly more males in the patient group than in the control group, brain volumes were corrected for sex and sex chromosomal SNPs were removed in all analyses. As expected, patients showed on average smaller brain volumes. They also received fewer years of education, most likely due to their illness. The level of parental education, an estimate of socioeconomic status, was not different between the patient and control groups. There was a good prediction of disease status in the target sample by the PSS based on the PGC sample (with a maximum R2=0.046 (increase in R2 after adding PSS to the model), p=0.001, at Pcutoff-SZ 0.1 (14,751 mostly independent SNPs, see figure 1), indicating that the polygenic score is a reliable measure of schizophrenia risk. Testing this association in the full GWAS sample from Utrecht (n= 1,342 subjects, adding all subjects for which no MRI data was available) resulted in a similar variance explained (R2=0.035), as expected with much smaller p-values (4.0×10−7, data not shown).
Table 1.
Demographic information.
| Schizophrenia patients | Healthy controls | Significance | |
|---|---|---|---|
| n | 152 | 142 | |
| Gender (m/f) | 121/31 | 79/63 | p=1×10−5 |
| Age in years (sd) | 32.0 (10.9) | 32.3 (12.2) | ns |
| Handedness (r/l/ambidexter/unknown) | 120/13/5/14 | 115/14/4/9 | ns |
| Level of education in years | 12.2 (2.5) | 13.9 (2.9) | p=1.1×10−7 |
| Level of parental education in years | 13.2 (3.1) | 13.2 (3.2) | ns |
| Age of first psychotic symptoms in years (sd) | 22.5 (5.4) | na | |
| Duration of illness in years (sd) | 8.5 (9.6) | na | |
| Duration of untreated illness in years (sd) | 1.4 (2.7) | na | |
| Duration of treatment with medication (sd) | 7.1 (10.1) | na | |
| Intracranial volume in ml (sd)** | 1671.3 (116.1) | 1683.6 (118.0) | ns |
| Total brain volume in ml (sd)* | 1492.0 (55.0) | 1513.0 (43.4) | p=3.5×10−4 |
| Gray matter volume in ml (sd) * | 617.0 (27.6) | 623.5 (24.6) | p=0.035 |
| White matter volume in ml (sd) * | 502.6 (34.3) | 515.72 (32.0) | p=0.001 |
sd=standard deviation, ns=not significant, na = not applicable. Level of parental education is a measure of socioeconomic level.
= corrected for age, sex and intracranial volume.
= corrected for age and sex.
Figure 1. The variance explained of different phenotypes by PSS for different Pcutoff-SZ SNP sets.
SZ= schizophrenia, TB= total brain volume, WM= white matter volume, GM= gray matter volume, IC= intracranial volume. y-axis = explained variance by the PSS of this phenotype. For dichotomous traits Nagelkerke’s pseudo R2 was compared between a model with only covariates and a model including covariates and the PSS. For continuous traits the difference in R2 was used. Intracranial volume and sex were included as negative controls. For more information see supplementary table 1.
The polygenic schizophrenia score was significantly associated with total brain volume at different Pcutoff-SZ, irrespective of disease status (with a maximum at Pcutoff-SZ 0.006: R2=0.048, p=1.6×10−4, see figure 1 and Table S1 in Supplement 1). The association was in the expected direction: higher genetic risk scores were associated with smaller total brain volumes. When including disease status in the analysis, the effect of PSS on brain volume remained significant (R2=0.038, p=0.001) and there was no significant interaction between PSS and disease status, indicating that the association was similar in the patient and control groups. PSS was specifically associated with reduced white matter volume (R2=0.051, p=8.6×10−5 at Pcutoff-SZ 0.006) and did not explain variance in gray matter volume (R2=0.002, p=0.232 at Pcutoff-SZ 0.006). As expected, no association was found between PSS and either sex or intracranial volume, which can be regarded as negative controls. From graphical inspection of the data we concluded that there were no outliers in brain volume or in PSS that could significantly influence the results.
While the 14,751 SNPs with p-values up to 0.1 (for effect on schizophrenia) substantially contributed in explaining variation in disease status, a much smaller subset of 2,020 schizophrenia risk variants explained most variance in white matter volume (with Pcutoff-SZ 0.01). Within this set of 2,020 mostly independent SNPs, which were selected based on their association to schizophrenia, we explored which SNPs contributed most to the effect on white matter volume. SNPs were tested for their individual effect on white matter volume (in the same target sample). With a ‘white matter polygenic score’ analysis we selected an optimal p-value cutoff. White matter p-values were used to construct sets of SNPs with p-values below different cutoffs (p-value cutoff for white matter, or Pcutoff-WM): 0.06; 0.08; 0.1; 0.2; and 0.3. Of these sets of SNPs, the set with Pcutoff-WM 0.1 jointly explained most variance in white matter volume compared to the other Pcutoff-WM. Therefore we included SNPs with white matter p-value < 0.1 for further analyses. This small subset of 186 SNPs explaining variance in both schizophrenia and white matter volume thus represents the most important SNPs in determining the genetic overlap between the two phenotypes. Together these 186 SNPs explain 0.7% of the variance in disease status, which is a relatively large part (i.e. 9.2% of 2,020 SNPs explain 35% (0.007/0.02) of the variance explained by these 2,020 SNPs. This set included one of the ten SNPs with genome-wide significance in the PGC sample (rs17662626 near PCGEM1).
Next, we investigated the type and function of the 375 genes located within 100kb on either side of these 186 SNPs. An overview of these genes can be found in Table S2 in Supplement 1. Genes can be grouped in 7 functional clusters (table 2): immunoglobulin-like cell-adhesion molecules, signal peptides, zinc fingers, ion channels, (neurotransmitter) receptors, WD repeat proteins and transcription factors. The cluster of immunoglobulin-like cell-adhesion molecules is enriched compared to the reference database, having enrichment scores of 1.42 (an enrichment score of >1.3 is regarded as significant enrichment (22)). The most prevalent gene ontology (GO) categorie is cell-cell adhesion (GO:0016337, p=0.0007). However, after correction for testing multiple GO categories, this is not significantly enriched compared to the reference database. Similar results were found with flanking regions of 10kb (data not shown).
Table 2. Clusters of genes selected for variance explained in both schizophrenia and white matter volume.
An enrichment score of >1.3 is considered a significant enrichment compared to a reference database.
| Clustername | Genes | Enrichment score |
|---|---|---|
| Immunoglobulin-like cell-adhesion molecules | LSAMP, SIGLEC1, BOC, CNTN4, IGSF8, IGSF9, SLAMF9, CEACAM16 | 1.42 |
| Signal peptides | TMEM18, TMEM59L, TMEM74, TMEM139, TMC2, TMC5, GPR157, CEACAM16, SLAMF9, SLC35F1, TMCC3, SLC37A2, CHODL, KIAA0319, LRTM1, TAS2R40, C16orf62, C7orf44, GPM6A, LRP3, NKAIN2, CEACAM19 | 1.23 |
| Zinc fingers | ZDHHC7, MTF2, SMYD2, TRIM10, TRIM15, TRIM26, TRIM31, TRIM40, RNF39, DEF8, ZC3H14 | 1.02 |
| Ion channels | CACNA2D3, KCNJ9, KCNJ10, KCNAB2, TRPM3, ACCN2 | 0.93 |
| (Neurotransmitter) receptors | GRM3, GABRB2, HTR1F, ADCYAP1R1, TAS2R40, PTGER1, GPR157 | 0.91 |
| WD repeat proteins | WDR17, WDR52, WDR88, EML5, PPP2R2C, KIAA1239 | 0.82 |
| Transcription factors | ASXL3, TCF4, TCF20, HIVEP2, FOXI1, CRTC1, MTF2, EBF4, ZNF23, ZNF100, ZNF707, ESRRG, GLIS3 | 0.55 |
Discussion
We investigated the combined effect of schizophrenia-associated SNPs on brain volume, a highly heritable phenotype associated with this disease. Schizophrenia-associated SNPs explained around 5% of the variance in total brain and white matter volume, in patients as well as in healthy controls. This effect was largely exerted by only a fraction (n=2,020) of all SNPs with effect on disease status (n=14,751). Our data not only suggest that a relatively small subset of all schizophrenia-associated variants is related to white matter development, but also that disruptions in white matter development contribute to schizophrenia in susceptible individuals. Disease status was successfully predicted by the PSS, indicating that the genetic risk variants do indeed reflect schizophrenia risk in this smaller sample. Our finding that genes regulating white matter development are relevant to schizophrenia is consistent with our earlier report that the phenotypic overlap between schizophrenia and white matter volume is almost completely explained by genetic factors (11). Indeed, diffusion tensor imaging (DTI) studies show that abnormalities in white matter integrity are present before the onset of schizophrenia (23), while increased density and altered distribution of white matter neurons (24) and a reduction in oligodendrocyte numbers have repeatedly been found in post mortem brains of schizophrenia patients (23). Taken together, these data are consistent with a model in which genetic risk variants increase schizophrenia vulnerability through aberrant development of brain connectivity (25). Transition to the disease proper most likely occurs in interaction with other genetic or environmental risk factors.
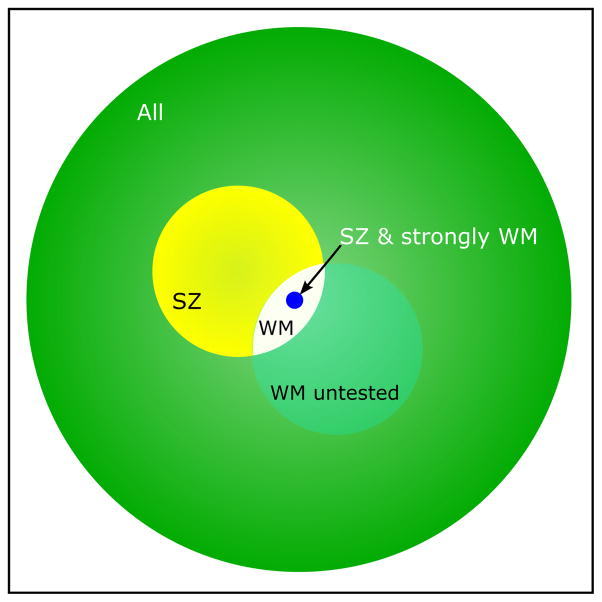
Another important observation is that a much smaller number of common variants appears to determine the overlap between schizophrenia and brain volume than the entire set that modulates schizophrenia. While almost 15,000 SNPs with Pcutoff-SZ up to 0.1 substantially contributed to the variance explained in disease status, only a little over 2,000 SNPs with Pcutoff-SZ up to 0.01 contributed to the variance in white matter volume. In fact, the variance explained in white matter volume decreased when adding more SNPs (with higher p-values for association to schizophrenia). This is in agreement with a model in which only a subset of the schizophrenia-associated SNPs influences brain volume. In this case, adding extra SNPs with effect on schizophrenia, but without a substantial effect on brain volume (OR>1, logOR>1) changes the PSS so that the variance explained in brain volume is diminished at higher cutoffs. These results suggest a specific subset of the disease-associated SNPs that is related to brain volume. A schematic representation of these subsets is shown in figure 2. Perhaps there are other SNPs with more subtle effects on white matter volume, also outside the 2,020 SNPs set, but their effect might easily be overshadowed by the relatively large amount of SNPs without effect on white matter volume. Since only 2% of the 2,020 SNPs were located in the HLA region, these are unlikely to have confounded the analyses. Applying the same strategy to other phenotypes related to schizophrenia, such as cognitive dysfunction, dopamine receptor binding, or other suitable endophenotypes, could similarly result in subsets of candidate variants that help elucidate the biology of the disorder. The same method can obviously be applied to other diseases and their endophenotypes.
Figure 2. Schematic representation of the SNPs involved in different phenotypes.
The large green circle represents all 117,924 SNPs included after quality control and removing SNPs in LD. The yellow circle represents the 14,751 SNPs having the largest effect on schizophrenia in the target sample. The small white area stands for the subset of these SNPs (n=2,020) that do also explain variance in white matter volume. There could be more SNPs influencing white matter volume (represented by the translucent white circle), but these were not investigated. The small blue circle represents the 186 SNPs who are likely to contribute most to both schizophrenia and white matter volume. The size of the circles represents the number of SNPs in that group.
Several other points are important to address. First, only a small amount of the total variance in brain volume and disease status is captured by the PSS. This is comparable with data reported previously (3) and can be explained by incomplete capture of rare variants and gene-gene interactions, among others (4). Using a polygenic score method based on common variants, we cannot aim to explain the majority of the variance in brain volume. Second, since reductions in gray matter volume are commonly reported in schizophrenia patients (6;7), it might seem remarkable that the PSS is not associated with gray matter volume. However, a recent study indicates that in schizophrenia patients gray matter volume is mostly determined by unique environmental factors (explaining 43% of the variance) (11). This is in contrast to white matter volume, which has a much stronger genetic component (63%). Our data provide genetic support for the observation in this twin study. Furthermore, brain volume can be influenced by environmental factors such as the use of psychotropic medication (26). The association between the PSS and brain volume was found not only in the total group, but also within the control group. Since psychotropic medication is used by patients only, it is unlikely that the observed effect could be explained for a substantial part by the use of medication. Because medication use is unreliably assessed in retrospect, we did not include it in the analyses. Lastly, the results do not imply that brain volume is influenced by a smaller number of common variants than schizophrenia. The effects of SNPs that do influence brain volume, but not schizophrenia, are not captured by the PSS.
Among the 2,020 independent schizophrenia risk variants explaining most variance in white matter volume, 186 SNPs were shown to affect white matter volume most strongly. We functionally clustered the 375 genes located near these 186 SNPs in order to generate hypotheses about the biological processes affected by these variants. Functional clustering of the genes should be regarded as explorative, and should be interpreted with caution, since these analyses are biased towards the effect of larger genes and well-investigated pathways (27). Besides, functional relationships between SNPs and nearby genes are often assumed, but not necessarily present. Still, the identified gene clusters have highly relevant functions. The significantly overrepresented cluster of immunoglobulin-like cell adhesion molecules contains proteins that are involved in axon guidance or cell-cell interactions (28). LSAMP is expressed at dendrites of neurons in cortical and subcortical regions of the limbic system and mediates neurite outgrowth (29). A significant increase in LSAMP expression was found in the dorsolateral prefrontal cortex of patients with schizophrenia and bipolar disorder (29). Contactin 4 (CNTN4) regulates neural network formation and has previously been implicated in autism and anorexia nervosa (30;31). Also other clusters contain many genes with relevant, neuronal functions, such as the receptors for glutamate, GABA and serotonin, which have been previously implicated in schizophrenia (32). Each of these genes may make relevant targets for further research.
To conclude, these results indicate that a relatively small subset of schizophrenia genetic risk variants is related to the (normal) development of white matter. This in turn suggests that disruptions in white matter growth increase the susceptibility to develop schizophrenia.
Supplementary Material
Acknowledgments
This work was supported by a Veni grant from Zorg Onderzoek Nederland, Medische Wetenschappen to SB (ZON-MW, the Dutch organization for health research and development, project number: 91686137), by a grant from the Dutch Brain Foundation to SB (grant 14F06(2)-34) and by Top Institute Pharma (project T5-203). The GROUP project was supported by a grant from ZON-MW, within the Mental Health program (project number: 10.000.1001). Genome-wide association study of this sample was funded by the National Institute of Mental Health (grant RO1 MG078075 to RAO).
The Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium (for a list of affiliations, see Supplement 2):
Stephan Ripke, Alan R Sanders, Kenneth S Kendler, Douglas F Levinson, Pamela Sklar, Peter A Holmans, Dan-Yu Lin, Jubao Duan, Roel A Ophoff, Ole A Andreassen, Edward Scolnick, Sven Cichon, David St. Clair, Aiden Corvin, Hugh Gurling, Thomas Werge, Dan Rujescu, Douglas H R Blackwood, Carlos N Pato, Anil K Malhotra, Shaun Purcell, Frank Dudbridge, Benjamin M Neale, Lizzy Rossin, Peter M Visscher, Danielle Posthuma, Douglas M Ruderfer, Ayman Fanous, Hreinn Stefansson, Stacy Steinberg, Bryan J Mowry, Vera Golimbet, Marc De Hert, Erik G Jönsson, István Bitter, Olli P H Pietiläinen, David A Collier, Sarah Tosato, Ingrid Agartz, Margot Albus, Madeline Alexander, Richard L Amdur, Farooq Amin, Nicholas Bass, Sarah E Bergen, Donald W Black, Anders D Børglum, Matthew A Brown, Richard Bruggeman, Nancy G Buccola, William F Byerley, Wiepke Cahn, Rita M Cantor, Vaughan J Carr, Stanley V Catts, Khalid Choudhury, C Robert Cloninger, Paul Cormican, Nicholas Craddock, Patrick A Danoy, Susmita Datta, Lieuwe de Haan, Ditte Demontis, Dimitris Dikeos, Srdjan Djurovic, Peter Donnelly, Gary Donohoe, Linh Duong, Sarah Dwyer, Anders Fink-Jensen, Robert Freedman, Nelson B Freimer, Marion Friedl, Lyudmila Georgieva, Ina Giegling, Michael Gill, Birte Glenthøj, Stephanie Godard, Marian Hamshere, Mark Hansen, Thomas Hansen, Annette M Hartmann, Frans A Henskens, David M Hougaard, Christina M Hultman, Andrés Ingason, Assen V Jablensky, Klaus D Jakobsen, Maurice Jay, Gesche Jürgens, René S Kahn, Matthew C Keller, Gunter Kenis, Elaine Kenny, Yunjung Kim, George K Kirov, Heike Konnerth, Bettina Konte, Lydia Krabbendam, Robert Krasucki, Virginia K Lasseter, Claudine Laurent, Jacob Lawrence, Todd Lencz, F Bernard Lerer, Kung-Yee Liang, Paul Lichtenstein, Jeffrey A Lieberman, Don H Linszen, Jouko Lönnqvist, Carmel M Loughland, Alan W Maclean, Brion S Maher, Wolfgang Maier, Jacques Mallet, Pat Malloy, Manuel Mattheisen, Morten Mattingsdal, Kevin A McGhee, John J McGrath, Andrew McIntosh, Duncan E McLean, Andrew McQuillin, Ingrid Melle, Patricia T Michie, Vihra Milanova, Derek W Morris, Ole Mors, Preben B Mortensen, Valentina Moskvina, Pierandrea Muglia, Inez Myin-Germeys, Deborah A Nertney, Gerald Nestadt, Jimmi Nielsen, Ivan Nikolov, Merete Nordentoft, Nadine Norton, Markus M Nöthen, Colm T O’Dushlaine, Ann Olincy, Line Olsen, F Anthony O’Neill, Torben F Ørntoft, Michael J Owen, Christos Pantelis, George Papadimitriou, Michele T Pato, Leena Peltonen, Hannes Petursson, Ben Pickard, Jonathan Pimm, Ann E Pulver, Vinay Puri, Digby Quested, Emma M Quinn, Henrik B Rasmussen, János M Réthelyi, Robert Ribble, Marcella Rietschel, Brien P Riley, Mirella Ruggeri, Ulrich Schall, Thomas G Schulze, Sibylle G Schwab, Rodney J Scott, Jianxin Shi, Engilbert Sigurdsson, Jeremy M Silverman, Chris C A Spencer, Kari Stefansson, Amy Strange, Eric Strengman, T Scott Stroup, Jaana Suvisaari, Lars Terenius, Srinivasa Thirumalai, Johan H Thygesen, Sally Timm, Draga Toncheva, Edwin van den Oord, Jim van Os, Ruud van Winkel, Jan Veldink, Dermot Walsh, August G Wang, Durk Wiersma, Dieter B Wildenauer, Hywel J Williams, Nigel M Williams, Brandon Wormley, Stan Zammit, Patrick F Sullivan, Michael C O’Donovan, Mark J Daly & Pablo V Gejman
Footnotes
Disclosures
All individual authors on this paper declare no biomedical financial interests or potential conflicts of interest. The following are financial disclosures for the PGC Consortium: Eli Lilly funded portions of the genotyping for CATIE and TOP. P.F.S. received research funding from Eli Lilly in connection with CATIE. T.S.S. received research funding from Eli Lilly and consulting fees from Janssen Pharmaceutica, GlaxoSmithKline and Bristol-Myers Squibb. J.A.L. received research funding from AstraZeneca Pharmaceuticals, Bristol-Myers Squibb, GlaxoSmithKline, Janssen Pharmaceutica and Pfizer and consulting and educational fees from AstraZeneca Pharmaceuticals, Bristol-Myers Squibb, Eli Lilly, Forest Pharmaceuticals, GlaxoSmithKline, Janssen Pharmaceutica, Novartis, Pfizer and Solvay. D.St.C. received research funding from GlaxoSmithKline and Generation Scotland, Genetics Health Initiative. F.A. received funds from Pfizer, Organon and the Foundation for the National Institutes of Health. D.W.B. has received research support from Shire and Forest, has been on the speakers’ bureau for Pfizer and has received consulting honoraria from Forest and Jazz. T.W. has received consulting and lecture fees from H. Lundbeck A/S. O.A.A. has received Speaker’s honorarium from AstraZeneca, Janssen, Bristol-Myers Squibb and GlaxoSmithKline. I.M. has received a Speaker’s honorarium from Janssen and AstraZeneca. A.K.M. has received consulting fees or honoraria from Eli Lilly & Company, Janssen Pharmaceutica, Merck, Bristol-Meyers Squibb, Pfizer, PGxHealth (a division of Clinical Data, Inc.), Roche Diagnostics and Vanda Pharmaceuticals and has received research support from Eli Lilly & Company. T.L. has received consulting fees or honoraria from Merck, Eli Lilly & Company, Golden Helix, Inc., InforMed Insights and PGxHealth (a division of Clinical Data, Inc.). I.B. has been an advisory board member, consultant and lecturer for AstraZeneca, Bristol-Myers Squibb, Eli Lilly, EGIS, Janssen, H. Lundbeck A/S, Novartis, Pfizer, Richter and Schering-Plough and received a grant for an investigator-initiated study from H. Lundbeck A/S. J.J.M. has received consulting and speaker’s fees from Johnson & Johnson, Schering-Plough and Eli Lilly. C.P. has received grant support from Janssen-Cilag, Eli Lilly, Hospira (Mayne) and AstraZeneca, provided consultancy to Janssen-Cilag, Eli Lilly, Hospira (Mayne), AstraZeneca, Pfizer and Schering-Plough and has undertaken investigator-initiated studies supported by Eli Lilly, Hospira, Janssen Cilag and AstraZeneca. The Denmark-Aarhus group (The GEMS Stud with principal investigators A.D.B., O.M. and P.B.M.) received research funding from H. Lundbeck A/S. E.G.J. has served as an unpaid consultant for Eli Lilly.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
AF Terwisscha van Scheltinga, Email: aterwiss@umcutrecht.nl.
Steven C. Bakker, Email: S.Bakker-2@umcutrecht.nl.
Neeltje E.M. van Haren, Email: N.E.M.vanHaren@umcutrecht.nl.
Eske M. Derks, Email: E.M.Derks@amc.uva.nl.
Jacobine E. Buizer-Voskamp, Email: J.E.Buizer@umcutrecht.nl.
Heleen B.M. Boos, Email: H.B.M.Boos@umcutrecht.nl.
Wiepke Cahn, Email: W.Cahn@umcutrecht.nl.
HE Hulshoff Pol, Email: H.E.Hulshoff@umcutrecht.nl.
Stephan Ripke, Email: ripke@atgu.mgh.harvard.edu.
Roel A. Ophoff, Email: rophoff@mednet.ucla.edu.
RS Kahn, Email: R.Kahn@umcutrecht.nl.
Reference List
- 1.Cardno AG, Marshall EJ, Coid B, Macdonald AM, Ribchester TR, Davies NJ, et al. Heritability estimates for psychotic disorders: the Maudsley twin psychosis series. Archives of General Psychiatry. 1999;56:162–168. doi: 10.1001/archpsyc.56.2.162. [DOI] [PubMed] [Google Scholar]
- 2.Ripke S, Sanders AR, Kendler KS, Levinson DF, Sklar P, Holmans PA, et al. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;10:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Purcell SM, Wray NR, Stone JL, Visscher PM, O’donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee SH, Decandia TR, Ripke S, Yang J, Sullivan PF, Goddard ME, et al. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat Genet. 2012 doi: 10.1038/ng.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 6.Hulshoff Pol HE, Schnack HG, Bertens MG, van Haren NEM, van der Tweel I, Staal WG, et al. Volume changes in gray matter in patients with schizophrenia. American Journal of Psychiatry. 2002;159:244–250. doi: 10.1176/appi.ajp.159.2.244. [DOI] [PubMed] [Google Scholar]
- 7.Wright IC, Rabe-Hesketh S, Woodruff PW, David AS, Murray RM, Bullmore ET. Meta-analysis of regional brain volumes in schizophrenia. American Journal of Psychiatry. 2000;157:16–25. doi: 10.1176/ajp.157.1.16. [DOI] [PubMed] [Google Scholar]
- 8.Baare WF, Hulshoff Pol HE, Boomsma DI, Posthuma D, De Geus EJ, Schnack HG, et al. Quantitative genetic modeling of variation in human brain morphology. Cerebral Cortex. 2001;11:816–824. doi: 10.1093/cercor/11.9.816. [DOI] [PubMed] [Google Scholar]
- 9.Cannon TD, Thompson PM, van Erp TG, Toga AW, Poutanen VP, Huttunen M, et al. Cortex mapping reveals regionally specific patterns of genetic and disease-specific gray-matter deficits in twins discordant for schizophrenia. Proc Natl Acad Sci U S A. 2002;99:3228–3233. doi: 10.1073/pnas.052023499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldman AL, Pezawas L, Mattay VS, Fischl B, Verchinski BA, Zoltick B, et al. Heritability of brain morphology related to schizophrenia: a large-scale automated magnetic resonance imaging segmentation study. Biol Psychiatry. 2008;63:475–483. doi: 10.1016/j.biopsych.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 11.Haren NEMv, Rijsdijk F, Schnack HG, Picchioni MM, Toulopoulou T, Weisbrod M, et al. The Genetic and Environmental Determinants of the Association Between Brain Abnormalities and Schizophrenia: The Schizophrenia Twins and Relatives Consortium. Biol Psychiatry. 2012 doi: 10.1016/j.biopsych.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boos HB, Aleman A, Cahn W, Hulshoff Pol HE, Kahn RS. Brain volumes in relatives of patients with schizophrenia: a meta-analysis. Arch Gen Psychiatry. 2007;64:297–304. doi: 10.1001/archpsyc.64.3.297. [DOI] [PubMed] [Google Scholar]
- 13.Genetic Risk and Outcome in Psychosis (GROUP) investigators. Evidence that familial liability for psychosis is expressed as differential sensitivity to cannabis: an analysis of patient-sibling and sibling-control pairs. Arch Gen Psychiatry. 2011;68:138–147. doi: 10.1001/archgenpsychiatry.2010.132. [DOI] [PubMed] [Google Scholar]
- 14.Haren NEMv, Hulshoff Pol HE, Schnack HG, Cahn W, Brans R, Carati I, et al. Progressive brain volume loss in schizophrenia over the course of the illness: evidence of maturational abnormalities in early adulthood. Biological Psychiatry. 2008;63:106–113. doi: 10.1016/j.biopsych.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 15.Andreasen NC, Flaum M, Arndt S. The Comprehensive Assessment of Symptoms and History (CASH). An instrument for assessing diagnosis and psychopathology. Archives of General Psychiatry. 1992;49:615–623. doi: 10.1001/archpsyc.1992.01820080023004. [DOI] [PubMed] [Google Scholar]
- 16.Boos HB, Cahn W, van Haren NE, Derks EM, Brouwer RM, Schnack HG, et al. Focal And Global Brain Measurements in Siblings of Patients With Schizophrenia. Schizophr Bull. 2011 doi: 10.1093/schbul/sbq147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Transactions on Medical Imaging. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- 18.Brouwer RM, Hulshoff Pol HE, Schnack HG. Segmentation of MRI brain scans using non-uniform partial volume densities. Neuroimage. 2010;49:467–477. doi: 10.1016/j.neuroimage.2009.07.041. [DOI] [PubMed] [Google Scholar]
- 19.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Courchesne E, Chisum HJ, Townsend J, Cowles A, Covington J, Egaas B, et al. Normal brain development and aging: quantitative analysis at in vivo MR imaging in healthy volunteers. Radiology. 2000;216:672–682. doi: 10.1148/radiology.216.3.r00au37672. [DOI] [PubMed] [Google Scholar]
- 21.Buizer-Voskamp JE, Muntjewerff JW, Strengman E, Sabatti C, Stefansson H, Vorstman JA, et al. Genome-Wide Analysis Shows Increased Frequency of Copy Number Variation Deletions in Dutch Schizophrenia Patients. Biological Psychiatry. 2011 doi: 10.1016/j.biopsych.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei Huang D, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi N, Sakurai T, Davis KL, Buxbaum JD. Linking oligodendrocyte and myelin dysfunction to neurocircuitry abnormalities in schizophrenia. Prog Neurobiol. 2011;93:13–24. doi: 10.1016/j.pneurobio.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Connor CM, Crawford BC, Akbarian S. White matter neuron alterations in schizophrenia and related disorders. Int J Dev Neurosci. 2011;29:325–334. doi: 10.1016/j.ijdevneu.2010.07.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heuvel MPvd, Kahn RS. Abnormal brain wiring as a pathogenetic mechanism in schizophrenia. Biol Psychiatry. 2011;70:1107–1108. doi: 10.1016/j.biopsych.2011.10.020. [DOI] [PubMed] [Google Scholar]
- 26.Moncrieff J, Leo J. A systematic review of the effects of antipsychotic drugs on brain volume. Psychol Med. 2010;40:1409–1422. doi: 10.1017/S0033291709992297. [DOI] [PubMed] [Google Scholar]
- 27.Elbers CC, van Eijk KR, Franke L, Mulder F, van der Schouw YT, Wijmenga C, et al. Using genome-wide pathway analysis to unravel the etiology of complex diseases. Genet Epidemiol. 2009;33:419–431. doi: 10.1002/gepi.20395. [DOI] [PubMed] [Google Scholar]
- 28.Fabre PJ, Shimogori T, Charron F. Segregation of ipsilateral retinal ganglion cell axons at the optic chiasm requires the Shh receptor Boc. J Neurosci. 2010;30:266–275. doi: 10.1523/JNEUROSCI.3778-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Behan AT, Byrne C, Dunn MJ, Cagney G, Cotter DR. Proteomic analysis of membrane microdomain-associated proteins in the dorsolateral prefrontal cortex in schizophrenia and bipolar disorder reveals alterations in LAMP, STXBP1 and BASP1 protein expression. Mol Psychiatry. 2009;14:601–613. doi: 10.1038/mp.2008.7. [DOI] [PubMed] [Google Scholar]
- 30.Cottrell CE, Bir N, Varga E, Alvarez CE, Bouyain S, Zernzach R, et al. Contactin 4 as an autism susceptibility locus. Autism Res. 2011;4:189–199. doi: 10.1002/aur.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang K, Zhang H, Bloss CS, Duvvuri V, Kaye W, Schork NJ, et al. A genome-wide association study on common SNPs and rare CNVs in anorexia nervosa. Mol Psychiatry. 2011;16:949–959. doi: 10.1038/mp.2010.107. [DOI] [PubMed] [Google Scholar]
- 32.Kegeles LS, Mao X, Stanford AD, Girgis R, Ojeil N, Xu X, et al. Elevated Prefrontal Cortex gamma-Aminobutyric Acid and Glutamate-Glutamine Levels in Schizophrenia Measured In Vivo With Proton Magnetic Resonance Spectroscopy. Arch Gen Psychiatry. 2012 doi: 10.1001/archgenpsychiatry.2011.1519. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.