Abstract
INTRODUCTION
ALK gene rearrangements occur in ~5% of lung adenocarcinomas (ACA), leading to ALK overexpression and predicting response to targeted therapy. Fluorescence in situ hybridization (FISH) is the gold standard for detection of ALK rearrangements in lung ACA but requires specialized equipment and expertise. Immunohistochemistry (IHC) for ALK protein overexpression is a promising screening modality, with reports of newer antibodies showing excellent sensitivity and specificity for ALK-rearranged lung ACA.
METHODS
In this study, we analyze ALK IHC (5A4 clone) in 186 cases from our clinical service and compare with ALK FISH and EGFR and KRAS mutation status.
RESULTS
Twelve cases had concordant ALK protein overexpression and ALK rearrangement by FISH. Three ALK-rearranged cases lacked ALK protein expression. Of these discrepant cases, one had a coexisting EGFR mutation and a subtle “atypical” ALK rearrangement with a break in the 5’ centromeric portion of the FISH probe. One case had a concurrent BRAF mutation; followup testing on a metastasis revealed absence of the ALK-rearrangement with persistent BRAF mutation. In one ALK-rearranged, protein negative case, very limited tissue remained for ALK IHC, raising the possibility of false negativity due to protein expression heterogeneity. Importantly, ALK protein expression was detected in one case initially thought not to have an ALK rearrangement. In this case, FISH was falsely negative due to interference by benign reactive nuclei. After correcting for these cases, ALK IHC was 93% sensitive and 100% specific as compared to FISH.
CONCLUSIONS
ALK IHC improves the detection of ALK rearrangements when used together with FISH, and its use in lung adenocarcinoma genetic testing algorithms should be considered.
Keywords: ALK, lung adenocarcinoma, FISH, immunohistochemistry
INTRODUCTION
Rearrangement of the anaplastic lymphoma kinase (ALK) gene occurs in ~5% of lung adenocarcinomas and predicts response to the targeted inhibitor crizotinib. 1 In most cases, ALK fuses with EML4 via a small intrachromosomal inversion event; however, other translocation events such as TFG-ALK and KIF5B-ALK have also been described. 2 Fluorescence in situ hybridization (FISH)-based analysis detecting chromosome 2 inversions and other ALK translocations represents the current standard for diagnosis of ALK-rearranged lung adenocarcinomas.
The National Comprehensive Cancer Network recommends testing for EGFR mutations (by sequence-level analysis) and ALK rearrangement by FISH on all lung adenocarcinomas from patients with advanced disease. 3EGFR activating mutations are detected in ~20% of lung adenocarcinomas and predict response to EGFR tyrosine kinase inhibitors. 4 As a result of these recommendations, pathologists have seen a dramatic increase in requests for both EGFR mutation analysis and ALK FISH on lung adenocarcinoma specimens.
With FISH as the mainstay for detection of ALK rearrangements, the ALK Break Apart FISH Probe Kit (Abbott Molecular, Des Plaines, IL) has become an FDA-approved companion diagnostic for targeted therapy with the ALK inhibitor crizotinib in lung cancers. (http://www.accessdata.fda.gov/cdrh_docs/pdf11/p110012a.pdf) However, FISH can be expensive and time-consuming and requires specialized fluorescence microscopy equipment and expertise. The ALK FISH assay in particular can be difficult to interpret because the most common alteration, the intrachromosomal inversion, leads to a subtle (>2 probe diameter) separation in the 5’ and 3’ signals. 5 In addition, cells without a rearrangement can not uncommonly have some nonspecific signal separation. As a result, the assay is prone both to false negatives and false positives and has significant interobserver variability. 6
EML4-ALK fusions drive ALK transcriptional upregulation and protein expression. 7 Immunohistochemistry (IHC) for ALK protein expression has been available for many years for use in the diagnosis of anaplastic large cell lymphoma, but the traditional antibodies for use in lymphoma (i.e. CD246) are insufficiently sensitive for detection at the level at which it is expressed in ALK-rearranged lung cancers. 8 However, several recent studies have demonstrated that a relatively new ALK clone, 5A4, can accurately identify ALK-rearranged lung ACA as compared to FISH. Published studies from France and Korea comparing the 5A4 antibody to ALK FISH demonstrated a sensitivity and specificity of 95 to 100%.9,10 In contrast, a series from the United States (published in abstract form) suggested that the sensitivity of the 5A4 antibody is only 82%.11 According to these studies, ALK expression can be variable; while strong staining appears to be 100% specific for the presence of rearrangement by FISH, weak-to-intermediate staining has been reported in FISH negative tumors. The basis for the discrepancies between ALK FISH and IHC is unclear.
In this study, we compare ALK FISH and IHC in a cohort of 186 cases derived from our clinical workflow, which includes concurrent mutational analysis of EGFR and KRAS. We demonstrate that ALK IHC correlates well with FISH. However, several discordant cases were identified, including two cases which occurred in patients with concurrent oncogene mutations, both with rearrangements detected by FISH and negative ALK IHC. In addition, we identified two discrepant cases without known concurrent oncogenic mutations: one with strong positive ALK protein expression and negative FISH results, and one with absent ALK protein expression and positive FISH results. We herein examine the basis for these discrepancies and determine that, in most cases, the discrepancies can be resolved either with repeat testing or closer analysis of the FISH results to exclude atypical rearrangements unlikely to have functional consequences. As a result of our findings, we propose a clinical testing algorithm that incorporates both ALK IHC and FISH to maximize the sensitivity and specificity of detection of ALK-rearranged lung adenocarcinomas.
METHODS
Cases were clinically selected for ALK analysis based on tumor type, patient characteristics, and tumor stage from September 2010 to April 2012. Clinical histories were derived from clinic charts and the electronic medical record following approval by the Brigham and Women's Hospital and/or Dana Farber Cancer Institute Institutional Review Boards.
FISH
Four μm -thick formalin-fixed, paraffin-embedded tissue sections were used for evaluation of ALK genetic status by FISH with the commercial LSI ALK dual color, break-apart rearrangement probe (Abbott Molecular, Abbott Park, IL). Briefly, tissue sections were mounted on positively charged slides and air dried. Targeted tumor areas were circled with a diamond pen following review of the corresponding hematoxylin and eosin (H&E) slide with a pathologist. Slides were deparaffinized, dehydrated, immersed in 0.2N HCl for 20 min then washed and incubated in Pretreatment Solution at 80°C for 30 min. Slides were washed then incubated in 0.5 mg/ml Protease solution (Paraffin Pretreatment Kit I, Abbott Molecular) at 37°C for 35 minutes, washed again and dried at 40-50°C on slide warmer for 2-5 min. The tissue was then fixed in 10% buffered formalin at room temperature (RT) for 10 min, washed in Wash Buffer and dried on slide warmer as described above. Cellular DNA was denatured in 70% formamide in 2X SSC pH 7.0 at 72°C for 5 min and slides were dehydrated at RT in 70%, 85%, 100% ethanol for 1 minute each. ALK probe was denatured at 73°C for 5 min. Hybridization was carried out overnight at 37°C. Post-hybridization wash was performed in 2X SSC/0.3%NP-40, pH 7.0-7.5 at72°C for 2 minutes. Slides were counterstained with DAPI and stored in dark at -20°C before microscope examination.
Results were analyzed with a fluorescence Zeiss Axiophot microscope. A minimum of 50 nuclei from two separate areas of tumor were independently scored by two technologists. Representative images were captured using Leica Microsystem Imaging (Leica Microsystems Inc., Buffalo Grove, IL).
Samples were classified as positive for ALK rearrangement when ≥15% of nuclei showed split signals (i.e. red and green signals were separated by ≥2 signal diameters) or single red signals (3′ ALK) were observed. H&E and FISH slides for all cases were reviewed by a pathologist to confirm that scoring was carried out in the tumor cell population.
Immunohistochemistry
Immunohistochemistry for ALK was performed on 4 μm -thick formalin-fixed, paraffin-embedded tissue sections using clone 5A4 (Novocastra, Newcastle, UK). Briefly, slides were deparaffinized, then treated with Peroxidase Block (DAKO, Carpinteria, CA) for 15 minutes to quench endogenous peroxidase activity. Antigen retrieval was carried out in citrate buffer (pH 6) in a pressure cooker at 122°C for 30-45 minutes. The sections were then incubated with the primary mouse monoclonal anti-ALK antibody at a 1:50 dilution for 40 minutes, washed in 50 mM Tris-HCl (pH 7.4), and incubated with horseradish peroxidase–conjugated secondary antibodies (Envision Plus detection kit, DAKO).
Staining was developed through incubation with diaminobenzidine (DAB), and sections were counterstained.
The stained slides were reviewed by two pathologists (LMS and JLH) blinded to the FISH results. Staining was graded as semiquantitatively as follows: 0 for absent or barely perceptible expression in rare cells, 1 (low) for weak to moderate multifocal expression and 2 (high) for strong staining in most cells. All positive cases demonstrated a granular, cytoplasmic expression pattern. Focal, weak rimming of intracellular mucin droplets was considered negative.
Mutation analysis
For mutation analysis, DNA was extracted from dissected formalin-fixed, paraffin-embedded 5μm tissue sections containing more than 50% tumor cells. The EGFR kinase domain (exons 18 through 21) and KRAS exons 2 and 3 were amplified using nested PCR, as previously described.12,13 PCR products underwent direct bidirectional sequencing by dye terminator sequencing. Sequence analysis was performed by using Mutation Surveyor (SoftGenetics, State College, PA) and confirmed by qualified molecular pathologists (N.I.L., L.M.S.).
RESULTS
ALK FISH
From September 2010 to April 2012, 830 cases underwent ALK FISH testing in our laboratory, of which 25 (3%) demonstrated an ALK rearrangement. Of these, 186, including 15 FISH positive cases, 161 FISH negative cases, and 10 cases that failed by FISH were tested by ALK IHC. The FISH positive cases included seven balanced rearrangements, characterized by a split signal, and eight unbalanced rearrangements, characterized by a loss of the 5’ probe.
ALK Immunohistochemistry
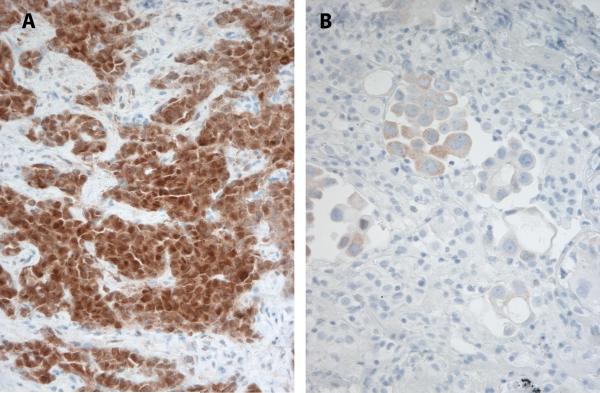
ALK protein expression was detected in 13 cases. ALK IHC was negative in 170 cases. Three cases had insufficient tumor tissue remaining for IHC interpretation. ALK protein expression in positive cases ranged from weak and multifocal (1+) to strong and diffuse (2+) (Figure 1a). In three cases, we noted significant intratumoral heterogeneity, ranging from absent to moderate ALK expression (Figure 1b).
Figure 1.
ALK immunohistochemistry in lung adenocarcinoma reveals variable levels of protein expression in ALK rearranged lung tumors. (A) Strong and diffuse reactivity (2+) and (B) heterogeneous expression ranging from absent to low (1+) in two cases with ALK rearrangement by FISH.
ALK FISH and IHC Correlation
Twelve IHC positive cases showed an ALK rearrangement by FISH. The intensity of ALK protein expression did not correlate with type of ALK rearrangement (data not shown). One IHC positive case did not show an ALK rearrangement at the time of initial clinical review. In three IHC-negative cases, an ALK rearrangement was detected by FISH. Of the remaining cases, 160 were negative by both IHC and FISH, including six cases that were noted to have ALK rearrangements detected below the 15% cutoff for positivity (range of 6-14% abnormal cells). Eight cases that were deemed insufficient for FISH for technical reasons (i.e. high background, poor hybridization, etc.) were scored negative by IHC. Two cases failed both FISH and IHC.
Analysis of cases with intratumoral protein expression heterogeneity
In the three cases with heterogenous ALK protein expression, the percentage of tumor cells with rearrangement by FISH ranged from 86 to 94%. Although in most cases only selected fields are scored by FISH, two cases with ALK IHC heterogeneity contained only a small number of tumor cells (one was a cytology cell block and one was a needle core biopsy) and were scored in their entirety. This observation indicates that the ALK rearrangement is a consistent finding in the tumor cell population despite variable levels of intratumoral ALK protein expression.
Mutation analysis
EGFR mutation results were available from the tumors of all but one patient, who had insufficient material. 36 (19%) cases were EGFR mutated and 151 were wild type. KRAS mutation results were available for 146 patients. 41 (22%) were KRAS mutated and 127 cases were wild type, 19 were not tested, and 1 was insufficient. One case contained both an ALK rearrangement by FISH and an EGFR c.2573T>G (p.Leu858Arg) mutation. None of the ALK IHC positive cases had a concurrent oncogenic mutation.
Examination of discrepant cases
Four cases had discrepant ALK FISH and ALK IHC results. The discrepancies are categorized below as follows: cases with presumed “atypical” or nonfunctional ALK rearrangements (false positive FISH results), cases with false negative FISH results, and cases with false negative IHC results.
Cases with false positive FISH results
In two cases, additional clinical history was useful in interpreting the discrepant results (Table 2): Case 1 was a 61 year old man with a 45 pack year smoking history and right large hilar mass, mediastinal lymphadenopathy and bony metastases at the time of presentation. Cervical mediastinoscopy revealed metastatic poorly differentiated lung adenocarcinoma to level 4 lymph nodes. Because the patient had aggressive disease and significant symptoms at presentation, palliative chemotherapy was initiated prior to the completion of genomic testing. Subsequent FISH on the level 4 lymph node specimen showed a rearrangement involving the ALK probe in 32% of tumor cells; however, ALK IHC was negative. Mutational analysis on that same sample revealed wild type EGFR and KRAS; however, as part of a research protocol at the Dana Farber Cancer Institute, the patient's tumor was tested for BRAF mutations and was found to have an exon 15 mutation (c.1799T>A (p.Val600Glu)). At the time of progression of disease (approximately 5 months after diagnosis), the patient was started on a phase one trial of crizotinib (DFCI 06-068). Five days after initiating crizotinib, the patient was urgently admitted for bowel perforation secondary to visceral metastases. He required an emergency resection of a small bowel metastasis; ALK FISH performed on this specimen was negative. ALK IHC was negative. A BRAF Val600Glu mutation was again detected. He died one month later while under the care of hospice.
Table 2.
Cases with false negative ALK FISH or ALK IHC results.
| Discrepancy type | Sample type | ALK FISH | ALK IHC | Other mutations | Followup | Reason for discrepancy |
|---|---|---|---|---|---|---|
| FISH false negative | Lung wedge biopsy | No translocation detected | Positive (2+) | WT | Repeat FISH on original sample: unbalanced translocation in 86% of tumor cells | Small area of tumor in inflamed/reactive lung; first FISH analysis did not include tumor |
| IHC false negative | Lymph node biopsy | Unbalanced translocation in 38% of cells | Negative | N/A: Insufficient material | None available | Extremely limited specimen-interpret negative results with caution |
Case 2 was a 65 year old nonsmoking woman with multiple bilateral pulmonary nodules and right pleural effusion at the time of presentation. Diagnostic thoracentesis revealed lung adenocarcinoma. Genotyping of tumor revealed an EGFR exon 21 (c.2573T>G (p.Leu858Arg)) kinase activating mutation. The patient was started on erlotinib, and she had a partial response with dramatic clinical benefit. Unfortunately, her disease progressed after 9 months of erlotinib at 100 mg as a single agent. Pemetrexed was added to the regimen. During one of the therapeutic thoracentesis, ALK FISH analysis revealed ALK rearrangement detected in 32/50 nuclei. The patient elected not to enroll in any clinical trials at that time. However, after 3 cycles of Pemetrexed/erlotinib therapy, the patient decided to stop all treatment for best supportive care. After 8 months of best supportive care, the patient decided to be re-evaluated for possible additional treatment. Another diagnostic thoracentesis was performed. ALK FISH was again performed. Careful analysis of the FISH results in the tumor recurrence revealed an atypical ALK rearrangement that involved an asymmetrically split green signal, with a bright single green signal in addition to a small green signal fused to a red signal (Figure 2). While the EGFR sequencing data was pending, the patient was started on erlotinib at 100 mg daily and crizotinib at 200 mg twice daily. Unfortunately, the patient clinically worsened despite 2 weeks of combination treatment. The patient was taken off all therapy and died 2 days after being under the care of hospice. The EGFR sequencing data confirmed the presence of a p.Thr790Met acquired resistance mutation.
Figure 2.
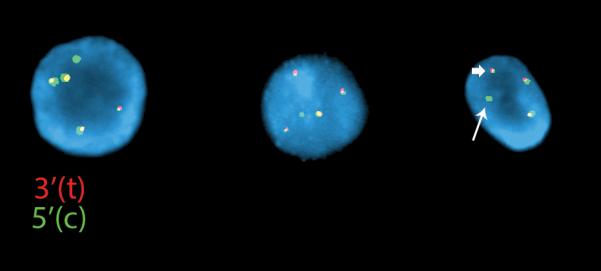
Atypical rearrangement by FISH using the LSI ALK dual color probes (Abbott Molecular) in a patient with a coexisting EGFR L858R mutation. The tumor cells show 2-3 three normal fused signals and an asymmetrically split green (5’ centromeric) signal with a bright single green signal (arrow) in addition to a small green signal fused to a red signal (arrowhead).
Case with false negative FISH results
A lung wedge biopsy containing adenocarcinoma underwent ALK FISH testing and was negative for a rearrangement. Mutation analysis was also negative. ALK IHC revealed 2+ staining in tumor cells; however, the cells were present only as scattered small clusters in a background of inflamed and reactive lung tissue. Re-review of the original FISH specimen revealed that the counts had been performed in an area with exuberant type II pneumocyte hyperplasia but without tumor cells. Repeat FISH analysis revealed an ALK rearrangement in 86% of tumor cells. (Table 2)
Case with false negative IHC results
A lymph node biopsy containing metastatic adenocarcinoma performed at a referring hospital was received for mutational analysis and ALK FISH. There was insufficient tumor to perform mutation analysis; however, ALK FISH was carried out and revealed an ALK rearrangement in 38% of cells. ALK IHC was negative (score of 0) on this specimen; however, <50 tumor cells remained for examination. Clinical followup is limited on this case. (Table 2)
Sensitivity and specificity of ALK FISH and IHC
Taking the above observations into account, in the end we considered 14 cases to contain “true” ALK-rearrangements, of which 13 showed IHC positivity, for a sensitivity of 93%. The specificity of ALK protein expression for the presence of a rearrangement by FISH was 100%. (Table 3) In our experience, the specificity of FISH in clinical practice was 98.5%, due to the detection of nonfunctional or atypical ALK rearrangements.
Table 3.
ALK Immunohistochemistry as compared to ALK FISH results following comprehensive molecular and clinical review of discrepant cases.
| ALK FISH | ||||
|---|---|---|---|---|
| ALK Immunohistochemistry | Positive | Negative | Total | |
| Negative | 1 | 162 | 163 | |
| Low (1+) | 4 | 0 | 4 | |
| High (2+) | 9 | 0 | 9 | |
| Total Positive | 13 | 0 | 13 | |
| Total | 14 | 162 | 176* | |
Total case tally does not include 10 cases that were insufficient either by FISH and/or IHC.
DISCUSSION
In this study, we examined a cohort of 186 lung adenocarcinoma cases taken directly from our clinical ALK FISH workflow and retrospectively compared the FISH results to ALK immunohistochemistry. On initial analysis, ALK IHC appeared to have limited sensitivity for detection of ALK rearranged lung cancers. Of the 15 cases originally identified as containing an ALK rearrangement, only 12 demonstrated clear positive ALK expression (sensitivity of 80%). However, careful analysis of the clinical, genetic, and FISH studies in these discrepant cases indicate that this conclusion was unwarranted. In two discrepant cases, simultaneous oncogenic mutations were identified, and most literature supports the observation that ALK rearrangements occur at best rarely with oncogenic mutations. 14 In one such case of a patient with a concurrent BRAF activating mutation, the ALK-rearranged clone was detected only at the time of diagnosis and was absent in a distant metastasis. This observation, together with the absent ALK protein expression, would argue that the detected rearrangement was either a technical artifact or, if truly present, was likely not transcribed or translated and was lost during disease progression. 15
In another case of a patient with an ALK rearrangement and EGFR activating mutation, careful analysis of the FISH results in the tumor recurrence revealed an atypical rearrangement characterized by an asymmetric splitting of the 5’ probe. The significance of this alteration is unclear, although this cytogenetic finding has not been associated with ALK activation. Importantly, such asymmetric split signals could easily lead to false positive interpretation, especially in suboptimal specimens with weak green probe signals, as was likely the case in the original specimen examined from this patient. It will be important to determine if these atypical rearrangements can produce an ALK fusion product that is not detectable by IHC (such as through RNA analysis); unfortunately this type of analysis was not possible in this case due to the limited size of the tumor sample. Clinically, this patient's course was consistent with having a driver mutation in the EGFR gene, as she originally responded to EGFR tyrosine kinase inhibitor treatment, and at the time of relapse was found to have the p.Thr790Met EGFR TKI resistance mutation, described in approximately 50% of patients who progress through targeted therapy. 16
Importantly, IHC also detected an additional ALK-rearranged case that would have been missed by relying on FISH alone. In this case, a large section of lung tissue in which the tumor was present only as isolated nests in a background of reactive pneumocyte hyperplasia was originally scored as FISH negative for ALK rearrangement. Even on H&E stained slides, reactive pneumocyte hyperplasia can sometimes be difficult to distinguish from neoplasia. Identification of small nests of tumors in a reactive background is significantly more difficult with fluorescence microscopy. Prompted by the positive ALK IHC, the original slide was reexamined, and an ALK rearrangement was identified in the tumor cells.
In the final discrepant case, ALK FISH was positive and IHC was negative. However, tumor tissue was extremely limited in this case, no mutation data was available, and, because the slide was sent from a referring hospital, the specifics of fixation and tissue handling were unclear. In the absence of corresponding mutation analysis or additional material on which to confirm the results, we cannot entirely exclude the possibility that the FISH result represents a false positive. Alternatively, this discrepancy may indeed reflect a falsely negative IHC. We have detected ALK protein expression heterogeneity in a subset of cases stained; in the absence of obvious heterogeneity at the chromosomal level, one may conclude that ALK protein expression heterogeneity likely reflects intratumoral differences in transcription and protein processing. These observations suggest that negative results on very limited tumor tissue, as with other immunohistochemical and genetic studies, should be interpreted with caution.
These findings argue for a combined FISH and IHC approach to maximize the sensitivity and specificity of detection of ALK-rearranged lung cancers. Because of its reliance on specialized equipment and personnel, the use of FISH in any diagnostic algorithm frequently introduces some delay and may be uninterpretable in cases with nuclear overlapping, crush artifact, or technical limitations. IHC can be readily incorporated into the surgical pathology workflow, has less than a one day turnaround time, and is a robust technique that may deliver results even when the FISH fails.
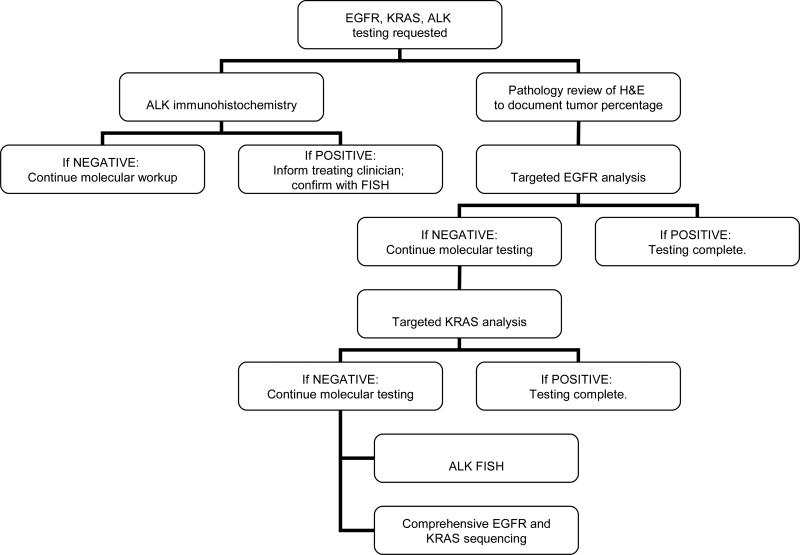
Keeping in mind that an individual institution's testing practice depends on local resources, expertise, and reimbursement, we propose that an algorithmic approach to molecular diagnosis in lung carcinoma can help to control costs, eliminate unnecessary testing, and improve turnaround time. In our hospital, we have instituted a diagnostic algorithm that combines ALK IHC, EGFR and KRAS mutation analysis, and ALK FISH. This algorithm operates under the assumption that these alterations essentially occur in a mutually exclusive fashion. Although KRAS alterations are not targetable, the presence of a KRAS mutation identifies a substantial number of cases that do not require ALK FISH analysis. This algorithm employs sequential testing as follows: (1) targeted analysis of EGFR exons 19 and 21; if negative, move to (2) targeted analysis of KRAS codons 12 and 13; and if negative move to (3) ALK FISH and Sanger sequencing of EGFR exons 18-21 and KRAS exons 2 and 3. (Figure 3; Additional details regarding targeted mutation analysis are available upon request.) ALK IHC is ordered up front; if this returns positive, the treating oncologist will immediately be informed, thereby accelerating the initiation of targeted therapy. ALK IHC positive case will undergo confirmatory ALK FISH analysis using the FDA-approved kit. Even as next generation sequencing becomes more pervasive in clinical laboratories, ALK IHC can complement molecular analysis to more rapidly triage cases for ALK FISH testing.
Figure 3.
Immunohistochemical, molecular, and cytogenetic algorithm for lung adenocarcinoma testing.
Although FISH is considered the gold standard for diagnosis of ALK rearrangement, this review of our clinical experience with ALK FISH testing indicates that it is prone both to false negative and false positive results. In the absence of comprehensive outcome data on this population, we cannot draw conclusions about the true sensitivity and specificity of FISH and IHC in predicting response to crizotinib therapy. However, institution of prospective IHC and FISH analysis in clinical diagnostics will help to address this issue.
Table 1.
Cases with false positive ALK FISH results.
| Case | Sample type | Testing date | Clnical ALK FISH report | ALK IHC | Other mutations | Therapy | Followup |
|---|---|---|---|---|---|---|---|
| 1 | Lung biopsy | September, 2010 | Balanced translocation in 32% of tumor cells | Negative | BRAF c.1799T>A (p.Val600Glu) | Chemotherapy, Crizotinib | progression |
| Small bowel metastasis, resection | February, 2011 | No rearrangement detected | Negative | BRAF c.1799T>A (p.Val600Glu) | - | death | |
| 2 | Pleural fluid | November, 2010 | Balanced translocation in 64% of tumor cells | Negative | EGFR c.2573T>G (p.Leu858Arg) | Erlotinib+ Pemetrexed | Partial response x 9 months |
| Lung core biopsy | February, 2012 | Atypical ALK rearrangement with split centromeric probe | Negative | EGFR c.2573T>G (p.Leu858Arg) and c.2369C>T (p.Thr790Met) | Erlotinib + Crizotinib | death |
Footnotes
Conflicts of interest and sources of funding relevant to this manuscript:
None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaw AT, Solomon B. Targeting anaplastic lymphoma kinase in lung cancer. Clin Cancer Res. 2011;17:2081–2086. doi: 10.1158/1078-0432.CCR-10-1591. [DOI] [PubMed] [Google Scholar]
- 3.Febbo PG, Ladanyi M, Aldape KD, et al. NCCN Task Force report: Evaluating the clinical utility of tumor markers in oncology. J Natl Compr Canc Netw. 2011;9(Suppl 5):S1–32. doi: 10.6004/jnccn.2011.0137. [DOI] [PubMed] [Google Scholar]
- 4.Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 5.Camidge DR, Kono SA, Flacco A, et al. Optimizing the detection of lung cancer patients harboring anaplastic lymphoma kinase (ALK) gene rearrangements potentially suitable for ALK inhibitor treatment. Clin Cancer Res. 2010;16:5581–5590. doi: 10.1158/1078-0432.CCR-10-0851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wallander ML, Geiersbach KB, Tripp SR, et al. Comparison of Reverse Transcription Polymerase Chain Reaction, Immunohistochemistry and Fluorescence in situ Hybridization Methodologies for Detection of Echinoderm Microtuble-Associated Protein-Like 4/Anaplastic Lymphoma Kinase Fusion Positive Non-Small Cell Lung Carcinoma: Implications for Optimal Clinical Testing. Archives of Pathology and Laboratory Medicine. 2012 doi: 10.5858/arpa.2011-0321-OA. In Press. [DOI] [PubMed] [Google Scholar]
- 7.Boland JM, Erdogan S, Vasmatzis G, et al. Anaplastic lymphoma kinase immunoreactivity correlates with ALK gene rearrangement and transcriptional up-regulation in non-small cell lung carcinomas. Hum Pathol. 2009;40:1152–1158. doi: 10.1016/j.humpath.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 8.Mino-Kenudson M, Chirieac LR, Law K, et al. A novel, highly sensitive antibody allows for the routine detection of ALK-rearranged lung adenocarcinomas by standard immunohistochemistry. Clin Cancer Res. 2010;16:1561–1571. doi: 10.1158/1078-0432.CCR-09-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McLeer-Florin A, Moro-Sibilot D, Melis A, et al. Dual IHC and FISH Testing for ALK Gene Rearrangement in Lung Adenocarcinomas in a Routine Practice: A French Study. J Thorac Oncol. 2012;7:348–354. doi: 10.1097/JTO.0b013e3182381535. [DOI] [PubMed] [Google Scholar]
- 10.Paik JH, Choe G, Kim H, et al. Screening of anaplastic lymphoma kinase rearrangement by immunohistochemistry in non-small cell lung cancer: correlation with fluorescence in situ hybridization. J Thorac Oncol. 2011;6:466–472. doi: 10.1097/JTO.0b013e31820b82e8. [DOI] [PubMed] [Google Scholar]
- 11.Glassco J, Kyshtoobayeva A, Bloom KJ. Assessment of the ALK Antibody, 5A4 in Detecting ALK Rearrangements in Non-Small Cell Lung Cancer Specimens. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2012:25. [Google Scholar]
- 12.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 13.Sequist LV, Joshi VA, Janne PA, et al. Epidermal growth factor receptor mutation testing in the care of lung cancer patients. Clin Cancer Res. 2006;12:4403s–4408s. doi: 10.1158/1078-0432.CCR-06-0099. [DOI] [PubMed] [Google Scholar]
- 14.Zhang X, Zhang S, Yang X, et al. Fusion of EML4 and ALK is associated with development of lung adenocarcinomas lacking EGFR and KRAS mutations and is correlated with ALK expression. Mol Cancer. 2010;9:188. doi: 10.1186/1476-4598-9-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung Y, Kim P, Jung Y, et al. Discovery of ALK-PTPN3 gene fusion from human non-small cell lung carcinoma cell line using next generation RNA sequencing. Genes Chromosomes Cancer. 2012;51:590–597. doi: 10.1002/gcc.21945. [DOI] [PubMed] [Google Scholar]
- 16.Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]