Abstract
Hematopoietic stem cells (HSCs) are defined by their ability to self-renew and reconstitute all elements of the hematopoietic system. Acute myeloid leukemia (AML) is thought to arise from, and be maintained by, leukemic stem cells (LSCs), which exhibit similar features to HSCs, including the abilities to self-renew and differentiate into non-self-renewing cells. Acquisition of stem-cell-like characteristics by the LSCs is likely mediated in part by molecular mechanisms that normally regulate HSC function. Thus, understanding the shared and unique aspects of the molecular regulation of these cell populations will be important to understanding the relationship between normal hematopoiesis and leukemogenesis. MicroRNAs (miRNAs) are small noncoding RNAs that act at the posttranscriptional level to regulate protein expression. Unfortunately, most investigations of the role of miRNAs in normal hematopoiesis have been restricted to studies of their effects on lineage commitment in progenitors and mature effector cell function, but not on HSCs. Recent studies have identified miRNAs that enhance HSC function, and an abundance of profiling studies using primary AML samples have identified dysregulated miRNAs that may target genes implicated in self-renewal (HOX genes, P53, and PTEN), thus providing a potential link between normal and malignant stem cells. While these studies as well as recent in vivo models of miRNA-induced leukemogenesis (e.g. miR-29a, miR-125b) suggest a role for miRNAs in the development of AML, future studies using serial transplantation of primary AML blasts, from both mouse models and primary human AML specimens, will be necessary to assess the roles of miRNAs in LSC biology.
Keywords: acute myeloid leukemia, experimental models of leukemia, hematopoiesis, hematopoietic stem cells, leukemia stem cell, microRNAs
Introduction
Hematopoietic stem cells (HSCs) exhibit the unique ability to give rise to all of the cells of the hematopoietic system as well as themselves during the lifetime of an organism over numerous cell divisions in a process referred to as self-renewal [Kondo et al. 2003; Weissman, 2000]. Acute myeloid leukemias (AMLs) include cells that exhibit similar features to HSCs, including the functional capacity to self-renew, as demonstrated through serial transplantation, as well as the ability to differentiate into non-self-renewing leukemic blasts; these self-renewing leukemic cells are referred to as leukemic stem cells (LSCs) [Bonnet and Dick, 1997; Lapidot et al. 1994]. Given their similarities in function, gene expression, surface immunophenotype, and cytologic features, many have hypothesized that AML is the result of a stepwise progression of genetic and/or epigenetic events accumulating in HSCs or immature hematopoietic progenitors, culminating in the aberrant acquisition of stem-cell-like characteristics, including self-renewal, in progenitor cells that typically do not possess the ability to self-renew [Jamieson et al. 2004; Passegue et al. 2003]. Given this possibility, it is likely that many of the molecular mechanisms regulating the various functions of HSCs are also responsible for the transformation of HSC/progenitors into LSCs. Thus, comparing these populations with one another may yield important insights into the shared and unique aspects of their molecular regulation.
MicroRNAs (miRNAs) are small non-protein-coding RNAs that regulate gene expression by inhibiting the translation or catalyzing the degradation of target mRNAs. Since the first miRNA, lin-4, was identified in 1993, miRNAs have been shown to play critical roles in the regulation of many biological processes including cell differentiation, proliferation, and apoptosis, with significant influences on normal and malignant hematopoiesis [Vasilatou et al. 2010; Kluiver et al. 2006]. MiRNAs are transcribed as primary precursors (pri-microRNA) before being further processed into shorter precursor forms (premicroRNA) by the nuclear RNase Drosha. They are subsequently exported to the cytoplasm where they undergo editing by the endoribonuclease Dicer into mature 19-24 nucleotide miRNA duplexes [Lee et al. 2003]. miRNAs then assemble into an RNA-induced silencing complex (RISC) to effect suppression of translation or degradation of a potentially large number of target mRNAs [Gregory et al. 2005]. Because target recognition by miRNAs is based on perfect (in plants) or imperfect base pair matching (mammals) with the 3'- or 5'-untranslated regions (UTRs) of mRNAs, each miRNA may target a large number of mRNAs [Doench and Sharp, 2004]. Interestingly, in some cases miRNAs may also regulate gene expression by means other than direct binding to the UTRs of mRNAs, such as by acting as decoys for RNA binding proteins [Eiring et al. 2010; Malecova and Morris, 2010]. Thus, changes in the expression of even a small number of miRNAs can have a profound influence on many biological processes. For more details regarding miRNA biogenesis, processing, and target recognition, we refer the reader to several excellent reviews [Newman and Hammond, 2010; Siomi and Siomi, 2010; Winter et al. 2009].
Investigators have demonstrated functional roles for miRNAs in various aspects of normal and malignant hematopoiesis, including lineage commitment and differentiation, apoptosis, and mature immune effector cell function. These topics have been previously summarized in several excellent reviews [Vasilatou et al. 2010; Havelange et al. 2009; Kluiver et al. 2006], and therefore we do not repeat these summaries here; however, it is noteworthy that the vast majority of existing studies concerning the role of miRNAs in lineage commitment have not comprehensively evaluated their effects in stem and multipotent/oligopotent progenitor populations. For example, studies on the role of miR-223 in granulopoiesis [Pulikkan et al. 2010; Johnnidis et al. 2008; Fukao et al. 2007; Fazi et al. 2005; Chen et al. 2004], miR-181 in B- and T-cell differentiation [Chen et al. 2004], miR-150 in B-cell and megakaryocytic differentiation [Lu et al. 2008; Xiao et al. 2007], and miR-221 in erythropoiesis [Bruchova et al. 2007; Felli et al. 2005] highlighted the effects of miRNAs on hematopoietic differentiation, but each was limited to studies of mature effector cell output or a particular committed progenitor population. This is partly due to the reliance on in vitro models of induced differentiation in leukemic cell lines or heterogeneous progenitor populations, rather than the use of more physiologic systems that include the evaluation of these miRNAs in maturing HSCs and lineage committed progenitors. Moreover, little is known about the function of these or other miRNAs in HSCs. Fortunately, emerging data reveal a major role for miRNAs in the earliest stages of hematopoiesis. The implication of these studies is that many of the same miRNAs that regulate HSC/progenitor function may also regulate the behavior of the development of their malignant counterparts, LSCs. Understanding the shared and unique utilization of these mechanisms will be important to unraveling the relationship between normal hematopoiesis and leukemogenesis.
MicroRNAs in hematopoietic stem cells
Until recently, most of the available data regarding miRNA function in HSCs was based on expression profiling studies and in silico bioinformatic predictions. However, a number of recent investigations have directly evaluated miRNA expression and function in HSCs, providing the first confirmation of the role of miRNAs in HSC function (Tables 1 and 2).
Table 1.
MicroRNAs with characterized roles in normal hematopoietic and/or leukemic stem cell function.
| Increased expression in HSCs (species) | Function/phenotype | Affected progenitor populations | Role in self-renewal assessed by secondary transplantation | Proposed mechanism(s) of action | References | |
|---|---|---|---|---|---|---|
| mi R-125a | Yes (mouse) | Overexpression in mouse bone marrow cells leads to an increase in HSC pool size as measured by competitive limiting dilution assays. | HSCs (functionally, but not immunophenotypically defined) | No | Reduced apoptosis by targeting the proapoptotic protein Bak1. | Guo et al. [2010] |
| miR-125b | Yes (mouse, human) | Overexpression in mouse bone marrow cells leads to a myeloproliferative disorder that progresses to AML. | Not determined | Yes: transplantation of leukemic mouse bone marrow into secondary immunodeficient recipients led to death within weeks. | None proposed | O'Connell et al. [2010] |
| Overexpression in human CD34+ progenitors increases engraftment in the xenograft setting. | ||||||
| miR-125b | Yes (mouse) | Overexpression in mouse HSCs leads to an expansion of lymphoid balanced and lymphoid biased HSCs as assayed by serial transplantation. | HSCs (self-renewal assayed by serial transplantation) | Yes: HSC function assayed by serial transplantation. | Anti-apoptotic effects at least partially mediated by target genes Klf13 and Bmf. | Ooi et al. [2010] |
| miR-125b | Yes (mouse) | Overexpression in mouse fetal liver cells leads to a myeloproliferative neoplasm, B-ALL, and T-ALL. Co-expression with Bcrabl in bulk 5-FU treated bone marrow leads to accelerated leukemogenesis. | Not determined | No | Targeting of the proapoptotic transcripts Bak1 and Bmf, as well as P53 (confirmed in human only). | Bousquet et al. [2010] |
| miR-125b | Yes (mouse, human) | Overexpression in HSPCs enriched mouse bone marrow cells and CD34+ human bone marrow cells leads to perturbed myeloid differentiation. Overexpression in mouse fetal liver megakaryocyte and megakaryocyte/erythroid progenitors (MkP/MEP) leads to increased proliferation and self renewal of these populations. | MkP/MEP (self renewal assayed by serial replating, but not serial transplantation) | No | Impaired global miRNA biogenesis via direct targeting of DICER1, and direct targeting of the tumor suppressor ST18. | Klusmann et al. [2010] |
| miR-29a | Yes (mouse, human) | Overexpression in mouse bone marrow enriched for HSC/progenitor cells leads to myeloid biased differentiation, and a myeloproliferative disorder that progresses to AML. | MPP, CMP, GMP | Yes: affected CMP and GMP populations, as well as leukemic blasts, were shown to serially transplant disease. | Regulation of the G1 to S phase transition, possibly by downregulation of HBP1, which may act in coordination with other targets yet to be defined. | Han et al. [2010] |
| miR-155 | No | Overexpression in HSC enriched mouse bone marrow leads to a myeloproliferative disorder with expansion of GM populations, and reduced numbers in the erythroid, lymphoid, and megakaryocyte lineages. | Unknown (heterogeneous populations transplanted) | No | Increased Akt activation by targeting the inositol phosphatase SHIP1 [O'Connell et al. 2009]. Other confirmed targets have been linked to AML (PU.1, Picalm) and myeloproliferative disorders (Cutl1, Csf1r). | O'Connell et al. [2008] |
HSC, hematopoietic stem cells; AML, acute myeloid leukemia; ALL, acute lymphoid leukemia; 5-FU, 5-Fluorouracil; HSPCs, hematopoietic stem and progenitor cells; MkP, megakaryocyte progenitors; MEP, megakaryocyte erythroid progenitors; MPP, multipotent progenitors; CMP, common myeloid progenitors; GMP, granulocyte macrophage progenitors; GM, granulocyte/monocyte.
Table 2.
MicroRNAs enriched in hematopoietic stem cells.
| HSC definition | Upregulated miRNAs | Reference |
|---|---|---|
| Lin-Kit+Sca+CD34-Flk2-(mouse) | miR-99b, −125a, let-7e, miR-130a, −10a, −10b, −125b, −31, −99a, −100, −146b, −425, −422b, −18a, −15b |
Guo et al. [2010] |
| Lin-Kit+Sca+CD150+CD48− | miR-125b, −125a, −155, −99a, −126, −196b, −130a, −542−5p, −181c, −193b, and let-7e |
O'Connell et al. [2008] |
| ES150+ (CD45+EPCR+CD48 −CD150+) ES150− (CD45+EPCR+CD48 −CD150−) SLAM HSC− (CD150+CD48−, CD45 enriched) LKS (Lin-Kit+Sca+) |
miR-29a (only in ES150+ and ES150-), −125b, −196a, −196b, −130a, −148b, −351, and let-7d |
Petriv et al. [2010] |
Although the earliest efforts to characterize miRNA expression and function in human HSCs did not utilize highly purified cells, these studies nonetheless had an important influence on subsequent studies, as they attempted to utilize complementary miRNA and mRNA data sets to predict miRNA function in human HSCs. One of the earliest efforts to characterize miRNAs in HSCs/progenitors was performed in human CD34+ cells, which are frequently utilized as surrogates for human HSCs. It is important to note, however, that the CD34+ compartment represents a heterogeneous mixture of hematopoietic progenitors, of which <1% are functionally defined HSCs [Majeti et al. 2009; Peault et al. 1993]. Georgantas and colleagues performed the first large scale profiling of miRNAs using human CD34+ cells from normal human bone marrow and mobilized peripheral blood CD34+ cell harvests [Georgantas et al. 2007]. Using a solid-state array, 33 miRNAs in total were found to be expressed in CD34+ cells. Using mRNA expression data previously generated from CD34+ cells, in silico algorithms for predicting mRNA targets of miRNAs, as well as taking into account the presumed developmental stage at which previously identified major regulators of erythroid, myeloid, or lymphoid differentiation act, the investigators made conclusions about the developmental stage-specific roles of miRNAs. While this represented an important early attempt to dissect the functional roles of miRNAs in early human hematopoiesis, the model generated relied on numerous assumptions about the developmental stage-specific roles of these mRNA targets. Thus, the functional roles of miRNAs in HSCs or specific lineage committed progenitors remained an open question.
Studies of miRNA function in HSCs are performed more easily in mice than in humans due to the ability to interrogate gene function genetically, as well as through the use of robust congenic transplantation systems to assess the function of prospectively isolated, highly purified HSCs and committed progenitor cell populations [Wilson et al. 2007]. Guo and colleagues demonstrated the importance of miRNAs in HSCs using a conditional Dicerlox/lox knockout (KO) mouse model in which Dicer was selectively deleted in the hematopoietic system using the Mx-Cre transgene. HSC/progenitor function was assessed by transplanting Dicer KO and wild-type (WT) bone marrow to reconstitute hematopoiesis in lethally irradiated recipient mice. Although Dicer KO HSCs persisted in recipient mice for months, there was a significant reduction in the primitive LKS (Lineage-cKit+Sca-1+) progenitor population as well as all mature lineages, particularly myeloid cells. Furthermore, while they found complete deletion of the floxed allele in Cre+Dicerlox/wt cells by genomic DNA analysis, in the Cre+Dicerlox/lox cells, undeleted alleles (functionally WT) persisted, likely suggesting positive selection for HSCs with intact Dicer function. In an in vitro methylcellulose assay the Dicer KO cells did not form any colonies. Together, these studies indicate that Dicer is required for normal HSC function. In addition, Dicer KO LKS cells exhibited increased rates of apoptosis, caspase-3 activity, and increased Ki-67 staining, consistent with an increase in cell death and proliferation. Thus, this study demonstrated that Dicer, and by extension, its mature miRNA products, are essential for maintaining the most immature hematopoietic cell pool [Guo et al. 2010].
In another study measuring miRNA expression in mouse HSCs, O'Connell and colleagues examined the expression of 137 miRNAs in LKS cells and total bone marrow from C57BL/6 mice [O'Connell et al. 2010]. Of these 137 miRNAs, 11 were found to be enriched in LKS progenitors compared with total bone marrow including miR-125a, miR-125b, miR-155, miR-99a, miR-126, miR-196b, miR130a, miR-542, miR-181, miR-193, and miR-let7e. Fractionation of the LKS compartment into more stringently defined HSC populations with either endothelial protein C receptor (EPCR) or SLAMf1/CD150 yielded two populations with further enrichment for these 11 miRNAs. With the exception of miR-193b, enrichment of the same set of miRNAs was found in an analysis of CD34+ human cord blood (CB) cells compared with CB CD34- cells, demonstrating that this pattern of miRNA expression in early hematopoiesis is evolutionarily conserved. This is one of the few studies to corroborate miRNA expression patterns in mouse and human hematopoiesis. As described later in this review, several miRNAs that enhance HSC self-renewal have also been found to be dysregulated in human AML, suggesting that their failure to downregulate appropriately in malignant hematopoiesis may be in part responsible for the impaired differentiation, apoptosis, or enhanced self-renewal that characterizes AML blasts [Garzon et al. 2008a].
In the most comprehensive evaluation of miRNA expression in the hematopoietic system to date, Petriv and colleagues et al. examined the expression levels of 288 miRNAs in 27 phenotypically distinct cell populations ranging from hematopoietic stem/progenitor cells to terminally differentiated mature blood cells [Petriv et al. 2010]. The mean number of miRNAs detected in each population was 96 (range 49 to 149). Analysis of miRNA expression patterns revealed several interesting trends. For example, miR-130a, miR-196a, and miR-196b were highly expressed in stem/progenitor cells and downregulated upon commitment to lymphoid lineages. A phylogenetic analysis of miRNA expression showed that these 27 cell populations could be grouped into six clusters including stem and progenitor cells, lymphoid cells, and four other branches corresponding with different myeloid lineage cells. These studies provide strong evidence for lineage-specific miRNA expression in hematopoiesis, and they set the stage for future studies aimed at confirming the hypothesized roles for these miRNAs. It will be interesting to see how integration of these data sets with mRNA expression and proteomic data from the same progenitor populations might promote the identification of interesting targets for further investigation. Interestingly, expression analysis of a subset of miRNAs in single cells found that populations such as granulocyte-macrophage progenitors (GMPs), thought to be relatively homogeneous with respect to function, displayed less cell-to-cell miRNA expression variation as compared with more functionally heterogeneous populations, such as CD45+CD48+ cells. Thus, these data suggest that miRNA expression variability in single cells may underlie the functional heterogeneity observed in such populations, providing a potential method to molecularly characterize distinct populations within heterogeneous progenitor populations.
HSCs are defined by their ability to self-renew and reconstitute all of the cells of the hematopoietic system by giving rise to downstream multipotent, oligopotent, and committed progenitor cells. As mentioned previously, early studies focused on identifying miRNAs enriched in immature hematopoietic subsets without generating data on their in vivo roles. Fortunately, several recent studies have begun to shed light on the function of miRNAs in HSCs. In the aforementioned study by O'Connell and colleagues the 11 miRNAs found to be upregulated in HSCs were retrovirally transduced into bone marrow cells and used in competitive repopulation assays. MiR-125b was chosen for further study, as it demonstrated the greatest increase in repopulation, and had also been shown to be overexpressed in AMLs harboring t(2;11)(p21;q23) [Klusmann et al. 2010; Bousquet et al. 2008]. Lethally irradiated mice transplanted with bone marrow cells retrovirally transduced to ectopically overexpress miR-125b developed a progressive expansion of immature myeloid cells in the PB, culminating in AML. Owing to the heterogeneous nature of the cells transduced to express miR-125b, it is unclear which progenitor population or populations served as the cell of origin for this expansion [O'Connell et al. 2010]; however, these data support a leukemogenic role for miR-125b in at least a subset of human AMLs.
In a separate study, Guo and colleagues found that a cluster of miRNAs including miR-125a, miR-99b, and let-7e is highly expressed in mouse HSCs/progenitor cells, thus choosing these miRNAs for further investigation. Retroviral transduction and overexpression of miR-125a alone, but not other members of the cluster, in immature mouse bone marrow cells was sufficient to enhance their reconstitution potential in lethally irradiated mice. Furthermore, serial replating assays in methylcellulose demonstrated decreased colony formation in the presence of an antagomir against miR-125a, and HSCs transduced with miR-125a exhibited preserved hematopoietic cell output through 18 days of ex vivo culture, suggesting that miR-125a enhances HSC activity in a cell autonomous manner. The cell surface immunophenotypes of reconstituted bone marrow cells overexpressing miR-125a were significantly altered and could not be used to prospectively isolate HSCs by conventional gating strategies using LKS, CD48 and CD150 specific antibodies; however, competitive limiting dilution assays demonstrated an eightfold increase in the HSC pool size. However, it is not clear whether the observed increase in HSC activity is solely attributable to this increase in HSC number. Attempts to identify target genes of miR-125a in human AML cell lines identified the proapoptotic protein Bak1 (Bcl-2 antagonist/killer1) as a potential target gene. Interestingly, bone marrow cells from Bak1−/− mice demonstrated no obvious immunophenotypic or functional derangements, suggesting that expansion of the HSC pool by miR-125a arises from multiple gene targets or different physiologic targets in mouse HSCs [Guo et al. 2010].
In our own studies, we have independently demonstrated high levels of miR-125b expression in both mouse and human HSCs. Transduction of highly purified mouse HSCs (Lin-ckit+Sca-1+CD150+CD34-FLK2-) with miR-125b expressing lentivirus induced enhanced engraftment as shown by serial transplantation with equal numbers of purified HSCs (miR-125b expressing and empty vector control) into secondary recipients. Similar to the previously described studies of miR-125 family members in HSCs, the mechanism of enhanced HSC function was due to induction of an anti-apoptotic state in mouse HSCs, most likely through down-regulation of Bmf and KLF13 mRNA expression. However, in contrast to the other studies demonstrating induction of AML or simply increased HSC engraftment capability, we observed an increase in the number of total HSCs with preferential rescue and expansion of lymphoid-biased HSCs [Beerman et al. 2010]. This expansion of lymphoid-biased HSCs was accompanied by increased numbers of both B and T progenitors and mature lymphoid cells in the peripheral blood [Ooi et al. 2010].
In a separate set of studies, we determined that human HSCs and AML LSCs share high expression of miR-29a. While the phenotype induced by overexpression of miR-29a in immature mouse bone marrow cells is discussed in the next section, it is important to note that prior to the development of AML, miR-29a induced the appearance of nonleukemic committed myeloid progenitor cells (common myeloid progenitors [CMPs] and GMPs) that were capable of long-term engraftment without loss of differentiation potential, consistent with the aberrant induction of self-renewal by miR-29a. While our studies did not directly address the issue of HSC function due to the rapid development of a myeloproliferative neoplasm-like phenotype, future studies will be aimed at evaluating HSC function in the setting of miR-29a overexpression and knockdown [Han et al. 2010]. We discuss the role of the miR-29 family in AML in a following section.
Together, these studies demonstrate clear roles for miRNAs in HSC biology, and in several cases, high levels of miRNA expression are shared between HSCs and AML and sufficient to induce AML in mouse models. Because these miRNAs can regulate self-renewal as well as lineage choice (at the level of the HSCs), these studies suggest that some HSC-relevant miRNAs may be important in myeloid leukemogenesis, possibly even in the self-renewing component of AML, the LSCs. For example, the fact that overexpression of miR-125b and miR-29a both enhance HSC or progenitor self-renewal capability and also induce AML suggests that these miRNAs may play key roles in conferring self-renewal properties to LSCs. A notable limitation of these studies is the reliance on overexpression systems to study miRNA function in HSCs. Sufficiency experiments will be essential for determining the necessity of these miRNAs for normal HSC function. In some cases, such studies may need to be performed in a conditional manner since constitutive knockdown may prevent proper development of HSCs. For example, we were unable to generate stably engrafting HSCs transduced with antisense miR-125b lentiviruses, suggesting a requirement for miR-125b in HSC function (data not shown); confirmation of this hypothesis could easily be performed in a conditional knockout or knockdown model.
MicroRNAs in acute myeloid leukemia
Numerous studies have established roles for miRNAs as tumor suppressors or oncogenes in hematologic malignancies. In some cases, the roles of specific miRNAs in cancer development have been inferred by their cancer-associated loss or gain [Croce, 2009]. Mechanistically, oncogenic miRNAs are thought be to expressed at high levels in cancer or precancerous cells with suppression of the translation of mRNAs encoding tumor suppressors, while tumor suppressor miRNAs are expressed at reduced levels, releasing an inhibitory signal and allowing for the increased translation of oncogenic mRNAs [Garzon et al. 2006a]. Examples of these include miR-15a and miR-16 −1 in chronic lymphocytic lymphoma (CLL) [Calin et al. 2002], the miR-17∼92 cluster in B-cell lymphomas [He et al. 2005], and miR-155 in Hodgkin and nonHodgkin lymphomas [Eis et al. 2005; Kluiver et al. 2005]. Although recent studies have described dysregulation of miRNAs in acute lymphoblastic lymphoma (ALL) [Schotte et al. 2010; Wang et al. 2010; Kaddar et al. 2009], functional studies have been limited to only a few miRNAs (miR-125b, miR-155, miR-196b) [Coskun et al. 2010; Gefen et al. 2010; Tassano et al. 2010; Costinean et al. 2006]. These studies suggest that miRNAs involved in AML pathogenesis may also be relevant to ALL. However, as the pathogenic relationship between the LSCs and the HSCs in ALL is not as well established as in AML, we focus this review on the role of miRNAs in the LSCs in AML.
In AML, the LSC may arise either directly from HSCs that have acquired multiple mutations or committed progenitors that have acquired the ability to self-renew [Jamieson et al. 2004; Passegue et al. 2003]. Mouse models of chronic myeloid leukemia (CML) and AML have provided evidence that the LSCs may arise from either population, with the specific population involved likely depending on the exact oncogenic event and disease context [Krivtsov et al. 2006; Passegue and Weisman, 2005; Cozzio et al. 2003]. Moreover, there is increasing evidence of a bidirectional interaction between LSCs and the bone marrow microenvironment that may influence lineage fate, survival, and sensitivity to treatment. As with HSCs, the gold standard for defining a LSC population is demonstrating self-renewal potential by serial transplantation in vivo. Although studies have demonstrated roles for miRNAs in established AMLs, miRNAs have not been characterized in primary AML LSCs in the xenograft setting. Thus, the majority of the evidence for functional roles of miRNAs in the experimental setting is from mouse models as described in the section entitled ‘MicroRNA induction of AML in experimental models’.
Two studies by Lu, Volinia, and colleagues were among the first to demonstrate that miRNA expression patterns reflect the lineage and differentiation state of multiple types of tumors [Volinia et al. 2006; Lu et al. 2005]. These studies were subsequently followed by a number of large-scale miRNA expression profiling studies in AML over the past several years. Cumulatively, these studies have identified miRNA expression signatures that strongly correlate with morphologic subtype, cytogenetic status, molecular abnormalities, and prognosis; (Table 3) the results of these studies are summarized in detail in a number of excellent reviews [Vasilatou et al. 2010; Havelange et al. 2009]. Notable findings included the identification of miRNAs associated with specific cytogenetic or molecular subtypes. For example, specific miRNA signatures were found to be associated with core binding factor (CBF), mixed lineage leukemia rearranged (MLL), trisomy 8, and cytogenetically normal (CN) AMLs [Jongen-Lavrencic et al. 2008; Marcucci et al. 2008a]. AMLs harboring fms-like tyrosine kinase with internal tandem duplication (FLT3-ITD) or cytoplasmic mutated nucleophosmin (NPMc+) were also found to be associated with specific miRNAs [Garzon et al. 2008b]. Interestingly, when 157 miRNAs were measured in 100 primary AML specimens representing common karyotypic abnormalities, specific cytogenetic groups such as t(15;17), CBF [t(8;21) and inv(16)], MLL, and CN-AML clustered into distinct groups, but differences in chromosomal number did not explain differences in miRNA expression. Thus, miRNA expression differences among these cytogenetic groups is not likely to be a direct result of the chromosomal abnormalities themselves, but rather a reflection of the unique biology of each karyotypic subtype. Similarly, AMLs with t(15;17) showed strong upregulation of six miRNAs, with most of these miRNAs located on chromosome 14, away from the t(15;17) translocation itself, again suggesting that miRNA dysregulation reflects the different biology of leukemic subtypes, rather than the structural genetic abnormalities themselves [Jongen-Lavrencic et al. 2008]. As the LSCs may arise from different progenitor populations in each AML subtype, the differential expression of miRNAs may reflect a context specific role in the maintenance of each particular LSC [Dixon-McIver et al. 2008].
Table 3.
MicroRNAs dysregulated in acute myeloid leukemia by cytogenetic and molecular subtype.
| Cytogenetic or molecular subtype | miRNAs increased (PUTATIVE TARGET) | miRNAs decreased | References |
|---|---|---|---|
| Normal karyotype | miR-10a (H0XA1), −10b, −26a, −30c, −16−2, −21 (PTEN), −181b (TLR3, IL1B), −368, and −192, and let-7a−2 | miR-126, −203, −200c, −182, −204, −196b, −193, −191, −199a, −194, −183, −299, and −145 | Garzon et al. [2008b] |
| t(15;17) | miR-382, −134, −376a, −127 (BCL6), −299-5p, −323, −154, −154*, −299, −368, −370, −224, and −382 | miR-17−3p, −185, −187, −194, −200a, −200b, −200c, −330, and −339 | Dixon-McIver et al. [2008]; Jongen-Lavrencic et al. [2008]; Li et al. [2008] |
| t(8;21) | miR-146a, −126 (PLK2), and −126* | let-7b, let-7c (RAS), miR-133a, −223 | Dixon-McIver et al. [2008]; Jongen-Lavrencic et al. [2008]; Li et al. [2008]; Fazi et al. [2007]; |
| lnv(16) | miR-99a, −100, −224, −126, and −126* | miR-127 (BCL6) | Dixon-McIver et al. [2008]; Jongen-Lavrencic et al. [2008]; Li et al. [2008] |
| 11q23 | miR-326, −219, −194, −301, −324, −339, −99b, −328, and the miR-17−92 cluster | miR-34b (CDK4, CCNE2, P53 target), −15a (BCL2), −29a, −29c, −372 (LATS2), −30a, −29b, −30e, −196a (HOXA7, HOXD8, HOXB8), −102, 331, −299, −193, let−7f (RAS) | Garzon et al. [2008b]; Li et al. [2008] |
| Trisomy 8 | miR-124a (CEBPa), −30d, −29b/c, let−7d, and 37 others (see ref) | None | Garzon et al. [2008b] |
| NPM1 mutated | miR-10a (HOXA1), −10b, −196a, and −196b (HOXB8), miR29s (a/b/c), let−7s (a/b/f), miR-15a, −16−1 | miR-204 (HOXA10, MEIS−1) | Garzon et al. [2008a]; Jongen−Lavrencic et al. [2008] |
| FLT3−ITD | miR-155, −10b | None | Garzon et al. [2008b]; Jongen−Lavrencic et al. [2008] |
| CEBPα mutated | miR-128, −181a, −181b, −181c (TLR4, IL1B), miR-219−1−3p, miR-355* | miR-34a (P53 target), miR-194 | Marcucci et al. [2008a] |
| All (compared with CD34+ normal bone marrow) All (compared with normal bone marrow mononuclear cells) | miR-21 (PTEN), −26a, −26b, −29b, 34a (P53 target), −221 (Kip1), −222 (c−kit) let−7e, 142−5p, −155, 181a/b/c, −195, −34a (P53 target), −221 (KipD, −222 (c−kit) | miR-23b, −126, −130a, −135, −93, −146, −106b, −125a miR-34c (P53 target), −26a, −199a (potential tumor suppressor), −103, −372 (potential oncogene), −26a, −26b, −29b, −23b | Garzon et al. [2008b]; Isken et al. [2008]; Jongen−Lavrencic et al. [2008] Isken et al. [2008]; Jongen−Lavrencic et al. [2008] |
| Improved Outcome | miR-181a, −181b (cytogenetically normal AML), miR-199a, −191 (all cytogenetic groups) | miR-124, −128−1, −194, −219−5p, −220a, −320 (cytogenetically normal AML) | Garzon et al. [2008b]; Marcucci et al. [2008b] |
| Better response to hypomethylating agent treatment | miR-29b | None | Blum et al. [2010] |
In parentheses next to particular miRNAs are notable putative targets, a subset of which have been functionally confirmed. In bold type are miRNAs that are variably dysregulated depending on the cell population of comparison, i.e. miRNAs that are decreased compared with normal CD34+ cells but increased relative to total bone marrow mononuclear cells. AML, acute myeloid leukemia.
is part of standard nomenclature for the minor passenger strand during miRNA processing that is usually degraded but occasionally remains viable and can have its own mRNA targets- i.e., miR-355* denotes a particular functional miRNA distinct from miR-355.
Major roles for many of these miRNAs have been postulated based on their predicted targets or their position within the genome, but overall there have been few functional studies of miRNA function in AML. Nevertheless, these studies support the notion that the LSCs in different cytogenetic and molecular subtypes of AML may be regulated by different miRNAs. In addition, they provide important information regarding miRNAs that have potential as therapeutic targets or markers of prognosis, as well as miRNAs that may have direct functional roles in AML. In the remainder of this section, we highlight findings from these studies that suggest roles for miRNAs in LSC function.
MicroRNA regulation of self-renewal pathways in acute myeloid leukemia
HOX genes are thought to regulate self-renewal in both HSCs and LSCs [Argiropoulos and Humphries, 2007], with overexpression of HOXA10 in mouse bone marrow cells leading to perturbed myeloid differentiation and progression to AML [Thorsteinsdottir et al. 1997]. In addition, HOXA9 overexpression alone and in combination with MEIS-1 both lead to a myeloproliferative disorder that progresses to AML [Kroon et al. 2001]. Thus, the finding that high expression of miR-10a and miR-10b, miRNAs predicted to target HOX family members, was associated with specific cytogenetic and molecular subtypes of AML suggests a potential role for these miRNAs in LSC function. In addition, Deberdini and colleagues measured miRNA and mRNA expression in total bone marrow cells from 30 patients with AML and normal cytogenetics (CN-AML) and identified miR-10a, miR-10b, and miR-196a, miRNAs located within the intergenic regions of HOX genes, as highly expressed in AML. They also demonstrated a statistically significant association with high expression of nearly all genes in the HOXA and HOXB clusters [Debernardi et al. 2007]. Similarly, miR-10a and miR-10b were shown to be associated with CN-AML harboring NPMc+ [Garzon et al. 2008a]. Notable down-regulated miRNAs included miR-204, confirmed to be a regulator of HOXA10 and MEIS-1. Given that HOXA1 is a putative target of miR-10a and miR-10b [Garzon et al. 2006b] and HOXB8 has been shown to be a target of miR-196a [Yekta et al. 2004], these studies suggest a complex regulatory network involving miRNAs and HOX genes.
P53 has been shown to promote HSC quiescence and self-renewal [Liu et al. 2009], with recent studies showing that deficiency of p53 likely promotes AML by eliminating its ability to limit aberrant self-renewal in hematopoietic progenitors [Zhao et al. 2010]. In a study comparing expression of 154 miRNAs in 50 primary AML samples with both normal bone marrow and CD34+ selected hematopoietic progenitors, miR-34a was found to be upregulated [Isken et al. 2008]. This is an intriguing observation, since miR-34a has been identified as a P53 target with proapoptotic function [Chang et al. 2007; Raver-Shapira et al. 2007; Tarasov et al. 2007]. Thus, it is interesting to speculate that miR-34a may regulate p53 activity, thereby contributing to LSC development, quiescence, or resistance to therapy.
Garzon and colleagues determined that MLL (11q23) rearranged AMLs are associated with a distinct miRNA expression signature that includes eight upregulated and six downregulated miRNAs [Garzon et al. 2008b]. Notable dysregulated miRNAs included miR-21, which was upregulated and targets the tumor suppressor PTEN [Meng et al. 2007], and miR-196, which was downregulated and targets several HOX genes. In a separate study, upregulation of miR-21 was identified across all subtypes of AML [Jongen-Lavrencic et al. 2008]. PTEN has been implicated in HSC biology, as PTEN deficient mice exhibit increased proliferation and depletion of HSCs, while promoting a skewed pattern of lineage differentiation and the emergence of AML containing a distinct LSC [Yilmaz et al. 2006; Zhang et al. 2006].
An analysis of 29 AML patients with FLT3-ITD mutations revealed upregulation of miR-155 when compared with FLT3-wt/NPMc+ patients [Garzon et al. 2008a]. As discussed above, overexpression of miR-155 alone has been shown to lead to a preleukemic B-cell proliferation and a myeloproliferative disorder [O'Connell et al. 2008; Costinean et al. 2006], suggesting a role in LSC formation. However, these studies do not identify the specific progenitor population in which miR-155 exerts its effects. In experiments blocking FLT3 signaling in FLT3-ITD cell lines there was no decrease in miR-155, suggesting that its increased expression is independent of FLT3 signaling. This may imply a cooperative leukemogenic role for miR-155 limited to LSCs that are dependent on constitutive FLT3 signaling.
Li and colleagues performed an analysis of 435 miRNAs in 54 AML samples (47 primary AML samples and 7 cell lines) harboring t(15;17), t(8;21), inv(16), or MLL rearrangements, along with three normal bone marrow samples (two CD34+ selected and one CD15+ selected) [Marcucci et al. 2008a]. Samples grouped into three discrete clusters, characterized by CBF [t(8;21) and inv(16)], t(15;17), and MLL rearrangements [Li et al. 2008]. The CBF driven cluster was characterized by overexpression of miR-126/126*, and the MLL rearranged cluster was characterized by overexpression of the miR-17-92 cluster, suggesting a pathogenic role for these miRNAs in each particular cytogenetic subtype. Forced expression of miR-126 in t(9;11) and inv(16) bearing cell lines significantly decreased apoptosis and increased cell viability, while transduction of mouse bone marrow progenitor cells led to increased replating efficiency, particularly in cooperation with the t(8;21) fusion gene (AML-ETO). These studies suggest a role for this miRNA in enhancing LSC self-renewal, but this remains to be validated in an in vivo transplantation model.
These studies highlight an important issue for gene profiling studies in AML. Control or reference populations in AML studies may be CD34+ cells, mononuclear cells, or total bone marrow cells. The choice of ‘normal’ reference populations may greatly influence the interpretation of gene expression data because each of these populations is highly heterogeneous, both with respect to lineage as well as differentiation state. In this particular study, comparison of AML blast populations to either CD34+ selected or total bone marrow controls yielded markedly different results for a subset of miRNAs, including miR-26a, −26b, and -29b (with each having intermediate levels of expression between CD34+ cells and total bone marrow cells). Thus, it is not surprising that other studies have resulted in discordant findings, with Dixon-McIver and colleagues showing that miR-26a is decreased in AML (in comparison with total bone marrow) and Cammarata and colleagues showing that miR-26a is increased in AML (in comparison with CD34+ cells) [Cammarata et al. 2010; Dixon-McIver et al. 2008]. These studies likely reflect the developmental stage-specific expression of miRNAs in normal hematopoiesis, as well as the pitfalls of using heterogeneous progenitor populations as normal controls. Given that the LSCs likely arise in the HSCs or more committed progenitors [Passegue et al. 2003], we believe these purified populations would serve as the most appropriate controls for such comparisons. MiRNAs associated with cytogenetically or molecularly defined subsets of AML may reflect cooperating lesions that act synergistically with their defined genetic lesions. As these lesions may accumulate in a specific temporal sequence, comparison of miRNA profiles in residual nonleukemic progenitor populations after treatment with their leukemic counterparts may help determine the order in which the lesions were acquired.
MicroRNA induction of acute myeloid leukemia in experimental models
Because self-renewal is one of the defining characteristics of the LSCs and the experimental gold standard for self-renewal is demonstration of serial transplantability of candidate stem cells, xenograft models of AML, preferably with primary patient samples, will be needed to establish the function of miRNAs in the LSCs. While such studies are currently lacking in the literature, several recent studies suggest important roles for miRNAs in LSC function due to their ability to induce AML or myeloid disorders in mice.
miR-196b is a transcriptional target of MLL that lies between HOXA9 and HOXA10, both of which are frequently overexpressed in MLL leukemias. As putative targets of miR-196b include members of the HOX cluster (HOXB8, HOXB7), it likely plays a role in fine-tuning levels of HOX gene expression. Expression of miR-196b, HOXA9, and HOXA10 appears to be higher in less differentiated cells, particularly ST-HSCs. Popovic and colleagues demonstrated that introduction of the MLL-AF9 fusion into mouse bone marrow progenitors, which increases proliferative capacity, also increased miR-196b expression. Treatment of these cells with an antagomir for miR-196b abrogated their replating potential in methylcellulose, demonstrating the central role of miR-196b in this phenotype. Overexpression of miR-196b in c-kit+ bone marrow cells also led to increased replating potential in methylcellulose and introduced a partial block in differentiation. Interestingly, predicted HOX gene targets did not show detectable expression in the presence or absence of miR-196b overexpression, leaving the mechanism of action of miR-196b unclear. Together, these results suggest that miR-196b may enhance self-renewal in hematopoietic progenitors, but in vivo transplantation studies will be necessary to determine whether increased expression of miR-196b is sufficient to induce AML alone, or in concert with MLL, and whether it regulates the LSCs in a such a model [Popovic et al. 2009].
Mi and colleagues demonstrated that the miR-17-92 cluster is overexpressed in MLL-rearranged AML and that enforced expression of miR-17-92 in Hela and 293T cells inhibits apoptosis [Mi et al. 2010]. Transfection of mouse bone marrow progenitor cells with a truncated form of the miR-17-92 cluster lacking miR-92 led to increased methylcellulose colony forming capacity, particularly when cotransfected with MLL-ELL. MLL-ELL transfected cells appeared morphologically less differentiated than those transfected with miR-17-92 alone, suggesting synergism between the two factors in enhancing proliferation and blocking differentiation. However, which progenitor population(s) these effects take place in remains unclear, with further studies necessary to determine whether they are sufficient for the development of LSCs.
Enforced expression of miR-155 in immature mouse bone marrow cells transplanted into lethally irradiated recipient mice led to the development of a myeloproliferative phenotype with expansion of granulocyte/monocyte (GM) cell populations at the expense of other hematopoietic cells including erythroid and lymphoid cells, as well as megakaryocytes and platelets [O'Connell et al. 2008]. Although many GM cells exhibited morphologic evidence of dysplasia, the myeloproliferative condition did not progress to acute leukemia, suggesting that miR-155 may be sufficient to induce a preleukemic state that requires additional mutations for full transformation. It would be interesting to perform serial transplants to determine whether there is an enhancement of self-renewal in the miR-155 expressing HSC.
Han and colleagues were the first to experimentally demonstrate miRNA function in myeloid leukemogenesis [Han et al. 2010]. After demonstrating that miR-29a is expressed at relatively high levels in human HSC and multipotent progenitors (MPPs) as well as primary AML blasts, they showed that ectopic expression of miR-29a in mouse HSCs/progenitors induced a myeloproliferative phenotype with expansion of myeloid progenitors and proliferation of more mature myeloid cells. Secondary transplants with CMPs and GMPs demonstrated aberrant acquisition of self-renewal properties, and MPPs showed a unique hyperproliferation, indicating developmental stage-specific effects of miR-29a. Secondary transplants with bulk spleen or bone marrow cells revealed that the myeloproliferative phenotype progresses to AML with a definable, c-kit+ LSC. Thus, while miR-29a was the first miRNA shown to induce AML in vivo following induction of self-renewal in committed progenitors, these studies did not directly address whether secondary mutations may be necessary for transformation of normal progenitors into self-renewing leukemic blasts (Figure 1). In contrast, recent studies have demonstrated a growth inhibitory and potential tumor suppressor role for miR-29 family members, as overexpression of both miR-29a and miR-29b in the K562 and Kasumi-1 AML cell lines resulted in decreased proliferation and increased apoptosis [Garzon et al. 2009]. Possible explanations for the different effects of miR-29 family members include the cell-context specific effects of these miRNAs, i.e. transformed human AML cell lines versus primary mouse HSCs/committed progenitors, as well as the potential unique mRNA targets of miR-29 family members due to their different subcellular locations, with miR-29b predominantly localized in the nucleus (via its hexanucleotide nuclear localization signal) as opposed to the predominantly cytoplasmic location of miR-29a [Hwang et al. 2007]. Thus, the overexpression of these two miR-29 family members might be expected to lead to very different phenotypes.
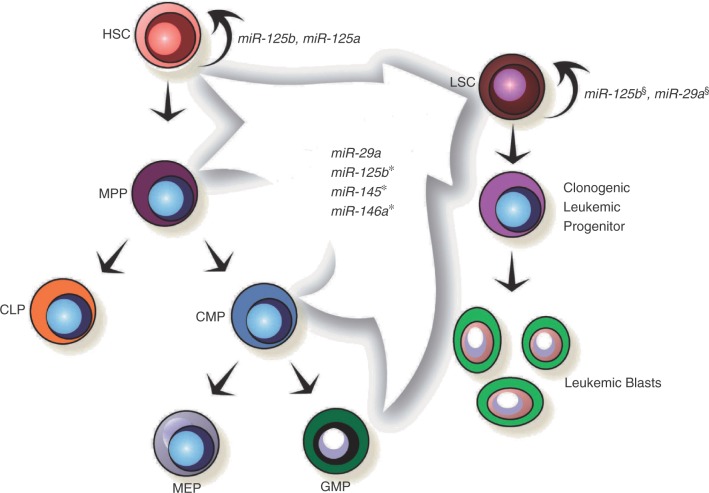
Figure 1.
MicroRNAs with demonstrated roles in hematopoietic stem cell (HSC) and leukemic stem cell (LSC) function. Each of the miRNAs contained within the arrow have demonstrated roles in inducing acute myeloid leukemia in the mouse hematopoietic system. Please see the text for details. *It remains unclear in which progenitor population(s) miR-125b, miR-145, and miR-146a exert their effects. §Both miR-125b and miR-29a have been demonstrated to induce myeloproliferative disorders that progress to a serially transplantable acute myeloid leukemia in mice. This suggests that they support self-renewal of the leukemic stem cell, but more formal experiments perturbing their function in the LSC itself are needed to come to a definitive conclusion. HSC, hematopoietic stem cell; LSC, leukemic stem cell; MPP, multipotent progenitor; CMP, common myeloid progenitor; MEP, megakaryocyte erythroid progenitor; GMP, granulocyte macrophage progenitor; CLP, common lymphoid progenitor.
As described earlier, O'Connell and colleagues showed that retroviral mediated overexpression of miR-125b in bone marrow cells followed by transplantation into lethally irradiated recipient mice leads to a myeloproliferative disorder that progresses to AML [O'Connell et al. 2008]. The early phase of this disease is characterized by an increase in monocytes and neutrophils and a decrease in lymphocytes, red blood cells, and platelets, indicating that an early effect of miR-125b may be the induction of a GM lineage bias. While this phenotype could be due to miR-125b-induced expansion of myeloid-biased HSCs, this possibility has not yet been investigated. In addition, because the bone marrow cells were not purified prior to retroviral transduction, sufficiency experiments with specific progenitor cells could not be performed. Transplantation of bone marrow from AML-bearing mice into secondary recipients led to their death from leukemia rapidly, indicating that the AML possesses a LSC population. Whether or not such AMLs depend on miR-125b for maintenance remains unclear.
The myelodysplastic syndromes (MDS) are associated with a risk for progression to AML, and recent studies have suggested a role for miRNA dysregulation in this process. A common subtype of MDS characterized by an interstitial deletion of chromosome 5q is characterized by refractory anemia, variable neutropenia, and a normal or high platelet count with megakaryocytic dysplasia. Starczynowski and colleagues recently evaluated the expression of miRNAs located on the common deleted region on chromosome 5q, and identified miR-145 and miR-146a as consistently downregulated in MDS with del(5q). Retroviral mediated knockdown of these miRNAs using a miR decoy followed by transplantation into lethally irradiated mice led to the development of thrombocytosis, megakaryocytic dysplasia, and variable neutropenia, but not anemia [Starczynowski et al. 2010a]. When the mice were followed for longer periods of time, a subset subsequently developed myeloid malignancies (bone marrow failure, a myeloproliferative disorder, or acute leukemia) with a latency of 8 months [Starczynowski et al. 2010b]. Retroviral-mediated overexpression of TRAF6, a target of miR-146a, in mice led to the same thrombocytosis, megakaryocytic dysplasia, and neutropenia, as well as the development of a transplantable AML in a subset of animals. Furthermore, miR-146a has since been demonstrated to be downregulated in primary AML samples with both a normal karyotype and del(5q) compared with normal CD34+ cells (miR-145 was significantly downregulated in samples with a normal karyotype, but not del(5q)). Retroviral-mediated overexpression of miR-145 and miR-146a in the human promyelocytic leukemia cell line HL60, known to express these miRNAs at low levels, led to impairment of growth and increased apoptosis, further implicating these miRNAs in the pathogenesis of leukemic progression.
Future prospects
It is now apparent that miRNAs such as miR-155, miR-125a/b, and miR-29a regulate HSC function. Given the many shared properties of HSCs and LSCs, perhaps it is not surprising that these miRNAs that regulate HSC function also likely play important roles in the AML LSC. It is important to note, however, that while overexpression of some of these miRNAs in the normal hematopoietic system induces AML or myeloproliferative-like disorders, none of the published studies to date directly address the role of miRNAs in the LSCs. The use of in vitro colony forming assays and overexpression/knockdown studies in leukemic cell lines are likely to identify miRNAs that regulate AML cell growth, survival, and response to chemotherapy, but they may be insufficient to identify those miRNAs that regulate functionally defined properties such as self-renewal. Thus, demonstrating that these miRNAs are required for LSC self-renewal will require their assessment in serial transplantation assays using primary AML samples and immunodeficient mouse strains. Ideally, such functional assays would assess the ability of human AML LSCs to serially transplant following manipulation of miRNA expression in xenografted hosts using antagomirs or other strategies [Ma et al. 2010; Zhang et al. 2010; Gumireddy et al. 2008; Krutzfeldt et al. 2005]. As these studies are already underway in numerous labs, we expect many more stories of miRNA function in AML LSCs to emerge in the coming years.
Footnotes
C.Y. Park is supported by an award from the NCI/NIH (award number 3P30CA008748-44S5) and is a Fellow Scholar in Basic Science of the American Society of Hematology.
The authors declare no conflicts of interest in preparing this article.
References
- Argiropoulos B., Humphries R.K. (2007) Hox genes in hematopoiesis and leukemogenesis. Oncogene 26: 6766–6776 [DOI] [PubMed] [Google Scholar]
- Beerman I., Bhattacharya D., Zandi S., Sigvardsson M., Weissman I.L., Bryder D., et al. (2010) Functionally distinct hematopoietic stem cells modulate hematopoietic lineage potential during aging by a mechanism of clonal expansion. Proc Natl Acad Sci USA 107: 5465–5470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum W., Garzon R., Klisovic R.B., Schwind S., Walker A., Geyer S., et al. (2010) Clinical response and miR-29b predictive significance in older AML patients treated with a 10-day schedule of decitabine. Proc Natl Acad Sci USA 107: 7473–7478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet D., Dick J.E. (1997) Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 3: 730–737 [DOI] [PubMed] [Google Scholar]
- Bousquet M., Harris M.H., Zhou B., Lodish H.F. (2010) MicroRNA miR-125b causes leukemia. Proc Natl Acad Sci USA, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet M., Quelen C., Rosati R., Mansat-De V. Mas, La Starza R., Bastard C., et al. (2008) Myeloid cell differentiation arrest by miR-125b-1 in myelodysplastic syndrome and acute myeloid leukemia with the t(2;11)(p21;q23) translocation. J Exp Med 205: 2499–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruchova H., Yoon D., Agarwal A.M., Mendell J., Prchal J.T. (2007) Regulated expression of microRNAs in normal and polycythemia vera erythropoiesis. Exp Hematol 35: 1657–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin G.A., Dumitru C.D., Shimizu M., Bichi R., Zupo S., Noch E., et al. (2002) Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA 99: 15524–15529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammarata G., Augugliaro L., Salemi D., Agueli C., La Rosa M., Dagnino L., et al. (2010) Differential expression of specific microRNA and their targets in acute myeloid leukemia. Am J Hematol 85: 331–339 [DOI] [PubMed] [Google Scholar]
- Chang T.C., Wentzel E.A., Kent O.A., Ramachandran K., Mullendore M., Lee K.H., et al. (2007) Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell 26(5): 745–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Z., Li L., Lodish H.F., Bartel D.P. (2004) MicroRNAs modulate hematopoietic lineage differentiation. Science 303: 83–86 [DOI] [PubMed] [Google Scholar]
- Coskun E., von der Heide E.K., Schlee C., Kuhnl A., Gokbuget N., Hoelzer D., et al. (2010) The role of microRNA-196a and microRNA-196b as ERG regulators in acute myeloid leukemia and acute T-lymphoblastic leukemia. Leuk Res, in press. [DOI] [PubMed] [Google Scholar]
- Costinean S., Zanesi N., Pekarsky Y., Tili E., Volinia S., Heerema N., et al. (2006) Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci USA 103: 7024–7029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzio A., Passegue E., Ayton P.M., Karsunky H., Cleary M.L., Weissman I.L. (2003) Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev 17: 3029–3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce C.M. (2009) Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 10: 704–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debernardi S., Skoulakis S., Molloy G., Chaplin T., Dixon-McIver A., Young B.D. (2007) MicroRNA miR-181a correlates with morphological sub-class of acute myeloid leukaemia and the expression of its target genes in global genome-wide analysis. Leukemia 21: 912–916 [DOI] [PubMed] [Google Scholar]
- Dixon-McIver A., East P., Mein C.A., Cazier J.B., Molloy G., Chaplin T., et al. (2008) Distinctive patterns of microRNA expression associated with karyotype in acute myeloid leukaemia. PLoS One 3: e2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J.G., Sharp P.A. (2004) Specificity of microRNA target selection in translational repression. Genes Dev 18: 504–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiring A.M., Harb J.G., Neviani P., Garton C., Oaks J.J., Spizzo R., et al. (2010) miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell 140: 652–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eis P.S., Tam W., Sun L., Chadburn A., Li Z., Gomez M.F., et al. (2005) Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA 102: 3627–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazi F., Racanicchi S., Zardo G., Starnes L.M., Mancini M., Travaglini L., et al. (2007) Epigenetic silencing of the myelopoiesis regulator microRNA-223 by the AML1/ETO oncoprotein. Cancer Cell 12: 457–466 [DOI] [PubMed] [Google Scholar]
- Fazi F., Rosa A., Fatica A., Gelmetti V., Marchis M.L.D., Nervi C., et al. (2005) A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell 123: 819–831 [DOI] [PubMed] [Google Scholar]
- Felli N., Fontana L., Pelosi E., Botta R., Bonci D., Facchiano F., et al. (2005) MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci USA 102: 18081–18086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T., Fukuda Y., Kiga K., Sharif J., Hino K., Enomoto Y., et al. (2007) An evolutionarily conserved mechanism for microRNA-223 expression revealed by microRNA gene profiling. Cell 129: 617–621 [DOI] [PubMed] [Google Scholar]
- Garzon R., Fabbri M., Cimmino A., Calin G.A., Croce C.M. (2006a) MicroRNA expression and function in cancer. Trends Mol Med 12: 580–587 [DOI] [PubMed] [Google Scholar]
- Garzon R., Garofalo M., Martelli M.P., Briesewitz R., Wang L., Fernandez-Cymering C., et al. (2008a) Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci USA 105: 3945–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R., Heaphy C.E., Havelange V., Fabbri M., Volinia S., Tsao T., et al. (2009) MicroRNA 29b functions in acute myeloid leukemia. Blood 114: 5331–5341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R., Pichiorri F., Palumbo T., Iuliano R., Cimmino A., Aqeilan R., et al. (2006b) MicroRNA fingerprints during human megakaryocytopoiesis. Proc Natl Acad Sci USA 103: 5078–5083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R., Volinia S., Liu C.G., Fernandez-Cymering C., Palumbo T., Pichiorri F., et al. (2008b) MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood 111: 3183–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gefen N., Binder V., Zaliova M., Linka Y., Morrow M., Novosel A., et al. (2010) Hsa-miR-125b-2 is highly expressed in childhood ETV6/RUNX1 (TEL/AML1) leukemias and confers survival advantage to growth inhibitory signals independent of p53. Leukemia 24: 89–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgantas R.W., III, Hildreth R., Morisot S., Alder J., Liu C.G., Heimfeld S., et al. (2007) CD34+ hematopoietic stem-progenitor cell microRNA expression and function: A circuit diagram of differentiation control. Proc Natl Acad Sci USA 104: 2750–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory R.I., Chendrimada T.P., Cooch N., Shiekhattar R. (2005) Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123: 631–640 [DOI] [PubMed] [Google Scholar]
- Gumireddy K., Young D.D., Xiong X., Hogenesch J.B., Huang Q., Deiters A. (2008) Small-molecule inhibitors of microrna miR-21 function. Angew Chem Int Ed Engl 47: 7482–7484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S., Lu J., Schlanger R., Zhang H., Wang J.Y., Fox M.C., et al. (2010) MicroRNA miR-125a controls hematopoietic stem cell number. Proc Natl Acad Sci U SA 107: 14229–14234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y.-C., Park C.Y., Bhagat G., Zhang J., Wang Y., Fan J.-B., et al. (2010) microRNA-29a induces aberrant self-renewal capacity in hematopoietic progenitors, biased myeloid development, and acute myeloid leukemia. J Exp Med 207: 475–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havelange V., Garzon R., Croce C.M. (2009) MicroRNAs: New players in acute myeloid leukaemia. Br J Cancer 101: 743–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Thomson J.M., Hemann M.T., Hernando-Monge E., Mu D., Goodson S., et al. (2005) A microRNA polycistron as a potential human oncogene. Nature 435: 828–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H.W., Wentzel E.A., Mendell J.T. (2007) A hexanucleotide element directs microRNA nuclear import. Science 315: 97–100 [DOI] [PubMed] [Google Scholar]
- Isken F., Steffen B., Merk S., Dugas M., Markus B., Tidow N., et al. (2008) Identification of acute myeloid leukaemia associated microRNA expression patterns. Br J Haematol 140: 153–161 [DOI] [PubMed] [Google Scholar]
- Jamieson C.H., Ailles L.E., Dylla S.J., Muijtjens M., Jones C., Zehnder J.L., et al. (2004) Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med 351: 657–667 [DOI] [PubMed] [Google Scholar]
- Johnnidis J.B., Harris M.H., Wheeler R.T., Stehling-Sun S., Lam M.H., Kirak O., et al. (2008) Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature 451: 1125–1129 [DOI] [PubMed] [Google Scholar]
- Jongen-Lavrencic M., Sun S.M., Dijkstra M.K., Valk P.J., Lowenberg B. (2008) MicroRNA expression profiling in relation to the genetic heterogeneity of acute myeloid leukemia. Blood 111: 5078–5085 [DOI] [PubMed] [Google Scholar]
- Kaddar T., Chien W.W., Bertrand Y., Pages M.P., Rouault J.P., Salles G., et al. (2009) Prognostic value of miR-16 expression in childhood acute lymphoblastic leukemia relationships to normal and malignant lymphocyte proliferation. Leuk Res 33: 1217–1223 [DOI] [PubMed] [Google Scholar]
- Kluiver J., Kroesen B.J., Poppema S., van den Berg A. (2006) The role of microRNAs in normal hematopoiesis and hematopoietic malignancies. Leukemia 20: 1931–1936 [DOI] [PubMed] [Google Scholar]
- Kluiver J., Poppema S., de Jong D., Blokzijl T., Harms G., Jacobs S., et al. (2005) BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol 207: 243–249 [DOI] [PubMed] [Google Scholar]
- Klusmann J.-H., Li Z., Bohmer K., Maroz A., Koch M.L., Emmrich S., et al. (2010) miR-125b-2 is a potential oncomiR on human chromosome 21 in megakaryoblastic leukemia. Genes Dev 24: 478–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M., Wagers A.J., Manz M.G., Prohaska S.S., Scherer D.C., Beilhack G.F., et al. (2003) Biology of hematopoietic stem cells and progenitors: Implications for clinical application. Annu Rev Immunol 21: 759–806 [DOI] [PubMed] [Google Scholar]
- Krivtsov A.V., Twomey D., Feng Z., Stubbs M.C., Wang Y., Faber J., et al. (2006) Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature 442: 818–822 [DOI] [PubMed] [Google Scholar]
- Kroon E., Thorsteinsdottir U., Mayotte N., Nakamura T., Sauvageau G. (2001) NUP98-HOXA9 expression in hemopoietic stem cells induces chronic and acute myeloid leukemias in mice. EMBO J 20: 350–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzfeldt J., Rajewsky N., Braich R., Rajeev K.G., Tuschl T., Manoharan M., et al. (2005) Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438: 685–689 [DOI] [PubMed] [Google Scholar]
- Lapidot T., Sirard C., Vormoor J., Murdoch B., Hoang T., Caceres-Cortes J., et al. (1994) A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 367: 645–648 [DOI] [PubMed] [Google Scholar]
- Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., et al. (2003) The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419 [DOI] [PubMed] [Google Scholar]
- Li Z., Lu J., Sun M., Mi S., Zhang H., Luo R.T., et al. (2008) Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc Natl Acad Sci USA 105: 15535–15540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Elf S.E., Miyata Y., Sashida G., Huang G., Di Giandomenico S., et al. (2009) p53 regulates hematopoietic stem cell quiescence. Cell Stem Cell 4: 37–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., et al. (2005) MicroRNA expression profiles classify human cancers. Nature 435: 834–838 [DOI] [PubMed] [Google Scholar]
- Lu J., Guo S., Ebert B.L., Zhang H., Peng X., Bosco J., et al. (2008) MicroRNA-mediated control of cell fate in megakaryocyte-erythrocyte progenitors. Dev Cell 14: 843–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Reinhardt F., Pan E., Soutschek J., Bhat B., Marcusson E.G., et al. (2010) Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol 28: 341–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeti R., Becker M.W., Tian Q., Lee T.L., Yan X., Liu R., et al. (2009) Dysregulated gene expression networks in human acute myelogenous leukemia stem cells. Proc Natl Acad Sci USA 106: 3396–3401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malecova B., Morris K.V. (2010) Transcriptional gene silencing through epigenetic changes mediated by non-coding RNAs. Curr Opin Mol Ther 12: 214–222 [PMC free article] [PubMed] [Google Scholar]
- Marcucci G., Maharry K., Radmacher M.D., Mrozek K., Vukosavljevic T., Paschka P., et al. (2008a) Prognostic significance of, and gene and microRNA expression signatures associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: A Cancer and Leukemia Group B Study. J Clin Oncol 26: 5078–5087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci G., Radmacher M.D., Maharry K., Mrozek K., Ruppert A.S., Paschka P., et al. (2008b) MicroRNA expression in cytogenetically normal acute myeloid leukemia. N Engl J Med 358: 1919–1928 [DOI] [PubMed] [Google Scholar]
- Meng F., Henson R., Wehbe-Janek H., Ghoshal K., Jacob S.T., Patel T. (2007) MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 133: 647–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S., Li Z., Chen P., He C., Cao D., Elkahloun A., et al. (2010) Aberrant overexpression and function of the miR-17-92 cluster in MLL-rearranged acute leukemia. Proc Natl Acad Sci USA 107: 3710–3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman M.A., Hammond S.M. (2010) Emerging paradigms of regulated microRNA processing. Genes Dev 24: 1086–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell R.M., Chaudhuri A.A., Rao D.S., Baltimore D. (2009) Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci USA 106: 7113–7118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell R.M., Chaudhuri A.A., Rao D.S., Gibson W.S.J., Balazs A.B., Baltimore D. (2010) MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. Proc Natl Acad Sci USA 107: 14235–14240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell R.M., Rao D.S., Chaudhuri A.A., Boldin M.P., Taganov K.D., Nicoll J., et al. (2008) Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med 205: 585–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi A.G., Sahoo D., Adorno M., Wang Y., Weissman I.L., Park C.Y. (2010) MicroRNA-125b expands hematopoietic stem cells and enriches for the lymphoid-balanced and lymphoid-biased subsets. Proc Natl Acad Sci USA, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passegue E., Jamieson C.H., Ailles L.E., Weissman I.L. (2003) Normal and leukemic hema-topoiesis: Are leukemias a stem cell disorder or a reacquisition of stem cell characteristics? Proc Natl Acad Sci USA 100(Suppl. 1): 11842–11849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passegue E., Weisman I.L. (2005) Leukemic stem cells: Where do they come from? Stem Cell Rev 1: 181–188 [DOI] [PubMed] [Google Scholar]
- Peault B., Weissman I.L., Buckle A.M., Tsukamoto A., Baum C. (1993) Thy-1-expressing CD34+ human cells express multiple hematopoietic potentialities in vitro and in SCID-hu mice. Nouv Rev Fr Hematol 35: 91–93 [PubMed] [Google Scholar]
- Petriv O.I., Kuchenbauer F., Delaney A.D., Lecault V., White A., Kent D., et al. (2010) Comprehensive microRNA expression profiling of the hematopoietic hierarchy. Proc Natl Acad Sci USA 107: 15443–15448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic R., Riesbeck L.E., Velu C.S., Chaubey A., Zhang J., Achille N.J., et al. (2009) Regulation of miR-196b by MLL and its overexpression by MLL fusions contributes to immortalization. Blood 113: 3314–3322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulikkan J.A., Dengler V., Peramangalam P.S., Peer A.A. Zada, Muller-Tidow C., Bohlander S.K., et al. (2010) Cell-cycle regulator E2F1 and microRNA-223 comprise an autoregulatory negative feedback loop in acute myeloid leukemia. Blood 115: 1768–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raver-Shapira N., Marciano E., Meiri E., Spector Y., Rosenfeld N., Moskovits N., et al. (2007) Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell 26: 731–743 [DOI] [PubMed] [Google Scholar]
- Schotte D., Lange-Turenhout E.A., Stumpel D.J., Stam R.W., Buijs-Gladdines J.G., Meijerink J.P., et al. (2010) Expression of miR-196b is not exclusively MLL-driven but is especially linked to activation of HOXA genes in pediatric acute lymphoblastic leukemia. Haematologica 95: 1675–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H., Siomi M.C. (2010) Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell 38: 323–332 [DOI] [PubMed] [Google Scholar]
- Starczynowski D.T., Kuchenbauer F., Argiropoulos B., Sung S., Morin R., Muranyi A., et al. (2010a) Identification of miR-145 and miR-146a as mediators of the 5q- syndrome phenotype. Nat Med 16: 49–58 [DOI] [PubMed] [Google Scholar]
- Starczynowski D.T., Morin R., McPherson A., Lam J., Chari R., Wegrzyn J., et al. (2010b) Genome-wide identification of human microRNAs located in leukemia-associated genomic alterations. Blood, in press. [DOI] [PubMed] [Google Scholar]
- Tarasov V., Jung P., Verdoodt B., Lodygin D., Epanchintsev A., Menssen A., et al. (2007) Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: Mir-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle 6: 1586–1593 [DOI] [PubMed] [Google Scholar]
- Tassano E., Acquila M., Tavella E., Micalizzi C., Panarello C., Morerio C. (2010) MicroRNA-125b-1 and BLID upregulation resulting from a novel IGH translocation in childhood B-Cell precursor acute lymphoblastic leukemia. Genes Chromosomes Cancer 49: 682–687 [DOI] [PubMed] [Google Scholar]
- Thorsteinsdottir U., Sauvageau G., Hough M.R., Dragowska W., Lansdorp P.M., Lawrence H.J., et al. (1997) Overexpression of HOXA10 in murine hematopoietic cells perturbs both myeloid and lymphoid differentiation and leads to acute myeloid leukemia. Mol Cell Biol 17: 495–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilatou D., Papageorgiou S., Pappa V., Papageorgiou E., Dervenoulas J. (2010) The role of microRNAs in normal and malignant hematopoiesis. Eur J Haematol 84: 1–16 [DOI] [PubMed] [Google Scholar]
- Volinia S., Calin G.A., Liu C.G., Ambs S., Cimmino A., Petrocca F., et al. (2006) A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 103: 2257–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li Z., He C., Wang D., Yuan X., Chen J., et al. (2010) MicroRNAs expression signatures are associated with lineage and survival in acute leukemias. Blood Cells Mol Dis 44: 191–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman I.L. (2000) Stem cells: Units of development, units of regeneration, and units in evolution. Cell 100(1): 157–168 [DOI] [PubMed] [Google Scholar]
- Wilson A., Oser G.M., Jaworski M., Blanco-Bose W.E., Laurenti E., Adolphe C., et al. (2007) Dormant and self-renewing hematopoietic stem cells and their niches. Ann NY Acad Sci 1106: 64–75 [DOI] [PubMed] [Google Scholar]
- Winter J., Jung S., Keller S., Gregory R.I., Diederichs S. (2009) Many roads to maturity: Microrna biogenesis pathways and their regulation. Nat Cell Biol 11: 228–234 [DOI] [PubMed] [Google Scholar]
- Xiao C., Calado D.P., Galler G., Thai T.H., Patterson H.C., Wang J., et al. (2007) miR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell 131: 146–159 [DOI] [PubMed] [Google Scholar]
- Yekta S., Shih I.H., Bartel D.P. (2004) MicroRNA-directed cleavage of HOXB8 mRNA. Science 304: 594–596 [DOI] [PubMed] [Google Scholar]
- Yilmaz O.H., Valdez R., Theisen B.K., Guo W., Ferguson D.O., Wu H., et al. (2006) PTEN dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature 441: 475–482 [DOI] [PubMed] [Google Scholar]
- Zhang J., Grindley J.C., Yin T., Jayasinghe S., He X.C., Ross J.T., et al. (2006) PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature 441: 518–522 [DOI] [PubMed] [Google Scholar]
- Zhang S., Chen L., Jung E.J., Calin G.A. (2010) Targeting microRNAs with small molecules: From dream to reality. Clin Pharmacol Ther 87: 754–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Zuber J., Diaz-Flores E., Lintault L., Kogan S.C., Shannon K., et al. (2010) p53 loss promotes acute myeloid leukemia by enabling aberrant self-renewal. Genes Dev 24: 1389–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]