Abstract
The zebrafish maxillary barbel is an integumentary organ containing skin, glands, pigment cells, taste buds, nerves, and endothelial vessels. The maxillary barbel can regenerate (LeClair & Topczewski, 2010); however, little is know about its molecular regulation. We have studied FGF-related signaling molecules during barbel regeneration, comparing these to a well-known regenerating appendage, the zebrafish caudal fin. Multiple FGF ligands (fgf20a, fgf24), receptors (fgfr1–4) and downstream targets (pea3, il17d) are expressed in normal and regenerating barbel tissue, confirming FGF activation. To test if specific FGF pathways were required for barbel regeneration, we performed simultaneous barbel and caudal fin amputations in two temperature-dependent zebrafish lines. Zebrafish homozygous for a point mutation in fgf20a, a factor essential for caudal fin blastema formation, regrew maxillary barbels normally, indicating that the requirement for this ligand is appendage-specific. Global overexpression of a dominant negative FGF receptor, Tg(hsp70l:dn-fgfr1:EGFP)pd1 completely blocked fin outgrowth but only partially inhibited barbel outgrowth, suggesting reduced requirements for FGFs in barbel tissue. Maxillary barbels expressing dn-fgfr1 regenerated peripheral nerves, dermal connective tissue, endothelial tubes, and a glandular epithelium; in contrast to a recent report in which dn-fgfr1 overexpression blocks pharyngeal taste bud formation in zebrafish larvae (Kapsimali et al., 2011), we observed robust formation of calretinin-positive tastebuds. These are the first experiments to explore the molecular mechanisms of maxillary barbel regeneration. Our results suggest heterogeneous requirements for FGF signaling in the regeneration of different zebrafish appendages (caudal fin vs. maxillary barbel) and taste buds of different embryonic origin (pharyngeal endoderm vs. barbel ectoderm).
Keywords / MeSH Headings: Zebrafish, Regeneration, Signal Transduction, Receptor, Fibroblast Growth Factor, Type 1, FGF20a protein, zebrafish, HSP70 Heat-Shock Proteins, Wound Healing, Taste Buds
Introduction
The zebrafish, Danio rerio, is a popular model for studying morphological and molecular aspects of regeneration. In contrast to mammals, which have limited regenerative abilities (Gurtner et al., 2008), zebrafish can regenerate the retina (Becker and Becker, 2007), optic nerve (Becker and Becker, 2007), heart (Poss, 2007), kidney (Diep et al., 2011), spinal cord (Becker and Becker, 2008), the lateral line sensory system (Dufourcq et al., 2006) and the caudal fin (reviewed by Tal et al., 2009). Investigating the molecular similarities and differences among regenerating species, and among organs within those species, may reveal certain molecular regulators that are widely used. Alternatively, comparative regeneration studies can reveal unique mechanisms that may be restricted to certain clades, species, or cell lineages. Understanding these phenomena at the molecular level may help to explain or augment the limited regenerative capacity of other species, including our own.
The zebrafish caudal fin is intensively studied in regeneration research, leading the way in the broader field of vertebrate limb regeneration (Akimenko et al., 2003). The caudal fin is covered by a glandular, pigmented epithelium and contains 16–18 bony fin rays surrounded by nerves, fibroblasts, and vascular tissue. After amputation, the cut surface of the fin responds in a morphologically stepwise manner with five recognized stages: wound healing, epithelium thickening, blastema formation, regenerative outgrowth and termination. In the first stage, an apical epidermal cap (AEC) forms over the wound. This epithelial tissue thickens and secretes growth factors, prompting the adjacent mesenchymal cells to dedifferentiate, migrate, and proliferate (Nechiporuk and Keating, 2002). In the third stage, a distally projecting blastema forms, representing a population of progenitor cells (Akimenko et al., 1995). Regenerative outgrowth includes the division, migration and re-differentiation of these cells to replace the missing tissue types. The final step is termination, when the fin reaches its original size. As recently demonstrated, the caudal fin can repeat this process as often as necessary, accurately restoring appendage size and shape even if periodically amputated dozens of times (Azevedo et al., 2011).
Fibroblast growth factor (FGF) signaling is ubiquitous in many physiological and developmental processes, including multiple stages of fin regeneration (Barrientos et al., 2008; Bouzaffour et al., 2009; Wills et al., 2008; Yin and Poss, 2008). Up to 27 zebrafish FGFs have been identified, most of which are secreted (review by Wakahara et al., 2007). These extracellular ligands bind and activate transmembrane FGF receptors, causing dimerization among multiple receptor subtypes (e.g., fgfr1a, 1b, 2, 3 and 4) and activating downstream signaling. In 2005, it was shown that a regeneration-defective zebrafish phenotype, devoid of blastema (dob), was caused by a temperature-sensitive null mutation in the ligand fgf20a (Whitehead et al., 2005). During caudal fin regeneration, fish homozygous for this mutation formed an abnormal regenerative epithelium, did not produce a blastema, and failed to express typical blastema markers. Remarkably, this mutation did not affect embryonic development or any other aspect of the adult phenotype, suggesting it was a regeneration-specific molecule.
Also in 2005, a transgenic zebrafish line was generated in which FGF signaling could be broadly repressed by the heat-induced overexpression of a dominant-negative form of fibroblast growth factor receptor 1a (Lee et al., 2005). Fgfr1a is considered critical for FGF signaling because it is the receptor most highly expressed in the early stages of caudal fin regeneration, beginning in blastema cells 18 hours post surgery (Poss et al., 2000). Expression continues within both the proximal and distal blastema at 48 hours post surgery, and is maintained during later regenerate outgrowth. Consistent with this spatial and temporal expression, studies perturbing Fgfr1a signaling using heat-shock transgenics, small-molecule drugs or antisense-mediated knockdown have confirmed an essential role in fin regeneration. Inhibiting FGF signaling early in regeneration prevents blastema formation and decreases the expression of downstream genes such as muscle segment homeobox (msx) genes, which are considered markers of blastema cells; blocking FGF signaling late in regeneration affects cell distribution and proliferation (Poss et al., 2000; Thummel et al., 2006). The requirement for FGFs in zebrafish caudal fin regeneration is thus well established, with multiple mutant and transgenic lines available to manipulate this pathway.
The zebrafish maxillary barbel is a cranial sensory structure that has recently been studied for its regenerative potential (LeClair and Topczewski, 2010; Moore et al., 2012). At approximately 30–40 days post-fertilization, barbel buds extend from the maxillae and eventually form adult appendages that are 2–4 millimeters long (Fig. 1A). Smaller nasal barbels develop near the olfactory pits. Barbel appendages of varying construction can be found in other fishes, amphibians and reptiles (Fox, 1999); however, in zebrafish, the maxillary barbel remains optically transparent throughout the life cycle, allowing its internal structure to be studied in detail. After surgical amputation, barbels heal rapidly and appear to follow the classic morphological stages of zebrafish fin regeneration, including blastema formation, elongation, and termination (Fig. 1B). Although barbels and fins differ in size, shape, location, and function, both are epithelial-mesenchymal appendages that contain similar cell types, including glands, pigment cells, nerves, blood vessels and connective tissues (Fig. 1C). Therefore barbels and fins may both be useful models to examine the regenerative potential of these cell lineages and to make comparisons between similar structures within the same species. However, our current knowledge of barbel regeneration at the molecular level is very limited. It is not known what genes are expressed during maxillary barbel regeneration, what regulatory networks control this gene expression, and how these networks act in the barbel as compared to the fin.
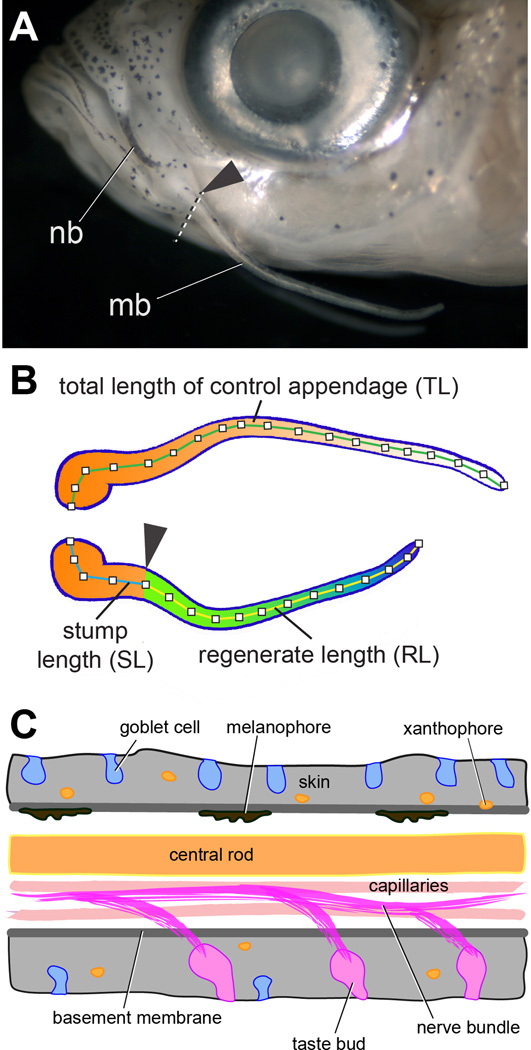
Figure 1. Overview of the zebrafish maxillary barbel (ZMB).
A) Position of the nasal barbel (nb) and maxillary barbel (mb) in a wild type adult zebrafish. The arrowhead and dotted line mark the approximate amputation plane for all barbel regeneration experiments.
B) Schematic of a matched pair of maxillary barbels after a regeneration experiment. Orange-white indicates original tissue of the contralateral control barbel (top) and the proximal stump of the amputated barbel (bottom). Green-blue indicates regenerated tissue. Segment labels correspond to the measurements obtained from digital images of the barbels, as described in the Methods.
C) Schematic diagram of cell types in the adult zebrafish maxillary barbel. Anterior is to the left. By convention, in this orientation the upper surface is ‘dorsal’ and the lower surface is ‘ventral’.
Given the well-established biological requirement for FGF signaling in zebrafish caudal fin regeneration, we hypothesized that this pathway would also be necessary for maxillary barbel regeneration. To support this hypothesis, we surveyed the expression of selected FGF ligands, receptors and downstream targets in normal and regenerating barbel tissue, demonstrating that many components of this pathway are active. To test the requirement for specific FGF signaling molecules during maxillary barbel regeneration, we used the fgf20azp3 mutant and dn-fgfr1-EGFPpd1 transgenic lines previously described, simultaneously administering both fin and barbel amputations under permissive and restrictive conditions. Homozygous fgf20a mutants failed to regenerate the caudal fin as predicted; however, barbel regeneration in these individuals was normal, indicating that the requirement for fgf20a is appendage-specific. Global heat shock of hemizygous dn-fgfr1 transgenics also abolished caudal fin regeneration, but only partially reduced barbel outgrowth in the same individuals, indicating that barbel regeneration may be less dependent on the FGF pathway. Finally, within the dn-fgfr1 line we examined the regeneration of maxillary barbel taste buds, which are abundant on the ventral surface and distal tip of this appendage. In contrast to larval pharyngeal taste buds, whose development was recently reported to be strongly FGF-dependent (Kapsimali et al., 2011), numerous calretinin-positive taste bud clusters regenerated equally well in maxillary barbels with or without induction of dn-fgfr1. These experiments provide the first molecular context for the regeneration of maxillary barbel tissue, and highlight substantial differences between the barbel and the caudal fin. The influence of FGFs also appears to diverge between larval taste bud differentiation and adult taste bud regeneration. These contexts may be used to explore why some tissues in an organism regenerate more readily than others, and how the molecular pathways of appendage regeneration have evolved.
Materials and Methods
Fish Husbandry
Zebrafish were reared in a laboratory colony on a 14 hr:10 hr light:dark cycle. Fish were crossed and reared at 28°C using standard methods (Westerfield, 2000). Routine adult fish density was kept below 1 fish/200 mL and all fish were fed live brine shrimp and/or commercial fish flakes once or twice daily for the duration of the study. All animal protocols were approved by the Institutional Animal Care and Use Committees (IACUCs) of DePaul University and/or Children’s Hospital of Chicago Research Center.
Surgical Procedures
All surgical procedures were performed under light anesthesia, using 0.015% buffered Tricaine (Sigma-Aldrich: E102521) in system water. For caudal fin clips, the distal half of the fin was removed with a razor blade. For barbel clips, the left maxillary barbel was removed with a pair of fine spring-scissors as previously described (LeClair and Topczewski, 2009); the right maxillary barbel was left intact as a control. After surgeries, fish were held in a crossing tank of system water plus methylene blue before being returned to the recirculating system or aquaria. For terminal tissue collection, fish were first anesthetized in Tricaine, then euthanized in ice water.
Fish Lines, DNA Extraction and Genotyping
Zebrafish of the wild type strain “AB” were obtained from stocks at Children’s Memorial Research Center. This strain was used for outcrossing and gene expression studies.
The fgf20azp3 zebrafish line was obtained from the Zebrafish International Resource Center (ZIRC #ZL1047) as a single clutch of ~100 embryos. 3–4 months later, selected adult fish were genotyped by caudal fin clips followed by observation of tail regeneration. Although 33°C is considered the optimal temperature for activating the regeneration deficiency in this line (fgf20azp3/zp3), we observed that many homozygous mutants had impaired tail regeneration at 28°C as previously reported (Whitehead et al., 2005). Genotypes were confirmed by PCR using a ZIRC-supplied thermal cycling protocol and primers specific for the fgf20azp3 mutation: forward primer (FGT_01d) = 5’ TTT GAG GAG AAT TGG AAC AAC ACT T 3’; reverse primer (FGT_02) = 5’ TTT TTG GGG TGG TTT TGA GTT T 3’. The forward primer (FGT_01d) contains a base pair mismatch (bold, underlined) adjacent to the fgf20azp3 mutation, creating an AloI restriction site only in amplicons from the mutated allele; in wild type PCR products, this site is not formed. Digestion of the PCR products with AloI yielded two fragments from mutants (207 and 39 base pairs long) or one fragment from wild types (246 base pairs long).
For DNA extraction, freshly dissected caudal fin tissue was placed in a PCR tube with 50 µL DNA extraction buffer (10 mM Tris pH 8.2, 10 mM EDTA, 200 mM NaCl, 0.5% SDS) and 200 µg/mL freshly added proteinase K. The tissue was digested at 55°C for 18 hours, heat-inactivated at 98°C for 10 minutes, and the DNA precipitated with ethanol. Pellets were resuspended in 20–50 µL Tris-EDTA (pH 8.0) and stored at −20°C.
The JumpStart REDTaq PCR Reaction kit (Sigma-Aldrich: P0982) was used in 20 µL reactions containing 10 µL RedTaq ReadyMix, 1 µL forward primer (0.5 µM), 1 µL reverse primer (0.5 µM), 1 µL tailfin DNA template (50 – 200 ng) and 7 µL nuclease-free distilled water. After thermal cycling, 8 µL of each PCR product was added to 2 µL (20 U) of AloI enzyme, 2 µL enzyme buffer and 8 µL distilled water. The reaction was incubated at 30°C overnight and 10 uL of the digest was run on a 50:50 3% agarose: 3% MetaPhor gel (Cambrex, Inc.) with ethidium bromide in TBE buffer for ~3 hours at 90 V. Products were visualized on a UV light box and captured as TIFF files. Confirmed homozygous mutant males were saved for later crosses (see “fgf20a Regeneration Experiments and Heat Treatments”, below).
The heat-shock inducible dominant negative fgfr1 line (Tg(hsp70l:dn-fgfr1-EGFP)pd1) was obtained from ZIRC (#ZL1476). These transgenic fish contain an insert with a heat-sensitive promoter (hsp70l) that is progressively activated at temperatures from 36–38°C (Shoji and Sato-Maeda, 2008), driving transcription of a modified FGFR1 that has the intracellular kinase domain replaced by enhanced green fluorescent protein (EGFP), allowing visual confirmation of expression. Adults were genotyped using the ZIRC-supplied PCR thermal cycling protocol and primers specific for EGFP: forward primer (EGFP6) = 5’ TTC TTC AAG TCC GCC ATG CCC G 3’; reverse primer (EGFP8) = 5’ GCA CGC TGC CGT CCT CGA TGT T 3’. Genomic DNA collection and PCR amplification were performed as above; amplified DNA was evaluated by gel electrophoresis on 0.8–1.2% TBE agarose gels with ethidium bromide. Surprisingly, 100% of the fish had a transgenic insertion confirmed by PCR (data not shown), therefore an in vivo heat-shock test was needed to identify those fish whose insertions were functional. dn-fgfr1 males of unknown genotype were outcrossed with wild type (AB) females and fertilized eggs were collected. After 3 days, a subsample of 10–12 embryos was transferred to a microcentrifuge tube of egg water and heat shocked at 37–38°C for 30 minutes in a water bath. Embryos were then incubated for 3 hours at 28°C in a Petri dish to allow for protein synthesis. Embryos were screened for EGFP using a Nikon Eclipse 50i fluorescent microscope. The tested males produced either 100% EGFP offspring, 50% EGFP offspring, or no EGFP offspring. Males that produced 50:50 offspring, indicating single, functional transgenic insertions, were separated to cross for future experiments.
RNA Extraction and Reverse Transcription Polymerase Chain Reaction (RT-PCR)
Total RNA isolation was performed for qualitative analyses of gene expression in wildtype tissues. Barbels or fin tissues were removed to a drop of RNALater (Invitrogen), trimmed, and then transferred in a minimal volume of fluid to the cap of a microfuge tube containing 250 µL RNA isolation reagent (RNAzol, Molecular Research Center). The tissue was pulse-spun to the bottom of this tube, then snap-frozen in liquid nitrogen. Tubes were then thawed on ice and the tissue was homogenized with a motorized mini-grinder (Kontes “Pellet Pestle”). Samples of similar tissues were then combined into larger aliquots and processed for total RNA using the Direct-zol RNA MiniPrep kit (Zymo Research). To avoid genomic contamination, RNA was treated with DNAse during this procedure. RNA concentrations were determined by measuring the absorbance at 260nm on a NanoDrop 1000c spectrophotometer. RNA aliquots were stored at −80°C until use. To visualize RNA integrity, 2–3 µL of each sample were run for 20 minutes at >200 V on a 1% lithium borate gel with ethidium bromide (Brody et al., 2004) and photographed on an AlphaInnotech gel imager.
RT-PCR primers were obtained from published articles or designed using NCBI Primer BLAST and Primer 3 bioinformatics programs. To confirm specificity, all primers were first tested on DNase-treated total RNA from wild type regenerating caudal fin; each reaction produced a predominant band of the expected size (not shown). RT-PCR reactions were assembled using the SuperScript III One-Step System with Platinum Taq (Invitrogen). A 10 uL reaction contained 50–100 ng total RNA, 5 µL SuperScript reaction mix, 0.4 µL SuperScript III RT/Platinum Taq Mix, 0.2 uL each of forward and reverse primers (10 mM), and RNase free water. Larger volumes were scaled up proportionately. The thermal cycler was set to perform a “touchdown PCR” from approximately 5°C above to −5°C below the Tm of each primer pair. Gel electrophoresis and imaging were performed as previously described.
fgf20a Regeneration Experiments and Heat Treatments
To test the necessity of fgf20a during maxillary barbel regeneration, we designed an experiment to challenge adult homozygous mutants (fgf20azp3/zp3) and heterozygous sibling controls (fgf20a+/zp3) to regenerate both maxillary barbels and caudal fins under either permissive or restrictive temperatures (Fig. 2). In this design, barbel regeneration was the desired response variable and caudal fin regeneration was an in vivo internal control. Confirmed homozygous mutant males (Fig. 2A) were outcrossed to wild type females (Fig. 2B) to create 100% heterozygous offspring. After 3–4 months, heterozygous females were backcrossed to their homozygous fathers to create 50:50 homozygous recessive: heterozygous clutches (Fig. 2C). These offspring clutches were not genotyped, keeping the experimenter blind; probabilistically, however, we expected half to be regeneration-deficient. 3–6 months post fertilization, offspring fish received simultaneous barbel and caudal fin amputations followed by 24 hours of recovery in 28°C water (Fig. 2D). These fish were then haphazardly split between a 33°C aquarium, the restrictive temperature for the fgf20azp3 allele (Whitehead et al., 2005), and a 28°C aquarium, representing standard zebrafish rearing temperatures (Fig. 2E). To maintain constant temperature, the 33°C tank was wrapped on three sides with foam insulation. Both aquaria were otherwise identical, and received the same daily feedings and periodic water changes. After 14 days, all of the fish were euthanized and their tissues collected for morphological analysis (Fig. 2F).
Figure 2. Diagram of the regeneration experiment using the temperature-sensitive fgf20azp3 mutant zebrafish line.
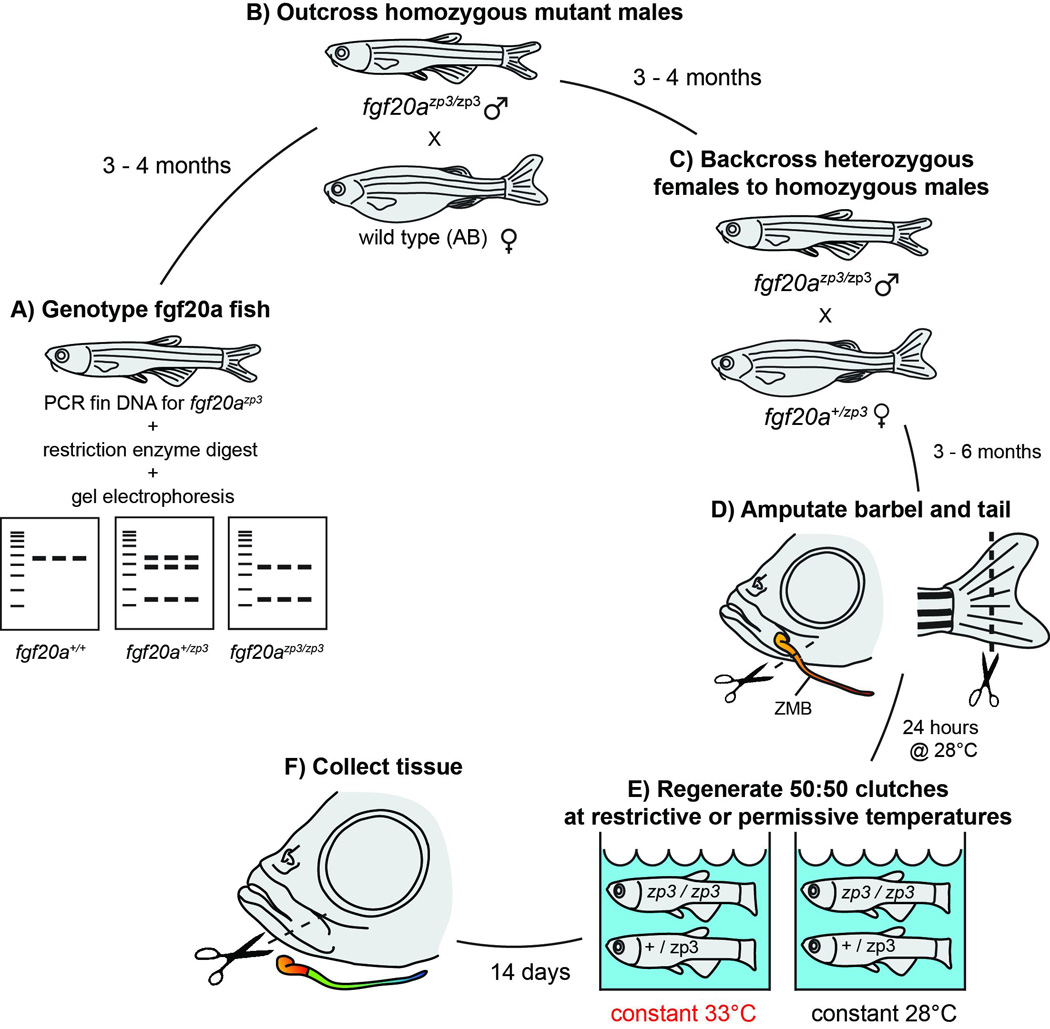
fgf20azp3 fish were genotyped by performing caudal fin amputations and collecting DNA for PCR. A restriction enzyme digest of the PCR products (A) produced smaller fragments in mutant carriers (for details see Methods). Homozygous recessive males were crossed to wild type females (B), generating 100% heterozygotes. Heterozygous female offspring were then back crossed to their fathers (C) to produce 50:50::homozygous: heterozygous clutches. Offspring were reared 3–6 months, after which the caudal fin and left maxillary barbel were removed (D); the right maxillary barbel was left intact as a control. After 24 hours at 28°C to allow for wound healing, fish were haphazardly split between 2 tanks, one at a constant 33°C and one at a constant 28°C (E). Barbels and caudal fins were allowed to regenerate for 14 days, then collected for analysis (F).
hsp70l:dn-fgfr1-EGFP Regeneration Experiments and Heat-Shock Apparatus
To test the necessity of fgfr1 during maxillary barbel regeneration, we designed a similar mixed-clutch, simultaneous-regeneration experiment (Fig. 3). Confirmed Tg(hsp70l:dn-fgfr1-EGFP)pd1 hemizygous males were outcrossed to wildtype females (Fig. 3A). A subsample of each clutch (8–10 larvae) was heat-shocked as described to confirm 50:50 transgenic:wildtype populations (Fig. 3B). The remaining embryos, which were not exposed to elevated water temperatures, were allowed to grow to 3–6 months old. 58 fish from multiple, pooled 50:50 clutches had simultaneous fin and barbel clips performed as described, and were allowed to recover for 24 hours post surgery in 28°C water (Fig. 3C). These fish were then haphazardly split into four 2.8L tanks on an Aquaneering housing rack, two with heat-shock treatment and two without (Fig. 3D). The dominant negative fgfr1-EGFP construct is most strongly activated when water temperatures are above 37°C (Lee et al., 2005); to achieve these temperatures, fish regenerating under heat shock (HS) conditions had their flow-through tanks supplemented with an in-rack heating system (Duszynski et al., 2011). In this system, an outlet timer turned on a calibrated submersible heating element for approximately 2 hours per day for 14 days: one hour to gradually heat the water, and another hour to hold peak temperature. Fish regenerating under “no heat shock” (NHS) conditions were kept in adjacent 2.8-liter tanks with no heaters; the system water temperature was constant at approximately 28°C. Flow rates in all four tanks were kept on a low drip (approximately 10 mL/min) for the duration of the experiment. The water temperatures in each heat-shock tank were recorded every 30–60 seconds by a submersible temperature probe connected to a battery-powered data logger (HOBO U12 Logger, Onset Computer Corporation). Four hours after the last heat shock cycle, all of the fish were euthanized and their tissues were collected for morphological analysis (Fig. 3E).
Figure 3. Diagram of the regeneration experiment using the dn-fgfr1 transgenic zebrafish line.
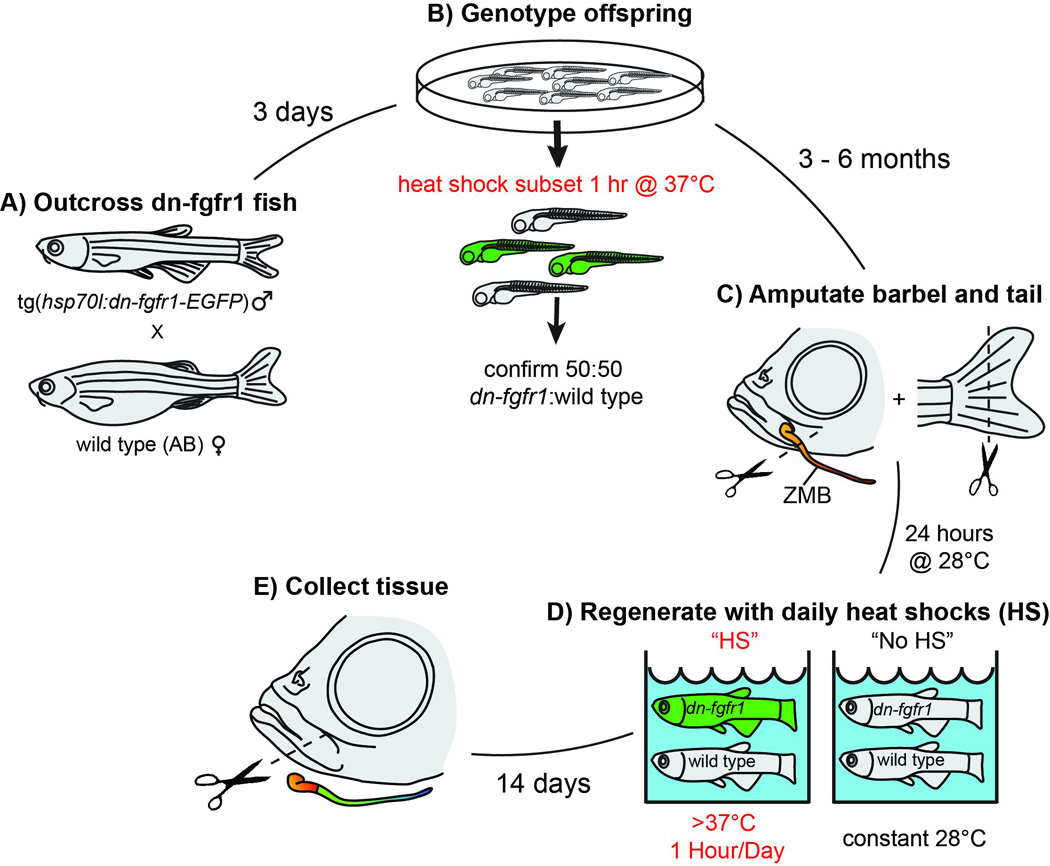
Tg(hsp70l:dn-fgfr1-EGFP) fish were genotyped by outcrossing adult males to wild type females (A) and screening a subsample of embryos for EGFP expression after a 1-hour heat-shock treatment (B). Clutches of 50:50::hemizygous:wild type fish were reared for 3–6 months, after which the caudal fin and left maxillary barbel were removed (C); the right maxillary barbel was left intact as a control. After 24 hours at 28°C to allow for wound healing, fish were haphazardly split between heat-shock tanks (HS = >37°C for one hour/day) and no heat-shock tanks (NHS = constant 28°C; D). Barbels and caudal fins were allowed to regenerate for 14 days, then collected for analysis (E).
Linear Measurements of Barbel Regeneration
As fish were collected, a small piece of fresh dorsal fin was removed and frozen for genotype confirmation, after which each fish was fixed whole in 4% paraformaldehyde-phosphate buffered saline (PF-PBS) overnight. Fixed tissue was rinsed in 1× PBS and stored in 70% ethanol until analysis. To minimize experimenter bias, we used a randomization and blinding protocol between tissue collection and measurement. As each fish was fixed, it was placed in a tube labeled only with an integer number. The treatment tank for each fish was recorded, and this information was kept secure by one investigator (EEL). The second investigator (RJD) received the numbered tubes and analyzed these samples in an order dictated by a random number generator (www.random.org); this investigator was thus blind to both treatment group and genotype during analysis. Only after all measurements were collected was the treatment group revealed.
The left and right maxillary barbels of adult zebrafish are highly symmetrical, differing by <2% in length, with a portion of this variation being human measurement error (unpublished data). The length of the right, unamputated barbel was thus assumed to represent the original length of the left, amputated barbel. Matched pairs of barbels were photographed in saline and digitized using Image J (National Institutes of Health, available at http://rsbweb.nih.gov/ij/). The amount of regeneration was measured as both an absolute distance (regenerate length, in millimeters past the amputation plane) and as a percent of the contralateral control using a previously published formula (LeClair and Topczewski, 2010). A schematic of these measurements is shown in Figure 1B. The segmented line tool in ImageJ was used to plot manually selected points (white squares) along the midline of each appendage. Total length (TL) was measured on the control barbel from the proximal end of the central rod to the barbel tip. Stump length (SL) was measured on the regenerated barbel from the proximal end of the central rod to the amputation plane (black arrowhead). Regeneration length (RL) was measured on the regenerated barbel from the amputation plane to the barbel tip. Raw data were exported to Prism 5 software (Graphpad) for graphing and statistical analysis. Between-group equality of variances was first assessed using the F-test; datasets passing these screens were analyzed parametrically using Student’s t. To account for multiple tests and maintain an overall Type I error rate of 0.05, the level required to declare statistical significance was revised downwards using the Bonferroni correction: p = 0.05/5 = 0.01, where 5 = the number of post hoc tests (Sokal and Rohlf, 2011).
Maxillary Barbel Taste Bud Regeneration Assay
To examine adult taste bud regeneration under heat-shock conditions, we performed maxillary barbel amputations on a second group of fish from the inducible dn-fgfr1:EGFP transgenic line. Fifty adult fish from a 50:50 wild type:hemizygous clutch had simultaneous barbel and tail amputations, an overnight recovery, and then 14 days of once-daily heat shock (>37.5°) using the semi-automated heat shock system. In this experiment, we omitted the non-heated controls. All of the fish survived in good health. Four hours after the last heat shock, the fish were euthanized and fixed in 4% paraformaldehyde/PBS overnight at 4°C. The following day, the fish were screened for the presence/absence of a regenerated caudal fin and global EGFP expression. 100% of the fish lacking tail regeneration (26/50) were also EGFP-positive, confirming both genotype and the heat-shock activation of the transgene (not shown). After sorting, the caudal fins, control barbel and regenerated barbel of each fish were removed into a Petri dish of saline and photographed under magnification with a micrometer scale. All same-genotype barbels (EGFP− or EGFP+) were then pooled and processed simultaneously for immunohistochemistry.
Barbel tissues were washed three times in phosphate-buffered saline + 0.5% Triton-X (PBS-TrX), blocked at least 2 hours in PBS-TrX + 10% goat serum, and incubated overnight with a rabbit anti-calretinin primary antibody (Millipore #AB5054) diluted 1:1,000 – 1:2,000 in incubation solution (PBS-TrX + 2% DMSO + 2% goat serum). After 5–6 rinses in PBS-TrX, a fluorescent red secondary was applied (anti-rabbit Cy3, Jackson ImmunoResearch) diluted 1:400 in incubation solution. This incubation was for 2 hours at room temperature. After 5–6 rinses in PBS-TrX, the tissues were mounted on glass slides in CFM (Electron Microscopy Sciences) with DAPI as a nuclear counterstain. Comparable image stacks from EGFP+ and EGFP− barbels (regenerates and controls) were acquired on a Leica SPE confocal microscope. To obtain maximum resolution of taste bud fine structure, selected scans were deconvoluted using Huygens© software (version 4.1, Scientific Volume Imaging).
Results
Multiple FGF signaling molecules are expressed in adult maxillary barbels and in the distal part of the regenerating barbel
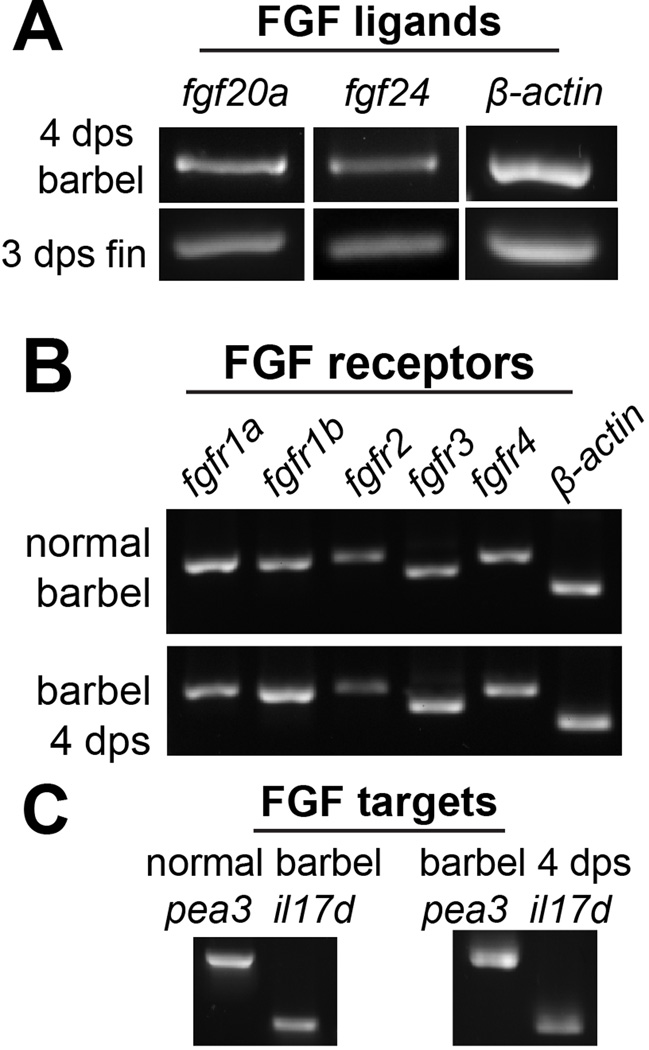
As previously reviewed, multiple FGF ligands, receptors, and downstream targets are expressed during caudal fin regeneration. However there is no published data on this pathway in barbel tissue. We therefore designed a set of polymerase chain reaction (PCR) primers targeting well-known components of FGF signaling (Table 1) and confirmed their specificity on reverse-transcribed total mRNA from regenerating caudal fin blastemae (not shown). The same reaction conditions were then used to screen for the expression of each mRNA in wild type adult maxillary barbel tissue, and in the distal portion of regenerating maxillary barbels at 4 days post surgery (4 dps; Fig. 4). All of the FGF-related genes were amplified, including the ligands fgf20a and fgf24, the FGF receptors fgfr1a, 1b, 2, 3 and 4, and downstream signaling targets pea3 (ETS-domain transcription factor pea3: Brown et al., 1998) and il17rd (interleukin 17 receptor D il17rd: Tsang et al., 2002). These detection experiments were repeated on two independent RNA extractions, with similar results (data not shown). Thus the maxillary barbel, like the caudal fin, expresses multiple FGF signaling members both constitutively and during the early stages of regeneration.
Table 1. Primers for reverse transcription polymerase chain reaction (RT-PCR) detection of FGF receptors, ligands and downstream targets.
Product sizes and specificity were predicted using NCBI’s Primer BLAST on zebrafish RefSeq RNA.
| Gene | ZBD-GENE | Primer | Sequence 5’-3’ | predicted size (bp) | |
|---|---|---|---|---|---|
| fgf20a | 060110-1 | fgf20a-F1 fgf20a-R1 |
GCAGATTTGGTATATTGGAATTCAT CTAGAACATCCTTGTAAAGCTCAGG (Stoick-Cooper et al., 2007) |
348 | |
| fgf24 | 030708-1 | fgf24-F1 fgf24-R1 |
GCAAGAAGATTAACGCCAATGG TTTAGGTCGACCCTTTCG |
272 | |
| fgfr1a | 980526-255 | fgfr1a-F1 fgfr1a-R1 |
AAACCGTGAAGTTTCGTTGC TCACCTCGATGTGTTTCAGC |
372 | |
| fgfr1b | 060503-14 | fgfr1b-F1 fgfr1b-R1 |
AACGTGTCCTTTGAGGATGC CTGGAGGAGAGACGTGAAGG |
365 | |
| fgfr2 | 030323-1 | fgfr2-F1 fgfr2-R1 |
TTGGCGGGTAACTCTATTGG TCAGGGAGGTCGTATTCTGG |
371 | |
| fgfr3 | 000816-1 | fgfr3-F1 fgfr3-R1 |
ACTGGACAGAGTCGGACAGC CAGACATCCTCAACCGTTCC |
321 | |
| fgfr4 | 980526-488 | fgfr4-F1 fgfr4-R1 |
GCTTACCAAGTGGCAAGAGG TGCAGTTGGAGGGTTTATCC |
367 | |
| il17rd | 020320-5 | il17rd-F3 il17rd-R3 |
CAGAGATTCGGGGATTTACG TGGCGCTGCGAGTGGAGCTG |
302 | |
| pea3 | 990415-71 | pea3-F1 pea3-R1 |
GCCTGGCTGCCCATCCATGT GCAATAGGTGCAGCGGGTGGT |
674 | |
Figure 4. FGF-related signaling molecules are expressed in normal and regenerating maxillary barbel tissue.
A) Reverse-transcription polymerase chain reaction (RT-PCR) on DNase-treated total RNA from distal regenerating maxillary barbels (4 days post surgery, dps) and distal regenerating caudal fins (3 days post surgery, dps) shows similar expression of fgf20a and fgf24 ligands. Normal and regenerating barbel tissues express similar FGF receptors (B) and downstream target molecules (C).
The ligand Fgf20a is essential for caudal fin, but not barbel regeneration
The fgf20a zebrafish line contains a temperature-sensitive null mutation that at 33°C abolishes caudal fin blastema formation, preventing regenerative outgrowth (Whitehead et al., 2005). To test if Fgf20a was also essential for maxillary barbel regeneration, we challenged 50:50 clutches of heterozygous (fgf20a+/zp3, hereafter called ‘wild types’) and homozygous recessive zebrafish (fgf20azp3/zp3, hereafter called ‘fgf20a mutants’) with simultaneous barbel and tail amputations and housed them under restrictive or control temperatures for 14 days (33°C vs. 28°C, Fig. 2). If fgf20a were essential for the regeneration of both types of appendages, we expected both to be equally inhibited at the restrictive temperature. In the original mutant screen, 72% of fgf20a mutants showed fin regeneration at 28°C; therefore, we also expected that fin and/or barbel regeneration in these animals might be partially inhibited at the control temperature. In contrast, wild type siblings were expected to regenerate normally at both temperatures, although possibly at slightly different rates.
At the restrictive temperature (33°C), all wildtype siblings regenerated caudal fins normally (n = 18; Fig. 5A). All fgf20a mutants held in the same tank, however, had substantial caudal fin defects (n = 13; Fig. 5B). Six out these 13 mutants (46%) had only a thin epithelium covering the fin wound, and there was no visible blastema or any outgrowth. The other seven mutants (54%) were very similar but had regenerated one or two short, abnormal fin rays. At the permissive temperature (28°C), all wild type siblings had normal tail regeneration (n = 20); however all fgf20a mutant tails were grossly abnormal (n = 14, data not shown). Four of the 14 mutants (29%) showed complete inhibition of fin regeneration, similar to the restrictive condition. The other 10 mutants (71%) partially regenerated, but the regenerated portion of the fin was opaque, disorganized, and only half the size of wild type regenerates (not shown). These results indicate that the conditions in our aquaria were able to completely (at 33°C) or partially (at 28°C) suppress Fgf20a signaling in 100% of homozygous recessive individuals, replicating previously described caudal fin defects in this mutant line.
Figure 5. The temperature sensitive mutation fgf20azp3 has no effect on maxillary barbel regeneration.
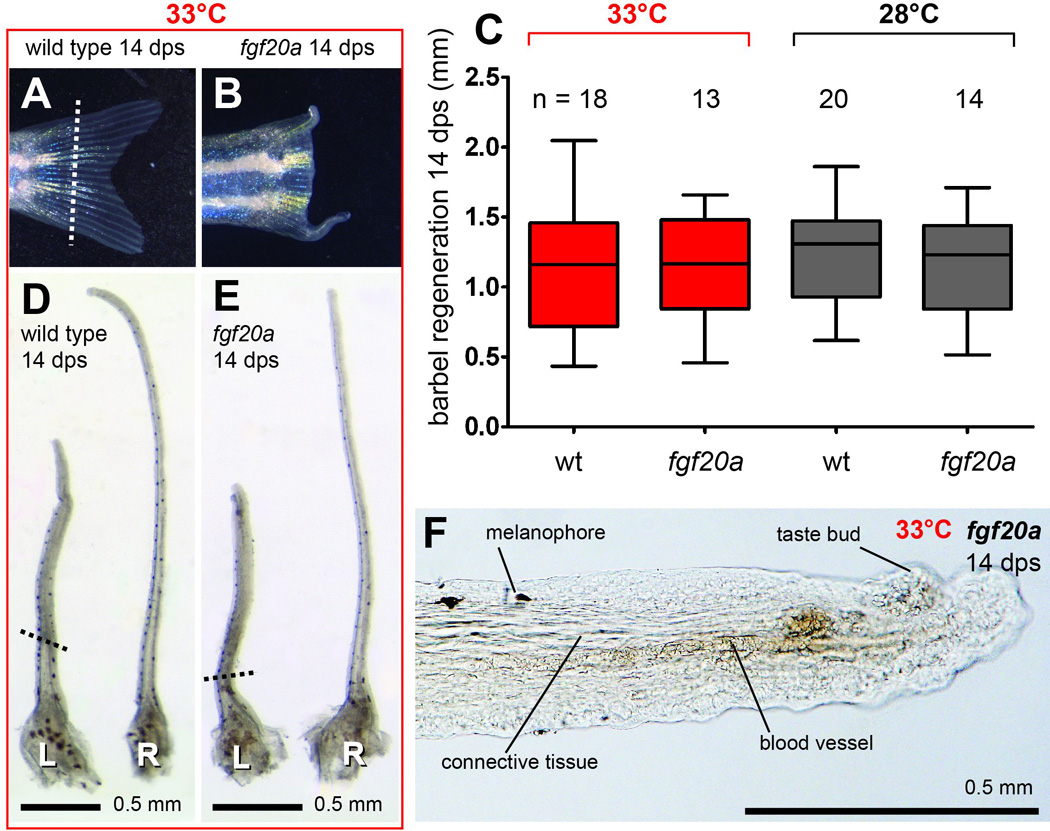
Wild type zebrafish (fgf20azp3/+) at 33°C regenerated caudal fins normally (A), while homozygous recessive siblings (fgf20azp3/zp3) held in the same tank lacked outgrowth (B). The white dotted line indicates the caudal fin amputation plane. C) 14 days post surgery (dps), there was no effect of genotype on maxillary barbel regeneration at either experimental temperature (33°C, red or 28°C, black). Regeneration was measured as millimeters past the amputation plane. D–E) Paired wild type (D) and homozygous recessive (E) maxillary barbels treated at 33°C for 14 days, showing similar gross morphology. L = left, amputated barbel; R = right, unamputated barbel. The black dotted lines indicate the barbel amputation planes. These barbels are from the same animals whose caudal fins are shown in panels A and B, respectively; both fish were of similar body size. F) Magnification of the distal end of the regenerated barbel in panel E, showing normally arranged connective tissue, blood vessels, melanophores and taste bud hillocks.
In contrast to the dramatic effects on caudal fin regeneration in fgf20a mutants, neither restrictive nor control temperature conditions had any effect on maxillary barbel regeneration. After 14 days at 33°C, the amputated maxillary barbels of fgf20a mutants produced on average approximately 1.2 mm of tissue distal to the amputation plane, corresponding to a recovery of 41.3% of the contralateral control appendage (Fig. 5C, Table 2A). Amputated maxillary barbels in wild type siblings produced on average 1.1 mm of distal tissue, or 42.9% of the contralateral control appendage. This was a highly non-significant difference (HS-fgf20a vs. HS-wt at 33°C; t = 0.22, df = 29, p = 0.82). Morphologically, the regenerated barbels of mutants and wild types held at 33°C were indistinguishable (Fig. 5D,E). Although shorter than the contralateral controls, all of the regenerated appendages had glandular skin, dorsally-located melanophores, abundant taste bud hillocks, erythrocyte-filled capillaries and extensive dermal connective tissue (Fig. 5F). Similar quantitative and qualitative results were observed at the permissive temperature (28°C). Although this temperature also inhibited caudal fin regeneration in 100% of the fgf20a mutant individuals, these same fish regrew barbels just as robustly as their wild type siblings (HS-fgf20a vs. HS-wt at 28°C; 1.1 m vs. 1.2 mm; t = 0.68; df = 32; p = 0.49). Although there were no significant differences in barbel regenerate length among groups, there was substantial individual variation, with up to a twofold difference among fish of the same genotype within a treatment. However, the levels of variation in each treatment group were similar (F-test for equality of variances; HS-wt vs. HS-fgf20a at 33°C: F = 1.5, df = (17,12), p = 0.44; HS-wt vs. HS-fgf20a at 28°C: F = 1.1, df = 13,19, p = 0.80). Finally, we observed that the absolute amount of barbel tissue regenerated at 28°C was almost identical to that regenerated at 33°C regardless of genotype, suggesting that this temperature differential does not strongly affect the amount of new barbel tissue obtained after 14 days.
Table 2. Morphological measurements of maxillary barbel regeneration.
After 14 days at the indicated treatments, barbel regeneration past the amputation plane was measured in millimeters (see Methods & Figure 1B). Boxplots of these data are graphed in Figures 5, 6 and 8, respectively.
| A. fgf20a regeneration experiments at restrictive (33°C) and ‘permissive’ (28°C) temperatures | |||||
|---|---|---|---|---|---|
| treatment | genotype | N | mean regenerated barbel length (mm) |
range (mm) | 95% CI of mean |
| 33°C | wild type | 18 | 1.1 | 0.43 – 2.00 | 0.91 – 1.40 |
| 33°C | fgf20azp3/zp3 | 13 | 1.2 | 0.46 – 1.70 | 0.95 – 1.40 |
| 28°C | wild type | 20 | 1.2 | 0.62 – 1.90 | 1.10 – 1.40 |
| 28°C | fgf20azp3/zp3 | 14 | 1.1 | 0.52 – 1.70 | 0.94 – 1.40 |
| B. dn-fgfr1 barbel regeneration experiments (HS = heat shock; NHS = no heat shock) | |||||
|---|---|---|---|---|---|
| treatment | genotype | N | mean regenerated barbel length (mm) |
range (mm) | 95% CI of mean |
| HS | wild type | 14 | 1.50 | 0.83 – 2.00 | 1.30 – 1.70 |
| HS | dn-fgfr1 | 14 | 0.70 | 0.08 – 1.40 | 0.46 – 0.95 |
| NHS | dn-fgfr1 + wild type | 30 | 1.70 | 0.92 – 2.10 | 1.60 – 1.80 |
| C. dn-fgfr1 barbel and taste bud regeneration experiments (HS = heat shock) | |||||
|---|---|---|---|---|---|
| treatment | genotype | N | mean regenerated barbel length (mm) |
range (mm) | 95% CI of mean |
| HS | wild type | 24 | 1.10 | 0.52 – 1.60 | 1.30 – 1.70 |
| HS | dn-fgfr1 | 26 | 0.57 | 0.10 – 1.10 | 0.47 – 0.67 |
Heat shock activation of hsp70l:dn-fgfr1 reduces, but does not prevent, maxillary barbel outgrowth
Having observed that maxillary barbels could regenerate independently of fgf20a, we next wished to see if the entire FGF pathway was dispensable for the regeneration of this appendage. To do this, we used a dominant-negative Fgfr1 transgenic zebrafish line (Tg(hsp70l:dn-fgfr1-EGFP)pd1) to globally restrict FGF signaling in vivo (Fig. 3). A once-daily heat shock treatment (1 hr of water temperatures >37°C) was expected to block all outgrowth of the caudal fin in hsp70l:dn-fgfr1 hemizygotes (hereafter called ‘dn-fgfr1’ fish) and simultaneously induce EGFP expression, in contrast to wild type sibling controls. If FGF signaling were necessary for barbel regeneration, we expected barbel outgrowth to be blocked in the same hemizygous individuals. Conversely, if FGF signaling were not necessary for barbel regeneration, we would expect to see fin regeneration halted, but barbels outgrow normally. Finally, we expected that all wild type fish receiving heat shocks, plus all non-heat shock controls (wild type + dn-fgfr1:EGFP) would regenerate normally both fin and barbel appendages.
As expected, heat-shocked hemizygous dn-fgfr1 zebrafish showed a complete block of caudal fin regeneration (Fig. 6A). 14 days after surgery, 100% of these fish (n =14) had only a thin layer of epithelium covering the amputated fin rays (< 0.1 mm), having failed to regenerate any structures past the amputation plane. These animals also showed strong whole-body EGFP expression, visually confirming transgene activation (Fig. 6B). In contrast, all of the wildtype fish receiving heat shocks, and all of the (wild type + dn-fgfr1) fish not receiving heat shocks regenerated the entire caudal fin normally (data not shown). Thus, we were able in our heat-shock tanks to maintain the conditions necessary to induce dn-fgfr1 expression and maintain a strong in vivo blockade of caudal fin regeneration for 14 days.
Figure 6. Heat-shock induction of dn-fgfr1 reduces, but does not prevent, maxillary barbel regeneration.
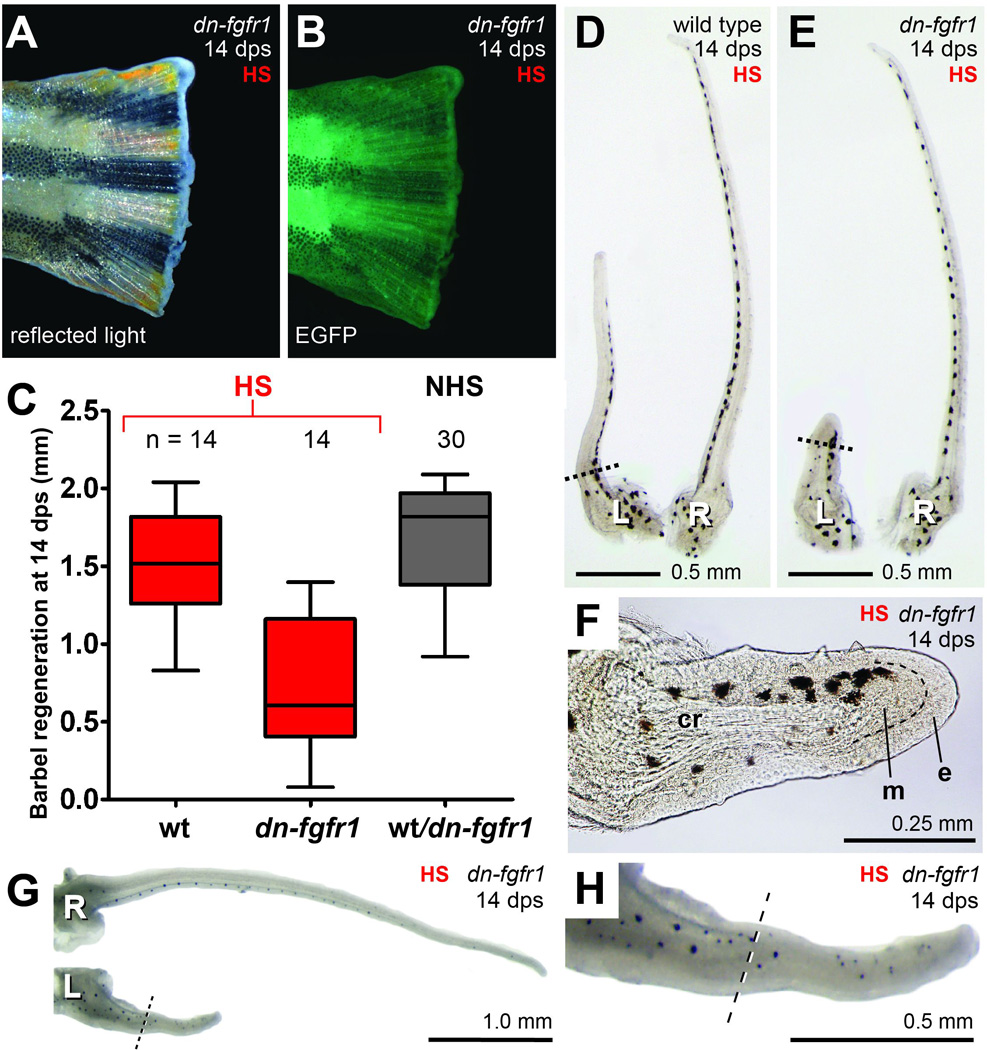
White light (A) and fluorescent (B) illumination of an amputated hemizygous dn-fgfr1:EGFP caudal fin after 14 days of daily heat shock (> 37°C). The fin tissue shows inhibition of regeneration and enhanced green fluorescent protein (EGFP) expression. C) Comparison of maxillary barbel regeneration among heat-shocked wild type (red, wt), heat-shocked hemizygous (red, dn-fgfr1) and non-heat-shocked mixed zebrafish (black, wt & dn-fgfr1). In the absence of heat shock, wild type and hemizygous fish of this line cannot be distinguished and are therefore grouped together. Matched pairs of wild type (D) and hemizygous dn-fgfr1 (E) barbels after 14 days of daily heat shock. The transgenic animal in this example had strongly inhibited barbel regeneration. L = left, amputated barbel; R = right, unamputated barbel. F) Magnification of the regenerated barbel shown in E; although there is little regrowth, a small mesenchymal bulge (m) projects past the amputation plane. e = epithelial cap. G). The average amount of barbel regeneration observed in heat-shocked dn-fgfr1 animals (~0.5 mm). These barbels had more robust tissue outgrowths, including melanophores, a vascular loop, and abundant taste bud hillocks well past the amputation plane (H).
Under these conditions, maxillary barbel regeneration in dn-fgfr1 fish was not blocked; however, the regenerates were reduced in length (Fig. 6C, Table 2B). dn-fgfr1 maxillary barbels produced on average only 0. 7 mm of new tissue past the amputation plane, compared to 1.5 mm of outgrowth for heat-shocked wild type barbels– a highly significant result (HS-dn-fgfr1 vs. HS-wt; t = 5.3; df = 26; p < 0.0001). As previously noted, within each treatment group there was considerable variability in the amount of barbel tissue produced. Subjectively, we divided the dn-fgfr1 barbels into three morphological categories. The first category (3/14 fish = 21%) accomplished little or no appendage elongation (<10% regrowth). These barbels had an intact epithelium covering the amputation plane, indicating wound healing, but few or no underlying mesenchymal cells (Fig. 6D–F). Most dn-fgfr1 barbels grew much more successfully, forming a second category (7/14 fish = 50%) that regenerated 16–25% of the contralateral control. These barbels all had differentiated structures well past the amputation plane, including extensive connective tissue, a complete blood vessel loop, protruding taste bud hillocks and scattered melanophores (Fig. 6G–H). The third category of regenerates (4/14 fish = 29%) was similar in morphology to the previous group, but had the longest regenerate lengths, nearly comparable to wild types (42–50% of the contralateral control). The variable response of maxillary barbel regeneration differed from the ‘all-or-nothing’ response of fin regeneration, which was completely blocked in all of the heat-shocked dn-fgfr1 individuals. Finally, there was no significant difference in the mean barbel regrowth of heat-shocked wild types and a mixed clutch of (wild type + dn-fgfr1) fish that received no heat shocks (HS-wt = 1.5 mm vs. HS-wt+dn-fgfr1 = 1.7 mm; t = 2.1; df=42, p = 0.045), suggesting that heat shocks by themselves do not dramatically affect barbel regeneration.
EGFP expression is maintained in maxillary barbel tissue at least 24 hours after heat shock in the hsp70l:dn-fgfr1:EGFP zebrafish line
Although one could interpret the absence of caudal fin regeneration and presence of whole-body EGFP expression as strong internal controls that the dominant negative FGFR1 construct was successfully expressed in all of the transgenic animals, we decided to examine maxillary barbel tissue more closely for the proper activation of this construct under the conditions of our experiment. The activation of a heat-shock promoter depends not only on temperature, but also on endogenous heat-shock proteins, which can vary in type and quantity among tissues (Basu et al., 2002; Krone et al., 1997). We therefore considered that maxillary barbel regeneration was not completely blocked in dn-fgfr1 zebrafish because the EGFP-tagged FGF receptor was not being adequately expressed or localized in barbel cells, an appendage which has not been examined previously in this line.
The onset and duration of maxillary barbel EGFP activation was assessed semiquantitatively by performing unilateral barbectomies and tail fin amputations on a group of adult dn-fgfr1 zebrafish, holding them overnight, and then heat shocking them once for one hour at approximately 37.5°C. Four to five fish were collected at the start of the experiment (0 hours = after surgery, but before heat shock) and similar subgroups were collected 1, 2, 4, 8 and 24 hours after heat shock. To eliminate autofluorescence due to aldehyde fixation, we examined freshly collected maxillary barbel and caudal fin tissues rinsed in PBS, mounted in glycerol and imaged immediately. Both the unamputated control barbel and amputated, regenerating barbel were examined on the same microscope slide.
EGFP expression first became faintly visible in dn-fgfr1 maxillary barbel tissue 2 hours post heat shock, and was obvious at 4 hours, with intense membrane localization in surface cells (Fig. 7A). The EGFP signals in dn-fgfr1 caudal fin tissues collected at the same time points were similar in intensity, as assessed by fixed manual exposures under the same fluorescent microscope (not shown). Using confocal microscopy to image through the normal barbel shaft, we were also able to confirm EGFP expression in the full thickness of the epithelium as well as deep cells adjacent to the acellular central rod (Fig. 7A’). 24 hours after heat shock, unamputated dn-fgfr1 maxillary barbels had persistent strong EGFP expression in cell types throughout the appendage, including the stratified squamous epithelium, taste bud clusters, peripheral nerve fibers, and the flattened endothelial cells of the capillary walls (Fig. 7B, B’). Amputated barbel stumps showed similar deep EGFP expression, notably in the mesenchymal layer just under the wound epithelium (Fig. 7C). These EGFP signals in the barbels of dn-fgfr1 fish were comparable to those observed at the healed edges of the amputated caudal fin rays (Fig. 7D). We conclude that the dn-fgfr1 construct is active in all cell types of the zebrafish maxillary barbel and that a one-hour heat shock (>37°C) is sufficient to maintain expression in this appendage for at least 24 hours. To more closely examine EGFP expression in the full thickness of barbel tissue, a second group of heat-shocked barbels were minimally fixed (2 hours at room temperature), rinsed, sliced crosswise and mounted in agarose on the cut end. These views also showed intense EGFP expression in superficial and deep barbel cells (Fig. 7E,F). We conclude that the maxillary barbel, like the fin, expresses abundant membrane-bound dn-fgfr1 for up to 24 hours after global heat shock in vivo.
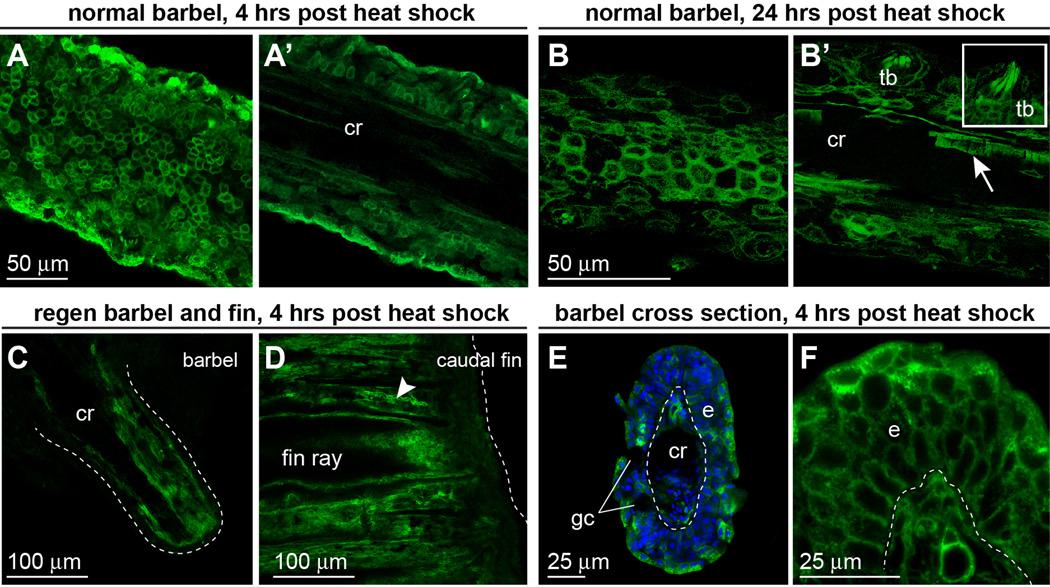
Figure 7. dn-fgfr1:EGFP is expressed throughout the maxillary barbel up to 24 hours after heat shock treatment.
A) Confocal image of an unfixed dn-fgfr1 maxillary barbel collected 4 hours after a one-hour heat shock. EGFP is prominent in epithelial cell membranes. A’) A deeper slice from the same z-stack shows EGFP in all cells surrounding the acellular central rod (cr). B). 24 hours after heat shock, membrane localization of EGFP is maintained in the barbel epithelium. B’) A deeper slice through the same z-stack shows persistent EGFP in basal epithelial cells, taste buds (inset), nerve fibers, and individual endothelial cells (arrow). C) Confocal slice through the center of an unfixed, amputated maxillary barbel 24 hours after heat shock; the distal end is to the lower right. EGFP is expressed throughout the full thickness of the stump, particularly in dermal tissues surrounding the central rod. Similar levels of EGFP expression were observed in amputated caudal fins at the same time point (D); the dotted line indicates the distal margin of each appendage. E) Fixed maxillary barbel tissue collected 4 hours after heat shock and mounted ‘on end’ in low-melt agarose. EGFP is visible in both epidermal (e) and deeper layers. Regions without EGFP include the epidermal goblet cells (gc) and the central rod (cr). F) ‘On end’ view of a second maxillary barbel showing membrane-localized EGFP in similar areas.
The maxillary barbel epithelium of hsp70l:dn-fgfr1 transgenic zebrafish regenerates well-organized calretinin-positive taste cells under heat-shock conditions
While this study was in progress, it was reported that the activation of dn-fgfr1 in larval zebrafish suppresses the formation of calretinin-positive (Calb2b+) gustatory cells in the developing pharyngeal taste buds (Kapsimali et al., 2011). Because the maxillary barbel epithelium is also well supplied with taste buds, we wished to examine if the regeneration of adult calretinin-positive cells could be similarly affected. We therefore repeated unilateral barbectomies and fin amputations on dn-fgfr1 animals and wild type siblings and treated both with once-daily heat shocks (one hour at >37.5°C) for 14 days. All fish were then analyzed for the presence/absence of caudal fin regeneration (to confirm in vivo transgene activation), the partial inhibition of dn-fgfr1 barbel regeneration (replicating our previous findings) and calretinin immunostaining of both normal and regenerating barbels (to assay the number, location and organization of taste buds). In addition to its effects on regeneration, prolonged blockade of FGF signaling can also impair the maintenance of normal adult zebrafish tissues, including fins (Wills et al., 2008) and retinal photoreceptors (Hochmann et al., 2012; Qin et al., 2011). We thus anticipated that heat-shock conditions might cause disruption or disorganization of barbel taste buds even in the normal, non-regenerating barbel epithelium of dn-fgfr1 animals.
After 14 days of heat-shock at 37.5°– 37.9°C (Fig. 8A), all wild type animals (n = 24) regenerated a normal-sized caudal fin (not shown), while all dn-fgfr1 individuals (n = 26) showed little caudal fin repair beyond wound healing (Fig. 8B). In some specimens, the tissue between the fin rays was retracted and filled with loose, necrotic cells. Ten of these caudal fins were analyzed by measuring the total tissue thickness (mesoderm + ectoderm) over the third most ventral fin ray; on average, less than 0.1 mm of tissue was present. Despite these strong in vivo effects on caudal fin outgrowth, the same dn-fgfr1 fish produced, on average, approximately 0.57 mm of barbel tissue past the amputation plane. This was about half of the maxillary barbel regrowth (1.1. mm) observed in heat-shocked wild types (Fig. 8C–D, Table 2C). Within each treatment group, the amount of barbel regeneration was again highly variable. However, the reduction in barbel outgrowth was similar in direction, magnitude and significance to the previous experiment (t = 8.5; df = 48; p < 0.0001; Table 2B vs. Table 2C).
Figure 8. The maintenance and regeneration of adult maxillary barbel taste buds are unaffected by 14 days of heat shock in the dn-fgfr1 line.
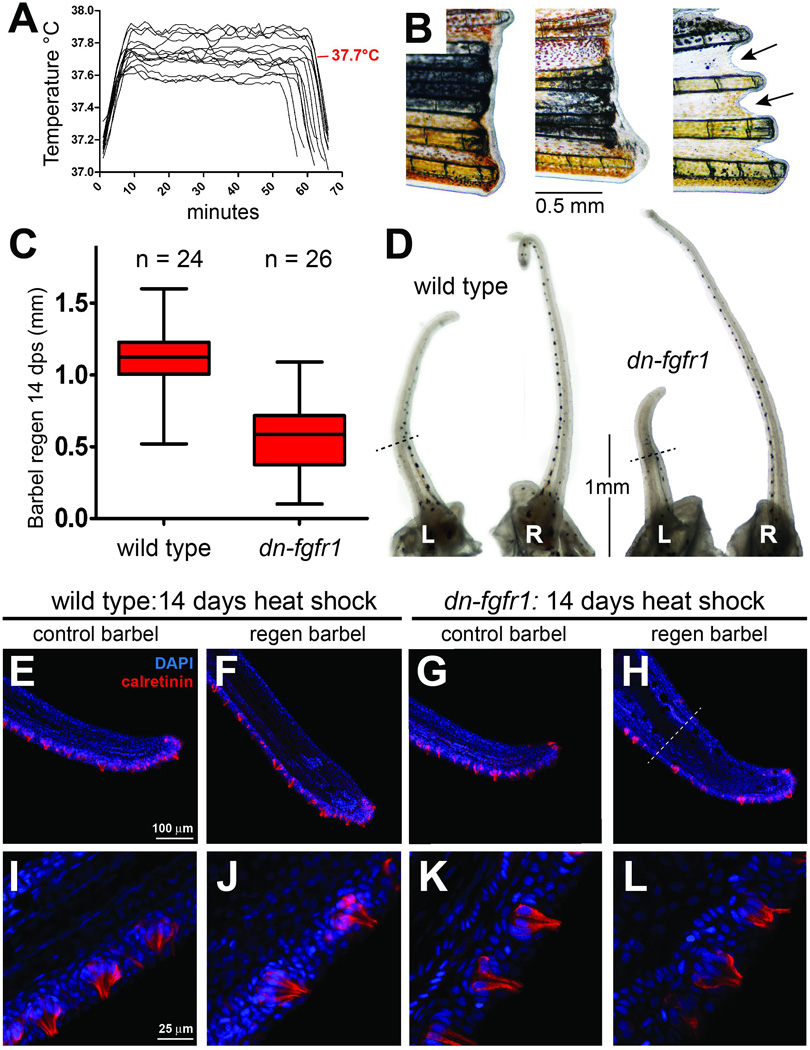
A) Representative in-tank temperature logs for the 14-day taste bud regeneration experiment. The heat-shock trace for each day is superimposed on the same temperature and time scale. The average peak temperature is shown in red (37.7°C). B) Gross morphology of dn-fgfr1 caudal fins at the 14-day endpoint. Caudal fin outgrowth was abolished; in some individuals, the fin margin shrank (arrows, right). All heat-shocked wild type siblings regenerated caudal fins normally (not shown). C) Heat-shock treatment reduced maxillary barbel regeneration approximately 50% compared to wild type controls (p < 0.0001) D) Gross morphology of wild type and dn-fgfr1 barbels at the 14 day endpoint. Black dotted lines indicate amputation planes. L = left, amputated barbel; R = right, unamputated barbel. E–L) To assay taste bud regeneration, all of the heat-shocked wild type and dn-fgfr1 barbels (amputated and unamputated) were stained for calretinin immunoreactivity; DAPI was used to mark nuclei. There was no difference in either the arrangement (E–H) or fine structure of calretinin-positive gustatory cells in the barbel epithelium (I–L) among any of the treatment groups.
Although this experiment effectively reproduced the blockade of caudal fin regeneration and the reduction of maxillary barbel regeneration in adult dn-fgfr1 zebrafish we had observed previously, there was no effect on the number, location or organization of calretinin-positive cells in the barbel epithelium (Fig. 8E–L). In heat-shocked wild types, the taste buds of both unamputated and regenerating barbels were normally arranged along two densely packed, roughly parallel ventral rows, with isolated buds dorsally (Fig. 8E,F). Each taste bud was a compact, onion-shaped cluster containing 8–10 well-defined, elongated calretinin-positive cells (Fig 8I,J). Multiple solitary chemosensory cells, a normal sub-population within the epithelium (Kotrschal, 2000; Kotrschal et al., 1997), were also detected. In unamputated heat-shocked dn-fgfr1 barbels, we saw the same normal taste bud pattern and morphology (Fig. 8G,K). Amputated dn-fgfr1 barbels regrew appendages shorter than wild type controls, but successfully regenerated numerous calretinin-positive cell clusters in the expected areas of the ventral epithelium and distal tip (Fig. 8 H,L). Magnification of these regenerated taste buds confirmed a typical radial arrangement of bottle-shaped cells with apical projections and basal nuclei; we also observed the expected solitary chemosensory cells. We conclude that, over the time period studied, daily heat shocks to adult dn-fgfr1 zebrafish do not disrupt the maintenance, regeneration or patterning of calretinin-positive cells in the maxillary barbel epithelium.
Discussion
A major goal of modern molecular biology is to discover what signaling pathways are active in particular biological contexts and to manipulate those pathways to alter cell differentiation and function. Only a few gene networks control many regenerative processes; however, the modulation of these networks can differ among species, organs and tissues (Ausoni and Sartore, 2009; Bely and Nyberg, 2010; Brockes and Kumar, 2008; Garza-Garcia et al., 2010). The zebrafish maxillary barbel provides a novel system for investigating appendage regeneration within the context of a model organism, providing an intra-specific complement to the well-studied caudal fin.
In this study, we confirmed that multiple FGF pathway members are actively transcribed in both normal and regenerating maxillary barbel tissue. Using well-characterized zebrafish lines with temperature-inducible defects in fgf20a and fgfr1, we tested how these signals contribute to maxillary barbel regeneration in vivo, using caudal fin regeneration as a simultaneous internal control. Under temperature regimes where fin outgrowth was completely abolished, maxillary barbel regeneration was either unaffected (in fgf20a homozygous recessives) or only partially inhibited (in dn-fgfr1 hemizygotes). 14 days of heat shock also produced no effect on adult maxillary barbel taste bud maintenance or regeneration. Our results demonstrate regional heterogeneity in the role of FGF ligands and receptors in zebrafish appendage regeneration and taste bud differentiation. We discuss the implications of this heterogeneity, considering both the limitations of our experiments and the ontogenetic and evolutionary diversity of zebrafish appendages.
Fgf20a: a fin-specific factor?
Our first target was fgf20a, previously shown to be essential for caudal fin regeneration (Bouzaffour et al., 2009; Shao et al., 2011; Whitehead et al., 2005) and fin maintenance (Wills et al., 2008). Dual-amputation of caudal fins and maxillary barbels in fgf20a homozygous recessives, followed by regeneration at the restrictive temperature (33°C) confirmed that disrupting this ligand blocks all caudal fin outgrowth. However, this genetic defect had no effect on maxillary barbel regeneration in the same animals. This indicates that the regenerative requirement for fgf20a is not universal, but appendage-specific. Although we readily detected fgf20a mRNA in wild type extracts of both normal and regenerating maxillary barbels, the role of this gene in barbel tissue is currently unclear, as all of our fgf20azp3/zp3 zebrafish raised at 28°C developed normal barbels and had no barbel regeneration defect. The zebrafish paralog of fgf20a, namely fgf20b, is therefore of interest to see if it is also present in barbel tissue, and if simultaneously disrupting both fgf20a and fgf20b would inhibit regeneration. Alternatively, other FGF ligands may substitute for the role of fgf20a in this appendage.
Activation of hsp70l:dn-fgfr1 partially, but not completely, inhibits barbel regeneration
The heat shock inducible dn-fgfr1:EGFP transgenic line is a widely used, well-validated and efficient way to inhibit FGF signaling in zebrafish embryos and adults (Ganz et al., 2010; Kaslin et al., 2009; Kikuchi et al., 2010; Lee et al., 2009; Lee et al., 2005; Martin and Kimelman, 2010; Yin et al., 2008). The ability of a dominant negative FGF receptor lacking the tyrosine kinase domain to dimerize with and thus block signaling downstream of multiple FGF receptors is based on the predicted structural similarities between those receptors, as well as early experimental work showing that adding a single such construct at sufficient concentration could block FGF-dependent processes in Xenopus oocytes and embryos (Amaya et al., 1991). In zebrafish, heat shock induction of dn-fgfr1:EGFP can phenocopy null mutations and morpholino knockdown of FGF ligands (Kapsimali et al., 2011; Neugebauer et al., 2009), downregulate FGF target genes (Martin and Kimelman, 2010) and mimic the chemical administration of ATP-competitive protein tyrosine kinase inhibitors (Lee et al., 2009; Nechiporuk et al., 2007; Nechiporuk and Raible, 2008; Poss et al., 2000) that are highly specific for Ffgr1 (Mohammadi et al., 1997). The activity of this construct can vary depending on the experimental context. Sources of variation include copy number (e.g., homozygous vs. hemizygous individuals, Hochmann et al., 2012; Qin et al., 2011) and the precise heat-shock temperature, with the strongest inhibitory effects seen close to 38°C (Lee et al., 2005). Cellular sources of variation may also exist, such as differential affinity of the fusion protein for each of the FGF receptors, and differential affinity of FGF ligands for specific receptors (Beenken et al., 2011). Because these effects are more difficult to measure, they are seldom investigated in vivo (but see Yamagishi and Okamoto, 2010).
By heat-shocking hemizygous dn-fgfr1 fish at temperatures between 37–38°C, we were able to reproduce a strong block in caudal fin regeneration, which was maintained for 14 days. In the same transgenic animals, most maxillary barbel regenerates grew out substantially, approaching half-normal lengths. Despite being shorter, these regenerated barbels had all of the cell types of a normal appendage, suggesting that cell proliferation, rather than differentiation, was most affected by this treatment. Although we did not measure the level of FGF suppression in barbel tissue directly, the absence of fin regeneration is assumed to represent effective activation of the dominant-negative transgene throughout the organism. Membrane-bound dnfgfr1:EGFP fusion protein also appeared ubiquitously and rather homogenously expressed in heat-shocked barbel tissue, consistent with what others have observed in caudal fin (Lee et al., 2005), heart (Lepilina et al., 2006), and retina (Qin et al., 2011) of the same transgenic line. This observation argues against inefficient or mosaic expression of the construct in maxillary barbel tissue, and supports a model in which the caudal fin and maxillary barbel respond differently to ‘equivalent’ levels of FGF suppression.
FGFs and their receptors are ubiquitous in both development and regeneration, making all tissues dependent on them to some degree. A more complex question is why some tissues appear more dependent on them than others. Other investigations have reported tissue-specific responses following global activation of the dn-fgfr1 construct. Heat shock of 18 hpf dn-fgfr1 embryos greatly reduced FGF target gene expression in the liver primordium, but not in adjacent tissues (Shin et al., 2007). Heat shock at 20 hpf reduced FGF target gene expression in embryonic hindbrain, but not in forebrain or spinal cord (Esain et al., 2010). In the adult zebrafish brain, a single heat shock significantly decreased cell proliferation in the ventral, but not dorsal, telencephalic stem cell populations, even though EGFP mRNA and fluorescent signal were equally expressed in both areas (Ganz et al., 2010). Most recently, heat shock treatments were found to affect the regeneration of different retinal cell populations (Hochmann et al., 2012)
What might explain these observations? If different adult cell populations have different endogenous levels of FGF receptors, the same quantity of dn-fgfr1 may not be sufficient to dimerize with, and thus block, all receptor activity. Similar-temperature heat shocks have greater repressive effects on distal, compared to proximal, amputations of the zebrafish caudal fin, suggesting a proximo-distal gradient of FGF activity (Lee et al., 2005). In addition to different baseline levels of FGFRs, tissues may differ in FGF receptor activation and efficiency, such that transduction continues even when receptor number is reduced. A comprehensive quantitative analysis of FGF receptor levels and downstream marker genes in the maxillary barbel might be done in a future study. Such mechanisms might explain why there seem to be tissue-specific “set points" for a particular pathway within an organism.
Another unresolved issue is why some regenerating tissues have consistent and robust responses to injury, while others have variable and limited responses. In all of our experiments, caudal fin regeneration was “all or nothing,” with almost no variation among individuals within a treatment group. Under the same conditions, maxillary barbel regeneration was highly variable, with individual regenerates varying several fold in length. This observation adds another level of biological variation, that of individual regenerative capacity, which is not yet fully explained. Beyond FGF signaling, many other pathways influence regeneration. The caudal fin regenerates using multiple interconnecting genetic networks, including Wnts (Stoick-Cooper et al., 2007), retinoic acid (Géraudie et al., 1995; Mathew et al., 2009), hedgehogs (Laforest et al., 1998), stromal cell derived factor (Bouzaffour et al., 2009) and microRNA pathways (Thatcher et al., 2008; Yin et al., 2008). Much less is known about the maxillary barbel, which differs from the caudal fin in both developmental and evolutionary origin. Evolutionarily, caudal fins are ancient, primitive vertebrate structures, whereas barbels have been independently and repeatedly derived in many groups of cyprinids, including the genus Danio (Briolay et al., 1998; Fox, 1999; He et al., 2008; Li et al., 2008). It would not be surprising, therefore, if these two appendages had different genetic programs for growth, maintenance, and regeneration.
Spatial and temporal diversity of taste bud regeneration
Few studies address the molecular regulation of vertebrate taste buds, which, depending on the species, are distributed on epithelia of the head, trunk, oral cavity or gut lining. Current work indicates that taste buds are highly autonomous epithelial modules that can differentiate independently of both neural input (Thirumangalathu et al., 2009) and inductive signals from the underlying mesenchyme (Barlow and Northcutt, 1997). During early development, however, FGF signaling is required for regional taste bud formation in both fish and mice. Heat shock of dn-fgfr1 zebrafish embryos at either 36–48 or 52–54 hpf causes reduction and disruption of both calretinin- and serotonin-positive taste cells in the pharyngeal, but not the palatal epithelium (Kapsimali et al., 2011). A similar regional heterogeneity has been observed in mouse tongue development, wherein genetic manipulation of FGF10 alters caudal, but not rostral taste papillae (Petersen et al., 2011). We therefore sought to test if the maxillary barbel taste buds would be maintained and/or regenerated under conditions where FGF-dependent caudal fin regeneration was blocked.
After 14 days of heat shock, all wildtype and dn-fgfr1:EGFP zebrafish both maintained and regenerated taste buds normally. We expected that these superficial cells would receive a very effective heat shock, and this was confirmed by intense EGFP signal in transgenic taste buds up to 24 hours after a single treatment. Given that we saw no disruption of taste bud distribution or architecture at the endpoint of this experiment, we infer that maxillary barbel taste bud maintenance and regeneration are not strongly FGF-dependent.
Our observations are limited to calretinin-positive taste cells, a commonly used molecular marker for such studies (Germana et al., 2007; Levanti et al., 2008; Northcutt, 2005; Zachar and Jonz, 2011). Other specialized neuroepithelial cell types, such as serotonin-positive cells (5HT+), were not examined. Heat shock was applied for only 14 days; however, considering the relatively high cellular turnover of skin epithelia, this seemed to be a reasonable interval in which to observe some effects. Using radioactive labeling in channel catfish acclimated to warm water (30°C), it was estimated that 50% of taste buds cells were replaced every 12 days (Raderman-Little, 1979). Turnover rates for taste cells in adult zebrafish, however, have so far not been tested. Longer periods of heat shock, higher induction temperatures, or alternative methods of interfering with FGFs might be necessary to detect significant morphological effects.
Theme, Variation and Future Directions
Our results are the first to highlight the molecular mechanisms controlling zebrafish maxillary barbel regeneration, and demonstrate that these mechanisms differ from those of the caudal fin. Despite the expression of similar FGF ligands, receptors and downstream effectors in both appendages, we observed significant phenotypic differences in appendage regeneration when interfering with this pathway at the ligand or receptor level. Specifically, FGF signaling through fgf20a appears dispensable for barbel regeneration, the opposite of what has been observed in the fin. FGF signaling also appears less crucial for barbel regeneration, as barbels still partially regrew under heat-shock conditions that completely blocked caudal fin outgrowth in the same animals. Further molecular comparisons between these two appendages may highlight specific features that are shared, as well as those that confer tissue-specific and individual variability on the regenerative response. Studying simple, accessible structures such as the zebrafish caudal fin has greatly expanded our understanding of the developmental and regenerative mechanisms provided by vertebrate evolutionary history. The maxillary barbel is yet another accessible and productive context for these investigations.
Acknowledgements
This research was conducted in partial fulfillment of the requirements of the Master of Science program in the Department of Biological Sciences at DePaul University (RJD). Financial support was provided by the National Institutes of Health (R15HD064169 to EEL, R01DE016678 to JT), the DePaul University Research Council, the Department of Biological Sciences, and the DePaul Graduate Research Fund (to RJD). Resources from the Zebrafish International Resource Center were supported by grant P40-RR012546 from the National Institutes of Health’s National Center for Research Resources (NIH-NCRR). We thank Bill Gilliland for expert advice on confocal microscopy and deconvolution software, and Ann Hogan for bibliographic assistance.
Footnotes
Author Contributions:
Planned the study: RJD, JT, EEL
Performed experiments, analyzed data, and produced figures: RJD, EEL
Provided facilities and reagents: JT, EEL
Wrote and edited the paper: RJD, JT, EEL
References
- Akimenko M, Johnson S, Westerfield M, Ekker M. Differential induction of four msx homeobox genes during fin development and regeneration in zebrafish. Development. 1995;121:347–357. doi: 10.1242/dev.121.2.347. [DOI] [PubMed] [Google Scholar]
- Akimenko MA, Marí-Beffa M, Becerra J, Géraudie J. Old questions, new tools, and some answers to the mystery of fin regeneration. Dev Dyn. 2003;226:190–201. doi: 10.1002/dvdy.10248. [DOI] [PubMed] [Google Scholar]
- Amaya E, Musci TJ, Kirschner MW. Expression of a dominant negative mutant of the FGF receptor disrupts mesoderm formation in Xenopus embryos. Cell. 1991;66:257–270. doi: 10.1016/0092-8674(91)90616-7. [DOI] [PubMed] [Google Scholar]
- Ausoni S, Sartore S. From fish to amphibians to mammals: in search of novel strategies to optimize cardiac regeneration. J Cell Biol. 2009;184:357–364. doi: 10.1083/jcb.200810094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo AS, Grotek B, Jacinto A, Weidinger G, Saúde L. The regenerative capacity of the zebrafish caudal fin is not affected by repeated amputations. PLoS One. 2011;6:e22820. doi: 10.1371/journal.pone.0022820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow L, Northcutt R. Taste buds develop autonomously from endoderm without induction by cephalic neural crest or paraxial mesoderm. Development. 1997;124:949–957. doi: 10.1242/dev.124.5.949. [DOI] [PubMed] [Google Scholar]
- Barrientos S, Stojadinovic O, Golinko M, Brem H, Tomic-Canic M. Growth factors and cytokines in wound healing. Wound Repair Regen. 2008;16:585–601. doi: 10.1111/j.1524-475X.2008.00410.x. [DOI] [PubMed] [Google Scholar]
- Basu N, Todgham AE, Ackerman PA, Bibeau MR, Nakano K, Schulte PM, Iwama GK. Heat shock protein genes and their functional significance in fish. Gene. 2002;295:173–183. doi: 10.1016/s0378-1119(02)00687-x. [DOI] [PubMed] [Google Scholar]
- Becker C, Becker T. Growth and pathfinding of regenerating axons in the optic projection of adult fish. J Neurosci Res. 2007;85:2793–2799. doi: 10.1002/jnr.21121. [DOI] [PubMed] [Google Scholar]
- Becker CG, Becker T. Adult zebrafish as a model for successful central nervous system regeneration. Restor Neurol Neurosci. 2008;26:71–80. [PubMed] [Google Scholar]
- Beenken A, Eliseenkova AV, Ibrahimi OA, Olsen SK, Mohammadi M. Plasticity in the interactions of the N-terminus of fibroblast growth factor (FGF)-1 with FGF receptors underlies FGF1's promiscuity. J Biol Chem. 2011 doi: 10.1074/jbc.M111.275891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bely AE, Nyberg KG. Evolution of animal regeneration: re-emergence of a field. Trends Ecol Evol. 2010;25:161–170. doi: 10.1016/j.tree.2009.08.005. [DOI] [PubMed] [Google Scholar]
- Bouzaffour M, Dufourcq P, Lecaudey V, Haas P, Vriz S. Fgf and Sdf-1 pathways interact during zebrafish fin regeneration. PLoS One. 2009;4:e5824. doi: 10.1371/journal.pone.0005824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briolay J, Galtier N, Brito R, Bouvet Y. Molecular phylogeny of Cyprinidae inferred from cytochrome b DNA sequences. Mol Phylogenet Evol. 1998;9:100–108. doi: 10.1006/mpev.1997.0441. [DOI] [PubMed] [Google Scholar]
- Brockes JP, Kumar A. Comparative aspects of animal regeneration. Annu Rev Cell Dev Biol. 2008;24:525–549. doi: 10.1146/annurev.cellbio.24.110707.175336. [DOI] [PubMed] [Google Scholar]
- Brody J, Calhoun E, Gallmeier E, Creavalle T, Kern S. Ultra-fast high-resolution agarose electrophoresis of DNA and RNA using low-molarity conductive media. BioTechniques. 2004;37:598–602. doi: 10.2144/04374ST04. [DOI] [PubMed] [Google Scholar]
- Brown LA, Amores A, Schilling TF, Jowett T, Baert JL, de Launoit Y, Sharrocks AD. Molecular characterization of the zebrafish PEA3 ETS-domain transcription factor. Oncogene. 1998;17:93–104. doi: 10.1038/sj.onc.1201911. [DOI] [PubMed] [Google Scholar]
- Diep CQ, Ma D, Deo RC, Holm TM, Naylor RW, Arora N, Wingert RA, Bollig F, Djordjevic G, Lichman B, Zhu H, Ikenaga T, Ono F, Englert C, Cowan CA, Hukriede NA, Handin RI, Davidson AJ. Identification of adult nephron progenitors capable of kidney regeneration in zebrafish. Nature. 2011;470:95–100. doi: 10.1038/nature09669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufourcq P, Roussigné M, Blader P, Rosa F, Peyrieras N, Vriz S. Mechano-sensory organ regeneration in adults: the zebrafish lateral line as a model. Mol Cell Neurosci. 2006;33:180–187. doi: 10.1016/j.mcn.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Duszynski RJ, Topczewski J, LeClair EE. Simple, Economical Heat-Shock Devices for Zebrafish Housing Racks. Zebrafish. 2011;8:211–219. doi: 10.1089/zeb.2011.0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esain V, Postlethwait JH, Charnay P, Ghislain J. FGF-receptor signalling controls neural cell diversity in the zebrafish hindbrain by regulating olig2 and sox9. Development. 2010;137:33–42. doi: 10.1242/dev.038026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox H. Barbels and barbel-like tentacular structures in sub-mammalian vertebrates: A review. Hydrobiologia. 1999;403:153–193. [Google Scholar]
- Ganz J, Kaslin J, Hochmann S, Freudenreich D, Brand M. Heterogeneity and Fgf dependence of adult neural progenitors in the zebrafish telencephalon. Glia. 2010;58:1345–1363. doi: 10.1002/glia.21012. [DOI] [PubMed] [Google Scholar]
- Garza-Garcia AA, Driscoll PC, Brockes JP. Evidence for the local evolution of mechanisms underlying limb regeneration in salamanders. Integr Comp Biol. 2010;50:528–535. doi: 10.1093/icb/icq022. [DOI] [PubMed] [Google Scholar]
- Germana A, Paruta S, Germana GP, Ochoa-Erena FJ, Montalbano G, Cobo J, Vega JA. Differential distribution of S100 protein and calretinin in mechanosensory and chemosensory cells of adult zebrafish (Danio rerio) Brain Res. 2007;1162:48–55. doi: 10.1016/j.brainres.2007.05.070. [DOI] [PubMed] [Google Scholar]
- Gurtner GC, Werner S, Barrandon Y, Longaker MT. Wound repair and regeneration. Nature. 2008;453:314–321. doi: 10.1038/nature07039. [DOI] [PubMed] [Google Scholar]
- Géraudie J, Monnot MJ, Brulfert A, Ferretti P. Caudal fin regeneration in wild type and long-fin mutant zebrafish is affected by retinoic acid. Int J Dev Biol. 1995;39:373–381. [PubMed] [Google Scholar]
- He S, Mayden RL, Wang X, Wang W, Tang KL, Chen WJ, Chen Y. Molecular phylogenetics of the family Cyprinidae (Actinopterygii: Cypriniformes) as evidenced by sequence variation in the first intron of S7 ribosomal protein-coding gene: further evidence from a nuclear gene of the systematic chaos in the family. Mol Phylogenet Evol. 2008;46:818–829. doi: 10.1016/j.ympev.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Hochmann S, Kaslin J, Hans S, Weber A, Machate A, Geffarth M, Funk RH, Brand M. Fgf Signaling is Required for Photoreceptor Maintenance in the Adult Zebrafish Retina. PLoS One. 2012;7:e30365. doi: 10.1371/journal.pone.0030365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapsimali M, Kaushik AL, Gibon G, Dirian L, Ernest S, Rosa FM. Fgf signaling controls pharyngeal taste bud formation through miR-200 and Delta-Notch activity. Development. 2011;138:3473–3484. doi: 10.1242/dev.058669. [DOI] [PubMed] [Google Scholar]
- Kaslin J, Ganz J, Geffarth M, Grandel H, Hans S, Brand M. Stem cells in the adult zebrafish cerebellum: initiation and maintenance of a novel stem cell niche. J Neurosci. 2009;29:6142–6153. doi: 10.1523/JNEUROSCI.0072-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, Macrae CA, Stainier DY, Poss KD. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotrschal K. Taste(s) and olfaction(s) in fish: a review of specialized sub-systems and central integration. Pflugers Arch. 2000;439:R178–R180. [PubMed] [Google Scholar]
- Kotrschal K, Krautgartner WD, Hansen A. Ontogeny of the solitary chemosensory cells in the zebrafish, Danio rerio. Chem Senses. 1997;22:111–118. doi: 10.1093/chemse/22.2.111. [DOI] [PubMed] [Google Scholar]
- Krone PH, Lele Z, Sass JB. Heat shock genes and the heat shock response in zebrafish embryos. Biochem Cell Biol. 1997;75:487–497. [PubMed] [Google Scholar]
- Laforest L, Brown CW, Poleo G, Geraudie J, Tada M, Ekker M, Akimenko MA. Involvement of the sonic hedgehog, patched 1 and bmp2 genes in patterning of the zebrafish dermal fin rays. Development. 1998;125:4175–4184. doi: 10.1242/dev.125.21.4175. [DOI] [PubMed] [Google Scholar]
- LeClair EE, Topczewski J. Methods for the study of the zebrafish maxillary barbel. J Vis. 2009 doi: 10.3791/1558. Exp, http://www.jove.com/index/details.stp?id=1558, [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClair EE, Topczewski J. Development and regeneration of the zebrafish maxillary barbel: a novel study system for vertebrate tissue growth and repair. PLoS One. 2010;5:e8737. doi: 10.1371/journal.pone.0008737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, Tseng WA, Lo FY, Liu TM, Tsai HJ. FoxD5 mediates anterior-posterior polarity through upstream modulator Fgf signaling during zebrafish somitogenesis. Dev Biol. 2009;336:232–245. doi: 10.1016/j.ydbio.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Lee Y, Grill S, Sanchez A, Murphy-Ryan M, Poss KD. Fgf signaling instructs position-dependent growth rate during zebrafish fin regeneration. Development. 2005;132:5173–5183. doi: 10.1242/dev.02101. [DOI] [PubMed] [Google Scholar]
- Lepilina A, Coon A, Kikuchi K, Holdway J, Roberts R, Burns C, Poss K. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- Levanti MB, Montalbano G, Laurà R, Ciriaco E, Cobo T, García-Suarez O, Germanà A, Vega JA. Calretinin in the peripheral nervous system of the adult zebrafish. J Anat. 2008;212:67–71. doi: 10.1111/j.1469-7580.2007.00836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang X, Kong X, Zhao K, He S, Mayden RL. Variation patterns of the mitochondrial 16S rRNA gene with secondary structure constraints and their application to phylogeny of cyprinine fishes (Teleostei: Cypriniformes) Mol Phylogenet Evol. 2008;47:472–487. doi: 10.1016/j.ympev.2007.09.012. [DOI] [PubMed] [Google Scholar]
- Martin BL, Kimelman D. Brachyury establishes the embryonic mesodermal progenitor niche. Genes Dev. 2010;24:2778–2783. doi: 10.1101/gad.1962910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew L, Sengupta S, Franzosa J, Perry J, La Du J, Andreasen E, Tanguay R. Comparative expression profiling reveals an essential role for raldh2 in epimorphic regeneration. J Biol Chem. 2009;284:33642–33653. doi: 10.1074/jbc.M109.011668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi M, McMahon G, Sun L, Tang C, Hirth P, Yeh BK, Hubbard SR, Schlessinger J. Structures of the tyrosine kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science. 1997;276:955–960. doi: 10.1126/science.276.5314.955. [DOI] [PubMed] [Google Scholar]
- Moore A, Mark T, Hogan A, Topczewski J, LeClair EE. Peripheral axons of the adult zebrafish maxillary barbel extensively remyelinate during sensory appendage regeneration. J Comp Neurol. 2012 doi: 10.1002/cne.23147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechiporuk A, Keating M. A proliferation gradient between proximal and msxb-expressing distal blastema directs zebrafish fin regeneration. Development. 2002;129:2607–2617. doi: 10.1242/dev.129.11.2607. [DOI] [PubMed] [Google Scholar]
- Nechiporuk A, Linbo T, Poss KD, Raible DW. Specification of epibranchial placodes in zebrafish. Development. 2007;134:611–623. doi: 10.1242/dev.02749. [DOI] [PubMed] [Google Scholar]
- Nechiporuk A, Raible DW. FGF-dependent mechanosensory organ patterning in zebrafish. Science. 2008;320:1774–1777. doi: 10.1126/science.1156547. [DOI] [PubMed] [Google Scholar]
- Neugebauer JM, Amack JD, Peterson AG, Bisgrove BW, Yost HJ. FGF signalling during embryo development regulates cilia length in diverse epithelia. Nature. 2009;458:651–654. doi: 10.1038/nature07753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcutt RG. Taste bud development in the channel catfish. Journal of Comparative Neurology. 2005;482:1–16. doi: 10.1002/cne.20425. [DOI] [PubMed] [Google Scholar]
- Petersen CI, Jheon AH, Mostowfi P, Charles C, Ching S, Thirumangalathu S, Barlow LA, Klein OD. FGF signaling regulates the number of posterior taste papillae by controlling progenitor field size. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002098. e1002098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss KD. Getting to the heart of regeneration in zebrafish. Semin Cell Dev Biol. 2007;18:36–45. doi: 10.1016/j.semcdb.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Poss KD, Shen J, Nechiporuk A, McMahon G, Thisse B, Thisse C, Keating MT. Roles for Fgf signaling during zebrafish fin regeneration. Dev Biol. 2000;222:347–358. doi: 10.1006/dbio.2000.9722. [DOI] [PubMed] [Google Scholar]
- Qin Z, Kidd AR, Thomas JL, Poss KD, Hyde DR, Raymond PA, Thummel R. FGF signaling regulates rod photoreceptor cell maintenance and regeneration in zebrafish. Exp Eye Res. 2011;93:726–734. doi: 10.1016/j.exer.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raderman-Little R. The effect of temperature on the turnover of taste bud cells in catfish. Cell Tissue Kinet. 1979;12:269–280. doi: 10.1111/j.1365-2184.1979.tb00149.x. [DOI] [PubMed] [Google Scholar]
- Shao J, Chen D, Ye Q, Cui J, Li Y, Li L. Tissue regeneration after injury in adult zebrafish: the regenerative potential of the caudal fin. Dev Dyn. 2011;240:1271–1277. doi: 10.1002/dvdy.22603. [DOI] [PubMed] [Google Scholar]
- Shin D, Shin CH, Tucker J, Ober EA, Rentzsch F, Poss KD, Hammerschmidt M, Mullins MC, Stainier DY. Bmp and Fgf signaling are essential for liver specification in zebrafish. Development. 2007;134:2041–2050. doi: 10.1242/dev.000281. [DOI] [PubMed] [Google Scholar]
- Shoji W, Sato-Maeda M. Application of heat shock promoter in transgenic zebrafish. Develop. Growth Differ. 2008;50:401–406. doi: 10.1111/j.1440-169X.2008.01038.x. [DOI] [PubMed] [Google Scholar]
- Sokal R, Rohlf F. Biometry. 4th ed. W. H. Freeman; 2011. [Google Scholar]
- Stoick-Cooper CL, Weidinger G, Riehle KJ, Hubbert C, Major MB, Fausto N, Moon RT. Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development. 2007;134:479–489. doi: 10.1242/dev.001123. [DOI] [PubMed] [Google Scholar]
- Tal TL, Franzosa JA, Tanguay RL. Molecular signaling networks that choreograph epimorphic fin regeneration in zebrafish - a mini-review. Gerontology. 2010;56:231–240. doi: 10.1159/000259327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher EJ, Paydar I, Anderson KK, Patton JG. Regulation of zebrafish fin regeneration by microRNAs. Proc Natl Acad Sci U S A. 2008;105:18384–18389. doi: 10.1073/pnas.0803713105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirumangalathu S, Harlow DE, Driskell AL, Krimm RF, Barlow LA. Fate mapping of mammalian embryonic taste bud progenitors. Development. 2009;136:1519–1528. doi: 10.1242/dev.029090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel R, Bai S, Sarras MP, Song P, McDermott J, Brewer J, Perry M, Zhang X, Hyde DR, Godwin AR. Inhibition of zebrafish fin regeneration using in vivo electroporation of morpholinos against fgfr1 and msxb. Dev Dyn. 2006;235:336–346. doi: 10.1002/dvdy.20630. [DOI] [PubMed] [Google Scholar]
- Tsang M, Friesel R, Kudoh T, Dawid IB. Identification of Sef, a novel modulator of FGF signalling. Nat Cell Biol. 2002;4:165–169. doi: 10.1038/ncb749. [DOI] [PubMed] [Google Scholar]
- Wakahara T, Kusu N, Yamauchi H, Kimura I, Konishi M, Miyake A, Itoh N. Fibin, a novel secreted lateral plate mesoderm signal, is essential for pectoral fin bud initiation in zebrafish. Dev Biol. 2007;303:527–535. doi: 10.1016/j.ydbio.2006.11.041. [DOI] [PubMed] [Google Scholar]
- Westerfield M. Ch. 1 General Methods for Zebrafish Care, The zebrafish book: a guide for the laboratory use of zebrafish (Danio rerio) Eugene, OR: Univ. of Oregon Press; 2000. [Google Scholar]
- Whitehead GG, Makino S, Lien CL, Keating MT. fgf20 is essential for initiating zebrafish fin regeneration. Science. 2005;310:1957–1960. doi: 10.1126/science.1117637. [DOI] [PubMed] [Google Scholar]
- Wills AA, Kidd AR, Lepilina A, Poss KD. Fgfs control homeostatic regeneration in adult zebrafish fins. Development. 2008;135:3063–3070. doi: 10.1242/dev.024588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi M, Okamoto H. Competition for ligands between FGFR1 and FGFR4 regulates Xenopus neural development. Int J Dev Biol. 2010;54:93–104. doi: 10.1387/ijdb.092849my. [DOI] [PubMed] [Google Scholar]
- Yin VP, Poss KD. New regulators of vertebrate appendage regeneration. Curr Opin Genet Dev. 2008;18:381–386. doi: 10.1016/j.gde.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin VP, Thomson JM, Thummel R, Hyde DR, Hammond SM, Poss KD. Fgf-dependent depletion of microRNA-133 promotes appendage regeneration in zebrafish. Genes Dev. 2008;22:728–733. doi: 10.1101/gad.1641808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachar PC, Jonz MG. Confocal imaging of Merkel-like basal cells in the taste buds of zebrafish. Acta Histochem. 2011 doi: 10.1016/j.acthis.2011.03.006. [DOI] [PubMed] [Google Scholar]