Abstract
Background & objectives:
Chittoor virus (CHITV) belongs to genus Orthobunyavirus, family Bunyaviridae. It has been isolated from various species of mosquitoes and pig from different parts of India. Five isolates of CHITV were characterized at the molecular level and compared with other Batai viruses (BATV) to find out any kind of reassortment in their genome.
Methods:
Complete nucelocapsid (S), glycoprotein (M) and partial RNA polymerase (L) segments of CHITV were amplified and sequenced. These sequences were compared with those of Batai viruses, isolated from different geographical locations in Asia, Africa and Europe.
Results:
Phylogenetic analysis revealed CHITV as a variant of BATV. High level of conservation was seen among the CHITV isolates studied. The CHITV sequences showed clustering in one lineage with the sequences from Japan and Malaysia, however, BATV sequences from Europe and Africa formed a separate phylogenetic lineage.
Interpretation & conclusions:
The study indicates the presence of a single genotype of CHITV circulating in India, despite the involvement of different hosts in the natural cycle by this virus. Analysis of the sequences of the S, M and L segments of genome indicated that the virus has not undergone any reassortment. This virus has not caused any epidemic involving humans, however, replication of the virus in different mosquito and vertebrate hosts species suggests that it is a cause of concern.
Keywords: Batai virus, Bunyaviridae, Chittoor virus, glycoprotein, molecular characterization
Chittoor virus (CHITV) was first isolated in 1957 from Anopheles barbirostris collected from Brahmanpalli, Chittoor district, Andhra Pradesh, India. This virus was placed in Bunyamwera group based on serological characterization1. It was found antigenically related to Calovo virus and Batai virus (BATV) isolated from Slovakia and Malaysia, respectively2. Subsequently, several isolates of this virus were obtained from Anopheles and Culex mosquitoes as well as from a piglet in India3,4. BATV, an important member of the Bunyamwera serogroup, genus Orthobunyavirus, family Bunyaviridae5. This virus has a worldwide distribution2,3,6–10. BATV, which causes mild febrile illness in humans and animals, has also been isolated from the blood of a suspected malaria patient in Sudan11. Reassortant has been found in Ngari virus (NRIV), one of the members of this group, which wa s found to be associated with haemorrhagic fever outbreaks in East Africa12. NRIV was reassortant with S and L RNA segments from Bunyamwera virus and an M RNA segment from BATV5,10. Based on molecular and serological studies, CHITV was characterized as a variant of BATV.
The genus Orthobunyavirus comprises 18 serogroups and is composed of segmented single stranded negative sense RNA viruses13. The virus genome is composed of three segments viz., small (S), medium (M) and large (L). The S segment encodes the nucleocapsid (N) and the non-structural (NSs) proteins, while the M segment encodes the virion surface glycoproteins (Gn, Gc) and non-structural proteins (NSm). The L segment encodes for the replicase/transcriptase L protein. The nonstructural proteins NSm participate in virus assembly14 and NSs is involved in counteracting the host immune response by blocking alpha/beta interferon induction14–16.
Repeated isolations of CHITV and seroprevalence in several States suggest that it has been circulating in India for a long time17. However, this virus has not caused any outbreak involving humans in India, its ability to replicate in vertebrates and mosquitoes may be cause of concern for public health. Therefore, to determine its distribution across different taxonomic entity and molecular variations, attempts were made to characterize CHITV isolates at molecular level which were obtained from different regions in India.
Material & Methods
Propagation of virus: Details of five CHITV isolates used in the study are provided in the Table. The isolates were procured from virus repository of National Institute of Virology, Pune. The lyophilized viruses were reconstituted in sterile distilled water and dilutions were made in minimum essential medium (MEM). Vero E6 cells grown in 150 cm2 bottles up to 90% confluency were infected with 1 multiplicity of infection (MOI) of each isolate. The cultures were observed daily for cytopathic effects (CPE). When more than 75 per cent cells showed CPE, virus was harvested by repeated freezing and thawing of the cultures followed by centrifugation at 3000 g for 30 min at 4°C to remove the cell lysate. The supernatant was collected, aliquoted and stored at -70°C, until use.
Table.
Details of Chittoor virus isolation from India
Amplification of the complete M, S and partial L segments of CHITV: Total RNA was extracted from 250 μl CHITV infected tissue culture fluid using Tripure (Roche, USA) and chloroform: isoamyl alcohol (29:1 v/v), these were purified by RNAid kit (Bio 101, USA) according to the manufacturer's instructions. The RNA was dissolved in 50μl of diethylpyrocarbonate treated water. Primers for M segment amplification were designed based on sequences of bunyaviruses available in GenBank, using conserved sequences of 5’ and 3’ end of glycoprotein gene. The amplification primers for M segment were (IngMF) 5’AGT AGT GTA CTA CCRA3’ and (IngMR) 5’AGT AGT GTG CTA CCG ATA ACA A3’. The complete S and partial L segments were amplified and sequenced with primers BUNYA1 and BUNYA2 and primers M13CBUNL1C and BUNL605R as described earlier12. The M segment reverse transcription (RT) reaction was carried out at 50°C for 30 minute and polymerase chain reaction (PCR) was performed at 94°C for 2 min, followed by 40 cycles of 94°C for 15 sec, 49°C for 30 sec, 68°C for 5 min, and a final extension at 68°C for 10 min. The PCRs were carried out using Superscript III single step RT-PCR system with Platinum Taq High fidelity (Invitrogen, USA), according to manufacturer's instructions. Amplified products were analyzed on agarose gel of appropriate concentration and amplicon of the desired size was purified using QIAquick gel extraction kit (Qiagen, USA), as prescribed by the manufacturer. Since M segment of CHITV isolate G 20217 is available in GenBank (DQ341311), only S and partial L segments were amplified for this isolate, while for the remaining four isolates (804986, 804988, 804992 and 8627-11) all the three segments were amplified.
Sequencing and phylogenetic analysis: Sequencing was done using different sets of primers for S and M segments as defined above. In both the cases, internal primers were designed to get complete sequence while for L gene, amplification primers were used. Cyclic sequencing was carried out at PCR conditions of 96°C for 1 min, 96°C for 10 sec, 45°C for 5 sec and 60°C for 4 min for 25 cycles, using ABI Big-Dye 3.1 dye chemistry (Applied Biosystems, USA). These products were purified using Dyex 2.0 kit (Qiagen) according to manufacturer's instructions and sequencing was done using the ABI 3100 automated DNA sequencer. The sequences obtained were cleaned and edited using KODON 2.01 software, USA for both the reads from the two ends. The nucleotide sequences were converted into amino acid (aa) for determining the open reading frames (ORFs) and molecular weight of the putative proteins using KODON 2.01 software, USA. The clean sequences of each segment were aligned using program Clustal W and phylogenetic tree was constructed using neighbor- joining (NJ) algorithm with 1000-bootstrap replicates as implemented in Mega v 4.0 software18.
BATV sequences (S, M and L segments) and a few representatives of Orthobunyavirus Ilesha (ILE), Germistone (GER), Maguri (MAG) viruses from GenBank were compared with CHITV. Distance analysis of all the three segments was performed by software MEGA v 4.0. Bunyamwera virus (BUNV) was used as outgroup for the phylogenetic analysis of all the three segments of CHITV. In addition, analysis was also performed with partial M segment (420 bp and 157 aa) of CHITV, including West Ukraine BATV sequences (accession number GU320299 and GU320330).
Results
All the five isolates of CHITV could be propagated in Vero-E6 cell line and CPE was observed on 3rd day post-infection. Single step RT-PCR could amplify 958-nucleotide (nt) of the S segment, 605 - nt of the L segment and 4436- nt of M segment. It was observed in CHITV isolates, M gene was encoded for 162 kDa protein of 1434 amino acid (aa) ORF. In spite of complete M gene amplification of CHITV, a few M gene sequences (FJ436798 and FJ436800) could not generate good quality of sequences to complete ORF and encoded for 1401 and 1405 aa only. N gene was encoded for 35.8kDa proteins of 233 aa while NSs gene was encoded for 101 aa.
Phylogenetic analysis showed that S segment of all BATV formed a monophyletic tree of CHITV isolates (FJ436802-6) with other BATV from Japan, Malaysia and Germany. BATV from Australia [AF325122] and MAGV were out of this group (Fig. 1). Comparison of nucleocapsid gene of ten sequences of BATV including the five CHITV isolates showed that these have 84.0 and 89.0 per cent nt and aa identity, respectively. Among CHITV sequences 97.0-100.0 per cent identity was found at nucleotide level. Phylogenetic analysis of aa sequences of NSs gene of all the BATVs showed that CHITV had 100.0 per cent similarity, while isolates from Malaysia and Japan were in a separate lineage with a similarity of 98.6 per cent (data not shown). ILEV [AM709779] clustered with BUNV and NRIV [AY593729].
Fig. 1.
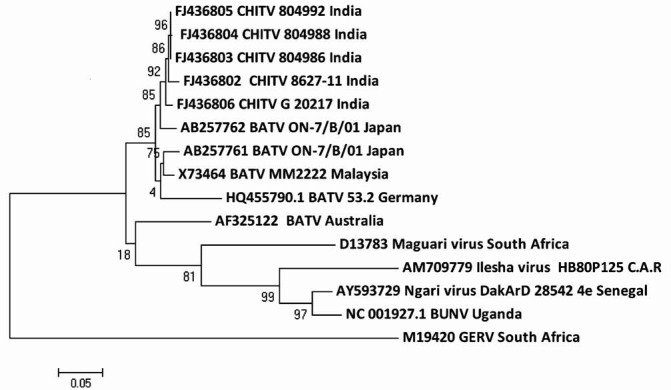
Phylogenetic tree of nucleotide sequences of CHITV and representative other BATV for complete S segment.
In the phylogenetic analysis of complete M segment, 13 BATV isolates were compared which showed formation of three phylogenetic lineages. First lineage (Asian) represented India, Malaysia (AY772534) and Japan (AB257765, AB257764) isolates with a difference of 8.0 and 3.4 per cent nt and aa, respectively. Second lineage (African) represented African isolates (Uganda DQ436460, Sudan DQ375393 and Kenya AY593725) with a difference of 5.2 and 2.4 per cent for nt and aa. Third lineage (European) was represented by Germany (HQ455791) and Slovakia (DQ334335) (Fig. 2). As reported earlier, NRIV is a reassortant virus which carries M segment of BATV and L and S segments of BUNV5. Similar result was obtained in this study, where NRIV (AY593725) clustered together with African isolates but not with BUNV as in the case of S segment. All the CHITVs showed similarity of up to 97.7-99.0 per cent and 98.9-99.8 per cent for nt and aa, respectively and clustered with Asian BATV. NRIV sequences showed a similarity of 88.3 and 95.4 per cent for nt and aa, respectively with Asian BATV and 94.8 and 97.8 per cent for nt and aa, respectively with African BATV, whereas MAGV and ILEV made a distant branch.
Fig. 2.
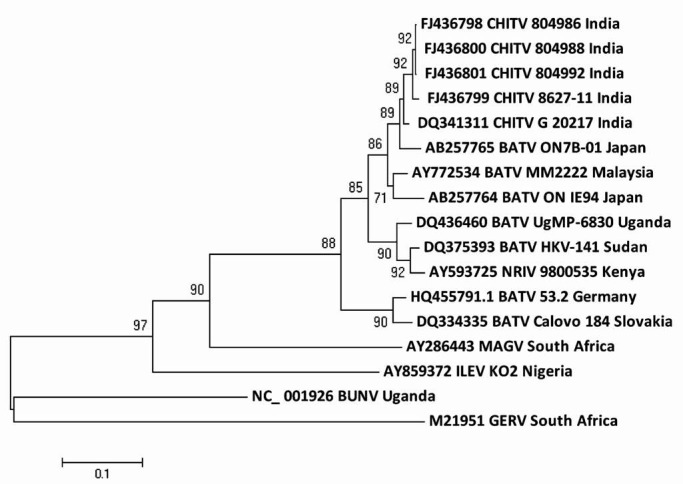
Phylogenetic tree of nucleotide sequences of CHITV and representative other BATV for complete M segment.
Phylogenetic analysis performed with partial M segment of BATV sequences including West Ukraine and Slovakia BATV sequences showed 14.8-17.4 per cent nt and 1.9-4.6 per cent aa difference, respectively with CHITV. This analysis suggests that three genotypes of BATV are circulating globally (Fig. 3).
Fig. 3.
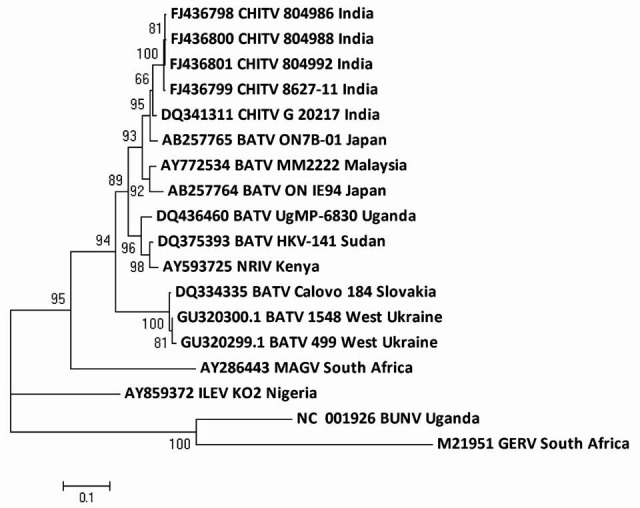
Phylogenetic tree of nucleotide sequences of CHITV and representative other BATV using neighbor joining method for partial M segment including West Ukraine isolates were constructed Bunyamwera virus was used as a outgroup.
RNA polymerase gene phylogenetic tree revealed the presence of two phylogenetic lineages (Asian and European). Similarly, all the Asian isolates clubbed together and made one lineage including isolates from India (FJ436793-7), Japan (AB257766, AB257767) and Malaysia (AY288469, AB257766). European lineage included sequence from Russia and west Ukraine. Among CHITV isolates, difference was only 0.0-2.6 per cent and 0.0-2.1 per cent, while with other BATV isolates, it was 4.7-7.5 per cent and 1.1-2.8 per cent at nt and aa level. With NRIV, CHITV isolates showed a difference of 29.3 per cent at nt and 11.0 per cent for aa level. The ILEV showed a difference of 29.9 and 20.0 per cent with CHITV isolates at nt and aa level, respectively. Reassortant virus NRIV showed high diversity to BATV and clustered with ILEV, MAGV and BUNV (NC_001925) with a similarity of 80.5 and 100.0 per cent at nt and aa level (Fig. 4).
Fig. 4.
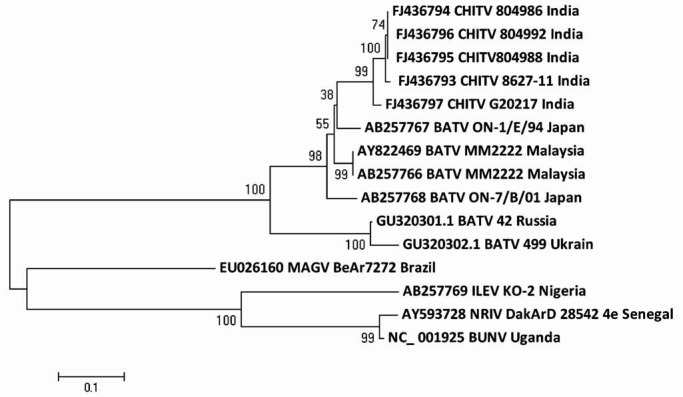
Phylogenetic tree of nucleotide sequences of CHITV and representative other BATV for partial L segment.
Discussion
During the present study, complete M and S segments and partial L segment were amplified and sequenced to characterize the five isolates of CHITV from India. It seems that genetically uniform strain of CHITV is circulating in India, despite being isolated from different hosts, i.e. mosquitoes and pigs. Perhaps, the origin of CHITV was a single strain, which was recently invaded from other region. The strains isolated in the 1960s and 1980s have not changed, indicating the conserved nature of the virus even after several years of circulation in nature. In this study not only S and L but M segment of CHITV also showed high conservation for nucleotide and amino acid level. This kind of conservation has been reported in S, M and L segments of BATV5,9.
The existence of three different genotypes of BATV circulating globally is known. Studies carried out recently19,20 also reported similar finding during the characterization of BATV isolates based on partial M segment. These studies also confirmed that despite geographical demarcation BATV is highly conserved and none of the CHITV isolates has undergone any reassortment.
In the past, a large number of bunyaviruses have been isolated in India. Among these, some of the bunyaviruses like Ingwavuma, Sathuperi, Thottapalaym (TPM), Umbre (UMB), sand fly fever viruses, Ganjam virus were characterized only at the serological level21–26. Recently, UMBV, TPMV, Ganjam virus were characterized by sequencing, which has helped in the designing of new primers for diagnosis27–29.
More studies are required in a larger set of bunyavirus isolates, including BATV to throw light on the origin of the virus and its maintenance in nature to understand their public health importance in India.32
References
- 1.Casals J, Whitman L. A new antigenic group of arthropod-borne viruses. Am J Trop Med Hyg. 1960;9:73–7. doi: 10.4269/ajtmh.1960.9.73. [DOI] [PubMed] [Google Scholar]
- 2.Bardos V, Cupkova E. The Calovo virus - the second virus isolated from mosquitoes in Czechoslovakia. J Hyg Epidemiol Microbiol Immunol. 1962;6:86–192. [PubMed] [Google Scholar]
- 3.Singh KRP, Pavri KM. Isolation of Chittoor virus from mosquitoes and demonstration of serological conversions in sera of domestic animals at Manjri, Poona, India. Indian J Med Res. 1966;54:220–4. [PubMed] [Google Scholar]
- 4.Geevarghese G, Prasanna NY, Jacob PG, Hanumaiah, Bhat HR. Isolation of Batai virus from sentinel domestic pig from Kolar district in Karnataka State, India. Acta Virol. 1994;38:239–40. [PubMed] [Google Scholar]
- 5.Briese T, Bird B, Kapoor K, Nichol ST, Lipkin WI. Batai and Ngari viruses, M segment reassortment and association with severe febrile disease outbreaks in East Africa. J Virol. 2006;80:5627–30. doi: 10.1128/JVI.02448-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaidamovich SY, Obukhova V R, Vinograd AI, Klisenko GA, Melnikova EE. Olkya - an arbovirus of the Bunyamwera group in the USSR. Acta Virol. 1973;17:444. [PubMed] [Google Scholar]
- 7.Klimas RA, Uclerx-van Haastev CM, Bishop DHL. Radioimmune assays and molecular studies that place Anopheles B and Turlock serogroup viruses in the Bunyavirus genus (Bunyaviridae) Am J Trop Med Hyg. 1981;30:876–87. doi: 10.4269/ajtmh.1981.30.876. [DOI] [PubMed] [Google Scholar]
- 8.Nashed NW, Olson JG, el-Tigani A. Isolation of Batai virus (Bunyaviridae:Bunyavirus) from the blood of suspected malaria patients in Sudan. Am J Trop Med Hyg. 1993;48:676–81. doi: 10.4269/ajtmh.1993.48.676. [DOI] [PubMed] [Google Scholar]
- 9.Wang F, Lv Z, Wang J, Fu S, Zhang H, Wang Z, et al. Sequencing and analysis of the full coding sequence of Batai virus isolated in China. Chinese J Virol. 2009;25:83–7. [PubMed] [Google Scholar]
- 10.Yanase T, Kato T, Yamakawa M, Takayoshi K, Nakamura K, Kokuba T, et al. Genetic characterization of Batai virus indicates a genomic reassortment between orthobunyaviruses in nature. Arch Virol. 2006;151:2253–60. doi: 10.1007/s00705-006-0808-x. [DOI] [PubMed] [Google Scholar]
- 11.Hubalek Z. Mosquito-borne viruses in Europe. Parasitol Res. 2008;103:S29–S43. doi: 10.1007/s00436-008-1064-7. [DOI] [PubMed] [Google Scholar]
- 12.Bowen MD, Trappier SG, Sanchez AJ, Meyer RF, Goldsmith CS, Zaki SR, et al. RVF Task Force: A reassortant bunyavirus isolated from acute hemorrhagic fever cases in Kenya and Somalia. Virology. 2001;291:185–90. doi: 10.1006/viro.2001.1201. [DOI] [PubMed] [Google Scholar]
- 13.Soldan SS, González-Scarano F. Emerging infectious diseases: the Bunyaviridae. J Neurovirol. 2005;11:412–23. doi: 10.1080/13550280591002496. [DOI] [PubMed] [Google Scholar]
- 14.Shi X, Kohl A, Léonard V, Li P, McLees A, Elliott RM. Requirement of the N-terminal region of the orthobunyavirus non-structural protein NSm for virus assembly and morphogenesis. J Virol. 2006;80:8089–99. doi: 10.1128/JVI.00579-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bridgen A, Weber F, Fazakerley JK, Elliot RM. Bunyamwera bunyaviruses nonstructural protein NSs is a nonessential gene product that contributes to viral pathogensis. Proc Natl Acad Sci USA. 2001;98:664–9. doi: 10.1073/pnas.98.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weber F, Bridgen A, Fazakerley JK, Streitenefeld H, Kessler N, Randall RE, et al. Bunyamwera bunyaviruses nonstructural protein NSs counteracts the induction of alpha/beta interferon. J Virol. 2002;76:7949–55. doi: 10.1128/JVI.76.16.7949-7955.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pavri KM, Sheikh BH. Distribution of antibodies reacting with Chittoor virus in humans and domestic ungulates of India. lndian J Med Res. 1966;54:225–8. [PubMed] [Google Scholar]
- 18.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 19.Terekhin SA, Grebennikova TV, Khutoretskaya NV, Butenko AM. Molecular genetic analysis of strains of Batai virus isolated from mosquitos in volgograd Region of Russian Federation, West Ukraine, and the Czech Republic. Mol Genet Microbiol Virol. 2010;25:31–3. [PubMed] [Google Scholar]
- 20.Jöst H, Bialonski A, Schmetz C, Günther S, Becker N, Schmidt-Chanasit J. Isolation and phylogenetic analysis of Batai virus, Germany. Am J Trop Med Hyg. 2011;84:241–3. doi: 10.4269/ajtmh.2011.10-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pavri KM, Sheikh BH, Singh KRP, Rajagopalan PK, Casals J. Balagodu virus, a new arbovirus isolated from Ardeola grayii (Sykes) in Mysore state, south India. Indian J Med Res. 1969;57:758–74. [PubMed] [Google Scholar]
- 22.Carey DE, Reuben R, Panicker KN, Shope RE, Myers RM. Thottapalayam virus: A presumptive arbovirus isolated from a shrew in India. Indian J Med Res. 1971;59:1758–60. [PubMed] [Google Scholar]
- 23.Dandawate CN, Rajagopalan PK, Pavri KM, Work TH. Virus isolations from mosquitoes collected in North Arcot District, Madras State & Chittoor district, Andhra Pradesh between November 1955 & October 1957. Indian J Med Res. 1969;57:1420–6. [PubMed] [Google Scholar]
- 24.Dandawate CN, Shah KV. Ganjam virus: a new arbovirus isolated from ticks Haemaphysalis intermedia Warburton and Nuttall, 1909 in Orissa, India. Indian J Med Res. 1969;57:799–804. [PubMed] [Google Scholar]
- 25.Govardhan MK, Dhanda V, Modi GB, Bhatt PN, Bhagwat RB, Dandawate CN, et al. Isolation of Phlebotomus (Sandfly) fever virus from sandflies & humans during the same season in Aurangabad Distract, Mahatrashtra State, India. Indian J Med Res. 1976;64:57–63. [PubMed] [Google Scholar]
- 26.Joshi MV, Elankumaran S, Joshi GD, Albert A, Padbidri VS, Manohar MB, et al. A post-epizootic survey of Rift Valley Fever-like illness among sheep at Veerapuram, Chennai, Tamil Nadu. Indian J Virol. 1998;14:155–7. [Google Scholar]
- 27.Yadav PD, Mishra AC, Mourya DT. Molecular characterization of Umbre virus (Bunyaviridae) Virol J. 2008;5:115–7. doi: 10.1186/1743-422X-5-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yadav PD, Vincent MJ, Nichol ST. Thottapalayam virus is genetically distant to the rodent-borne hantaviruses, consistent with its isolation from the Asian house shrew (Suncus murinus) Virol J. 2007;4:80–2. doi: 10.1186/1743-422X-4-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yadav PD, Vincent MJ, Khristova M, Kale C, Nichol ST, Mishra AC, et al. Genomic analysis reveals Nairobi sheep disease virus to be highly diverse and present in both Africa, and in India in the form of the Ganjam virus variant. Infect Genet Evol. 2011;11:1111–20. doi: 10.1016/j.meegid.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 30.Singh KRP, Pavri KM. Isolation of Chittoor virus from mosquitoes and demonstration of serological conversions in sera of domestic animals at Manjri, Poona, India. Indian J Med Res. 1966;54:220–4. [PubMed] [Google Scholar]
- 31.Jacob PG, Hanumaiah, Bhat HR. Isolation of Batai virus from sentinel domestic pig from Kolar district in Karnataka State, India. Acta Virol. 1994;38:239–40. [PubMed] [Google Scholar]
- 32.Dandawate CN, Rajagopalan PK, Pavri KM, Work TH. Virus isolations from mosquitoes collected in North Arcot District, Madras State & Chittor district, Andhra Pradesh between November 1955 & October 1957. Indian J Med Res. 1969;57:1420–6. [PubMed] [Google Scholar]


