Abstract
Background & objectives:
Haemophilus influenzae is an important cause of mortality and morbidity among young children in developing countries. Increasing incidence of antibiotic resistance especially production of extended spectrum beta lactamase (ESBL) has made treatment and management of H. influenzae infection more difficult. Nasopharyngeal H. influenzae isolates are excellent surrogate for determination of antibiotic resistance prevalent among invasive H. influenzae isolates. In this study, we characterized nasopharyngeal H. influenzae isolates obtained from healthy school going children in Delhi.
Methods:
Nasopharyngeal H. influenzae isolates were collected from healthy school going children and subjected to serotyping, fimbrial typing and antibiogram profiling. ESBL production was recorded using phenotypic as well as molecular methods. Multi locus sequence typing (MLST) of 13 representative nasopharyngeal H. influenzae isolates was performed as per guidelines.
Results:
A significant proportion (26 of 80, 32.5%) of nasopharyngeal isolates of H. influenzae were identified as serotype b. Fimbrial gene (hifA) was detected in 23 (28.75%) isolates. Resistance against commonly prescribed antibiotics (Amp, Tet, Chloro, Septran, Cephalexin) were observed to be high among the nasopharyngeal commensal H. influenzae. Extended spectrum beta lactamase (ESBL) production was observed in a five (6.25%) isolates by both double disk diffusion and molecular typing. MLST identified several novel alleles as well as novel sequence types.
Interpretation & conclusions:
Our findings showed high resistance against common antibiotics and detection of ESBL in nasopharyngeal H. influenzae isolates collected from normal healthy school going children in Delhi. Detection of H. influenzae type b capsular gene and the presence of fimbrial gene (hif A) suggest virulence potential of these isolates. Discovery of novel alleles and presence of new sequence types (STs) among nasopharyngeal H. influenzae isolates may suggest wider genetic diversity.
Keywords: ESBL, Haemophilus influenzae, MLST, nasopharyngeal isolates, serotype b
Haemophilus influenzae is an important aetiological agent of respiratory tract infection in young children in developing countries. An estimated 3 million cases of meningitis and severe pneumonia and approximately 386,000 deaths occur every year worldwide in children below the age of five years due to type b H. influenzae infection (Hib)1. The organism has been classified on the basis of capsular polysaccharide into six serotypes (a to f) as well as unencapsulated type2. In the developed countries, introduction of H. influenzae type b conjugate vaccine has almost eradicated the problem of invasive Hib diseases. However, the pathogen is still a major cause of concern in the developing countries, where incidence of invasive infection is ten times higher than that of the developed countries in prevaccination era3. In India, Invasive Bacterial Infections Surveillance (IBIS) study reported Hib to be a major cause of acute bacterial meningitis in addition to Streptococcus pneumoniae4. Similar Hib disease burden reports are also available from Pakistan5,6, Bangladesh7 and Nepal8. Resistance against multiple antibiotics and production of beta lactamases had been observed in many countries among H. influenzae9–12.
Pathogenic potential of commensal H. influenzae is unknown and unexplored. It is accepted that since the isolation of invasive Hib is difficult at times, the commensal Hib isolates are taken as surrogate to examine antibiotic resistance in invasive Hib isolates13. Type b H. influenzae is known to be carried in the nasopharynx of young children. Therefore, characterization of nasopharyngeal isolates can be taken as a surrogate for invasive isolates. The rates of carriage of type b H. influenzae vary from 0.6 to 13.2 per cent14–16. More importantly, it is thought that type b nasopharyngeal isolates may act as a source of infection among the siblings in the community. In this study, H. influenzae isolates obtained from nasopharynx of healthy school going children in the Delhi region were analysed for the presence of serotype b capsule, fimbriae, antibiotic resistance pattern, extended spectrum beta lactamase (ESBL) production and genetic diversity.
Material & Methods
Bacterial strains and culture conditions: This retrospective study was done in the Department of Microbiology, All India Institute of Medical Sciences (AIIMS), New Delhi, with 80 H. influenzae isolates through nasopharyngeal swabs from healthy school going children between the age of 5-14 yr in Delhi region during August, 2005 to July, 2007. Nasopahryngeal swabs were collected by a physician and transported to the laboratory in skim milk, tryptone, glucose, glycerol transport medium (STGG). The swabs were cultured on blood agar and chocolate agar medium for isolation of both Streptococcus pneumoniae and Haemophilus influenzae. Children studying in schools situated in both rural and urban areas of Delhi and National Capital Region (NCR) were screened after taking ethical clearance from Institute Ethical Committee. Among the 80 isolates 26 were type b. According to statistician this was a good number for making a statistically significant analysis. Informed consent was obtained from parents/ guardian of child before collecting samples. The study protocol was approved by institutional ethics committee of AIIMS. The nasopharyngeal swabs were cultured on Muller-Hinton chocolate agar at 37°C in an atmosphere of 5 per cent CO2 for 16-18 h for growth. Identification of H. influenzae with standard diagnostic assays like oxidase test, Gram stain, satellitism and their growth dependence on factors X and V was thought to be adequate for diagnosis7. In addition, since the incidence of invasive diseases caused by H. aegyptius and H. haemolyticus is not significant in comparison to Hib, tests for differentiating H. influenzae from these two species were not done. The main focus of the study was commensal Hib isolates. H. influenzae ATCC 49247, H. influenzae ATCC 49766 and Escherichia coli ATCC 35218 were maintained in the laboratory as reference strains.
Serotyping of H. influenzae: Serotyping was carried out using slide agglutination test (SAT) with monospecific antiserum (b) (Murex, UK) as per the manufacturer's instructions. As the main focus of this study was to characterize commensal Hib isolates with regard to their pathogenic potential, only type b antiserum was used. Serotyping results were confirmed using PCR based on type b capsule specific primers bl 5’GCGAAAGTGAACTCTTATCTCTC3’ and b2 5’GCTTACGCTTCTATCTCGGTGAA3’ as described earlier2. Non-typable isolates were not identified. Capsule-deficient mutants of serotype b (b-) were not characterized as all capB PCR positive isolates were confirmed by slide agglutination test.
None of the respiratory pathogens including Corynebacterium diphtheriae, Streptococcus pyogenes, Bordetella pertussis, carry bexA gene and since type b PCR is a standardized test as per published literature, bexA PCR was not carried out17,18. Positive and negative controls were used with each batch of PCR. However, internal controls were not used in this study.
Fimbrial typing: Commensal isolates of H. infuenzae are known to make use of fimbriae for initial colonization to host epithelial cells. PCR method was carried out for detection of presence of haemagglutinating fimbriae based on hifA gene that encodes for the major subunit of haemagglutinating fimbirae of H. influenazae. The gene hifA was amplified as described previously19. Primers were taken from the report of Geluk et al20.
Antimicrobial susceptibility testing: Antimicrobial susceptibility testing was carried out by disk diffusion method on Haemophilus test medium according to Clinical Laboratory Standards Institute (CLSI) guidelines, 200621. Susceptibility was tested against the following antibiotics (μg): ampicillin (10), amoxcillin-clavulinic acid (20/10), cefotaxime (30), cefepime (30), cefixime (5), azithromycin (15), tetracycline (30), ciprofloxacin (5), trimethoprim-sulphamethoxazole (1.25-23.75), chloramphenicol (30), and rifampin (30) by Kirby Bauer's disc diffusion method. H. influenzae ATCC 49247, H. influenzae ATCC 49766 and E. coli ATCC 35218 (when testing amoxcillin-clavulinic acid) were used as quality control bacteria.
ESBL screening and confirmation: ESBL screening was based upon chromogenic cephalosporins (nitrocephin) test and double disc diffusion method as per CLSI guidelines22. Isolates of H. influenzae producing ESBL were subjected to PCR for detection of TEM and SHV genes using the primers TEM: Forward 5’ CTTCCTGTTTTTGCTCACCCA 3’, Reverse 5’ ACGATACGGGAGGGCTTAC 3’ and SHV: Forward 5’ TCAGCGAAAAACACCTTG 3’ Reverse 5’ TCCCGCAGATAAATCACC 3’22. Although SHV is not known to play any role in ESBL production in H. influenzae, these plasmids can get transferred from other Gram negative bacteria. So the presence of SHV plasmid was also screened.
Multi locus sequence typing: Phylogenetic relationships among H. influenzae isolates were determined by multilocus sequence typing (MLST) of seven housekeeping genes adenylate kinase (adk), ATP synthase F1 subunit Gamma (atpG), fumerate reductase iron sulphur protein (frdB), fuculokinae (fucK), malate dehydrogenase (mdh), glucose -6- phosphate isomerase (pgi), recA protein (recA) as described in the MLST database (http://haemophilus.mlst.net). The primers used for the amplification of the housekeeping gene were as described elsewhere23. Due to economic constrains, MLST was carried out in only 13 isolates which were selected randomly. Out of these isolates 01, 08, 06, 13, 10, 03 and 12 were type b and isolates 02, 07, 11, 05 and 09 were non-type b isolates. Sequencing of all seven house keeping genes was carried out on both strands. Same primers used for both amplification and sequencing. High-fidelity enzyme AmpliTaq Gold DNA polymerase (Applied Biosystems, USA) was used for all PCR reactions. Big Dye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA) was used for cycle sequencing reactions. Sequencing was performed on Applied Biosystems 3130xl platform. Sequence analysis was done using SeqScape v2.6 software (Applied Biosystems). Phylogenetic analysis was performed using MEGA424. Dendogram was constructed for 13 isolates with combined sequences of seven housekeeping genes in the order adk, atpG, frdB, fucK, mdh, pgi and recA.
Results
Serotyping: Of the 80 H. influenzae isolates studied, 41 (51.25%) demonstrated presence of serotype b capsule production as tested by SAT. However, PCR amplification of capsular b gene (Fig. 1) revealed presence of capB gene in 26 (32.5%) isolates showing a discrepancy in 15 of 80 (18.75%) isolates. All the capB positive isolates by PCR were also positive by SAT.
Fig. 1.
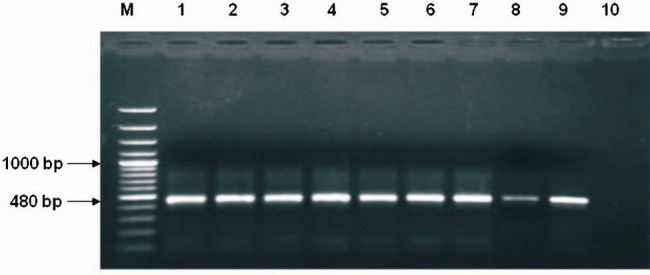
PCR amplified product (capB) in 1.5% agarose gel. Lane M: 100 bp Ladder, Lane 1: Positive control, Lanes 2-9: amplification of 480 bp from H. influenzae isolates, Lane 10: Negative control.
Fimbrial typing: For detection of fimbriae among the H. influnzae isolates, PCR amplification based on hifA gene was carried out (Fig. 2). Twenty three isolates (28.75%) demonstrated presence of fimbrial gene hifA. Majority of the nasopharyngeal H. influenzae isolates did not show presence of any haemagglutinating fimbriae. Of the 23 isolates carrying hifA gene, three were type b and 20 were of non type b serotype including possible non-typeable isolates (NTHi) which were not characterized.
Fig. 2.
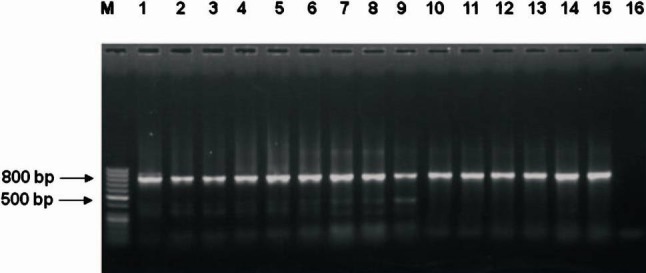
PCR amplified product (hifA) electrophoresed in 1.5% agarose gel. Lane M: 50 bp Ladder, Lane1: Positive control, Lanes 2-15: amplified 800 bp product H. influenzae isolates, Lane 16: Negative control.
Antimicrobial susceptibility testing: High resistance was observed against the commonly prescribed antibiotics. Resistance to ampicillin was shown by highest number of isolates 65 (81.25%) followed by tetracycline (45, 56.25%). Resistance to a β lactam- β lactamase inhibitor combination, combination of amoxicillin-clavilunic acid was exhibited by 13 (16.25%) isolates. Amongst the cephems (parenteral) antibiotics, 29 (36.25%) and 36 (45%) isolates showed resistance to cefotaxime and cefepime. In the cephems (oral), cefexime resistance was exhibited by 33 (41.25%) isolates. Only two isolates showed resistance against the macrolide antibiotic azithromycin, and seven showed ciprofloxacin resistance. Towards the foliate pathway inhibitors, TMP-SMZ resistance was shown by 27 (33.75%) isolates. Resistance against chloramphenicol and rifampin was observed in 31 (38.75%) and 18 (22.5%) isolates, respectively.
Screening of extended spectrum Beta-lactamases: Chromogenic nitrocephin test confirmed seven (8.75%) isolates to be β-lactamase producer. ESBL production was detected using double disc diffusion method in five (6.25%) isolates. Molecular detection of ESBL using PCR amplification of TEM and SHV plasmid, detected TEM gene in five (6.25%) isolates (Fig. 3).
Fig. 3.
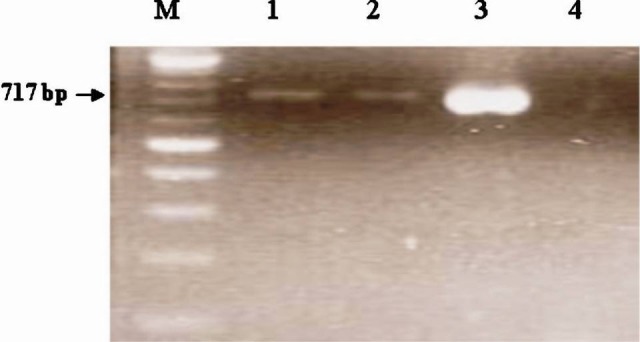
PCR amplified product (TEM) electrophoresed in 1.5% agarose gel. Lane M: 100 bp Ladder, Lanes 1-2: amplification of 717 bp from H. influenzae isolates, Lanes 3: Klebsiella pneumoniae ATCC 700603 and Lane 4: Negative control.
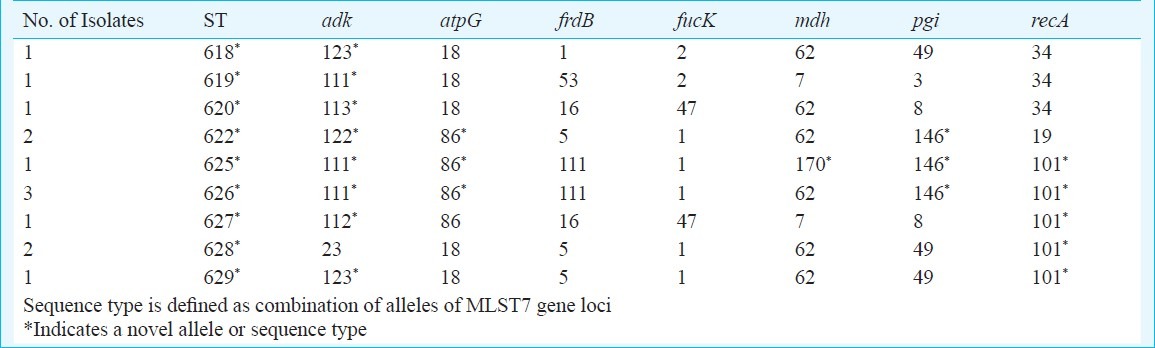
Multi locus sequence typing (MLST): Unique sequence types and high genetic variation were found to exist amongst the H. influenzae isolates obtained from the nasopharynx of healthy school going children in Delhi (Fig. 4). A total of 13 isolates were subjected to MLST. Nine new MLST alleles including adk alleles: 111, 112, 113, 122 and 123, atpG allele: 86, mdh allele: 170, pgi allele: 146, and recA allele: 101 were discovered. Nine novel, unique sequence types (STs) were discovered and the sequence information was submitted to the MLST database at http://haemophilus.mlst.net (STs 622, 618, 619, 620, 625, 626, 627, 628, and, 629). STs 625 and 626 are single locus variants differing at mdh locus. ST 628 and 629 are single locus variants differing at adk locus. Variations of MLST types (STs) at many loci indicated wider genetic diversity prevalent among nasopharyngeal isolates of H. influenzae (Table).
Fig. 4.
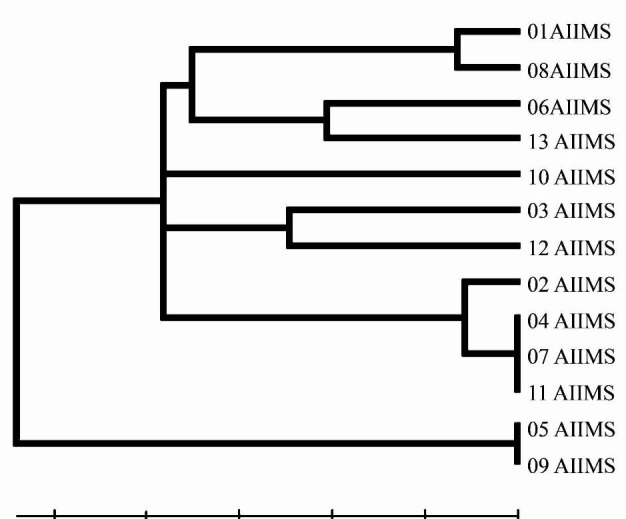
H. influenzae isolates UPGMA dendrogram based on MLST data for 13 type b.
Table.
MLST allelic profile generated from nasopharyngeal H. influenzae isolates from this study
Discussion
The carriage rates of H. influenzae type b among healthy individuals had been reported to vary from 0.6 to 13.2 per cent in different parts of the world14–16. The significance of invasive Hib disease as a major cause of morbidity and mortality in young children in India had been demonstrated by IBIS study4. Carriage of capsular type b H. infleunzae among healthy children in the community, may act as a source of infection for siblings. At the same time, carriage of the organism in the nasopharynx may also induce immunity among the younger population in the community through natural infection. Since almost all the isolates belonged to type III biotype, it was felt that it is not necessary to do further biotyping. The study mainly looked at the virulence markers and genomic relatedness of the isolates. Therefore, some of the phonotypic markers were not characterized. Studies reported increasing resistance against commonly prescribed antibiotics among invasive isolates of H. influenzae in India4,16,25. Appearance of ESBL production may indicate difficulties in management of H. influenzae related diseases in future. Only seven isolates were positive for beta lactamase as compared to higher number of isolates found to be resistant to third generation cephalosporins through disc diffusion testing. More than 30 different plasmids have been identified as responsible for resistance of Gram-negative bacteria to third generation cephalosporins26. Such plasmids are capable of getting transferred from one species to another through conjugation. TEM is reported as one of the major plasmid associated with ESBL production in H. influenzae27. Role of SHV in ESBL production in H. influenzae is not reported.
The disparty seen in this report is suggetive of some other mechanism of ESBL production in isolates circulating in Delhi region. Capsular serotyping by slide agglutination test, an easy method of identifying more virulent H. influenzae type b demonstrated false positive results, PCR serotyping method based on the presence or absence of capB gene loci, offered better results28. Carriage of type b H. influenzae varies in different geographical regions, which was observed to be high in this study indicating a potential source of infection for self and siblings. Thus it is important to determine the fimbrial presence. Reports suggest that fimbriae are abundantly present on the cell surface of hifA-positive strains29. Thus hifA positivity was considered to be indicative of presence of fimbriae. Presence of the fimbrial gene does not confirm expression of fimbriae. However, absence of the same in majority of the isolates is indicative of fimbriae independent mechanism of adherence. As the main focus of the study was type b H. influenzae, isolates were broadly categorized as type b and non-type b which also included non-typable isolates. Lack of haemagglutinating fimbiae among invasive type b H. influenzae has been reported earlier, presence of haemagglutinating fimbriae in lower number nasopharyngeal isolates in our study also suggest existence of alternative mechanism of adherence30.
MLST is a method used for both typeable and non typeable H. influenzae. In this study the genetic relatedness of type b and non-type b H. influenzae isolates circulating in Delhi was analyzed. A total of 774 different sequence types are listed in the MLST database till date. Sequence type diversity has been reported from various parts of the world (http://haemophilus.mlst.net). MLST data of nasopharyngeal H. influenzae isolates in our study revealed presence of many new alleles and sequence types (STs). Existence of wider genetic diversity among nasopharyngeal isolates, as shown by phylogenetic analysis, strongly suggest the possibility that non invasive nasopharyngeal H. influenzae isolates are under less evolutionary pressure to allow mutation in their genome. In comparison, phylogenetic analysis of invasive isolates (data not shown) reveal that invasive Hib isolates are more conserved in their genome probably indicating different lineage than that of nasopharyngeal H. influenzae isolates.
H. influenzae related diseases remain a major public health problem in developing countries, it is imperative to monitor drug resistance, virulence markers and genetic characteristics to understand the epidemiology of H. infleunzae infection. Appearance of non typeable H. influenzae (NTHi) as a major cause of locally invasive disease in many countries warrants surveillance of H. infleunzae infection. In our study, high resistance was observed against commonly used antibiotics including ESBL production. Presence of capB and hifA genes indicated virulence potential of these commensal isolates.
Acknowledgment
Authors acknowledge the Department of Science and Technology, Government of India, New Delhi, for financial support.
Footnotes
Conflict of Interest: None.
References
- 1.World Health Organization, Strategic advisory group of experts on immunization; The WHO position paper on Haemophilus influenzae type b conjugate vaccines. Wkly Epidemiol Rec. 2006;81:445–52. [PubMed] [Google Scholar]
- 2.Falla TJ, Crook DWM, Brophy LN, Makell D, Kroll JS, Moxon ER. PCR for capsular typing of Haemophilus iinfluenzae. J Clin Microbiol. 1994;32:2382–6. doi: 10.1128/jcm.32.10.2382-2386.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watt JP, Wolfson LJ, O’Brien KL, Henkle E, Deloria-Knoll M, McCall N, et al. Burden of disease caused by Haemophilus influenzae type b in children younger than 5 years: global estimates. Lancet. 2009;374:903–11. doi: 10.1016/S0140-6736(09)61203-4. [DOI] [PubMed] [Google Scholar]
- 4.Invasive Bacterial Infections Surveillance (IBIS) Group of the International Clinical Epidemiology Network. Are Haemophilus influenzae infections a significant problem in India. A prospective study and review? Clin Infect Dis. 2002;34:949–57. doi: 10.1086/339327. [DOI] [PubMed] [Google Scholar]
- 5.Weinberg GA, Ghafoor A, Ishaq Z, Nomani NK, Kabeer M, Anwar F, et al. Clonal analysis of Hemophilus influenzae isolated from children from Pakistan with lower respiratory tract infections. J Infect Dis. 1989;160:634–43. doi: 10.1093/infdis/160.4.634. [DOI] [PubMed] [Google Scholar]
- 6.Zaidi AK, Khan H, Lasi R, Mahesar W. Sindh Meningitis Group, Surveillance of pneumococcal meningitis among children in Sindh, southern Pakistan. Clin Infect Dis. 2009;48(Suppl 2):S129–35. doi: 10.1086/596491. [DOI] [PubMed] [Google Scholar]
- 7.Saha SK, Baqui AH, Darmstadt GL, Ruhulamin M, Hanif M, Arifeen S, et al. Invasive Haemophilus influenzae type b diseases in Bangladesh, with increased resistance to antibiotics. J Pediatr. 2005;146:227–33. doi: 10.1016/j.jpeds.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 8.Sharma PR, Adhikari RK, Joshi MP, Lal M, Chodon T, Pokhrel BM, et al. Intravenous chloramphenicol plus penicillin versus intramuscular ceftriaxone for the treatment of pyogenic meningitis in Nepalese children. Trop Doct. 1996;26:84–5. doi: 10.1177/004947559602600215. [DOI] [PubMed] [Google Scholar]
- 9.Wang A, Yu S, Yao K, Zhang W, Yuan L, Wang Y, et al. Antimicrobial susceptibility of Haemophilus influenzae strains and antibiotics usage patterns in pediatric outpatients: results from a children's hospital in China (2000-2004) Pediatr Pulmonol. 2008;43:457–62. doi: 10.1002/ppul.20789. [DOI] [PubMed] [Google Scholar]
- 10.The PROTEKT surveillance study: antimicrobial susceptibility of Haemophilus influenzae and Moraxella catarrhalis from community-acquired respiratory tract infections. J Antimicrob Chemother. 2002;50(Suppl S1):49–59. doi: 10.1093/jac/dkf810. [DOI] [PubMed] [Google Scholar]
- 11.Sahm DF, Brown NP, Thornsberry C, Jones ME. Antimicrobial susceptibility profiles among common respiratory tract pathogens: a GLOBAL perspective. Postgrad Med. 2008;120(3 Suppl 1):16–24. doi: 10.3810/pgm.2008.09.suppl52.280. [DOI] [PubMed] [Google Scholar]
- 12.Jansen WT, Verel A, Beitsma M, Verhoef J, Milatovic D. Surveillance study of the susceptibility of Haemophilus influenzae to various antibacterial agents in Europe and Canada. Curr Med Res Opin. 2008;24:2853–61. doi: 10.1185/03007990802381505. [DOI] [PubMed] [Google Scholar]
- 13.Das BK, Arora NK, Mathur P, Ostwal P, Mandal S, Kabra SK, et al. Nasopharyngeal carriage of Haemophilus influenzae. Indian J Pediatr. 2002;69:775–7. doi: 10.1007/BF02723690. [DOI] [PubMed] [Google Scholar]
- 14.Wang SR, Lo WT, Chou CY, Chen YY, Tsai SY, Chu ML, et al. Low rate of nasopharyngeal carriage and high rate of ampicillin resistance for Haemophilus influenzae among healthy children younger than 5 years old in northern Taiwan. Microbiol Immunol Infect. 2008;41:32–40. [PubMed] [Google Scholar]
- 15.Factor SH, LaClaire L, Bronsdon M, Suleymanova F, Altynbaeva G, Kadirov BA, et al. Streptococcus pneumoniae and Haemophilus influenzae type B Carriage, Central Asia. Emerg Infect Dis. 2005;11:1476–9. doi: 10.3201/eid1109.040798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jain A, Kumar P, Awasthi S. High ampicillin resistance in different biotypes and serotypes of Haemophilus influenzae colonizing the nasopharynx of healthy school-going Indian children. J Med Microbiol. 2006;55:133–7. doi: 10.1099/jmm.0.46249-0. [DOI] [PubMed] [Google Scholar]
- 17.Sam IC, Smith M. Failure to detect capsule gene bexA in Haemophilus influenzae types e and f by real-time PCR due to sequence variation within probe binding sites. J Med Microbiol. 2005;54:453–5. doi: 10.1099/jmm.0.45836-0. [DOI] [PubMed] [Google Scholar]
- 18.Kapogiannis BG, Satola S, Keyserling HL, Farley MM. Invasive infections with Haemophilus influenzae serotype a containing an IS1016-bexA partial deletion: possible association with virulence. Clin Infect Dis. 2005;41:e97–03. doi: 10.1086/498028. [DOI] [PubMed] [Google Scholar]
- 19.van Ham SM, van Alphen L, Mooi FR, van Putten JP. The fimbrial gene cluster of Haemophilus influenzae type b. Mol Microbiol. 1994;13:673–84. doi: 10.1111/j.1365-2958.1994.tb00461.x. [DOI] [PubMed] [Google Scholar]
- 20.Geluk F, Euk PP, Ham SMV, Jansen HM, Alphen LV. The fimbria gene cluster of nonencapsulated Haemophilus influenzae. Infect Immun. 1998;66:406–17. doi: 10.1128/iai.66.2.406-417.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Performance standards for antimicrobial susceptibility testing. Wayne, PA, USA: CLSI; 2006. Clinical and Laboratory Standards Institute (CLSI) M100-S16. [Google Scholar]
- 22.Lal P, Kapil A, Das BK, Sood S. Occurrence of TEM & SHV gene in extended spectrum beta-lactamases (ESBLs) producing Klebsiella sp. isolated from a tertiary care hospital. Indian J Med Res. 2007;125:173–8. [PubMed] [Google Scholar]
- 23.Meats E, Feil EJ, Stringer S, Cody AJ, Goldstein R, Kroll JS, et al. Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J Clin Microbiol. 2003;41:1623–36. doi: 10.1128/JCM.41.4.1623-1636.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 25.Nag VL, Ayyagari A, Venkatesh V, Ghar M, Yadav V, Prasad KN. Drug resistant Haemophilus influenzae from respiratory tract infection in a tertiary care hospital in North India. Indian J Chest Dis Allied Sci. 2001;43:13–7. [PubMed] [Google Scholar]
- 26.Paterson DL, Bonomo RA. Extended-spectrum β-lactamases: a clinical update. Clin Microbiol Rev. 2005;18:657–86. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farrell DJ, Morrissey I, Bakker S, Buckridge S, Felmingham D. Global distribution of TEM-1 and ROB-1 b-lactamases in Haemophilus influenzae. J Antimicrob Chemother. 2005;56:773–6. doi: 10.1093/jac/dki281. [DOI] [PubMed] [Google Scholar]
- 28.LaClaire LL, Tondella ML, Beall DS, Noble CA, Raghunathan PL, Rosenstein NE, et al. Identification of Haemophilu influenzae serotypes by standard slide agglutination serotyping and PCR-based capsule typing. J Clin Microbiol. 2003;41:393–6. doi: 10.1128/JCM.41.1.393-396.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamura M, Asaka T, Kirita A, Miyazaki H, Senda Y, Fujita S, et al. Occurrence of the fimbria gene hifA in clinical isolates of nonencapsulated Haemophilus influenzae. Microbiol Immunol. 2006;50:327–9. doi: 10.1111/j.1348-0421.2006.tb03800.x. [DOI] [PubMed] [Google Scholar]
- 30.Gilsdorf JR, Mccrea KW, Marrs CF. Role of pili in Haemophilus influenzae adherence and colonization. Infect Immun. 1997;65:2997–3002. doi: 10.1128/iai.65.8.2997-3002.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]


