Sir,
Increase in number of fever cases with joint pains, rashes as common symptoms and respiratory, gastrointestinal symptoms and mortality among a few cases1,2 were reported during July- August 2011 by the district health authorities in Jamshedpur (Latitude: 22°48’N, Longitude: 86°11’E), East Singhbhoom district, Jharkhand. This is a major industrial centre of East India with population of 11,04,713 (Census of India 2001)3. About 30,000 people were reported with fever (on an average daily 500 patients) and visited the outpatient departments of various hospitals in the city4. Tata Main Hospital in Jamshedpur, Jharkhand sent 10 blood samples from hospitalized patients suspected to have viral fever to National Institute of Virology (NIV), Pune on July 25, 2011. In the third week of July, district health authorities also sent serum samples from 10 patients with viral fever to National Center for Disease Control, New Delhi and anti-chikungunya virus (CHIKV) immunoglobulin M (IgM) antibodies were detected in three cases (District Surveillance Officer, Jamshedpur, personal communication). Serum samples from another 10 viral fever cases were also sent to School of Tropical Medicine, Kolkata, and anti-CHIKV IgM antibodies were detected in three cases and anti-dengue virus (DENV) IgM antibodies were detected in two cases suggesting co-circulation. Mortality reported in two fever cases admitted in the hospitals in the city created a panic situation among the citizens, of Jamshedpur2,4. Therefore, NIV, Pune, attempted to investigate the viral fever cases reported from August 2-9, 2011, to establish the aetiology and to suggest the preventive and control measures.
A suspected case was defined as a person with acute history of fever with any one or more of the following symptoms: joint pain, rash, headache, myalgia, photophobia and arthralgia. A subset of suspected cases (n=220) visiting the outpatient department, and/or admitted in tertiary care hospitals (Tata Main Hospital, Mahatma Gandhi Memorial Medical College Hospital and TELCO Hospital) in the city were examined and blood samples (2-3 ml) were collected for diagnosis. Blood samples were also collected from line-listed suspected cases (n=78) by district health authorities from three localities in Jamshedpur i.e. Somaya Zopadi Ghageri South (n=29), Gulti Zopadi Ghageri North (n=18), and Refugee Colony Mango (n=31).
Clinical samples (n=220) [blood from fever cases and cerebrospinal fluid (CSF) [referred by Tata Main Hospital (n=5) from patients presented with fever and neurological involvement] were aseptically collected and transported on ice to National Institute of Virology, Pune. Detection of anti-CHIKV IgM antibodies (n=220) and anti-DENV IgM antibodies (n=220) was carried out by enzyme linked immuno sorbant assay5,6. Blood samples (n=92) collected in early post onset day (duration <7 days) were screened for the presence of CHIKV and DENV by real time polymerase chain reaction (RT-PCR)7.
Intracranial inoculation of suckling mice with suspected clinical specimen for initial isolation and amplification of arboviruses is a classical procedure used worldwide8. Therefore, in this study infant (24-48 h old) Swiss albino mice were used for virus isolation. Methods for extraction, amplification and sequencing of viral RNA have been described previously9,10. Partial gene sequences of non-structural protein 1 (NS1) of CHIKV were used for phylogenetic analysis. Phylogenetic analysis was performed using the freeware11 MEGA 5. Best fit model for nucleotide substitution was selected from 24 models available in MEGA 5 based on minimum Akaike Information Criterion value12. Tamura Nei + I model of nucleotide substitution, obtained in the model test, was used for constructing phylogenetic tree based on maximum likelihood method. Reliability of the phylogenetic tree was estimated using bootstrap values run for 1000 iterations. In the Reunion Island most CHIKV isolated from patients presented an amino-acid substitution in the E1 glycoprotein, from an alanine (E1-226A) to a valine (E1-226V)13. This mutation has selective advantage in Aedes albopictus13. Therefore, it was thought desirable to investigate the presence of the E1-A226V change in these isolates. The same samples were screened for A226V mutation from E1 region by RT-PCR followed by sequencing9.
A total of 13,228 suspected cases were reported by public health authorities at Jamshedpur (District Surveillance Officer, personal communication). Among the 220 suspected cases investigated, 99 were male and 51 per cent were in the age group of 20-39 yr and 89 required hospitalization. Duration of hospitalization ranged between 1 and 14 days (median 4 days). Common clinical feature observed among suspected cases were fever (100%), joint pain (83.2%), rash (52.6%), headache (34.5%), myalgia (33.6%), retro-orbital pain (21%), nausea (20%) and vomiting (20%), diarrhoea (19%), itching (3.2%), oral ulcers (2.3%), cough (1.8%) and sore throat (0.9%). Neurological manifestations included altered sensorium (4.5%), unconsciousness (4.1%) and convulsions (4.1%). Rash was non pruritic, maculopapular and erythematous in character and started appearing 1-3 days after the initial symptom (fever/joint pain) and distributed on face, limbs and trunk of the body. Pre- and post-auricular lymphadenopathy was also noted among four patients.
Clinical samples collected from 220 suspected cases included 220 serum samples, 2 convalescent serum samples and 5 cerebrospinal fluids samples. Anti-CHIKV IgM antibodies were detected in 61.4 per cent (135/220), viral RNA in 45.6 per cent (42/92 i.e., the only available early POD samples) cases [serum collected during early post onset days (duration <7 days)] and both anti-CHIK IgM antibodies and CHIKV RNA detected in two cases. Anti-CHIKV IgM antibodies in serum and CSF sample were also detected in a 45 days old male suspect case presented with febrile convulsions. Anti-DENV IgM antibodies were not detected in any suspected cases, however, two serum samples were indeterminate for anti DENV IgM antibodies. Dengue viral RNA was not detected in any samples (n=92). Thus chikungunya infection was confirmed in 81.4 per cent (179/220) suspected cases. Laboratory findings in chikungunya cases were as follows; total leukocyte count (n=59), less then 4000/ ul was in two cases; platelet count (n=52), less then 1,50,000/ul in 29 cases; serum creatinine (n=37) more then 1.5 mg/dl in four cases; blood urea (n=37) more than 40 mg per cent in eight cases; serum glutamic oxaloacetic transaminase (n=25) more then 40 U/l in 15 cases; serum glutamic pyruvic transaminase (n=35) more than 40 U/l in 19 cases. Malarial parasite was not detected any of the suspected cases.
Mortality was reported in three laboratory confirmed chikungunya cases which presented with co-morbidity; a 45 yr old female with hypertension, a 60 yr old male with ischaemic heart disease and a 68 yr old male with diabetes mellitus. These three patients died due to multi organ failure. Febrile convulsions among children were also reported in two chikungunya positive cases.
CHIKV was isolated from 14 of 20 randomly selected from RTPCR positive serum samples collected (early POD) during the outbreak from suspected chikungunya cases and were RT-PCR positive for CHIKV. Sequencing based on ns1 region revealed homology with the African genotype. A226V mutation from E1 region was not observed in five isolates investigated. For phylogenetic analysis, ns1 sequences were obtained for five chikungunya cases. A phylogenetic tree was constructed based on partial ns1 region of 540 bp, over nucleotide positions 1161 to 1700, numbered according to the CHIKV prototype S27. The five isolates from the current Jamshedpur outbreak matched with recent Central/East African isolates from 2005 to 2008, sharing 98-99 per cent nucleotide identity (Figure). Sequencing of E1 gene of these five chikungunya isolates reveled that there is no amino-acid substitution in the E1 glycoprotein, from an alanine (E1-226A) to a valine (E1-226V).
Fig.
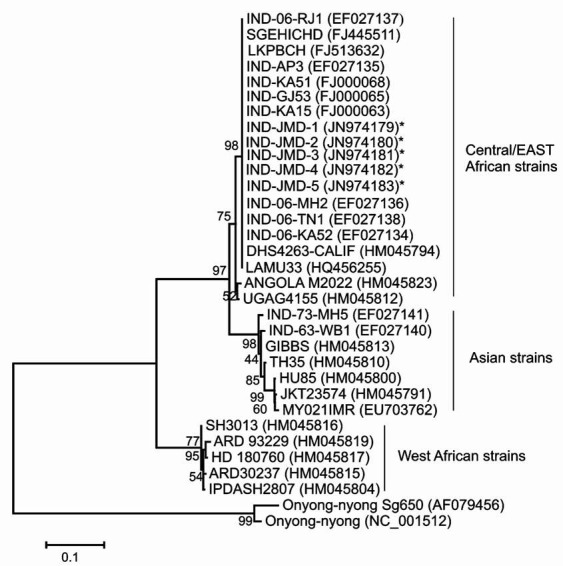
Phylogenetic relationships of chikungunya virus isolates from the 2011 Jamshetpur outbreak, India. The maximum likely hood tree was constructed using nucleic acid sequences of the ns1 gene (1160-1700 bp), with O’nyong nyong virus as an out-group virus. *Denotes the strains of present study (Accession Numbers JN974179, JN974180, JN974181, JN974182 and JN974183). The scale represents genetic distance.
Earlier, during May-September 1954, fever cases with increase in hospital admissions were also reported in Jamshedpur14. However, no conclusive aetiology was established15. The presence of CHIKV antibodies in human serum was also reported in 1964 from Jamshedpur16. Important preventive and control measures suggested to district health authorities include increasing awareness among people about how to avoid mosquito bites and strengthening vector surveillance for Aedes mosquito together with biological, chemical and environmental control measures against mosquitoes. Important observation noted in this outbreak was mortality associated with chikungunya infection in three suspected cases. Mortality due to severe chikungunya infection has been previously reported from Ahmedabad17,18 and Pune17,19.
Acknowledgment
Author thank Dr T.P. Madhusudanan [(AVM) Retd], General Manager, Health Services, Tata Main Hospital, Jamshedpur; Dr Swaran Singh, District Surveillance Officer, Dr Vibha Saran, Civil Surgeon, Jamshedpur East Singhbhoom District; health authorities from Mahatama Gandhi Memorial Hospital and TELCO Hospital Jamshedpur for their support during the investigation.
References
- 1.Viral outbreak puts hospitals on alert. Times of India. 2011. Jul 29, [accessed on August 30, 2011]. Available from: http://articles.timesofindia.indiatimes.com/2011-07-29/ranchi/29828705_1_ viral-fever- hospitalopd .
- 2.Fevered district in virus combat. The Telegraph Calcutta India. 2011. Aug 1, [accessed on August 30, 2011]. Available from: http://www. telegraphindia.com/1110801/jsp/jharkhand/story_14315495.jsp .
- 3.Census of India: List of towns. [accessed on August 30, 2011]. Available from: http://www.censusindia.gov.in/towns/jha_towns.pdf .
- 4.New scare revs up combat- 2 confirmed cases of chikungunya add to viral menace News. The Telegraph Calcutta India. 2011. Aug 2, [accessed on August 30, 2011]. Available from: http://www.telegraphindia.com/1110802/jsp/jharkhand/story_14321967.jsp .
- 5.Hundekar SL, Thakare JP, Gokhale MD, Barde PV, Argade SV, Mourya DT. Development of monoclonal antibody based antigen capture ELISA to detect chikungunya virus antigen in mosquitoes. Indian J Med Res. 2002;115:144–8. [PubMed] [Google Scholar]
- 6.Gadkari DA, Shaikh BH. IgM antibody capture ELISA in the diagnosis of Japanese encephalitis, West Nile and dengue virus infections. Indian J Med Res. 1984;80:613–9. [PubMed] [Google Scholar]
- 7.Gurukumar KR, Priyadarshini D, Patil JA, Bhagat A, Singh A, Shah PS, et al. Development of real time PCR for detection and quantitation of Dengue viruses. Virol J. 2009;6:1–8. doi: 10.1186/1743-422X-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moore CG, McLean RG, Mitchell CJ, Nasci RS, Tsai TF, Calisher CH, et al. Guidelines for arbovirus surveillance programs in the United States. Published by Fort Collins. Colorado: Centers for Disease Control and Prevention, Public Health Service, U.S. Department of Health and Human Services; 1993. [Google Scholar]
- 9.Arankalle VA, Shrivastava S, Cherian S, Gunjikar RS, Walimbe AM, Jadhav SM, et al. Genetic divergence of Chikungunya viruses in India (1963–2006) with special reference to the 2005-2006 explosive epidemic. J Gen Virol. 2007;88:1967–76. doi: 10.1099/vir.0.82714-0. [DOI] [PubMed] [Google Scholar]
- 10.Yergolkar PN, Tandale BV, Arankalle VA, Sathe PS, Sudeep A, Gandhe SS, et al. Chikungunya outbreaks caused by African genotype, India. Emerg Infect Dis. 2006;12:1580–3. doi: 10.3201/eid1210.060529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Posada D, Crandall KA. Selecting the best-fit model of nucleotide substitution. Systems Biol. 2001;50:580–601. [PubMed] [Google Scholar]
- 13.Schuffenecker I, Iteman I, Michault A, Murri S, Frangeul L, Vaney MC, et al. Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS Med. 2006;3:e263. doi: 10.1371/journal.pmed.0030263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khan N. Jamdhedpur fever. Indian J Med Sci. 1954;8:597–607. [Google Scholar]
- 15.Chari MV, Swamy TV. Jamshedpur fever. BMJ. 1955;2:1298–303. doi: 10.1136/bmj.2.4951.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pavari KM. Presence of chikungunya antibodies in human sera collected from Calcutta and Jamshedpur before 1963. Indian J Med Res. 1964;52:698–702. [PubMed] [Google Scholar]
- 17.Tandale BV, Sathe PS, Arankalle VA, Wadia RS, Kulkarni R, Shah S, et al. Systemic involvement and fatalities during Chikungunya epidemic in India, 2006. J Clin Virol. 2009;46:145–9. doi: 10.1016/j.jcv.2009.06.027. [DOI] [PubMed] [Google Scholar]
- 18.Malvankar DV, Shastri P, Bandyopadhyay T, Parmar J, Ramani KV. Increased mortality rate associated with Chikungunya epidemic, Ahemdabad, India. Emerg Infect Dis. 2008;14:412–5. doi: 10.3201/eid1403.070720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wadia RS. A neurotropic virus (chikungunya) and a neuropathic aminoacid (homocystein) Ann Indian Acad Neurol. 2007;10:198–213. [Google Scholar]


