ABSTRACT
The recent emergence of a novel human coronavirus (HCoV-EMC) in the Middle East raised considerable concerns, as it is associated with severe acute pneumonia, renal failure, and fatal outcome and thus resembles the clinical presentation of severe acute respiratory syndrome (SARS) observed in 2002 and 2003. Like SARS-CoV, HCoV-EMC is of zoonotic origin and closely related to bat coronaviruses. The human airway epithelium (HAE) represents the entry point and primary target tissue for respiratory viruses and is highly relevant for assessing the zoonotic potential of emerging respiratory viruses, such as HCoV-EMC. Here, we show that pseudostratified HAE cultures derived from different donors are highly permissive to HCoV-EMC infection, and by using reverse transcription (RT)-PCR and RNAseq data, we experimentally determined the identity of seven HCoV-EMC subgenomic mRNAs. Although the HAE cells were readily responsive to type I and type III interferon (IFN), we observed neither a pronounced inflammatory cytokine nor any detectable IFN responses following HCoV-EMC, SARS-CoV, or HCoV-229E infection, suggesting that innate immune evasion mechanisms and putative IFN antagonists of HCoV-EMC are operational in the new host. Importantly, however, we demonstrate that both type I and type III IFN can efficiently reduce HCoV-EMC replication in HAE cultures, providing a possible treatment option in cases of suspected HCoV-EMC infection.
IMPORTANCE
A novel human coronavirus, HCoV-EMC, has recently been described to be associated with severe respiratory tract infection and fatalities, similar to severe acute respiratory syndrome (SARS) observed during the 2002-2003 epidemic. Closely related coronaviruses replicate in bats, suggesting that, like SARS-CoV, HCoV-EMC is of zoonotic origin. Since the animal reservoir and circumstances of zoonotic transmission are yet elusive, it is critically important to assess potential species barriers of HCoV-EMC infection. An important first barrier against invading respiratory pathogens is the epithelium, representing the entry point and primary target tissue of respiratory viruses. We show that human bronchial epithelia are highly susceptible to HCoV-EMC infection. Furthermore, HCoV-EMC, like other coronaviruses, evades innate immune recognition, reflected by the lack of interferon and minimal inflammatory cytokine expression following infection. Importantly, type I and type III interferon treatment can efficiently reduce HCoV-EMC replication in the human airway epithelium, providing a possible avenue for treatment of emerging virus infections.
Observation
Coronaviruses are enveloped positive-stranded RNA viruses of veterinary and medical importance that are associated mainly with respiratory and enteric infections (1, 2). Some animal coronaviruses have long been known to cause severe diseases. In humans, however, it was long believed that coronaviruses cause mainly less severe respiratory infections known as the common cold. This changed with the appearance of the severe acute respiratory syndrome-associated coronavirus (SARS-CoV) which caused for the first time a coronavirus-induced life-threatening disease in humans and was responsible for the 2002-2003 epidemic involving more than 8,000 reported cases and almost 800 fatalities (1). The emergence of a novel coronavirus, HCoV-EMC, raises concerns that we may again face an epidemic caused by a zoonotic coronavirus (3). HCoV-EMC is associated with severe respiratory tract infection, renal failure, and fatalities (4, 5) and is, like SARS-CoV, closely related to bat coronaviruses (3). Since the HCoV-EMC animal reservoir and circumstances of zoonotic transmission are yet elusive, it is critically important to assess which barriers of HCoV-EMC host switching and human-to-human transmission are operational.
An important first barrier against invading respiratory pathogens is the respiratory epithelium, which represents the entry point and primary target tissue of respiratory viruses. To assess the zoonotic potential of HCoV-EMC, it is therefore critically important to determine if the human respiratory epithelium is susceptible to HCoV-EMC infection. To address this question, we used human airway epithelium (HAE) cultures that morphologically and functionally resemble the upper conducting airways in vivo (6). The HAE culture system is based on primary human bronchial epithelial cells obtained by biopsy, brushing, surgery, or lung transplant. Isolated bronchial epithelial cells are manipulated with chemically defined medium to initiate their differentiation into a pseudostratified human airway epithelial culture. When differentiation is complete, the pseudostratified HAE cell layer (i) contains basal, secretory, columnar, and ciliated cell populations and (ii) will generate mucus (6, 7). Therefore, this in vitro system recapitulates many aspects of the human airway epithelium, namely, the presence of well-defined cell types of the human airway epithelium, and physical barriers, such as the mucous layer.
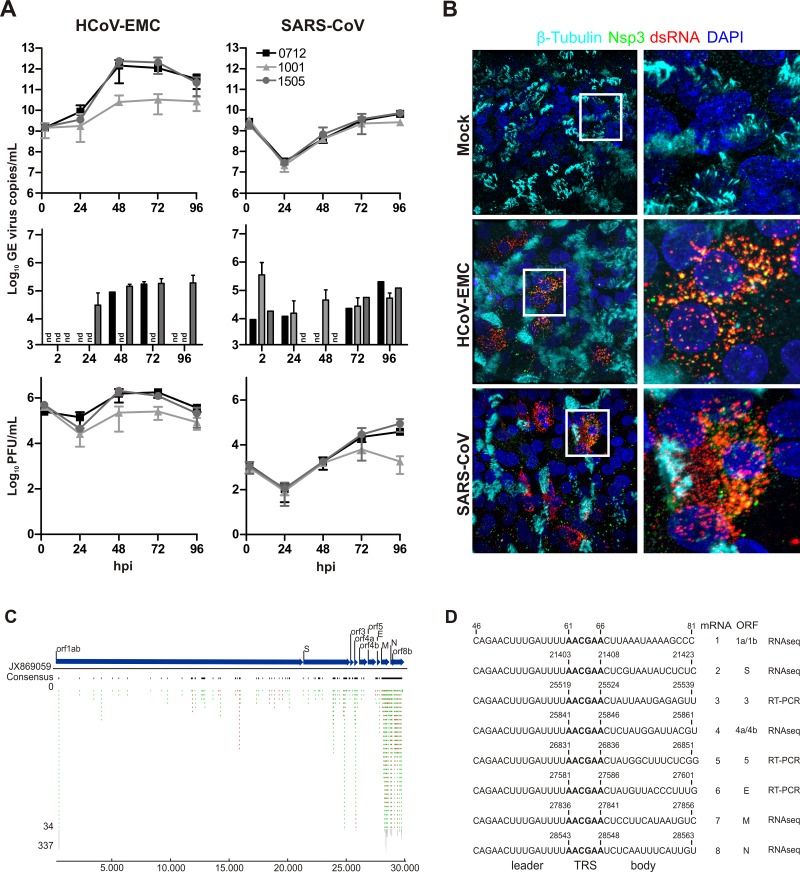
To this end, we have infected fully differentiated HAE cultures derived from three different donors with HCoV-EMC (3, 5) or SARS-CoV (strain Frankfurt-1) at a multiplicity of infection (MOI) of 0.1 and assessed viral growth kinetics. As shown in Fig. 1A, HAE cells are highly susceptible to HCoV-EMC infection, with peak virus production already at 48 h postinfection (hpi). In contrast, replication of SARS-CoV appeared slower and reached peak virus production later at 72 to 96 hpi (Fig. 1A). For both viruses, the vast majority of progeny virus was released at the apical side of HAE cultures, i.e., toward the mucous layer (Fig. 1A, top and bottom), compared to the lower and not always detectable virus release at the basolateral side, i.e., toward the medium (Fig. 1A, middle). We further analyzed HCoV-EMC and SARS-CoV infection of HAE cultures by immunofluorescence microscopy and stained for ciliated cells (β-tubulin), double-stranded (ds) RNA (as a marker for infected cells), and replicase gene-encoded nonstructural protein 3 (Nsp3; as a marker for coronavirus replicase-transcriptase complexes). As shown in Fig. 1B, HCoV-EMC-infected cells were readily identified with a cross-reacting polyclonal antiserum directed against SARS-CoV Nsp3 or a monoclonal antibody directed against dsRNA, which both visualized the characteristic punctuated perinuclear staining pattern for coronavirus replicase-transcriptase complexes. HCoV-EMC infected predominantly nonciliated cells, suggesting that the putative receptor for HCoV-EMC host cell entry is likely to be expressed on nonciliated cells of the human bronchial epithelium. We also analyzed intracellular HCoV-EMC-derived mRNAs by using an RNAseq approach. Total RNA from HCoV-EMC-infected HAE cultures was isolated at 6 hpi using Qiagen’s RNeasy kit followed by mRNA subtraction according to the manufacturer’s protocols. RNA-Seq libraries for an indexed Illumina sequencing run were established using ScriptSeq mRNA-Seq library preparation kit (Epicenter, WI) started from 1 ng mRNA. Quality-proven RNA-Seq libraries were analyzed using Illumina’s HiSeq2500 system according to Illumina’s TruSeq protocols for single reads (TruSeq SBS kit version 3-HS; 50 cycles). Data analysis was performed using CLC Genomics workbench 5.5 (CLC bio, Denmark). Before single-read mapping, raw reads were trimmed to eliminate ambiguous or remaining adapter sequences. We used all reads collected from 3 donors in duplicate experiments (total of 6 datasets) that failed to map to the human genome (25,053,494 out of 195,541,919 reads) for an alignment against the published HCoV-EMC genome sequence (GenBank accession no. JX869059.2). A total of 1,616 out of 25,053,494 (0.006%) reads could be assigned to the HCoV-EMC genome, and we observed a genome coverage reflecting the characteristic mRNA replication and transcription pattern expected for the coronavirus nested set of viral mRNAs (Fig. 1C). Indeed, we could identify several reads representing leader-body fusion sequences of predicted HCoV-EMC mRNAs 2, 4, 7, and 8 (Fig. 1D) (3). In addition, we experimentally determined by reverse transcription (RT)-PCR using total RNA from HCoV-EMC-infected HAE cells the leader-body fusion sequences of predicted mRNAs 3, 5, and 6 that were not represented in the RNAseq data (Fig. 1D; see also Table S1 in the supplemental material). Collectively, our data show that the human bronchial epithelium is highly permissive to HCoV-EMC infection and, accordingly, that all cellular factors required for cell entry (e.g., receptor), replication, and transcription of viral mRNAs, virus assembly, and release are available in the human host.
FIG 1 .
Replication of HCoV-EMC and SARS-CoV on HAE cultures. (A) HAE cultures from three donors (0712, black; 1001, light gray; 1505, dark gray) were prepared as described previously (7) and infected with HCoV-EMC or SARS-CoV (MOI = 0.1). Progeny virus release at the apical (top and bottom) and basolateral (middle) surfaces of HCoV-EMC- or SARS-CoV-infected HAE cultures was determined as genome equivalents (GE) or plaque-forming units (PFU) per ml at the indicated hpi by using quantitative real-time reverse transcription-PCR (qRT-PCR) specific for HCoV-EMC (16) and SARS-CoV (17) or titration of infectious particles on Vero cells. Experiments were performed in triplicate for each donor. Data are depicted as mean values ± standard deviations (SD); nd, not detected. (B) HCoV-EMC- and SARS-CoV-infected (MOI = 0.1) or mock-treated HAE cell cultures were fixed 48 hpi with 6% PFA and immunostained using the procedure as described (18). Rabbit polyclonal antiserum directed against SARS-CoV Nsp3 (green; anti-SARS-CoV antibody; Rockland) and mouse monoclonal antibody directed against dsRNA (red; J2; English & Scientific Consulting Bt.) were used as primary antibodies. Dylight 488-labeled anti-mouse IgG (H+L) and Dylight 647-labeled anti-rabbit IgG (H+L) (Jackson Immunoresearch) were applied as secondary antibodies, followed by two separate incubation steps with Cy3-conjugated mouse anti-β-tubulin antibody (light blue; Sigma) for staining of ciliated cells and DAPI (4',6-diamidino-2-phenylindole; Invitrogen) for staining nuclei (dark blue). Images were acquired using an EC, Plan-Neofluor 63×/1.40 oil differential inference contrast (DIC) M27 objective on a Zeiss LSM 710 confocal microscope. Image capture, analysis, and processing were performed using the ZEN 2010 (Zeiss) and Imaris (Bitplane Scientific Software) software packages. Representative images are shown from one (1505) of three donors. (C) Schematic representation of sequence reads of an RNAseq analysis of poly(A)-containing RNA derived from HCoV-EMC-infected HAE cultures (MOI = 1; 6 hpi). Single reads are depicted in green (sense) and red (antisense). The density of reads exceeding 34 for particular regions are shown condensed in gray. Blue arrows depict HCoV-EMC genes and open reading frames (ORFs). (D) Summary of detected HCoV-EMC mRNAs. Leader-body junctions of HCoV-EMC mRNAs are shown with 15 nucleotides upstream and downstream of the transcription regulatory sequence (TRS; bold). Numbers depict corresponding nucleotide positions in the HCoV-EMC genome. For all 8 viral mRNAs, the ORFs residing in the unique region and the method used for identification (RT-PCR or RNAseq) are indicated.
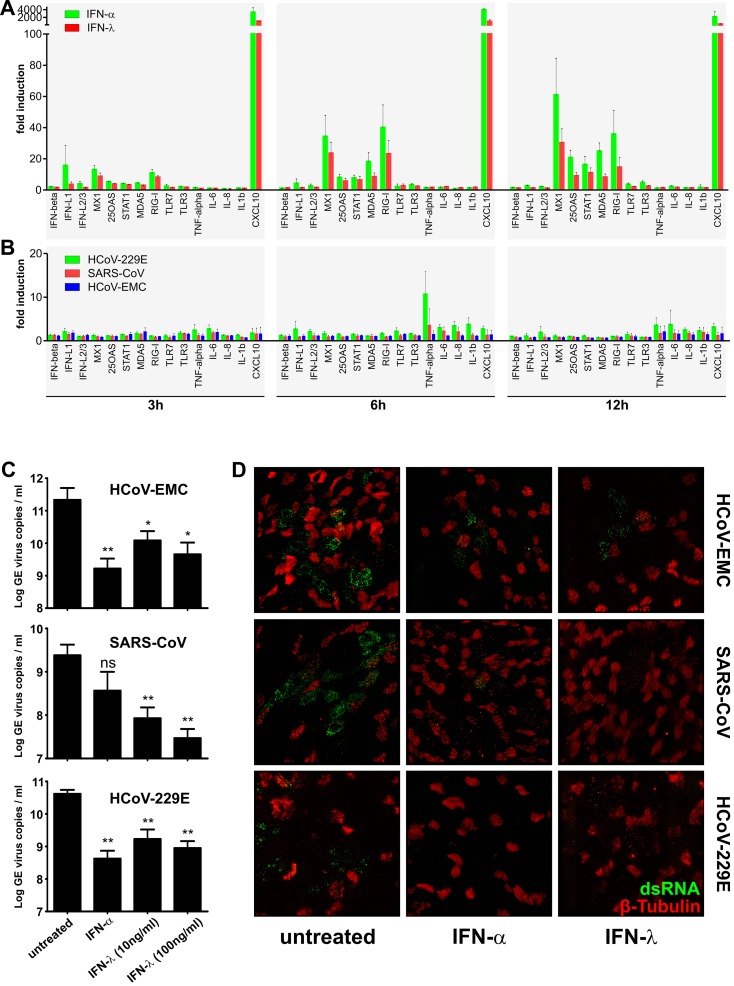
Next we assessed HAE host cell responses to HCoV-EMC infection on the transcriptional level and compared them to responses to SARS-CoV and HCoV-229E infection (MOI = 1). We chose to analyze the expression of a set of 15 cellular mRNAs (see Table S2 in the supplemental material) representing type I IFN, type III IFN, endosomal and cytoplasmic RNA sensor molecules, IFN-stimulated genes (ISGs), chemokines, and inflammatory cytokines, at 3, 6, and 12 h following type I IFN (100 IU of IFN-α) and type III IFN (10 ng/ml of IFN-λ3) (8) treatment or virus infection. As shown in Fig. 2A, HAE cultures respond swiftly to type I and type III IFN treatment with upregulation of ISG expression (i.e., Mx1, 2′-5′-OAS, Stat1, Mda5, Rig-I; Fig. 2A). Notably, the response of HAE cultures to IFN-λ treatment supports previous studies showing high expression of the IFN-λ receptor α-subunit (IFNLR1) in lungs and in epithelial cells (9). In contrast to IFN treatment, the HAE cultures displayed only limited early transcriptional response to coronavirus infection, and particularly, no induction of IFN-β was observed in HCoV-EMC-, SARS-CoV-, and HCoV-229E-infected cells (Fig. 2B). Also the expression of proinflammatory cytokines was only marginally induced, mainly in the common cold virus (HCoV-229E)-infected HAE cultures at 6 hpi. Thus, immediate host responses to HCoV-EMC infection of HAE cultures are very similar to those observed in SARS-CoV- and HCoV-229E-infected cells, suggesting that HCoV-EMC is already well adapted to replication in HAE cultures and that the human bronchial epithelium is not capable to mount a strong innate immune response in the absence of professional cytokine-producing cells, such as plasmacytoid dendritic cells, conventional dendritic cells, and macrophages (2, 10).
FIG 2 .
Human coronavirus-host interaction. (A) Gene expression analysis of IFN-treated HAE cultures. HAE cultures derived from three different donors were used untreated or were stimulated from the basolateral side with recombinant IFN-α (100 IU/ml; IFN-αA/D human; Sigma) or recombinant IFN-λ3 (10 ng/ml) (8) for 3, 6, and 12 h until total cellular RNA was extracted using RNeasy (Qiagen). Reverse transcription was performed with Moloney murine leukemia virus reverse transcriptase according to the manufacturer’s protocol (Invitrogen) using 1 µg of DNase-treated total RNA. Two microliters of diluted cDNA was amplified according to the manufacturer’s protocol, using primers targeting 15 different mRNA transcripts (see Table S1 in the supplemental material). Measurements and analysis were performed using a LightCycler 480 II instrument and software package (Roche). Cycle profile, 10 min at 95°C; 45 cycles of 10 s at 95°C, 20 s at 55°C, and 20 s at 72°C; followed by a melting curve step to confirm product specificity. Relative gene expression was calculated using the 2−ΔΔCt method (19) and is shown as fold induction of IFN-treated samples compared to that of untreated controls. (B) Gene expression analysis of virus-infected HAE cultures. HAE cell cultures were infected with HCoV-EMC, SARS-CoV, or HCoV-229E (MOI = 1), and total cellular RNA was isolated at 3, 6, and 12 hpi. Relative gene expression analysis was performed as described above. (C) Analysis of virus replication following IFN pretreatment. HAE cell cultures were left untreated or were treated from the basolateral side for 16 h with recombinant IFN-α (100 IU/ml; Sigma) or recombinant IFN-λ3 (10 ng/ml or 100 ng/ml) (8). The basolateral medium was replaced prior to infection with HCoV-EMC, SARS-CoV, and HCoV-229E (MOI = 0.1). Apical progeny virus release was determined at 48 hpi by qRT-PCR and is given as GE per ml. Each bar represents the mean ± SD from independent experiments performed in duplicate using HAE cultures derived from three different donors. ns, not significant (P > 0.05); *, P < 0.05; **, P < 0.01 (paired t test). (D) Immunofluorescence analysis of IFN-treated and virus-infected HAE cultures. HAE cultures were fixed with 6% PFA and immunostained using the procedure as described (18). Mouse monoclonal antibody directed against dsRNA (J2; English & Scientific Consulting Bt.) was applied as primary antibody and Dylight 488-labeled anti-mouse IgG (H+L) as secondary antibody (green; Jackson ImmunoResearch), followed by staining of cilia with Cy3-conjugated mouse anti-β-tubulin antibody (red; Sigma). Images were acquired using an EC, Plan-Neofluor 63×/1.40 oil DIC M27 objective on a Zeiss 710 confocal laser scanning microscope. Image capture, analysis, and processing were performed using the ZEN 2010 (Zeiss) and Imaris (Bitplane Scientific Software) software packages. Representative images are shown from one (0401) of three donors.
Since HAE cultures responded well to type I and type III IFN treatment, we addressed if these cytokines can reduce replication of HCoV-EMC, SARS-CoV, and HCoV-229E. HAE cultures derived from three different donors were left untreated or pretreated with IFN-α (100 IU) or IFN-λ3 (10 ng/ml and 100 ng/ml) (8) 16 h prior to infection (MOI = 0.1) with HCoV-EMC, SARS-CoV, or HCoV-229E, and apically released progeny virus genomes were determined by quantitative RT-PCR (qRT-PCR) at 48 hpi. As shown in Fig. 2C, pretreatment of HAE cultures with IFN-α reduced replication of HCoV-EMC and HCoV-229E for all three different donors and reduced replication of SARS-CoV for two of three donors. Accordingly, we observed a pronounced reduction of the number of dsRNA-positive cells in IFN-α-treated HAE cultures that had been infected with HCoV-EMC, SARS-CoV, or HCoV-229E (Fig. 2D). Notably, pretreatment of HAE cultures with IFN-λ3 also reduced replication of HCoV-EMC, SARS-CoV, and HCoV-229E for all three donors at both concentrations used (10 ng/ml and 100 ng/ml). Like for IFN-α treatment, we observed a pronounced reduction of the number of dsRNA-positive cells in IFN-λ3-treated and virus-infected HAE cultures, further corroborating the importance of type III IFN in epithelial antiviral host defense (9, 11).
In summary, we provide here conclusive evidence that the novel coronavirus HCoV-EMC can productively infect human bronchial epithelia cultures, suggesting that all necessary host cell factors for virus entry, RNA synthesis, and virus assembly and release are available in the human host. HCoV-EMC replication in HAE cultures was at least as efficient as replication of SARS-CoV (this study) and HCoV-229E (12). We conclude that HCoV-EMC is capable of infecting the primary target tissue, the human respiratory epithelium, which is in accordance to the reported clinical presentation of severe respiratory symptoms (4, 5). HCoV-EMC has been suggested to have a zoonotic origin, since closely related coronaviruses are known to replicate in bats. Considering that there is not yet any study reporting the successful isolation of a bat coronavirus, HCoV-EMC differs compared to known bat coronaviruses because it displays broad replication capability in diverse mammalian cell lines (13). Our data show that the highly pathogenic viruses HCoV-EMC and SARS-CoV can both replicate in HAE cultures similar to the common cold viruses HCoV-229E, HCoV-OC43, HCoV-NL63, and HCoV-HKU1 (12). Also, early host cell responses are very similar between high- and low-pathogenic human coronaviruses. Thus, although our data depict the zoonotic potential of HCoV-EMC by demonstrating efficient replication in the human respiratory epithelium, decisive factors that impact HCoV-EMC pathogenicity remain to be determined. Importantly, we could demonstrate that HCoV-EMC replication is equally vulnerable to the antiviral effects of type I and type III IFNs, suggesting a possibility to interfere with HCoV-EMC replication in the human respiratory tract. IFN-α treatment has indeed been explored as therapeutic strategy during the SARS epidemic and raised considerable promise (14). The critical importance of type III IFNs in epithelial host defense (11), recent reports that treatment of hepatitis C virus-infected patients with pegylated IFN-λ achieved rapid virological response, while adverse side effects were minimal (15), and our data concerning efficient inhibition of HCoV-EMC replication should encourage the further development of IFN-λ treatment options specifically for respiratory virus and emerging virus infections.
SUPPLEMENTAL MATERIAL
Primer sequences used for HCoV-EMC subgenomic mRNA analysis.
Primer sequences used for mRNA gene expression analysis.
ACKNOWLEDGMENTS
This study was supported by the Swiss National Science Foundation (project 31003A_132898; V.T.), the 3R Research Foundation Switzerland (project 128-11; V.T.), the German Ministry of Education and Research (BMBF project code SARS II; V.T., C.D., M.A.M.), the EU-FP7 project EMPERIE (contract number 223498; C.D.), Antigone (contract number 278976; C.D.), the German Research Foundation (DFG grants DR 772/3-1 and KA1241/18-1; C.D.), the Danish Cancer Society (fellowship to O.J.H.; grant support to R.H.), and the Danish Council for Independent Research, Medical Research (R.H.).
We are grateful to Bart Haagmans (Erasmus Medical Center, Rotterdam, The Netherlands) for helpful discussions.
Human bronchial epithelial cells were isolated from patients (>18 years old) who underwent bronchoscopy and/or surgical lung resection in their diagnostic pathway for any pulmonary disease and that gave informed consent. This was done in accordance with local regulation of the Kanton St. Gallen, Switzerland, as part of the St. Gallen Lung Biopsy Biobank (SGLBB) of the Kantonal Hospital, St. Gallen, which received approval by the ethics committee of the Kanton St. Gallen (EKSG 11/044, EKSG 11/103).
Footnotes
Citation Kindler E, Jónsdóttir HR, Muth D, Hamming OJ, Hartmann R, Rodriguez R, Geffers R, Fouchier RAM, Drosten C, Müller MA, Dijkman R, Thiel V. 2013. Efficient replication of the novel human betacoronavirus EMC on primary human epithelium highlights its zoonotic potential. mBio 4(1):e00611-12. doi:10.1128/mBio.00611-12.
REFERENCES
- 1. Perlman S, Netland J. 2009. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 7:439–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Weiss SR, Navas-Martin S. 2005. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 69:635–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van Boheemen S, de Graaf M, Lauber C, Bestebroer TM, Raj VS, Zaki AM, Osterhaus AD, Haagmans BL, Gorbalenya AE, Snijder EJ, Fouchier RAM. 2012. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio 3(6):e00473-12 http://dx.doi.org/10.1128/mBio.00473-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bermingham A, Chand MA, Brown CS, Aarons E, Tong C, Langrish C, Hoschler K, Brown K, Galiano M, Myers R, Pebody RG, Green HK, Boddington NL, Gopal R, Price N, Newsholme W, Drosten C, Fouchier RA, Zambon M. 2012. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro Surveill. 17:20290. [PubMed] [Google Scholar]
- 5. Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367:1814–1820 [DOI] [PubMed] [Google Scholar]
- 6. Fulcher ML, Gabriel S, Burns KA, Yankaskas JR, Randell SH. 2005. Well-differentiated human airway epithelial cell cultures. Methods Mol. Med. 107:183–206 [DOI] [PubMed] [Google Scholar]
- 7. Dijkman R, Koekkoek SM, Molenkamp R, Schildgen O, van der Hoek L. 2009. Human bocavirus can be cultured in differentiated human airway epithelial cells. J. Virol. 83:7739–7748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dellgren C, Gad HH, Hamming OJ, Melchjorsen J, Hartmann R. 2009. Human interferon-lambda3 is a potent member of the type III interferon family. Genes Immun. 10:125–131 [DOI] [PubMed] [Google Scholar]
- 9. Sommereyns C, Paul S, Staeheli P, Michiels T. 2008. IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 4:e1000017 http://dx.doi.org/10.1371/journal.ppat.1000017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cervantes-Barragan L, Züst R, Weber F, Spiegel M, Lang KS, Akira S, Thiel V, Ludewig B. 2007. Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood 109:1131–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pott J, Mahlakõiv T, Mordstein M, Duerr CU, Michiels T, Stockinger S, Staeheli P, Hornef MW. 2011. IFN-lambda determines the intestinal epithelial antiviral host defense. Proc. Natl. Acad. Sci. U. S. A. 108:7944–7949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dijkman R, Jebbink MF, Koekkoek SM, Deijs M, Jónsdóttir HR, Molenkamp R, Ieven M, Goossens H, Thiel V, van der Hoek L. Isolation and characterization of current human coronavirus strains in primary human epithelia cultures reveals differences in target cell tropism. Journal of Virology, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Muller MA, Raj VS, Muth D, Meyer B, Kallies S, Smits SL, Wollny R, Bestebroer TM, Specht S, Suliman T, Zimmerman K, Binger T, Eckerle I, Tschapka M, Zaki AM, Fouchier RAM, Haagams BL, Drosten C. 2012. Human coronavirus EMC does not require the SARS-Coronavirus receptor and maintains broad replicative capability in mammalian cell lines. mBio 3(6):e00515-12 http://dx.doi.org/10.1128/mBio.00515-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Loutfy MR, Blatt LM, Siminovitch KA, Ward S, Wolff B, Lho H, Pham DH, Deif H, LaMere EA, Chang M, Kain KC, Farcas GA, Ferguson P, Latchford M, Levy G, Dennis JW, Lai EK, Fish EN. 2003. Interferon alfacon-1 plus corticosteroids in severe acute respiratory syndrome: a preliminary study. JAMA 290:3222–3228 [DOI] [PubMed] [Google Scholar]
- 15. Muir AJ, Shiffman ML, Zaman A, Yoffe B, de la Torre A, Flamm S, Gordon SC, Marotta P, Vierling JM, Lopez-Talavera JC, Byrnes-Blake K, Fontana D, Freeman J, Gray T, Hausman D, Hunder NN, Lawitz E. 2010. Phase 1b study of pegylated interferon lambda 1 with or without ribavirin in patients with chronic genotype 1 hepatitis C virus infection. Hepatology 52:822–832 [DOI] [PubMed] [Google Scholar]
- 16. Corman VM, Eckerle I, Bleicker T, Zaki A, Landt O, Eschbach-Bludau M, van Boheemen S, Gopal R, Ballhause M, Bestebroer TM, Muth D, Muller MA, Drexler JF, Zambon M, Osterhaus AD, Fouchier RM, Drosten C. 2012. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveill. 17:pii =20285 [DOI] [PubMed] [Google Scholar]
- 17. Drosten C, Günther S, Preiser W, van der Werf S, Brodt HR, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RA, Berger A, Burguière AM, Cinatl J, Eickmann M, Escriou N, Grywna K, Kramme S, Manuguerra JC, Müller S, Rickerts V, Stürmer M, Vieth S, Klenk HD, Osterhaus AD, Schmitz H, Doerr HW. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967–1976 [DOI] [PubMed] [Google Scholar]
- 18. Dijkman R, Mulder HL, Rumping L, Kraaijvanger I, Deijs M, Jebbink MF, Verschoor EJ, van der Hoek L. 2009. Seroconversion to HCoV-NL63 in rhesus macaques. Viruses 1:647–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer sequences used for HCoV-EMC subgenomic mRNA analysis.
Primer sequences used for mRNA gene expression analysis.