Abstract
The twin-arginine translocation (Tat) pathway is one of two general protein transport systems found in the prokaryotic cytoplasmic membrane and is conserved in the thylakoid membrane of plant chloroplasts. The defining, and highly unusual, property of the Tat pathway is that it transports folded proteins, a task that must be achieved without allowing significant ion leakage across the membrane. The integral membrane TatC protein is the central component of the Tat pathway. TatC captures substrate proteins by binding their signal peptides. TatC then recruits TatA family proteins to form the active translocation complex. Here we report the crystal structure of TatC from the hyperthermophilic bacterium Aquifex aeolicus. This structure provides a molecular description of the core of the Tat translocation system and a framework for understanding the unique Tat transport mechanism.
The Sec and Tat1–3 protein transport pathways are responsible for protein export across the cytoplasmic membrane of bacteria and archaea, and for protein import into the thylakoids of choloroplasts. The Sec system threads unfolded proteins through a membrane-bound channel4. By contrast, the Tat pathway transports proteins that have already attained a folded state5, a more challenging task because folded proteins have a much larger cross-section than an unfolded peptide and so require a larger transport pathway. In addition, folded proteins have a diverse range of shapes and sizes, making it difficult to seal tightly around the protein during transport to preserve the membrane permeability barrier. How the Tat apparatus is able to accomplish this feat is unknown. In bacteria the Tat pathway has an important role in energy metabolism, nutrient acquisition and virulence1,5,6, whereas in plants the Tat pathway is essential for photosynthesis7.
Tat transport is carried out by the integral membrane proteins TatA (refs 8, 9), TatB (ref. 8) and TatC (ref. 10). TatA and TatB are structurally related and contain a single transmembrane helix followed by an amphipathic helix8,11, whereas TatC has multiple transmembrane helices 9. Proteins are targeted to the Tat pathway by amino-terminal signal peptides possessing a twin-arginine-containing sequence motif12–14 that is recognized by the TatC protein within a TatBC complex15–19. Binding of substrate proteins to the TatBC complex induces recruitment of TatA to assemble a transient TatABC-containing translocation site15,20,21, which is thought to facilitate transport by perturbing the membrane bilayer1,3,22.
TatC emerges as the core organizing component of the Tat pathway, directly and dynamically binding substrate, TatB and TatA23,24, and maintaining these interactions during the transport step16,20. A molecular-level understanding of TatC, the largest and most conserved element of the Tat translocation machinery, is required to begin to understand the remarkable Tat transport mechanism.
Structure of TatC
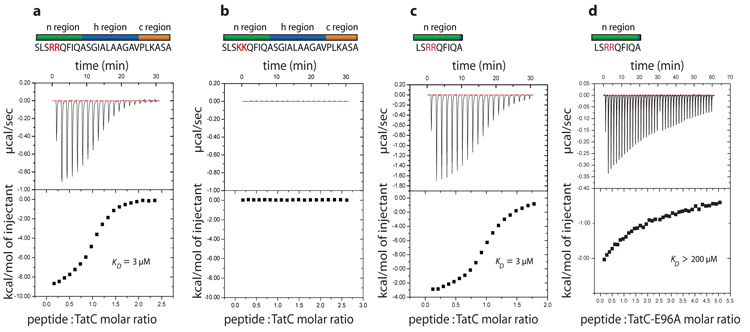
We targeted TatC proteins from thermophilic bacteria and archaea for structure determination. Aquifex aeolicus TatC (AaTatC) solubilized in the detergent lauryl maltose neopentyl glycol (LMNG)25 formed crystals that diffracted to high resolution. LMNG-solubilized AaTatC was able to bind a synthetic Tat signal peptide (dissociation constant (Kd) = 3 μM) at an approximately 1:1 protein:peptide ratio (Fig. 1a), but not a functionally inactive variant signal peptide15,16,26 (Fig. 1b). Thus the protein used for crystallization was functional for Tat signal peptide binding. Identical binding affinities were measured for the full-length signal peptide and for a truncated signal peptide comprising only the twin-arginine-containing n-region (Fig. 1c), consistent with crosslinking studies which indicate that TatC contacts the n-region of the signal peptide15,16.
Figure 1. Purified A. aeolicus TatC binds Tat signal peptides.
a–c, Isothermal titration calorimetry was used to assess interactions between AaTatC and the Tat signal peptide of the E. coli protein SufI (a), a functionally inactive variant SufI signal peptide in which the twin arginine residues have been substituted with twin lysine residues (b) and the n-domain of the SufI signal peptide (c). d, Interaction between the n-domain peptide and a AaTatC E96A variant.
Crystals of selenomethionine-labelled AaTatC (Supplementary Fig. 1) were produced and the structure of the protein solved at a resolution of 3.5 Å using anomalous dispersion (Supplementary Fig. 2a, b). The first four and last eleven amino acids of AaTatC are disordered and are not included in the structure (Supplementary Fig. 3). The final refined structure has an Rfree of 28.8% (Supplementary Table 1). The AaTatC fold is well-maintained in atomistic molecular dynamics (MD) simulations in a phospholipid bilayer, indicating that the structure would be stable in a membrane environment (Supplementary Fig. 4). AaTatC shares 40% sequence identity with Escherichia coli TatC (EcTatC; Supplementary Fig. 3), allowing functional data from the E. coli Tat system to be readily mapped to the AaTatC structure.
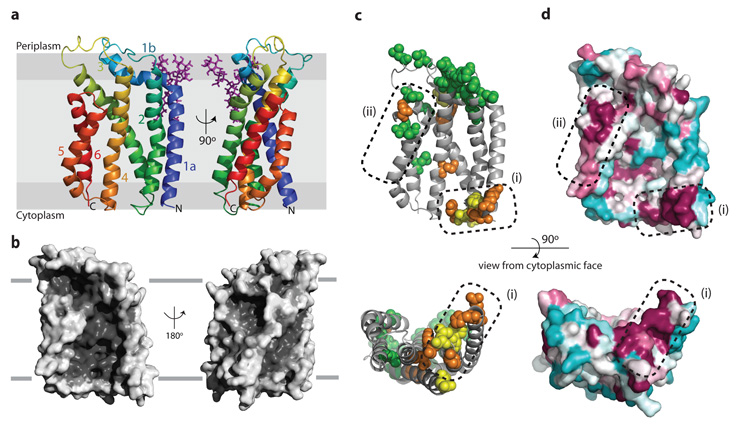
AaTatC contains six transmembrane helices (TM1–TM6), as anticipated9,27–29, which experimental evidence shows are oriented with the amino and carboxy termini of the protein at the cytoplasmic side of the membrane27–30. MD simulations of AaTatC in a phospholipid environment indicate that little of the structure protrudes outside the membrane bilayer (Fig. 2a and Supplementary Fig. 5). Three helices (TM2, TM3 and TM5) are strongly kinked within their transmembrane spans, whereas TM1 kinks abruptly towards the periplasmic side of the membrane to form an interfacial helix (helix 1b). The six transmembrane helices, with the exception of the C-terminal half of TM2 following the kink, are tilted at angles between 20° and 40° to the membrane normal. TM5 and TM6 are too short to span the membrane bilayer fully. Indeed, in MD simulations the membrane is distorted inwards at the ends of these helices (Supplementary Fig. 5). The restricted length of these helices is probably required, in part, to accommodate the TatB partner (see below) but it is also possible that the bilayer distortion caused by TM5 and TM6 may assist in forming the protein translocation pathway. The loops joining the transmembrane helices on the cytoplasmic face of AaTatC are short. However, at the periplasmic face of the membrane the loops are more elaborate, with the interfacial helix 1b and succeeding loop into TM2 making extensive interactions with the TM3–TM4 loop. The result is a structured periplasmic cap that stabilizes the relative positions and tilted orientations of the transmembrane helices. The presence of the cap structure explains why insertions and deletions in this region are not tolerated31,32 and why these loops are hotspots for inactivating mutations32 (Fig. 2c).
Figure 2. Structure of A. aeolicus TatC.
a, Cartoon representation of AaTatC coloured from blue at the N-terminus to red at the C-terminus. The position within the membrane bilayer is predicted by MD simulations (Supplementary Fig. 5). A semi-ordered molecule of LMNG detergent is present and is shown in purple. b, Surface representation of the AaTatC structure. The left hand views in a and b are the same orientation. c, Positions of inactivating substitutions in EcTatC identified by genetic screening32 (green), targeted substitution17,34 (orange), or both (yellow), mapped to AaTatC (full details in Supplementary Table 2). d, Sequence conservation at the surface of TatC displayed in the Consurf46 colouring scheme in which maroon represents high, and cyan low, sequence conservation. c, d, Dotted regions labelled (1) and (2) highlight conserved functional regions of the structure.
The overall shape of AaTatC resembles a cupped hand in which the transmembrane helices form a curved wall overhung on the concave face by the periplasmic cap (Fig. 2b). The result is a structure with a notably high surface area (~16,500 Å2) for the length of protein. A gap between the short TM5 and TM6 helices and the cap results in a prominent groove leading from the concave face of AaTatC to the periplasm. The AaTatC fold shows no similarity to previously described membrane protein structures. Importantly, the structure of TatC is unrelated to that of the Sec translocase even though the two proteins bind signal peptides of similar overall structure12. Crystallographic B-factors suggest that AaTatC has very restricted structural flexibility (Supplementary Fig. 6a). Similarly, MD simulations of AaTatC in a phospholipid bilayer show limited deformation modes (Supplementary Fig. 6b). It is, therefore, unlikely that TatC undergoes large conformational changes as typically found in small-molecule transporters33.
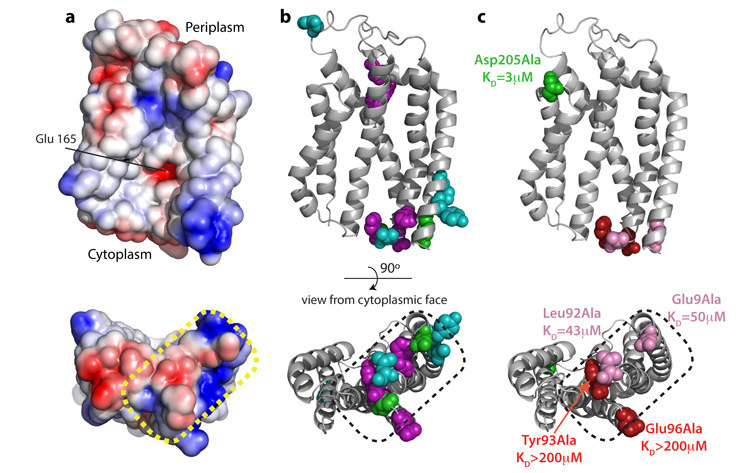
A notable feature of the surface of AaTatC is that the side chain of Glu 165 is exposed at the centre of the concave face, thereby placing an ionisable group in the hydrophobic interior of the bilayer (Fig. 3a). This residue is highly conserved as glutamate or glutamine. Substitution of the equivalent residue in EcTatC (Glu 170) with alanine does not affect signal peptide binding17, but severely compromises overall Tat transport activity17,34. In MD simulations Glu 165 is hydrated and perturbs the local bilayer environment by attracting a lipid head group. This suggests that burying Glu 165 in the membrane is highly unfavourable and that Glu 165 has an interaction partner in the cell.
Figure 3. Identification of the signal peptide binding site on TatC.
a, The electrostatic surface of TatC showing negatively charged regions as red and positively charged regions as blue. b, EcTatC residues implicated in signal peptide or substrate interactions mapped to AaTatC (full details in Supplementary Table 4). Substitutions that prevent signal peptide crosslinking to EcTatC (ref. 17) are shown in purple. Positions that give prominent photo-affinity crosslinks to substrate proteins36 are shown in green. Single amino acid substitutions that enable transport of substrates with defects in the signal peptide twin-arginine motif18,19,37 are shown in cyan. c, Effects of the indicated amino acid substitutions on the affinity of AaTatC for the SufI n-region peptide. Substitutions that affect signal peptide binding are coloured red or pink, and those that do not are coloured green. The corresponding experimental data are shown in Fig. 1d and Supplementary Fig. 9. The dotted region in a–c corresponds to region (1) in Fig. 2c, d.
Tat transport is energized by the proton electrochemical gradient35. However, no membrane surface-linked channels or transmembrane hydrogen bonding networks are apparent within AaTatC. Glu 165 is part of a short hydrogen-bonding network with Ser 107 and Trp 85 (Supplementary Fig. 7) but this is isolated from the membrane surface. Thus, if TatC participates in coupling transport to transmembrane proton movement, it does so in conjunction with other components.
To highlight functionally important regions of TatC we plotted amino acid substitutions that suppress the transport function of EcTatC (refs 17, 32, 34) on to the equivalent positions in the AaTatC structure (Fig. 2c). We also mapped sequence conservation on to the protein surface (Fig. 2d). There are two clusters of conserved and functionally important residues on the surface of TatC (labelled (1) and (2) in Fig. 2c, d). These are located at each end of the TatC molecule, on opposite sides of the membrane, and probably correspond to sites of interactions with partner proteins. A sequence covariance analysis shows that the strongest structural constraints in TatC are within those helix pairs (TM1/TM2 and TM5/TM6) that form the structural framework underlying the two functionally conserved surface sites (Supplementary Fig. 8 and Supplementary Table 3). Functionally important residues in the interior of the TatC molecule have structural roles mediating transmembrane kinks (conserved) or stabilizing the periplasmic cap (weakly conserved).
Signal peptide recognition
Residues in EcTatC that may be involved in signal peptide binding have been identified by crosslinking and genetic analysis17,19,36,37. The equivalent residues in AaTatC cluster on the cytoplasmic face of TM1–TM3 within conserved functional surface site (1) (Fig. 3b). Within this site the two negatively charged amino acids Glu 9 and Glu 96 would be appropriately positioned to coordinate the positively charged guanidinium groups of the sequential arginine residues of the signal peptide.
To directly test the involvement of this part of AaTatC in signal peptide interactions we measured the signal peptide binding affinities of single amino acid substitutions within the site (Figs 1d and 3c, and Supplementary Fig. 9). All the substitutions, including those of Glu 9 and Glu 96, resulted in large reductions in the affinity of AaTatC for the n-region peptide. By contrast, changing a functionally important34 residue in surface site (2) had no effect on signal peptide binding (Fig. 3c and Supplementary Fig. 9). This analysis confirms that the twin-arginine-containing region of Tat signal peptides bind to surface site (1).
Interactions with Tat components
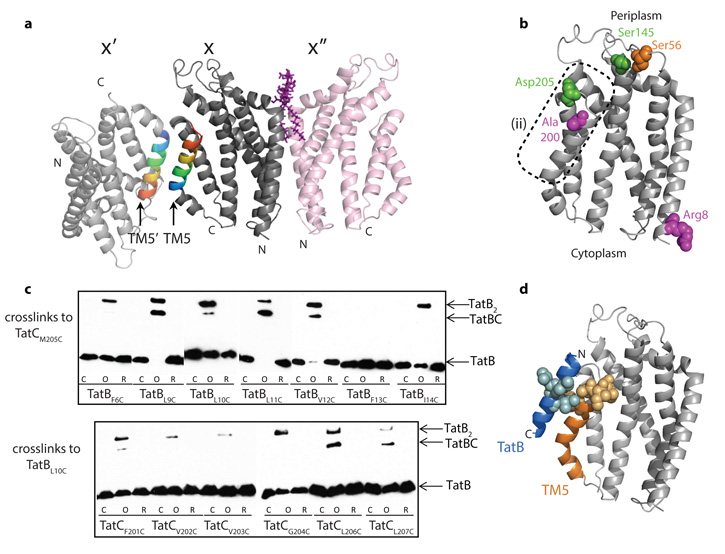
In E. coli, and in plant thylakoids, multiple copies of TatC are present in the TatBC complex26,34,38,39 and TatC has been reported to self-interact40, possibly as local TatC dimers41. We considered whether the protomer arrangement in the AaTatC crystal might reflect such interactions. One of two packing interactions in the crystals (between molecules X and X′′ in Fig. 4a) forms a topologically reasonable dimer (752 Å2 ×2 buried surface area; Supplementary Fig. 10), and biochemical characterization confirms that the LMNG-solubilized protein is a dimer in solution (Supplementary Fig. 11). Nevertheless, the low sequence conservation of the subunit interface (Fig. 2d and Supplementary Fig. 10), the results of sequence covariance analysis (Supplementary Fig. 8 and Supplementary Table 3), and inconsistency with photocrosslinking results (Fig. 4b)36 all argue against the biological relevance of this dimer. More generally, the lack of plausible inter-subunit contacts in the covariance analysis suggests that interactions between TatC protomers are not subject to specific structural or mechanistic constraints.
Figure 4. Sites of interaction with other Tat components.
a, AaTatC crystal contacts. The contact between molecules X and X′ involves a packing interaction between antiparallel TM5 helices, whereas that between molecules X and X′′ involves the semi-ordered LMNG molecule shown in purple. b, Positions in EcTatC that have been reported32,34,36 to interact with both TatA and TatB (green), with TatB (magenta), or with TatC (orange) mapped on to AaTatC (full details in Supplementary Table 5). The dotted region corresponds to region (2) in Fig. 2c, d. c, Disulphide crosslinking between E. coli TatB and TatC variants detected by immunoblotting with TatB antibodies. Lanes are untreated (C), oxidized with Cu(II)phenanthroline (O), or oxidized and then reduced with DTT (R). d, The complex between TatC and the transmembrane helix of TatB modelled on the AaTatC X–X′ crystal contact. Positions in EcTatC that form a disulphide bond to a EcTatB L10C variant (yellow), or positions in EcTatB that form a disulphide bond to a EcTatC M205C variant (blue), (data from c) are mapped on to the model.
In Gram-negative bacteria and plants TatC forms a tight complex with TatB. Known contact information for the two proteins in E. coli32,34,36 maps to the periplasmic end of conserved functional surface patch (2) in TatC (Fig. 4b) and to the periplasmic42 N terminus of the TatB transmembrane helix32 (TatB(TM)). To delineate further the proposed TatB–TatC contact site we undertook limited disulphide scanning mutagenesis of the E. coli TatB and TatC proteins in these areas (Fig. 4c). In the AaTatC crystals the contact between molecules X and X′ involves packing an inverted TM5 from one TatC molecule against the proposed TatB contact site in the other (Fig. 4a), showing that a transmembrane helix with the same orientation as TatB(TM) (N terminus at the periplasmic side of the membrane) is able to pack at this site. On the basis of this crystallographic contact we produced a model for the TatB(TM)–TatC complex consistent with the experimental interaction data (Fig. 4d). A notable feature of the model is that the C-terminal half of TM5 is well matched in length to the short TatB(TM) to which it binds.
Although our analyses localize the signal peptide and TatB binding sites to opposite ends of the TatC molecule, some evidence exists for functional and structural connections between TatB and signal peptides. First, photocrosslinks to TatB have been reported in the vicinity of the signal peptide binding site in TatC (ref. 36) (Fig. 4b). Second, the signal peptide h-region, which directly follows the n-region, photocrosslinks predominantly to TatB (refs 15, 16). Third, some substitutions that allow transport of substrates with defective Tat signal peptides map to the N terminus of TatB(TM)18,37. These data suggest that the amphipathic helix of TatB could run across the cytoplasmic face of TatC from the TatB(TM) binding site at TM5 to the signal peptide binding site at TM1 (Fig. 5). Alternatively, some of these interactions may relate to a stage in the transport cycle where the signal peptide is reaching across the membrane from the point of attachment of the twin-arginine motif at region (1) to the TatB(TM)-binding site in region (2). It is also possible that it is a TatB molecule bound to a neighbouring TatC molecule that is close to the signal peptide binding site.
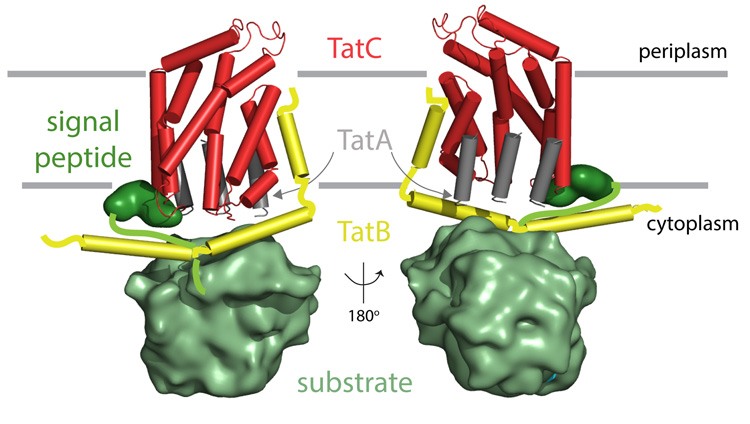
Figure 5. A conceptual model for the environment of TatC in the substrate-bound translocation site.
The model is viewed from the convex (left) and concave (right) sides of TatC. The interaction of TatC with the transmembrane helix of TatB is modelled as in Fig. 4d. The TatB amphipathic helices are positioned to allow TatB to be in the vicinity of the N terminus of TatC as shown by crosslinking36. The TatA transmembrane helix is shown bound to the concave face of TatC based on docking experiments (Supplementary Fig. 12). The position of the TatA amphipathic helix is unclear and is omitted from the conceptual model. TatA forms large polymers but only representative TatA molecules directly contacting TatC are shown. For simplicity, the model depicts a single TatC molecule but each TatC protein will be operating in the context of a larger TatBC complex.
The interaction of TatA with TatC is transient and may involve multiple TatA protomers binding to a single TatC protein. The only identified points of interaction between the two proteins are photocrosslinks36 at the periplasmic side of the membrane close to the modelled position of the TatB N terminus (Fig. 4b). This suggests that the periplasmically located N terminus42 of TatA can reach this region. In the absence of more detailed experimental information on the TatA binding site(s) we carried out in silico docking experiments and MD simulations using a homology model of the A. aeolicus TatA transmembrane helix (TatA(TM)). The results suggest that TatA(TM) binds within the concave face of TatC (Supplementary Fig. 12). This location might allow TatA to act as the interaction partner for exposed Glu 165, perhaps via interaction with the functionally essential conserved Gln/Glu in TatA(TM)21,43. The relatively large and concave surface of this region of TatC could promote a binding-induced conformational change in the bound TatA protomers that would allow them to act as the nucleation site for TatA polymerization.
The conceptual model shown in Fig. 5 summarizes the possible arrangement of the components of an active Tat translocation site indicated by the AaTatC structure.
Methods
Selection of clones, growth conditions and solubilizing detergent
A range of tatC genes from thermophilic bacteria and archaea were cloned into plasmid pWaldo-TEV-GFPe44 to allow expression in E. coli as N-terminal fusions to octahistidine-tagged GFP (TatC–GFP–8His). The resulting fusion proteins contain a TEV protease-cleavable linker between the TatC and GFP domains. The clones were cultured in a variety of background strains and growth conditions and TatC–GFP expression assessed by whole-cell fluorescence44,47. This screen identified Aquifex aeolicus TatC as suitable for high-level protein production. The A. aeolicus tatC gene48 contains two possible initiation codons. Although sequence comparisons between TatC proteins from within the phylum Aquificae indicate that the first of these codons is the authentic translation start site, the construct used here starts from the second methionine codon, thereby removing eight poorly conserved amino acids (sequence MSQEKLPE) from the N terminus of the protein. Fluorescence size exclusion chromatography (FSEC)49 was used to identify detergents that were able to solubilize the A. aeolicus TatC–GFP fusion protein in a monodisperse state. Subsequently size exclusion chromatography - multi-angle laser light scattering (SEC-MALLS) was used to assess whether these detergents were able to maintain TatC in a monodisperse state following proteolytic removal of the GFP domain. Detergents meeting this selection criterion were used to purify A. aeolicus TatC for crystallization trials.
Protein purification and crystallization
The A. aeolicus TatC–GFP construct was recombinantly expressed in E. coli Lemo56(DE3)47 cells cultured at 37 °C in Terrific Broth supplemented with 50 μg ml−1 kanamycin. When the cells attained OD600 = 5.0, 0.1 mM IPTG was added and the cells cultured for a further 16 h at 24 °C. Cells were disrupted by homogenization (Emulsifex C5, Avestin) and membranes recovered by differential centrifugation. Solubilization was carried out at 4 °C for 2 h in 1% w/v lauryl maltose neopentyl glycol (LMNG; Anatrace) in PBS containing 20 mM imidazole. The soluble fraction was loaded on to a 5 ml Ni-NTA column (Qiagen). The column was washed with five column volumes of PBS containing 0.02% LMNG and 25 mM imidazole, then with two column volumes of the same buffer containing 40 mM imidazole, and eluted with the same buffer containing 250 mM imidazole. TatC–GFP-containing fractions were pooled and incubated with 2 mg His-tagged TEV protease, 5 mM sodium citrate pH 6.3, 0.3 mM oxidized glutathione and 3 mM reduced glutathione (final concentrations). This mixture was dialysed in a 10 kDa molecular mass cutoff membrane (‘snakeskin tubing’, Fisher scientific) against size exclusion (SEC) buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.02% w/v LMNG) at 4 °C overnight. Passage over a 5 ml Ni-NTA column removed the His-tagged GFP and His-tagged TEV protease. TatC was concentrated in a centrifugal concentrator device (100 kDa molecular mass cutoff membrane, Millipore) to 5 ml, and loaded on to a Superdex 200 16/60 gel filtration column (GE Healthcare) pre-equilibrated in SEC buffer. Final peak fractions were concentrated to ~8 mg ml−1.
Selenomethionine-substituted TatC was expressed in E. coli L56 cells grown in M9 minimal medium, using the feedback inhibition of methionine biosynthesis method50. Cells were grown at 37 °C to an OD600 = 0.7 and induced as for the native protein. Purification was carried out as for the native protein but the final size exclusion buffer was supplemented with 5 mM β-mercaptoethanol. Full incorporation of selenomethionine was confirmed by mass spectrometry (Supplementary Fig. 1).
Crystallization was carried out using the sitting-drop vapour diffusion method at 21 °C with a drop size of 200 nl. The best crystals were obtained in 0.2 M ammonium sulphate, 0.02 M NaCl, 0.02 M sodium acetate pH 4.0, 33% v/v PEG 200 at a 50:50 ratio of protein:mother liquor and a drop volume of 400 nl. Crystals of the selenomethionine protein were obtained in the same condition. These were flash frozen in liquid nitrogen in the mother liquor, diffracted to 3.5 Å and belonged to the space group H32.
Data collection, structure determination and refinement
Diffraction data were collected at the Diamond light source (Beamline I04-1), at 120K from four SeMet-labelled crystals (λ = 0.92 Å). The data were processed using xia251 and data sets were scaled together using autoSHARP52. The first two selenium sites were located using Phenix Hyss53, and the final two positions identified in SHARP LLG gradient maps54. The hand was determined by SOLOMON55 solvent flattening (CC for observed versus solvent flattened Es was 45% versus 51%). The main chain was traced by hand as a continuous stretch of 220 residues in the experimental map with the SeMet sites allowing unambiguous determination of the chain direction and docking of sequence in the C-terminal portion of the protein. After initial refinement in autoBUSTER56, Buccaneer57 was used to autobuild a model which had the correct sequence throughout. Further rounds of building using COOT58 and refinement in autoBUSTERfollowed to give a final model for residues 5–229 with R = 25.2% and Rfree = 28.8%. 92% of residues lie in the favoured regions of the Ramachandran plot and the structure has a Molprobity45 score of 3.1 (84th percentile for structures 3.5 Å ± 0.25 Å). The quality of the electron density map is shown in Supplementary Fig. 2. There is one TatC protomer in the asymmetric unit and one molecule of LMNG located on a crystallographic two-fold (occupancy of all LMNG atoms set to 0.5).
The coordinates and experimental data have been deposited at the Protein Data Bank with accession code 4b4a.
Images for publication were generated using PyMol Version 1.5.0.4 (http://www.pymol.org; Schrödinger, LLC).
Protein analysis
Isothermal titration calorimetry was carried out using an iTC200 isothermal titration calorimeter (Microcal) using synthetic peptides (Generon Ltd) as the titrant. The n-region peptide was amidated on the C-terminus. Titrations were carried out at 25 °C using a reference power of 3 cal s−1 at a stirrer speed of 1,000 r.p.m. Isotherms were analysed and fitted in SEDPHAT59 using a hetero-association model (A + B↔AB) with ΔH and Kd as fitting parameters. To allow for inaccuracies in peptide concentrations an inactive peptide fraction was also included as a fitting parameter. Concentrations of TatC were measured by absorption at 280 nm in 6 M guanidinium chloride using an extinction coefficient of 24,870 M−1 cm−1 calculated from the sequence.
Disulphide-crosslinking experiments were carried out at 24 °C as described previously32,42.
Molecular dynamics simulations and in silico docking
All MD simulations were performed using GROMACS v4.5.4 at 323 K60. Coarse-grain MD simulations were run for 1 μs to permit the assembly and equilibration of a dipalmitoylphosphatidylcholine (DPPC) bilayer around the protein61,62 using a Martini force field63 (Supplementary Fig. 5). The systems were then converted to atomistic using the CG2AT-align method64. The atomistic systems used the Gromos 53a6 force field and were equilibrated for 1 ns with the protein restrained, before 100 ns of unrestrained atomistic MD.
Supplementary Material
Acknowledgements
We thank D. Byrne, G. Orriss and R. Owens for their contributions to the early stages of this project; R. Keller for advice; and J. Willem de Gier, D. Daley and R. Huber for providing strains and reagents. This work was supported by the Wellcome Trust (studentships to S.E.R., J.E.G. and M.A.M.; grant 083599, P.R.; grant 092970MA, M.S.P.S.), the Swedish Foundation for Strategic Research (‘Future research leaders 4’ to M.H.), the Swedish Research Council (grant 2010-5061 to M.H.), the E .P. Abrahams Cephalosporin Trust (M.K. and F.R.), the Biotechnology and Biological Sciences Research Council (studentship, M.J.L.; grant BB/E023347/1, S-M.L.; grant BB/1019855/1, P.J.S.), the Medical Research Council (grant G1001640, F.J.; grant G0900888, S.J.), and the European Research Council (Advanced Grant IMPRESS, J.M. and C.V.R.). Work in S.M.L.’s group is funded by the James Martin 21st Century School Vaccine Design Institute.
Footnotes
Supplementary Information is available in the online version of the paper.
Author Contributions The experiments were designed and manuscript written by S.E.R., M.J.T., B.C.B, S.M.L., M.H. and T.P. Experimental work was performed as follows: crystallization trials: S.E.R. and M.J.T.; structure determination: S.E.R., P.R. and S.M.L.; cloning and expression screening: J.E.G., M.J.T., M.J., S-M.L. and M.J.L.; homogeneity screening: M.J.T., M.J., S.E.R., M.A.M. and S.J.; MD simulations: P.J.S. and M.S.P.S; disulphide crosslinking: F.J. and T.P.; signal peptide binding and BN-PAGE: S.E.R., M.K. and F.R.; mass spectrometry: J.M. and C.V.R.
Author Information The coordinates and experimental data have been deposited at the Protein Data Bank with accession code 4b4a. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper.
References
- 1.Palmer T, Berks BC. The twin-arginine translocation (Tat) protein export pathway. Nature Rev. Microbiol. 2012;10:483–496. doi: 10.1038/nrmicro2814. [DOI] [PubMed] [Google Scholar]
- 2.Frobel J, Rose P, Muller M. Twin-arginine-dependent translocation of folded proteins. Phil. Trans. R. Soc. Lond. B. 2012;367:1029–1046. doi: 10.1098/rstb.2011.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Celedon JM, Cline K. Intra-plastid protein trafficking; how plant cells adapted prokaryotic mechanisms to the eukaryotic condition. Biochim. Biophys. Acta. 2012 doi: 10.1016/j.bbamcr.2012.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park E, Rapoport TA. Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Annu. Rev. Biophys. 2012;41:21–40. doi: 10.1146/annurev-biophys-050511-102312. [DOI] [PubMed] [Google Scholar]
- 5.Berks BC, Palmer T, Sargent F. The Tat protein translocation pathway and its role in microbial physiology. Adv. Microb. Physiol. 2003;47:187–254. doi: 10.1016/s0065-2911(03)47004-5. [DOI] [PubMed] [Google Scholar]
- 6.De Buck E, Lammertyn E, Anne J. The importance of the twin-arginine translocation pathway for bacterial virulence. Trends Microbiol. 2008;16:442–453. doi: 10.1016/j.tim.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Barkan A, Miles D, Taylor WC. Chloroplast gene expression in nuclear, photosynthetic mutants of maize. EMBO J. 1986;5:1421–1427. doi: 10.1002/j.1460-2075.1986.tb04378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Settles AM, et al. Sec-independent protein translocation by the maize Hcf106 protein. Science. 1997;278:1467–1470. doi: 10.1126/science.278.5342.1467. [DOI] [PubMed] [Google Scholar]
- 9.Sargent F, et al. Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J. 1998;17:3640–3650. doi: 10.1093/emboj/17.13.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bogsch EG, et al. An essential component of a novel bacterial protein export system with homologues in plastids and mitochondria. J. Biol. Chem. 1998;273:18003–18006. doi: 10.1074/jbc.273.29.18003. [DOI] [PubMed] [Google Scholar]
- 11.Hu Y, Zhao E, Li H, Xia B, Jin C. Solution NMR structure of the TatA component of the twin-arginine protein transport system from gram-positive bacterium Bacillus subtilis. J. Am. Chem. Soc. 2010;132:15942–15944. doi: 10.1021/ja1053785. [DOI] [PubMed] [Google Scholar]
- 12.Berks BC. A common export pathway for proteins binding complex redox cofactors? Mol. Microbiol. 1996;22:393–404. doi: 10.1046/j.1365-2958.1996.00114.x. [DOI] [PubMed] [Google Scholar]
- 13.Chaddock AM, et al. A new-type of signal peptide—central role of a twin-arginine motif in transfer signals for the ΔpH-dependent thylakoidal protein translocase. EMBO J. 1995;14:2715–2722. doi: 10.1002/j.1460-2075.1995.tb07272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stanley NR, Palmer T, Berks BC. The twin arginine consensus motif of Tat signal peptides is involved in Sec-independent protein targeting in Escherichia coli. J. Biol. Chem. 2000;275:11591–11596. doi: 10.1074/jbc.275.16.11591. [DOI] [PubMed] [Google Scholar]
- 15.Alami M, et al. Differential interactions between a twin-arginine signal peptide and its translocase in Escherichia coli. Mol. Cell. 2003;12:937–946. doi: 10.1016/s1097-2765(03)00398-8. [DOI] [PubMed] [Google Scholar]
- 16.Gerard F, Cline K. Efficient twin arginine translocation (Tat) pathway transport of a precursor protein covalently anchored to its initial cpTatC binding site. J. Biol. Chem. 2006;281:6130–6135. doi: 10.1074/jbc.M512733200. [DOI] [PubMed] [Google Scholar]
- 17.Holzapfel E, et al. The entire N-terminal half of TatC is involved in twin-arginine precursor binding. Biochemistry. 2007;46:2892–2898. doi: 10.1021/bi062205b. [DOI] [PubMed] [Google Scholar]
- 18.Kreutzenbeck P, et al. Escherichia coli twin arginine (Tat) mutant translocases possessing relaxed signal peptide recognition specificities. J. Biol. Chem. 2007;282:7903–7911. doi: 10.1074/jbc.M610126200. [DOI] [PubMed] [Google Scholar]
- 19.Strauch EM, Georgiou G. Escherichia coli tatC mutations that suppress defective twin-arginine transporter signal peptides. J. Mol. Biol. 2007;374:283–291. doi: 10.1016/j.jmb.2007.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mori H, Cline K. A twin arginine signal peptide and the pH gradient trigger reversible assembly of the thylakoid ΔpH/Tat translocase. J. Cell Biol. 2002;157:205–210. doi: 10.1083/jcb.200202048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dabney-Smith C, Mori H, Cline K. Oligomers of Tha4 organize at the thylakoid Tat translocase during protein transport. J. Biol. Chem. 2006;281:5476–5483. doi: 10.1074/jbc.M512453200. [DOI] [PubMed] [Google Scholar]
- 22.Cline K, McCaffery M. Evidence for a dynamic and transient pathway through the TAT protein transport machinery. EMBO J. 2007;26:3039–3049. doi: 10.1038/sj.emboj.7601759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frobel J, Rose P, Muller M. Early contacts between substrate proteins and TatA translocase component in twin-arginine translocation. J. Biol. Chem. 2011;286:43679–43689. doi: 10.1074/jbc.M111.292565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fritsch MJ, Krehenbrink M, Tarry MJ, Berks BC, Palmer T. Processing by rhomboid protease is required for Providencia stuartii TatA to interact with TatC and to form functional homo-oligomeric complexes. Mol. Microbiol. 2012;84:1108–1123. doi: 10.1111/j.1365-2958.2012.08080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chae PS, et al. Maltose-neopentyl glycol (MNG) amphiphiles for solubilization, stabilization and crystallization of membrane proteins. Nature Methods. 2010;7:1003–1008. doi: 10.1038/nmeth.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cline K, Mori H. Thylakoid ΔpH-dependent precursor proteins bind to a cpTatC–Hcf106 complex before Tha4-dependent transport. J. Cell Biol. 2001;154:719–730. doi: 10.1083/jcb.200105149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Punginelli C, et al. Cysteine scanning mutagenesis and topological mapping of the Escherichia coli twin-arginine translocase TatC Component. J. Bacteriol. 2007;189:5482–5494. doi: 10.1128/JB.00647-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeong KJ, et al. A periplasmic fluorescent reporter protein and its application in high-throughput membrane protein topology analysis. J. Mol. Biol. 2004;341:901–909. doi: 10.1016/j.jmb.2004.05.078. [DOI] [PubMed] [Google Scholar]
- 29.Behrendt J, Standar K, Lindenstrauss U, Bruser T. Topological studies on the twin-arginine translocase component TatC. FEMS Microbiol. Lett. 2004;234:303–308. doi: 10.1016/j.femsle.2004.03.048. [DOI] [PubMed] [Google Scholar]
- 30.Drew D, et al. Rapid topology mapping of Escherichia coli inner-membrane proteins by prediction and PhoA/GFP fusion analysis. Proc. Natl Acad. Sci. USA. 2002;99:2690–2695. doi: 10.1073/pnas.052018199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gouffi K, Santini CL, Wu LF. Topology determination and functional analysis of the Escherichia coli TatC protein. FEBS Lett. 2002;525:65–70. doi: 10.1016/s0014-5793(02)03069-7. [DOI] [PubMed] [Google Scholar]
- 32.Kneuper H, et al. Molecular dissection of TatC defines critical regions essential for protein transport and a TatB-TatC contact site. Mol. Microbiol. 2012 doi: 10.1111/j.1365-2958.2012.08151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forrest LR, Kramer R, Ziegler C. The structural basis of secondary active transport mechanisms. Biochim. Biophys. Acta. 2011;1807:167–188. doi: 10.1016/j.bbabio.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 34.Buchanan G, et al. Functional complexity of the twin-arginine translocase TatC component revealed by site-directed mutagenesis. Mol. Microbiol. 2002;43:1457–1470. doi: 10.1046/j.1365-2958.2002.02853.x. [DOI] [PubMed] [Google Scholar]
- 35.Mould RM, Robinson C. A proton gradient is required for the transport of two lumenal oxygen-evolving proteins across the thylakoid membrane. J. Biol. Chem. 1991;266:12189–12193. [PubMed] [Google Scholar]
- 36.Zoufaly S, et al. Mapping precursor-binding site on TatC subunit of twin arginine-specific protein translocase by site-specific photo cross-linking. J. Biol. Chem. 2012;287:13430–13441. doi: 10.1074/jbc.M112.343798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lausberg F, et al. Genetic evidence for a tight cooperation of TatB and TatC during productive recognition of twin-arginine (Tat) signal peptides in Escherichia coli. PLoS ONE. 2012;7:e39867. doi: 10.1371/journal.pone.0039867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bolhuis A, Mathers JE, Thomas JD, Barrett CM, Robinson C. TatB and TatC form a functional and structural unit of the twin-arginine translocase from Escherichia coli. J. Biol. Chem. 2001;276:20213–20219. doi: 10.1074/jbc.M100682200. [DOI] [PubMed] [Google Scholar]
- 39.Tarry MJ, et al. Structural analysis of substrate binding by the TatBC component of the twin-arginine protein transport system. Proc. Natl Acad. Sci. USA. 2009;106:13284–13289. doi: 10.1073/pnas.0901566106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orriss GL, et al. TatBC, TatB, and TatC form structurally autonomous units within the twin arginine protein transport system of Escherichia coli. FEBS Lett. 2007;581:4091–4097. doi: 10.1016/j.febslet.2007.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maldonado B, Buchanan G, Muller M, Berks BC, Palmer T. Genetic evidence for a TatC dimer at the core of the Escherichia coli twin arginine (Tat) protein translocase. J. Mol. Microbiol. Biotechnol. 2011;20:168–175. doi: 10.1159/000329076. [DOI] [PubMed] [Google Scholar]
- 42.Koch S, Fritsch MJ, Buchanan G, Palmer T. Escherichia coli TatA and TatB proteins have N-out, C-in topology in intact cells. J. Biol. Chem. 2012;287:14420–14431. doi: 10.1074/jbc.M112.354555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greene NP, et al. Cysteine scanning mutagenesis and disulfide mapping studies of the TatA component of the bacterial twin arginine translocase. J. Biol. Chem. 2007;282:23937–23945. doi: 10.1074/jbc.M702972200. [DOI] [PubMed] [Google Scholar]
- 44.Drew D, Lerch M, Kunji E, Slotboom DJ, de Gier JW. Optimization of membrane protein overexpression and purification using GFP fusions. Nature Methods. 2006;3:303–313. doi: 10.1038/nmeth0406-303. [DOI] [PubMed] [Google Scholar]
- 45.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tarry M, Skaar K, Heijne G, Draheim RR, Hogbom M. Production of human tetraspanin proteins in Escherichia coli. Protein Expr. Purif. 2012;82:373–379. doi: 10.1016/j.pep.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 48.Deckert G, et al. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 49.Kawate T, Gouaux E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 2006;14:673–681. doi: 10.1016/j.str.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 50.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 51.Winter G. xia2: an expert system for macromolecular crystallography data reduction. J. Appl. Cryst. 2010;43:186–190. [Google Scholar]
- 52.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol. Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 53.Adams PD, et al. The Phenix software for automated determination of macromolecular structures. Methods. 2011;55:94–106. doi: 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr. D. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- 55.Abrahams JP, Leslie AG. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D. 1996;52:30–42. doi: 10.1107/S0907444995008754. [DOI] [PubMed] [Google Scholar]
- 56.BUSTER 2.11.2. Global Phasing Ltd.; 2011. [Google Scholar]
- 57.Cowtan K. The Buccaneer software for automated model building. Tracing protein chains. Acta Crystallogr. D. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 58.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Houtman JC, et al. Studying multisite binary and ternary protein interactions by global analysis of isothermal titration calorimetry data in SEDPHAT: application to adaptor protein complexes in cell signaling. Protein Sci. 2007;16:30–42. doi: 10.1110/ps.062558507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 61.Monticelli L, Sorin EJ, Tieleman DP, Pande VS, Colombo G. Molecular simulation of multistate peptide dynamics: A comparison between microsecond timescale sampling and multiple shorter trajectories. J. Comput. Chem. 2008;29:1740–1752. doi: 10.1002/jcc.20935. [DOI] [PubMed] [Google Scholar]
- 62.Scott KA, et al. Coarse-grained MD simulations of membrane protein-bilayer self-assembly. Structure. 2008;16:621–630. doi: 10.1016/j.str.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 63.Monticelli L, Sorin EJ, Tieleman DP, Pande VS, Colombo G. Molecular simulation of multistate peptide dynamics: a comparison between microsecond timescale sampling and multiple shorter trajectories. J. Comput. Chem. 2008;29:1740–1752. doi: 10.1002/jcc.20935. [DOI] [PubMed] [Google Scholar]
- 64.Stansfeld PJ, Sansom MSP. From coarse grained to atomistic: a serial multiscale approach to membrane protein simulations. J. Chem. Theory Comput. 2011;7:1157–1166. doi: 10.1021/ct100569y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.