Abstract
The BRCT repeats of BRCA1 are essential for tumor suppression. Phospho-peptide affinity proteomic analysis identified a novel protein, Abraxas, that directly binds the BRCA1 BRCT repeats through a phospho-SXXF motif. Abraxas binds BRCA1 mutually exclusively with BACH1 and CTIP, forming a third Brca1 complex. Abraxas recruits the ubiquitin-interacting motif (UIM) containing protein Rap80 to BRCA1. Both Abraxas and Rap80 are required for DNA damage resistance, G2/M checkpoint control and DNA repair. Rap80 is required for a subset of Brca1-foci formation in response to IR and the UIM domains alone are capable of foci formation. The Rap80/Abraxas complex may help recruit Brca1 to DNA damage sites in part through recognition of ubiquitinated proteins.
The Brca1 tumor suppressor is associated with hereditary breast and ovarian cancer and has significant roles in DNA repair, cell cycle checkpoint control and maintenance of genomic stability (1, 2). Brca1 contains a N-terminal RING domain, a SQ cluster domain phosphorylated by DNA damage signaling kinases ATM/ATR(3), and two C-terminal BRCT repeats. Brca1 forms a heterodimeric complex with Bard1 (4) that exhibits E3 ubiqutin ligase activity(5–7). Many tumorigenic mutations in BRCA1 disrupt the BRCT repeats which constitute a phospho-peptide recognition domain that binds peptides containing a pSxxF motif (8–11).
To identify proteins that bind the BRCT domains of Brca1, we combined peptide affinity purification and stable isotope labeling with amino acids in cell culture (SILAC) (12–14) to identify phospho-peptides that directly bind to the BRCT domain of Brca1 and to quantify their abundance in the presence of DNA damage by mass spectrometry. Proteins from cells grown in heavy medium and cells grown in light medium treated with 10 Gy IR were prepared, mixed (1:1) and digested with Trypsin. Tryptic peptides bound to Gst-Brca1-BRCT were identified by mass spectrometry analysis. We searched for phospho-peptides that contained pSXXF motif and compared the peptides to the list of proteins we recently identified in an ATM/ATR substrate screen (Matsuoka et al., in preparation). In addition to the known Brca1 binding proteins Bach1 and CTIP, we identified peptides that represent a novel ATM/ATR substrate, FLJ13614 (Fig. 1B). We named this protein Abraxas for the Greek god and the gene Abra1. SILAC quantification analysis also indicated a doubly phosphorylated peptide, GEGFYS#RS#PTF containing p-S404 and p-S406, was enriched approximately 8-fold after DNA damage (Fig. S1). Both p-S406, and doubly, p-S404 p-S406, phosphorylated Abraxas peptides bound purified GST-Brca1-BRCT, while p-S404 and unphosphorylated peptides did not (Fig. 1C), a result confirmed using BIACORE analysis (Fig. S2A).
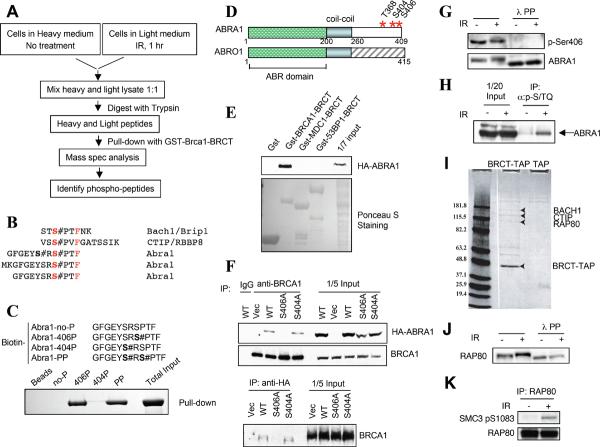
Fig. 1. Identification of Abraxas and Rap80 as Brca1-BRCT interacting proteins.
(A) A schematic view of experimental procedures for identifying phospho-peptides bound to Brca1-BRCT. (B) Identified phospho-peptides sequences. The # symbol indicates a phosphate resides on the previous residue (C). Phosphorylated Abra1 peptides bind to purified recombinant Brca1 BRCT domains. Biotinylated peptides on streptavidin beads were used to pull down purified recombinant GST-Brca1-BRCT and visualized by Comassie staining. (D) Diagram of Abra1 and its paralog Abro1 protein structures. (E) Abraxas specifically binds to Brca1 BRCT domains. HA-Abra1 was expressed in 293T cells and cell lysates were incubated with different purified GST-tagged BRCT domains. (F) HA-Abra1 association with endogenous Brca1 is dependent on S406 phosphorylation. HA-Abra1 wild type or mutant proteins were expressed in 293T cells. (G) Ser406 of Abra1 is phosphorylated in vivo. Lysates from 293T cells treated or not treated with IR and lambda phosphatase were resolved by SDS-PAGE. (H) Abraxas can be recognized by phospho-SQ/TQ antibodies against ATM/ATR substrates. Lysates from 293T cells were immunoprecipitated with anti-phospho-S/TQ antibodies and Western blots were probed with anti-Abra1 antibodies. (I) Rap80 was identified in TAP purification of Brca1-BRCT domain associated proteins. Retroviruses expressing either TAP only (TAP) or C-terminal TAP-tagged BRCT domain of Brca1 (BRCT-TAP) were introduced into Hela cells, and the infected cells were used for purification. A coomassie stained gel is shown. (J) Rap80 is phosphorylated in response to IR. (K) Rap80 can be recognized by phospho-antibodies against ATM/ATR substrates. Lysates from 293T cells were immunoprecipitated with anti-Rap80 antibody and probed with the indicated antibody.
Abraxas is well conserved in vertebrates (Fig. S3). Bioinformatics analysis also revealed KIAA0157, which is 39% identical to Abra1 in the N-terminal two-thirds of the protein (AA 1-260) we call the “ABR” domain. This protein, now named Abro1 (Abraxas Brother 1) (Fig. 1D), is also conserved in vertebrates (Fig. S3) but lacks the pSXXF motif and does not bind to Brca1 (data not shown).
Full length Abraxas binds the BRCT-repeats of Brca1 but not MDC1 and 53BP1 demonstrating specificity (Fig. 1E). A cancer predisposing Brca1 mutation, M1775R, known to disrupt BRCT integrity, abolished Abra1 binding (Fig. S2B). Abra1-Brca1 binding was phosphorylation-dependent as treatment of lysates with lambda protein phosphatase completely abolished binding (Fig. S2C), as did mutation of S406 (Fig. 1F and Fig. S2D). Abraxas S406 phosphorylation was confirmed using a phospho-specific antibody against p-S406 of Abraxas but was not increased by damage (Fig. 1G and Fig. S2E).
A search for additional Brca1-BRCT-binding proteins using TAP-tagged BRCA1-BRCT domains (15) expressed from a retrovirus in Hela cells identified Rap80, a ubiquitin-interacting motif (UIM), zinc-finger containing protein that interacts with retinoid-related testis-associated receptor (RTR) in vitro (Fig. 1I) (16). RAP80 caught our attention as it was also found to be phosphorylated on three sites, S140, S402, and S419, in response to IR in our ATM/ATR substrate screen (Matsuoka et al., in preparation). We confirmed that Rap80 bound Gst-Brca1-BRCT in vitro (Fig. S5), and that endogenous Rap80 bound Brca1 in vivo (Fig. S5 and see Fig. 4A). The binding of Rap80 to Brca1 was phosphorylation-dependent, as treatment with phosphatase abolished the binding (Fig S5, 4D).
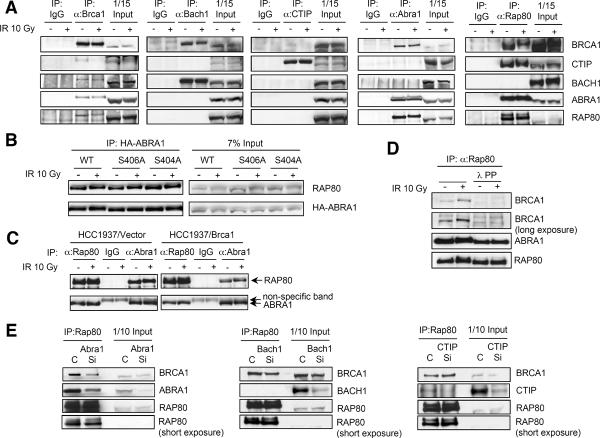
Fig. 4. Abra1 and Rap80 exist in a complex with Brca1.
(A) Brca1 forms distinct complexes with Abra1, Bach1 and CTIP. Immunoprecipitation with antibodies against Brca1, Bach1, CTIP, Abra1 and Rap80 was carried out with lysates from 293T cells treated or untreated with IR. (B) Rap80 interacts with the Abra1 mutant (S406A) that does not bind to Brca1. HA-tagged wild type or a mutant of Abra1 was expressed in 293T cells. Lysates of cells treated or untreated with IR were immunoprecipitated with anti-Rap80 antibodies. (C) TheRap80-Abra1 interaction is intact in HCC1937 cells that lack a functional Brca1. HCC1937 cells were either treated or untreated with IR. Cell lysates were immunoprecipitated with anti-Rap80, anti-Abra1 or control IgG. (D) The Rap80-Abra1 interaction is not phosphorylation dependent. Immunoprecipitation with Rap80 antibodies was carried out in cell lysates treated or not treated with lambda phosphatase. (E) The Rap80-Brca1 interaction decreases in cells treated with siRNA against Abra1. 293T cells were transfected with control oligo (C) or siRNA oligos against Abra1, Bach1 or CTIP (Si). 48 hrs later, cell lysates were prepared and immunoprecipitated with antibodies against Rap80. Immunoblotting was carried out with antibodies against Brca1, Abra1, Bach1, CTIP or Rap80.
The IR-induced phosphorylation of both Abraxas and Rap80 was confirmed as phosphatase-sensitive slower migrating forms of Abraxas and Rap80 appeared in IR treated cells (Fig. 1G, J). Furthermore, Abraxas was recognized by a pan-ATM/ATR substrate antibody after IR (Fig. 1H) and Rap80 was recognized by a phospho-SQ antibody to a known ATM site (Fig. 1K).
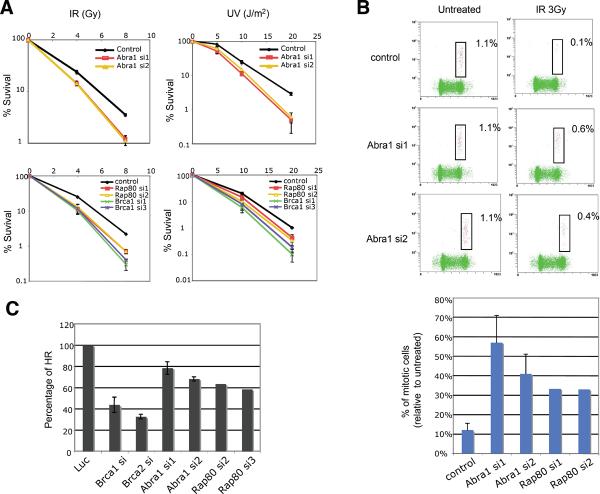
Cells depleted for Abraxas or Rap80 exhibited hypersensitivity to the killing effects of IR and UV compared with control cells (Fig. 2A) although less sensitive than Brca1-depleted cells (Fig. 2A), suggesting Abraxas and Rap80 mediate only a subset of Brca1 functions. Abraxas and Rap80 depleted cells also exhibit defects in G2/M checkpoint control in response to IR (Fig. 2B), again consistent with a role in control of Brca1 (17). Finally, Abraxas and Rap80 depletion reduces homologous recombination induced by DSBs induced by I-SceI cleavage (18), although to a lesser extent as reduction of Brca1 or Brca2 (19, 20) (Fig. 2C). These phenotypes were observed with multiple independent siRNAs for each gene.
Fig. 2. Abra1 and Rap80 are involved in DNA damage responses.
(A) Abra1-depleted or Rap80-depleted cells are sensitive to IR and UV. U2OS cells were treated with control oligos or siRNAs against Abra1, Rap80 or Brca1, incubated for 2 days, plated at low density, irradiated, and colonies counted after 14 days. (B) Analysis of the G2/M checkpoint. Cells were untreated or treated with 3 Gy IR as indicated, then incubated for 1 h at 37°C before fixation and p-H3 antibody staining. Three independent experiments were performed with siRNA oligos against Abra1. Two independent experiments were performed with siRNA oligos against Rap80 that yielded similar results. (C) Abra1-siRNA treated cells or Rap80-siRNA treated cells are defective for homologous recombination. DR-U2OS cells were transfected with siRNAs against luciferase, Brca1, Brca2, Abra1 or Rap80 separately. siRNAs against Brca1 or Brca2 were a mixture of three different siRNA oligos for each gene. Individual siRNA oligos were used for Abra1 or Rap80 genes. Three independent experiments were performed with siRNAs against luciferease, Brca1, Brca2 and Abra1.
Abraxas and Rap80 form foci that largely colocalize with Brca1 in a subpopulation of asynchronous U2OS cells (Fig. 3A,B) that represent S/G2 cells (21). They also form IR induced foci that overlap with Brca1 and Rad51 foci (Fig. 3A, B). When a UV laser was used to micro-irradiate cells, Abra1 relocalized to DNA damage sites as early as 15 min after UV, similar to Brca1 (Fig. 3C). Unlike Bach1 (22) and CTIP (7), neither Abra1 or Rap80 foci formation are dependent on Brca1. The GFP-tagged Abra1 S406A mutant, which does not bind Brca1, formed foci similar to wild type Abra1 in response to IR (Fig. S6A) and localized to UV laser-induced DNA damage as efficiently as wild-type GFP-Abra1 (Fig. 3C). In addition, stably integrated GFP-Abra1 (Fig. S6B) and Rap80 (data not shown) relocalized to DNA damage sites efficiently in Brca1-deficient HCC1937 cells after micro-irradiation with a UV laser.
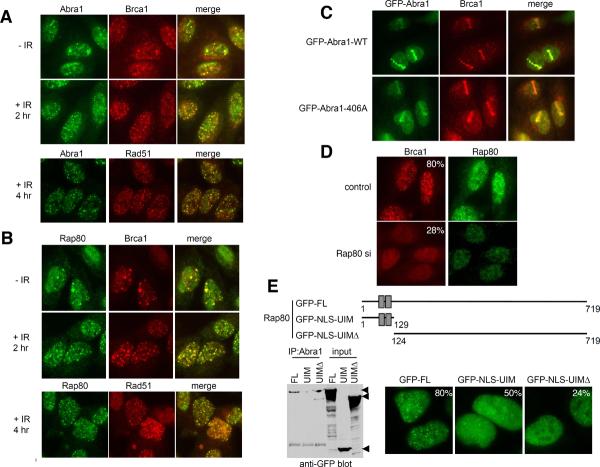
Fig. 3. Abra1 and Rap80 form DNA damage induced foci.
(A) and (B) Abra1 and Rap80 form DNA damage induced foci that colocalize with Brca1 and Rad51 foci. U2OS cells were either untreated or treated with IR 10 Gy for 2h, fixed and co-immunostatined with rabbit anti-Abra1 antibody, mouse anti-Brca1 antibody, or mouse anti-Rad51 antibody followed by proper Alex 488-conjugated or Cy3-conjuated antibodies. (C) Abra1 localizes to DNA damage regions as early as 15 min after damage, independent of binding to Brca1. Laser microirradiation was performed on U2OS cells carrying a stably integrated retroviral construct expressing GFP tagged wild type or mutant Abra1 (S406A). Cells were fixed and stained 15 min after laser treatment. (D) Brca1 foci formation is defective in Rap80-siRNA-treated cells. U2OS cells were transfected with control oligos or siRNAs against Rap80 for two days. Cells were then irradiated with 10 Gy IR, incubated for 2 h, fixed and immunostained with indicated antibodies. More than 400 cells were counted and cells containing more than 10 Brca1 foci were counted as positive. Three different oligos against Rap80 were used and similar results were obtained. (E) Rap80 IRIF formation is dependent on UIM domains independently of Abraxas binding. U2OS cells with stably integrated retroviral construct expressing GFP-WT, GFP-UIM or GFP-UIMΔ were irradiated with 10 Gy IR, 2h later, fixed and stained with antibodies against GFP followed with Alexa 488 secondary antibodies. More than 300 cells were counted to determine the percentage of cells forming foci for each cell line. Immunoprecipitation of Abraxas revealed binding to the C-terminal region of Rap80 lacking the UIM domain.
Importantly, depletion of Rap80 by siRNAs significantly reduced the foci formation of Brca1. For control oligos treated cells, around 80% of the cells form IR induced foci (IRIFs). However, when cells were depleted for Rap80, only 28% of the cells appeared to form IRIFs (Fig. 3D). Three non-overlapping siRNAs against Rap80 were used in multiple experiments and similar results were observed.
Rap80 contains two ubiquitin-interacting motif (UIM) domains at its N-terminus that might play a role in its response to DNA damage (Fig. 3E). To examine the role of UIMs in Rap80 foci formation, we made two deletion mutants. We found that the UIM domains of Rap80 alone could form IRIFs, although not as efficiently as full-length protein (only 50% of the total population). The truncated form of Rap80 lacking the UIM domains also retained the ability to form foci even less efficiently (Fig. 3E). However, in this case, the number of cells forming foci was not increased by IR. Thus, RAP80 appears to have two different means to form foci but only the UIM domain is responsive to IR.
Abraxas binds Brca1 in a manner that is mutually exclusive with Bach1 and CTIP. Bach1 or CTIP could not be detected in anti-Abra1 immunoprecipitates and reciprocally, Abra1 could not be detected in anti-Bach1 or anti-CTIP immunoprecipitates (Fig. 4A). This is consistent with each of these proteins associating with Brca1 through the same site on the BRCT motifs.
Unlike Abra1, Bach1 and CTIP, Rap80 does not possess a pSXXF motif suggesting it might associate with Brca1 via other known BRCT binding proteins. A significant portion of Rap80 could be co-immunoprecipitated with Abra1 (Fig. 4A) suggesting it might link Rap80 to Brca1. Rap80 lacking the UIM domains still retained the ability to associate with Abra1 (Fig. 3E) indicating the existence of a UIM-independent Abraxas-binding domain.
The Rap80-Abra1 interaction is independent of Brca1 because the S406A Abra1 mutant that does not bind to Brca1, maintained Rap80 binding (Fig. 4B). Furthermore, the Rap80-Abra1 interaction was intact in HCC1937 cells (Fig. 4C). While Rap80 interacts with Brca1 in a phosphorylation-dependent manner (Fig. 4D), the Rap80-Abra1 interaction is phosphorylation-independent (Fig. 4D). Therefore, Abra1 and Rap80 form a complex that interacts with Brca1.
CTIP could be detected in Rap80 immunoprecipitates (Fig. 4A) and anti-HA immunoprecipitates from cells expressing HA-Rap80 (Fig. S8). Thus, it is likely that Rap80 also resides in a complex with CTIP in vivo. To examine to what extent Abraxas and CTIP mediate Rap80 binding to Brca1, we immunoprecipitated Rap80 from cells treated with siRNAs against Abra1, Bach1 or CTIP. We found that Rap80-Brca1 interaction was significantly decreased when Abra1 levels were reduced by siRNAs (Fig. 4E). However, depleting Bach1 or CTIP did not affect the Rap80-Brca1 interaction significantly. Therefore, Rap80 interacts with Brca1 largely through binding to Abraxas. Bard1 is also present in the immunopreicipitated Abra1/Rap80/Brca1 complex (data not shown), suggesting the possibility that the Abra1/Rap80 complex might mediate the E3 ligase activity of Brca1/Bard1 heterodimers.
Our data together with previous studies (11, 23, 24), suggest that Brca1 BRCT domains directly interact with three different proteins, Abra1, Bach1 and CTIP through the p-SxxF motif, forming mutually exclusive complexes. Proteins that posses the p-SxxF motif may serve as adaptor proteins to recruit the Brca1/Bard1 E3 ubiquintin ligase to specific target proteins. This would be analogous to the role F-box proteins play in the SCF ubiqutin ligase ubiqutination pathway (25, 26). To distinguish these complexes, we propose to refer to these complexes as the Brca1 A-complex (Abra1), B-complex (Bach1), C-complex (CTIP). This will provide a framework for discussion of the individual functions of these and potentially novel Brca1 complexes yet to be discovered. Rap80 exists in the A complex and in some experiments was found in the C complex as well.
While Abraxas and its related paralog, Abro1, have no known functional motifs, Rap80 contains multiple motifs that interact with ubiquitin. Since the UIM domains of Rap80 form foci by themselves, it is likely that Rap80 localize to DNA damage sites through its UIM domains interacting with ubiquitinated proteins at the damaged sites. Furthermore, as Rap80 is required for at least a portion of Brca1 IRIFs, it may recruit Abraxas-Brca1 (and possibly CTIP-Brca1) complexes to DNA damage sites where they may ubiquitinate additional proteins, possibly amplifying ubiquitination in the same way Mdc1 amplifies H2AX phosphorylation by recruiting ATM (27). As ubiquitination seems central to Brca1's function and the DNA damage response in general, it is likely that Rap80 will play a critical role in this pathway.
The Brca1 A-complex is clearly involved in the DNA damage response. Besides forming damage-induced foci, depletion of Abra1 or Rap80 sensitizes cells to killing by IR and UV and disrupts G2/M checkpoint control and homologous recombination in response to DSBs. Neither depletion was as defective as reduction of Brca1 suggesting that the Brca1 A-complex controls only part of Brca1's role in these processes. The A-complex and B-complex are both required for homologous recombination (22). This leads to the interesting possibility that Brca1 promotes multiple distinct steps in various DNA damage responses. Furthermore, the A-complex and C-complex are both required for the G2-M checkpoint in response to IR suggesting they also perform different functions needed for cell cycle arrest. Complexes A and C are also implicated in transcription through their association with Rap80. The identification of three distinct Brca1 complexes will now allow us to specifically dissect the role of each in the DNA damage response and tumorigenesis.
Supplementary Material
Acknowledgements
We are grateful to J. Jin for retroviral expression constructs, B. Liu and S. Wu for help on using UV laser connected microscopy, E. Gillespie for assistance on the BIACORE analysis, K. Nakanishsi and M. Jasin for DR U2OS cells, F. Graham for the AdNGUS24i and AdCA36 adenoviruses. We are grateful to D.M. Livingston for exchanging information about Rap80 prior to submission. B.W. is a recipient of an NCI Howard Temin Award (1KO1, C A116275-01). A.S. is supported by T32CA09216 to the MGH Pathology Department at the Massachusetts General Hospital. This work was supported by grants from the NIH to S.J.E and S.G. S.J.E. is a Howard Hughes Medical Institute Investigator.
Footnotes
Supporting Online Material www.sciencemag.org
References
- 1.Venkitaraman AR. Cell. 2002;108:171. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 2.Narod SA, Foulkes WD. Nat Rev Cancer. 2004;4:665. doi: 10.1038/nrc1431. [DOI] [PubMed] [Google Scholar]
- 3.Cortez D, Wang Y, Qin J, Elledge SJ. Science. 1999;286:1162. doi: 10.1126/science.286.5442.1162. [DOI] [PubMed] [Google Scholar]
- 4.Wu LC, et al. Nat Genet. 1996;14:430. doi: 10.1038/ng1296-430. [DOI] [PubMed] [Google Scholar]
- 5.Baer R, Ludwig T. Curr Opin Genet Dev. 2002;12:86. doi: 10.1016/s0959-437x(01)00269-6. [DOI] [PubMed] [Google Scholar]
- 6.Hashizume R, et al. J Biol Chem. 2001;276:14537. doi: 10.1074/jbc.C000881200. [DOI] [PubMed] [Google Scholar]
- 7.Ruffner H, Joazeiro CA, Hemmati D, Hunter T, Verma IM. Proc Natl Acad Sci U S A. 2001;98:5134. doi: 10.1073/pnas.081068398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manke IA, Lowery DM, Nguyen A, Yaffe MB. Science. 2003;302:636. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- 9.Rodriguez M, Yu X, Chen J, Songyang Z. J Biol Chem. 2003;278:52914. doi: 10.1074/jbc.C300407200. [DOI] [PubMed] [Google Scholar]
- 10.Yu X, Chini CC, He M, Mer G, Chen J. Science. 2003;302:639. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 11.Glover JN, Williams RS, Lee MS. Trends Biochem Sci. 2004;29:579. doi: 10.1016/j.tibs.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 12.Everley PA, Krijgsveld J, Zetter BR, Gygi SP. Mol Cell Proteomics. 2004;3:729. doi: 10.1074/mcp.M400021-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Ong SE, Foster LJ, Mann M. Methods. 2003;29:124. doi: 10.1016/s1046-2023(02)00303-1. [DOI] [PubMed] [Google Scholar]
- 14.Amanchy R, Kalume DE, Pandey A. Sci STKE. 2005;2005:12. doi: 10.1126/stke.2672005pl2. [DOI] [PubMed] [Google Scholar]
- 15.Zhang D, Zaugg K, Mak TW, Elledge SJ. Cell. 2006;126:529. doi: 10.1016/j.cell.2006.06.039. [DOI] [PubMed] [Google Scholar]
- 16.Yan Z, Kim YS, Jetten AM. J Biol Chem. 2002;277:32379. doi: 10.1074/jbc.M203475200. [DOI] [PubMed] [Google Scholar]
- 17.Xu B, Kim ST, Lim DS, Kastan MB. Mol Cell Biol. 2002;22:1049. doi: 10.1128/MCB.22.4.1049-1059.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakanishi K, et al. Proc Natl Acad Sci U S A. 2005;102:1110. doi: 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moynahan ME, Pierce AJ, Jasin M. Mol Cell. 2001;7:263. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 20.Moynahan ME, Chiu JW, Koller BH, Jasin M. Mol Cell. 1999;4:511. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 21.Scully R, et al. Cell. 1997;90:425. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- 22.Cantor SB, et al. Cell. 2001;105:149. [Google Scholar]
- 23.Yu X, Chen J. Mol Cell Biol. 2004;24:9478. doi: 10.1128/MCB.24.21.9478-9486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenberg RA, et al. Genes Dev. 2006;20:34. doi: 10.1101/gad.1381306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. Cell. 1997;91:209. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 26.Bai C, et al. Cell. 1996;86:263. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 27.Lou Z, et al. Mol Cell. 2006;21:187. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.