Abstract
Rationale
One hypothesis for elevated Ca2+ leak through cardiac ryanodine receptors (RyR2) in heart failure (HF) is interdomain “unzipping” that can enhance aberrant channel activation. A peptide (DPc10) corresponding to RyR2 central domain 2460-2495 recapitulates this arrhythmogenic RyR2 leakiness by unzipping N- and central-domains. Calmodulin (CaM) and FK-506 binding protein (FKBP12.6) bind to RyR2 and stabilize the closed channel. Little is known about DPc10 binding to the RyR2 and how that may interact with binding (and effects) of CaM and FKBP12.6 with RyR2.
Objective
Measure, directly in cardiac myocytes, the kinetics and binding affinity of DPc10 to RyR2 and how that affects RyR2 interaction with FKBP12.6 and CaM.
Methods & Results
We used permeabilized rat ventricular myocytes, and fluorescently-labeled DPc10, FKBP12.6, and CaM. DPc10 access to its binding site is extremely slow in resting RyR2, but accelerated by promoting RyR opening or unzipping (by unlabeled DPc10). RyR2-bound CaM (but not FKBP12.6) drastically slowed DPc10 binding. Conversely, DPc10 binding significantly reduced CaM (but not FKBP12.6) binding to the RyR2. Fluorescence resonance energy transfer measurements indicate that DPc10 and CaM binding sites are separate and allow triangulation of the structural DPc10 binding locus on RyR2 vs. FKBP12.6 and CaM binding sites.
Conclusions
DPc10-RyR2 binding is sterically limited by the resting zipped RyR2 state. CaM binding to RyR2 stabilizes this zipped state, while RyR2 activation or prebound DPc10 enhances DPc10 access. DPc10 and CaM binding sites are distinct but allosterically interacting RyR2 sites. Neither DPc10 nor FKBP12.6 influences RyR2 binding of the other.
Keywords: Ryanodine receptor, DPc10, calmodulin, FKBP12.6, FRET
INTRODUCTION
The cardiac ryanodine receptor (RyR2) Ca2+ release channel in the sarcoplasmic reticulum (SR) membrane plays a central role in cardiac excitation-contraction (EC) coupling.1 Over the past ten years, diastolic Ca2+ leak through dysfunctional RyR2 has been recognized as an important factor contributing to altered Ca2+ homeostasis and arrhythmias in heart failure (HF). Evidence in several reports shows that RyR2 abnormality in HF causes increased diastolic Ca2+ leak, leading to contractile and relaxation dysfunction.2, 3, 4 Moreover, the abnormal Ca2+ leak through RyR2 provides a substrate for delayed afterdepolarization (DAD) that leads to lethal arrhythmias.5
One leading hypothesis explains the RyR2 dysfunction in HF and lethal arrhythmias, such as catecholaminergic polymorphic ventricular tachycardia (CPVT), by structural RyR2 changes that result in defective interaction (or zipping) between the N-terminal (N: 0-600) and the central (C: 2000-2500) domains.6 According to this concept, in the resting state, the N-terminal and central RyR2 domains interact with each other to act as a regulatory switch that influences RyR channel gating. This tight interdomain interaction, termed “domain zipping” seems to stabilize the closed channel. Weakening of these interdomain interactions may be caused by mutations in either the N-terminal or central regions of RyR2,7 or via competition by peptides derived from these two domains (domain unzipping),8, 9, 10 resulting in an increased opening probability of the RyR2 and leakiness of Ca2+. DPc10 is a synthetic peptide corresponding to a 36-residue stretch of the central domain (Gly2460-Pro2495) of RyR2.6 It has been shown that DPc10 can specifically and directly associate with the N-terminal domain11,12, and thus compete with its zipping to the central domain, and that the N-domain/DPc10 association can destabilize RyR2 (via domain unzipping), to increase Ca2+ leakiness.11 A single R2474S point mutation in DPc10 (DPc10-mut) inhibits all DPc10 effects, and a related human RyR2 mutation is associated with CPVT and RyR2 leakiness.
Tateishi et al.12 reported that a domain peptide (residues 163-195 of the N-terminal RyR2 domain, DP163-195) also induced Ca2+ leak from SR, presumably because it binds to the central domain and competes with the N-terminal/central zipping. Taken together, these data suggest that synthetic domain peptides bind to key subdomains of RyR2 and are capable of mimicking disease conditions of the RyR2 channel by interfering with interdomain interactions.
The FK506 binding proteins FKBP12 and FKBP12.6 are expressed in cardiac myocytes and can form tight complexes with RyR, at a stoichiometry of four FKBPs per tetrameric RyR channel. 1 As such, these FKBP isoforms are considered important RyR2 subunits, and have been reported to promote the closed channel state, but this role is controversial in myocytes from normal rat hearts.13 Indeed, we found that FKBP12 does not significantly alter Ca2+ sparks, whereas FKBP12.6 is slightly inhibitory, that PKA-dependent RyR2 phosphorylation does not alter FKBP binding and that only a small fraction of RyR2 in native myocytes has FKBP12.6 bound. 14 Two previous studies, in which RyR2 was treated with domain peptides to mimic pathologic Ca2+ leakage, found no direct effect of DPc10 on FKBP12.6 co-immunoprecipitation with RyR2. 11, 12 It is unknown whether FKBP12.6 influences binding of DPc10 to RyR2 or the ensuing increased Ca2+ leakage.
CaM is a ubiquitous Ca2+ binding protein that binds to the RyR2 and modulates its channel function.15 Binding of CaM within the cytosolic domain of RyR2 (at a site partly formed by residues 3583-3603) inhibits channel activity both at diastolic and elevated [Ca2+].16, 17 This indicates that CaM stabilizes the closed state of RyR2 in the resting state.18 Interestingly, concurrent addition of a high concentration of CaM with DPc10 in WT cardiomyocytes reduced the Ca2+ spark frequency compared to addition of DPc10 alone. Furthermore, in myocytes carrying a CPVT-linked RyR2 mutation (where •-adrenergic stimulation activates SR Ca leak) leads to defective interdomain interaction and reduced CaM binding to the RyR2 vs. WT myocytes.19 In addition, Ono et al.20 also reported that the CaM binding affinity to RyR2 in HF is significantly reduced compared to that of normal RyR2. Treatment of wild-type myocytes with DPc10 also inhibited CaM binding at the Z-line in the CPVT mutants.19
DPc10 and related RyR2 peptides may, therefore, serve as useful molecular probes to study the channel’s structure-function relationship. However, the details of DPc10 binding to RyR2, including affinity and kinetics, are still unknown. In the present study, our goal was to characterize the binding of DPc10 to the RyR2 in the relatively intact environment of saponin-permeabilized rat ventricular myocytes. We used fluorescent DPc10 to measure the affinity and kinetics of DPc10 binding to RyR2, and its influence on CaM and FKBP12.6 binding and function. Furthermore, we used fluorescence resonance energy transfer (FRET) between fluorescent FKBP12.6, DPc10, and CaM to determine how DPc10 alters CaM and FKBP12.6 binding and assess where the DPc10 binding site on RyR2 is in relation to CaM and FKBP12.6 binding sites.21, 22
METHODS
Rat ventricular myocytes were isolated and permeabilized as previously described.23 All procedures were performed according to the Guiding Principles in the Care and Use of the Animals approved by the Council of American Physiological Society. An expanded Methods section can be found in the Online Supplement. DPc10, FKBP12.6 and CaM were labeled at specific sites with small fluorescence probes similar to our previous studies.14,21,22 Competitive inhibition of fluorescent DPc10 (F-DPc10) binding to RyR2 by non-fluorescent DPc10 (NF-DPc10) showed that both bind to RyR2 at the same site and same affinity (Online Figure IA). NF-DPc10 and F-DPc10 produce similar effects on Ca2+ sparks and SR Ca2+ content (Online Figure IB), confirming that F-DPc10 exhibits the same functional effect as NF-DPc10.
RESULTS
Localization, and binding isotherms of F-DPc10 in permeabilized myocytes
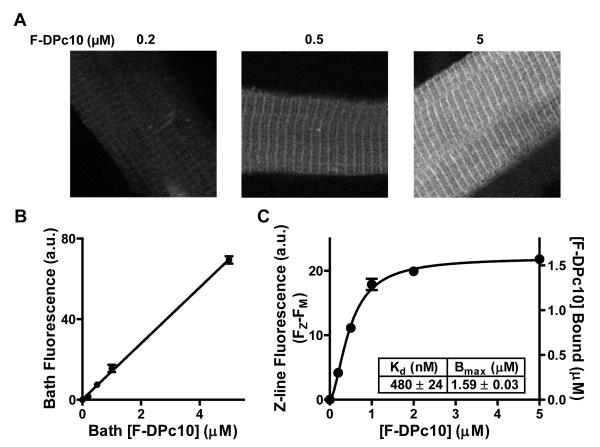
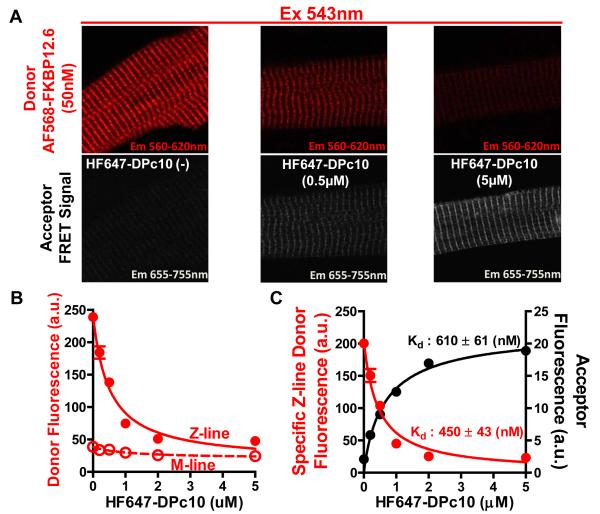
Figure 1A shows confocal images of saponin-permeabilized rat ventricular myocytes incubated with different concentrations of DPc10 labeled with 5-carboxyfluorescein at its N-terminus (F-DPc10). Myocytes were exposed to 0.2, 0.5, and 5 μmol/L F-DPc10, with intracellular [Ca2+] ([Ca2+]i) set at 50 nmol/L. F-DPc10 fluorescence is highest at the Z-lines, where RyR2 is concentrated, forming a typical cross-striated pattern. The difference between fluorescence intensity at the Z-line (FZ) and M-line (FM) is taken to represent [F-DPc10] specifically bound at the myocyte Z-line. We calibrated the bound [F-DPc10] in permeabilized myocytes using the linear relationship between F-DPc10 fluorescence and bath [F-DPc10] (Figure 1B). In-cell F-DPc10 binding isotherms indicate an apparent dissociation constant (Kd) for F-DPc10 binding at the Z-line of 480 ± 24 nmol/L; the maximal binding (Bmax), which reflects the concentration of F-DPc10 binding sites, was 1.59 ± 0.03 μmol/L (Figure 1C). This Bmax value for F-DPc10 is similar to our previous steady-state binding measurements of FKBP12.6 sites, which specifically binds to RyR2 in permeabilized myocytes with sub-nanomolar affinity.14 Thus, we infer that RyR2 is the main target for F-DPc10 (see also Discussion). To further test whether this Z-line associated F-DPc10 represents RyR2-bound F-DPc10, we measured FRET between FKBP12.6 (known to specifically bind to RyR2 with sub-nanomolar affinity)14 and F-DPc10. Figure 2A shows confocal images of FRET between FKBP12.6 labeled with Alexa Fluor 568 as a donor (AF568-FKBP12.6) and different concentrations of DPc10 labeled with HyLite Fluor 647 (HF647) as acceptor (HF647-DPc10). Donor (AF568-FKBP12.6) fluorescence at the Z-line was quenched by HF647-DPc10, but the M-line signal was not (Figure 2B). The apparent Kd calculated based on enhanced acceptor fluorescence (EAF) was 610 ± 61 nmol/L, and calculated based on donor fluorescence quench was 450 ± 43 nmol/L (Figure 2C). The donor quench measurement is less complicated (e.g., by donor bleed-through), consequently likely to be more accurate, yielding a Kd value that is remarkably similar to that obtain in our direct measurements of F-DPc10 at the Z-line (Figure 1C).
Figure 1. Localization and steady-state binding of F-DPc10 in permeabilized myocytes.
A, Confocal images of saponin-permeabilized myocytes incubated in internal solution containing 0.2 μmol/L, 0.5 μmol/L, 5 μmol/L F-DPc10. B, Dependence of bath F-DPc10 fluorescence on [F-DPc10]. C, Specific binding of F-DPc10 at the Z-lines (FZ-FM), calibrated based on the standard curve in B, and fit to single-site binding isotherm.
Data are reported as mean ± SE, from n = 10.
Figure 2. Steady-state Kd measurement of DPc10 by using FRET between FKBP12.6 labeled with AF568 (donor) and HF647-DPc10 (acceptor) in permeabilized cardiomyocytes.
A, Confocal images of FRET as the decrease of AF568-FKBP12.6 fluorescence (donor quench) upon addition of 0.5, 5μmol/L HF647-DPc10. B, Donor fluorescence intensity from Z-line and M-line plotted vs. [HF647-DPc10]. C, FRET due to HF647-DPc10 (acceptor) binding was detected at the Z-line (FZ-FM) either as decrease in donor fluorescence or as an enhancement in the acceptor fluorescence. Data are reported as mean ± SE.
Binding kinetics of F-DPc10 in permeabilized cardiac myocytes
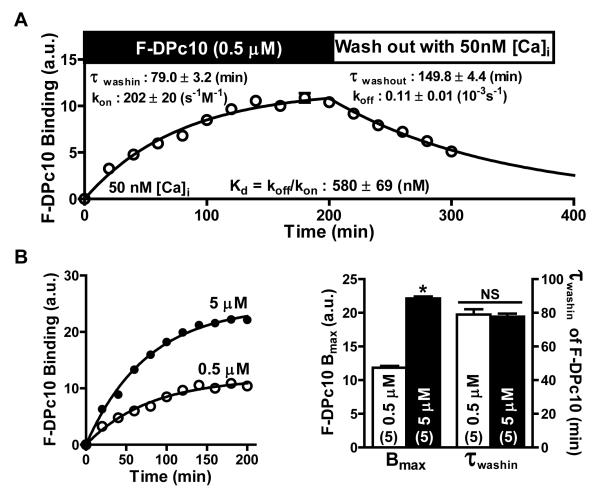
To characterize DPc10 binding kinetics at Z-lines, we performed F-DPc10 wash-in (500 nmol/L) and wash-out experiments in permeabilized myocytes (Figure 3A). Association (τwash-in = 79.0 ± 3.2 min) and dissociation (τwash-out = 149.8 ± 4.4 min) were very slow compared to similar FKBP12.6 measurements.14 From the wash-in/wash-out measurements, we calculated the association and dissociation rates constants, kon and koff, according to:
| (Eq.1) |
where kwash-out ≈ koff and k = 1/τ (s−1). Accordingly, kon = 202 ± 20 (s−1 M−1), and koff = 0.11 ± 0.01 (10−3 s−1). Based on these values and Kd = koff/kon, F-DPc10 binds at the Z-line with Kd = 580 ± 69 nmol/L, consistent with the steady-state Kd measurements (Figure 1). We repeated this kinetic analysis using FRET between FKBP12.6 and DPc10, thus assessing the RyR2-specific DPc10 binding (Online Figure IIA, B). Both methods of detecting FRET (EAF and donor quench) showed very similar, slow association and dissociation rates to those in Figure 3 for direct detection of F-DPc10 binding at the Z-line. Based on these kinetic and affinity analyses, we infer that most of the Z-line-specific DPc10 binding is to RyR2. That is also consistent with the Bmax, which would imply ~1 DPc10 per RyR2 monomer.
Figure 3. Kinetics of F-DPc10 binding at the myocyte Z-line.
A, Time course of F-DPc10 (0.5 μmol/L) wash-in and wash-out. B, Effect of [F-DPc10] (0.5 and 5 μmol/L) on τwash-in and Bmax. Data are reported as mean ± SE.
We were intrigued by the slow kwash-in, and conducted measurements to further understand the basis of this slow association. We tested the hypothesis that, at resting [Ca2+]i, DPc10 access to its RyR2 binding site is sterically hindered. If the N-terminal and central domains are tightly “zipped” to each other, this interaction may occlude the DPc10 binding site on the RyR2, thus limiting the DPc10 kon. Alternatively, a limiting factor may be the rate at which DPc10 adopts a conformation that can bind to RyR2. To discern between these mechanisms, we determined the effect of [F-DPc10] on τwash-in. If the small fraction of DPc10 in the right conformation limits binding rate, then τwash-in should be faster at higher [F-DPc10], according to Eq 1. Figure 3B shows that this was not the case. Instead, a 10-fold increase in [F-DPc10] had no significant effect on the τwash-in, although it did increase Bmax (Figure 3B, C). The same was seen when using FKBP12.6-DPc10 FRET to assess τwash-in with 0.5 vs. 5 μmol/L HF647-DPc10 (Online Figure IIIA, B). These results indicate that F-DPc10 association at its RyR2 binding site exhibits restricted access by a factor residing on RyR2, e.g. binding site opening or transitions from zipped to unzipped state.
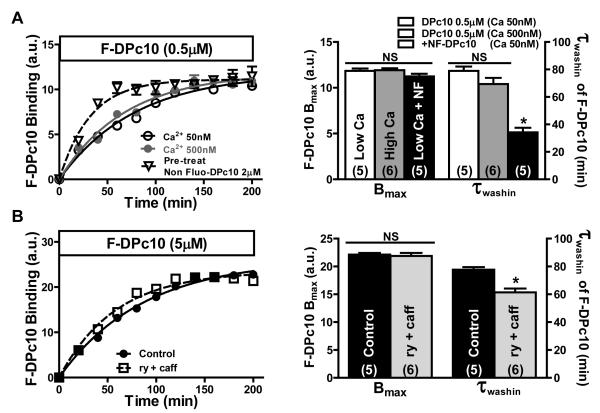
In our working model, under resting conditions, the RyR2 closed state may be stabilized by the interaction between the N-terminal and central domain in the “zipped” state. We hypothesized that conditions that promote RyR2 opening might enhance the rate of unzipping, and accelerate F-DPc10 τwash-in. To test this, we first monitored F-DPc10 wash-in at elevated Ca2+ (500 nmol/L). However, the 13% faster mean τwash-in was not significant (Figure 4A). While 500 nmol/L Ca2+ can increase RyR2 opening, it does not prolong open time appreciably, and the latter might be important in the propensity for unzipping. Thus, we pre-incubated myocytes with ryanodine (100 μmol/L) plus caffeine (5 mmol/L), which are known to favor long RyR2 openings and were reported to cause RyR2 domain unzipping in HEK293 cells.24 Ryanodine +caffeine produced a 21% faster τwash-in (p=0.002; Figure 4B). However, the most significant effect was seen after pre-saturating RyR2 with NF-DPc10 (and then NF-DPc10 washout with F-DPc10 present; Figure 4A). This treatment significantly accelerated F-DPc10 association by a factor of ~2 (Figure 4A). None of these treatments significantly altered Bmax (Figure 4A, B). Assuming that koff of NF-DPc10 is the same as for F-DPc10 and using Eq 1, the kon is increased by 3.2-fold by unzipping due to prebinding of NF-DPc10 to the RyR2. Our working hypothesis is that the RyR2 open state may increase the probability of an RyR2 shifting to the unzipped state and allow faster F-DPc10 wash-in. It also seems that the RyR2 open state (favored by caffeine-ryanodine) differs from the unzipped state (with DPc10 bound).
Figure 4. Effect of RyR2 channel modulators on the kinetics of F-DPc10 Z-line association.
A, Time course of F-DPc10 (0.5 μmol/L) Z-line binding in internal solution containing low [Ca2+]i, high [Ca2+]i, or after a 3 hr pre-equilibration with saturating [NF-DPc10] (2 μmol/L) in low [Ca2+]i (triangles). Data are reported as mean ± SE. B, Time course of F-DPc10 (5 μmol/L) Z-line binding after a 3 hr pre-equilibration in internal solution containing ryanodine (100 μmol/L) and caffeine (5 mmol/L). Data are reported as mean ± SE. (n values on bars).
Cross-talk between F-DPc10 and CaM or FKBP12.6 binding
1. Effect of CaM and FKBP12.6 on DPc10 binding at the myocyte Z-line
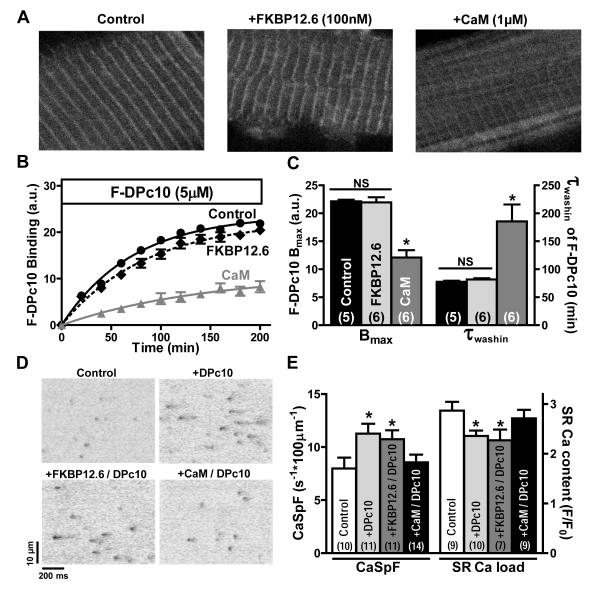
Both FKBP12.6 and CaM bind to the RyR2 and can reduce channel opening, which might alter DPc10 binding. Figure 5A shows representative confocal images of FKBP12.6 (100 nmol/L) and CaM (1 μmol/L) effects on F-DPc10 binding, as detected after a 200 min incubation with F-DPc10. While pre-equilibration with saturating CaM (1 μmol/L) greatly reduced F-DPc10 binding, pre-treatment with FKBP12.6 (100 nmol/L) did not alter F-DPc10 binding in permeabilized myocytes. Neither CaM nor FKBP12.6 pretreatment altered M-line F-DPc10 fluorescence (Online Figure IV). Figure 5B shows the time course of F-DPc10 wash-in with or without pretreatment with FKBP12.6 or CaM. Saturation of RyR2 with FKBP12.6 (100 nmol/L) did not alter either F-DPc10 maximal binding (Bmax) or τwash-in. In contrast, saturation of RyR2 with CaM dramatically reduced F-DPc10 Bmax and slowed DPc10 access to its binding site, as indicated by the large increase in τwash-in (Figure 5C). We infer that CaM stabilizes the domain interaction between N-terminal and central domains in the “zipped” state, and may thereby reduce DPc10 access to its binding site. To test for direct CaM-DPc10 interaction, we performed control FRET measurements between donor-labeled CaM and acceptor-labeled DPc10 in solution in the absence of RyR. The maximal FRET efficiency (<1%) ruled out direct CaM-DPc10 interaction.
Figure 5. Effects of FKBP12.6 and CaM on F-DPc10 binding at the Z-line, and on local Ca2+ release events in permeabilized cardiomyocytes.
A, Representative confocal images illustrating the effect of FKBP12.6 (100 nmol/L), and CaM (1 μmol/L) on the F-DPc10 (5 μmol/L) binding at the Z-lines. B, Time course of F-DPc10 (5 μmol/L) wash-in (circles), and in the presence of FKBP12.6 (100 nmol/L, diamonds) or CaM(1 μmol/L, triangles). C, Summary of fitting parameters (Bmax and τwash-in) for the data in panel B. Data are reported as mean ± SE. D, Ca2+ sparks measured using Fluo-4 as Ca2+ indicator. Representative line-scan images acquired after addition of DPc10 (5 μmol/L), DPc10 (5 μmol/L) plus FKBP12.6 (100 nmol/L), and DPc10 (5 μmol/L) plus CaM (1 μmol/L). [Ca2+]i = 50 nmol/L, buffered by 0.5 mmol/L EGTA. E, Summary of Ca2+ spark frequency and SR Ca2+ content. SR Ca2+ content was measured by addition of 15 mmol/L caffeine. Data are reported as mean ± SE (n values on bars).
Next, we asked whether RyR2 is activated by DPc10 and if FKBP12.6 or CaM can prevent this. We assessed Ca2+ sparks in permeabilized myocytes perfused with internal solution containing 50 nmol/L free Ca2+ plus 1 μmol/L AIP (to inhibit CaMKII activity). Line-scan images were recorded after 3 hr incubations under control conditions, and in the presence of 5 μmol/L DPc10, with or without 1 μmol/L CaM or 100 nmol/L FKBP12.6 (Figure 5D). DPc10 robustly increased Ca2+ spark frequency (CaSpF) vs. control, an effect almost completely blocked by CaM (Figure 5E). However, CaSpF activation by DPc10 was only slightly decreased by FKBP12.6 (Figure 5E), and not at all when normalized to SR Ca2+ content (Online Figure V). In DPc10-treated permeabilized myocytes, Ca2+ spark full width at half maximum (FWHM) and full duration at half maximum (FDHM) were significantly increased compared to control, and decreased when pre-treated with CaM (Online Table I).
Since Ca2+ spark frequency strongly depends on the SR Ca2+ content, we also measured SR Ca2+ content as the amplitude of caffeine-induced Ca2+ release (Figure 5E, right). In cells treated with DPc10 with or without FKBP12.6 the SR Ca2+ was significantly lower than under control conditions. In contrast, treatment with CaM plus DPc10 resulted in no significant decrease in SR Ca2+ content vs. control. Thus, the increased CaSpF in the presence of FKBP12.6 plus DPc10 cannot be secondary to increased SR Ca2+ content (which was in fact decreased). These results are consistent with a DPc10-induced increase in RyR2 channel activity resulting from defective interaction between N-terminal and central domains. This also agrees with the lack of FKBP12.6 effect on F-DPc10 binding kinetics (Figure 5B) and the potent inhibition of DPc10 binding by CaM (which may promote the “zipped” state and inhibit DPc10 access).
2. Effect of DPc10 on FKBP12.6 and CaM binding in permeabilized myocytes
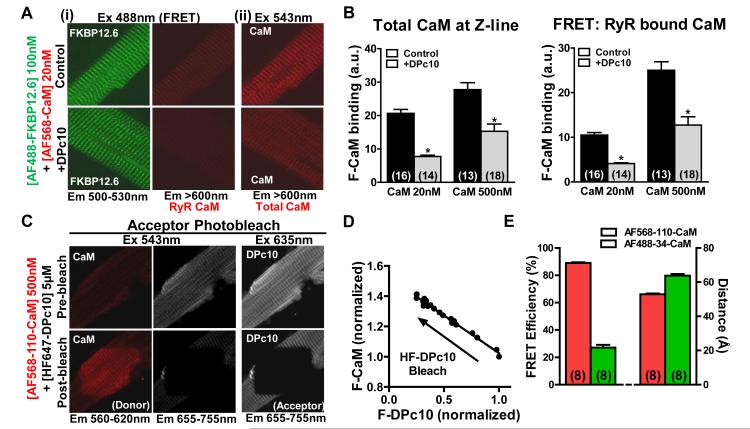
To examine the converse influence that DPc10 may have on FKBP12.6 and CaM binding to RyR2 in situ, we used fluorescent FKBP12.6 and CaM variants labeled with Alexa Fluor 488 or 568 (AF488 and AF568, respectively). These fluorescent proteins were added to saponin permeabilized myocytes with or without pre-equilibration with saturating DPc10 concentration. First, we found that AF488-FKBP12.6 at 1 nmol/L (near its Kd)14 forms a striated pattern that is not affected by pre-incubation with 5 μmol/L DPc10. (Online Figure VIA, B). Thus, DPc10 does not influence FKBP12.6 binding to RyR2. To measure CaM that is specifically RyR2-bound, we measured FRET between AF488-FKBP12.6 (donor) and AF568-34-CaM (acceptor in the N-domain)21 at a [CaM] near the Kd (20 nmol/L; Figure 6A (i)).22,25,26 Using direct excitation at 543 nm (emission at >600 nm) we detected total CaM at the Z-lines (Figure 6A (ii)). We also did this with high [CaM] (500 nmol/L) which saturates RyR2 with CaM under control conditions (without DPc10; Online Figure VIC). Figure 6B shows that pre-treatment with DPc10 significantly reduced CaM binding (at 20 nmol/L CaM) both at the RyR2 and overall at the Z-line, and by similar proportions. Even at high AF568-34-CaM levels (500 nmol/L) DPc10-treated myocytes exhibited reduced CaM binding at the RyR2 (FRET) and at the Z-lines (Total) vs. control. Thus, once F-DPc10 binds to the RyR2 and decreases N-terminal-central domain interactions, it reduces the CaM affinity for RyR2. Taken together, these results show that DPc10 and CaM binding to RyR2 are mutually inhibitory. To test whether DPc10 and CaM bind at the same or nearby RyR2 sites we measured FRET between CaM and DPc10.
Figure 6. The effect of DPc10 on CaM and FKBP12.6 binding to RyR2 in cardiac myocytes.
A, Representative confocal image of the effect of DPc10 on AF568-CaM binding at the Z-lines (ii, Ex =543 nm) and at the RyR2 detected by FRET between AF488-FKBP12.6 (donor) and AF568-CaM (acceptor) (i, Ex =488 nm). Myocytes were incubated with 5 μmol/L DPc10 (3 hrs, 25°C) before adding CaM. B, Quantitative analysis of panel A data for 20 and 500 nmol/L CaM. Data are reported as mean ± SE. C, Confocal images illustrating FRET between AF568-110-CaM (donor) and HF647-DPc10 (acceptor) measured using the acceptor photobleaching method. Photobleached area is clearly delineated in the middle of the myocyte image. D, Dependence of AF568-110-CaM fluorescence intensity on the extent of HF647-DPc10 photobleach. Data is best fitted by a linear function (R2 = 0.986), indicating that each donor participates in FRET with only one acceptor. E, Summary of FRET efficiency E and distances between AF568-110-CaM and HF647-DPc10, and between AF488-34-CaM and HF647-DPc10 derived from FRET. Data are reported as mean ± SE.
FRET between CaM and DPc10
We used a fluorescence donor probe (AF568) at the C-lobe of CaM21 (AF568-110-CaM) and HiLyte Fluor 647 (HF647) as acceptor probe on the N-terminus of DPc10 (HF647-DPc10). We utilized the acceptor photobleach approach with measurement of the resultant increase in donor (AF568-110-CaM) fluorescence in saponin permeabilized myocytes (Figure 6C). To use this approach quantitatively, all acceptor (DPc10) sites must be loaded, so that all donors can participate in FRET.
Our results above show that it is impractical to saturate RyR2 with both CaM and DPc10 (Figure 5B and 6B). To overcome this challenge, we pre-equilibrated the myocytes with saturating HF647-DPc10, thus loading all DPc10 binding sites on RyR2. Then when we add AF568-110-CaM (500 nmol/L) ~50% of RyR2s have donor, but all have acceptor, allowing quantitative analysis of enhanced donor fluorescence upon acceptor photobleach. Figure 6C shows selective photobleach of HF647-DPc10 (at 635 nm) in only the central region of the myocyte, and donor fluorescence was enhanced only in that region (lower left panel), indicating that donors and acceptors are within FRET range.
To rule out the possibility that there is energy transfer between a donor and multiple acceptors, we measured the relationship between donor fluorescence enhancement and acceptor photobleach, and found a linear relationship (Figure 6D), which indicates a 1:1 stoichiometry for CaM-DPc10 FRET. We interpret this result as clear evidence that the FRET efficiency (E) between AF568-110-CaM and HF647-DPc10 reflects the proximity of one CaM to one DPc10. E and donor-acceptor distance calculations are described in the Online Methods.
FRET efficiency between AF568-110-CaM and HF647-DPc10 upon 98.2 ± 0.2 % acceptor photobleach was 0.89 ± 0.01 (n=8). This corresponds to a distance of 53 ± 1 Å (Figure 6E), based on R0 = 75 Å for the AF568-HF647 donor-acceptor pair. With an alternative donor probe (AF488), this time attached at the N-lobe of CaM, and the same acceptor (HF647) on DPc10, we measured E= 0.27 ± 0.02 which corresponds to an interprobe distance of 63 ± 1 Å (Figure 6E). Thus, this result shows that the donor probes on CaM are 53-63 Å from the acceptor on DPc10, suggesting that CaM and DPc10 can simultaneously bind at distinct, yet nearby sites within the RyR2 structure. This again favors an allosteric rather than competitive basis for the mutual inhibition seen between CaM and DPc10 binding to the RyR2.
FRET between FKBP12.6 and DPc10
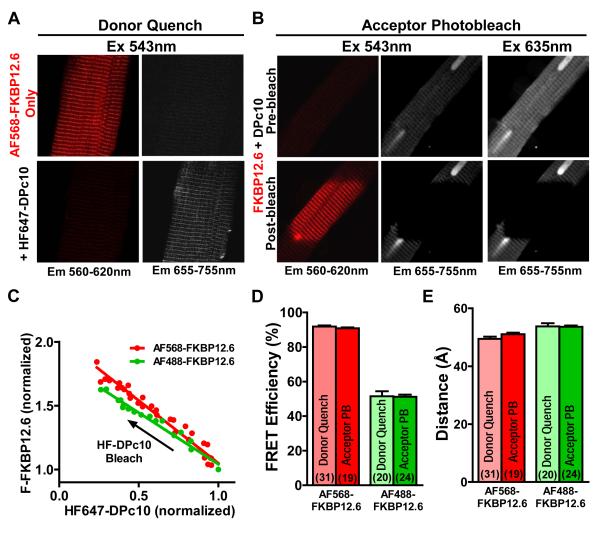
To gain further information about the topology of the DPc10 binding site on RyR2, we used the location of FKBP12.6 as a reference point.22, 27, 28 FKBP12.6 was labeled at position 1421 with the fluorescent donors AF488 (AF488-FKBP12.6) or AF568 (AF568-FKBP12.6), while DPc10 was labeled with the acceptor HF647. We used the same two methods as above to measure FRET in permeabilized myocytes. Figure 7A shows that when HF647-DPc10 (5 μmol/L; bottom) was added to myocytes equilibrated with donor (50 nmol/L AF568-FKBP12.6), there was very strong reduction in donor emission (560-620 nm) and simultaneous appearance of FRET in the acceptor emission channel (655-755 nm). Next, we monitored the increase in donor fluorescence after acceptor photobleach when both donor (AF568-FKBP12.6) and acceptor (HF647-DPc10) were pre-equilibrated (Figure 7B). The upper panel is before bleach, and the lower panel is after acceptor photobleach in only part of the myocyte, resulting in locally enhanced donor fluorescence (red). FRET between AF568-FKBP12.6 and HF647-DPc10 was almost complete (Figure7A-B), indicating close proximity between the donor and acceptor probes. To better gauge the FKBP12.6-DPc10 distance, we used an alternative donor probe, AF488-FKBP12.6 and the same HF647 acceptor on DPc10 (to reduce R0 for the FRET pair). Online Figure VIIA-B show representative confocal images of donor quench and acceptor photobleach using AF488-FKBP12.6 as a donor.
Figure 7. FRET between FKBP12.6 labeled with AF568 or AF488 (donor) and HF647-DPc10 (acceptor) in permeabilized cardiomyocytes.
A, Confocal images showing FRET as the decrease in AF568-FKBP12.6 fluorescence (donor quench) upon addition of HF647-DPc10. B, Confocal images illustrating FRET as the increase in AF568-FKBP12.6 fluorescence after photobleaching HF647-DPc10. Acceptor photobleach is clear in the center of the confocal myocyte image. C, Dependence of AF568- and AF488-FKBP12.6 fluorescence intensity on the extent of HF647-DPc10 photobleaching. Data is best fit by a linear function (R2 = 0.966 for AF568-FKBP12.6, R2 = 0.972 for AF488-FKBP12.6) indicating that each donor participates in FRET with only one acceptor. D, E, Summary of E and distances between AF568-/488-FKBP12.6 and HF647-DPc10 based on FRET measured by donor quench and acceptor photobleach. Data are reported as mean ± SE.
To ensure that FRET between FKBP12.6 and DPc10 accurately reflects interprobe distance, we performed several controls: (1) As shown in Online Figure VIIC, there was no significant difference in direct acceptor fluorescence intensity with or without equilibrated donors; (2) Online Figure VIID indicates that photobleach of the acceptor was essentially complete in both cases (AF488-FKBP12.6: 98.9 ± 0.3 %, AF568-FKBP12.6: 99.4 ± 0.4 %). (3) We also checked the stoichiometry of donor and acceptor using the method shown in Figure 6D. Figure 7C shows that fluorescence of AF568- and AF488-FKBP12.6 depended linearly on HF647-DPc10 fluorescence during progressive bleach, indicating that each donor is coupled to a single acceptor.
The average FRET efficiency between AF568/488-FKBP12.6 and HF647-DPc10 was used to estimate the distance between FKBP12.6 and DPc10. The FRET efficiency between AF568-FKBP12.6 and HF647-DPc10 measured by the donor quench method was E= 0.92 ± 0.01 (n=31), while measured by acceptor photobleach method E= 0.91 ± 0.01 (n=19) (Figure 7D), corresponding to a distance of 50 ± 1 Å and 51 ± 1 Å. For the shorter R0 pair (AF488-FKBP12.6 and HF647-DPc10) FRET E by the donor quench was 0.52 ± 0.03 (n=20) and by acceptor photobleach was 0.51 ± 0.01 (n=24) (Figure 7D right), corresponding to distances of 53 ± 1 Å and 54 ± 1 Å, respectively. Thus, remarkably similar results were obtained with two different donor-acceptor pairs, and two different methods for measuring FRET (Figure 7E). According to our FRET results, bound DPc10 is near both FKBP and CaM, which implies that reciprocal inhibition of CaM and DPc10 binding to RyR2 occurs through an allosteric mechanism rather than competition for the same binding site. Combining information from CaM-DPc10 and FKBP12.6-DPc10 FRET allows triangulation of relative positions on the RyR2 (see Discussion).
DISCUSSION
We used fluorescent DPc10, FKBP12.6, CaM and confocal microscopy of permeabilized cardiomyocytes and found that (1) DPc10 access to its binding site is sterically hindered in resting (zipped) RyR2, (2) F-DPc10 wash-in kinetics provide a sensitive measure of the RyR2 unzipped state in permeabilized myocytes, (3) DPc10 and CaM binding to RyR2 are mutually inhibitory (via allosteric rather than competitive interaction), and (4) DPc10, CaM and FKBP12.6 are physically 50-60 Å from each other as vertices of a roughly equilateral triangle on RyR2.
RyR2 is the main target of DPc10 binding at Z-lines
To assess DPc10 binding affinity and concentration at Z-lines, we used equilibrium and kinetic binding methods. Both methods (Figures 1C, 2C and 3A) yielded similar Kd values (~500 nmol/L), and a Bmax value of 1.6 μmol/L, which agrees with the concentration of RyR2 monomers and FKBP12.6 at the Z-line in rat ventricular myocytes.14,29 This Bmax value is higher than our previous measurements of Bmax of FKBP12.6 (~1 μmol/L) which binds very specifically (~1 nM Kd) to RyR2.14 The reason for this difference is that for DPc10 (vs. FKBP12.6) the fluorescence between Z-lines is a higher fraction of that at the Z-line (Online Figure VIII), in part because of the much higher DPc10 concentration required to saturate RyR2. For this reason we used the difference in Z- vs. M-line fluorescence (Fz-FM) to assess specific binding of F-DPc10 at the Z-lines. For FKBP12.6 we used cell average fluorescence,14 to measure Bmax in myocytes. If we re-analyze F-FKBP12.6 binding as we did F-DPc10 (using Fz-FM), the Bmax for FKBP12.6 was 1.3 μmol/L, consistent with F-DPc10 Bmax. Furthermore, the kinetics and affinity of Z-line associated DPc10 were almost the same as that of RyR2-specifically bound DPc10 (Figure 1-3 and Online Figure II and III). We conclude that RyR2 is the main specific Z-line target for F-DPc10.
Access of DPc10 to its RyR2 binding site is restricted
We found both the wash-in and wash-out kinetics of F-DPc10 binding (kwash-in and kwash-out) are extremely slow (Figure 3A). The calculated kon for F-DPc10 is about 1800-fold slower than that we measured for FKBP12.6 under similar conditions.14 This suggests either that DPc10 very slowly adopts a conformation that can bind RyR2, or that the DPc10 binding site on RyR2 becomes available only very slowly. The insensitivity of kwash-in to 10-fold higher [F-DPc10] (Figure 3B), is most consistent with the latter interpretation, indicating that kon is limited by RyR2 properties that restrict the access of DPc10 to its binding site. Further supporting this hypothesis, pretreatment with NF-DPc10 (Figure 4A) robustly increased in kon (~320%). We infer that the bound NF-DPc10 shifted RyR2 to the unzipped state allowing better access and exchange with F-DPc10. The simplest explanation for this is that the unzipped state relaxes back to the zipped state slowly with respect to F-DPc10 binding, so that when a NF-DPc10 dissociates it is more rapidly replaced by F-DPc10 (before re-zipping and greater steric hindrance returns). A second related possibility is that one DPc10 molecule may bind at 2 sites to RyR2 (one with higher affinity than the other). When saturated by NF-DPc10 in the unzipped state F-DPc10 may gain access and compete with NF-DPc10 at the low affinity site. Then when NF-DPc10 slowly dissociates from the high affinity site, F-DPc10 is already local and can reach steady state more rapidly (as observed). These are not mutually exclusive or unique possibilities.
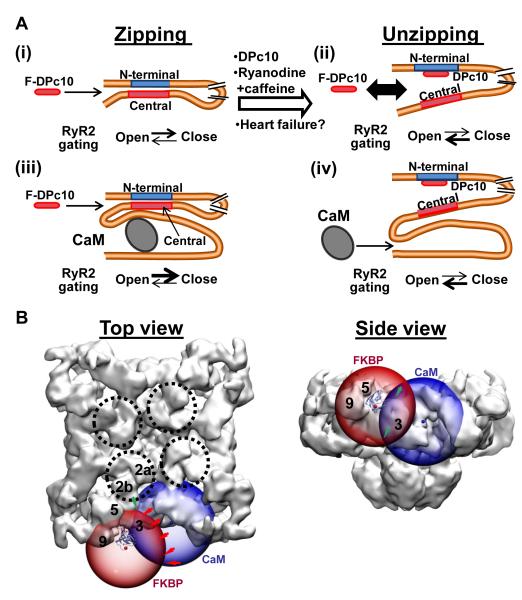
We also found that enhancing RyR2 open state (by caffeine +ryanodine) hastened the F-DPc10 association (Figure 4B). However, these effects on F-DPc10 kwash-in were small compared to that of pre-binding NF-DPc10, despite the very much stronger RyR2 channel opening expected. This agrees with Liu et al24 who reported that DPc10 more strongly unzips the N-terminal and central domains than did ryanodine + caffeine. Thus, we suggest that the unzipped and open states differ, although unzipping may increase RyR2 opening and that the open state may enhance the unzipping transition (and DPc10 access; Figure 8A).
Figure 8. Proposed model of the interaction between N-terminal and central domains, and localization of the DPc10 binding site in the 3D structure of RyR2.
A, Kinetics results (Figures 2-4) suggest that F-DPc10 access to its binding site is controlled by inter-domain interaction within RyR2. (i) The F-DPc10 access is sterically hindered in resting normal RyR2 (“zipped” state). (ii) Pre-treatment of RyR2 with physiological, pharmacological, or disease-mimetic agents that promote unzipping increase the F-DPc10 association rate. (iii) CaM inhibits the F-DPc10 binding to RyR2. (iv) DPc10 binding to RyR2 inhibits CaM binding to RyR2. B, Localization of DPc10 in the 3D structure of RyR2. FRET data between CaM and DPc10 (Figure 5), and between FKBP and DPc10 (Figure 6) suggest that DPc10 binds near to, or within the, RyR handle domain, between FKBP12.6 and CaM. The transparent blue sphere is centered at the surface projection (opaque blue ball) of the mass center of the cryo-EM CaM density37 and has a radius of 58 Å (Figure 5E). The transparent red sphere is centered at position 14 of FKBP27 (indicated by the opaque red ball) and has a radius of 53 Å (Figure 6E). The intersection of a sphere skin with the RyR surface defines possible locations of the DPc10 acceptor within the RyR 3D structure. In top view, we note that the FKBP sphere intersects the clamp domain. In side view, the intersection continues through the clamp but also through domain 3. The locus of the DPc10 should be approximately at the intersections between the spheres (green arrows) and the RyR surface.
Relationship between FKBP12.6 and DPc10 binding to RyR2
FKBP12.6 has been found to quiet RyR2 channel opening,30 but this is an intensely controversial issue,13,31 and FKBP12.6 may only inhibit pathologically leaky RyRs.11 Since more than 80% of the RyRs in the cardiomyocytes have no natively-bound FKBP12.6,14 adding saturating concentrations of exogenous FKBP12.6 ought to decrease Ca2+ leak caused by DPc10-induced unzipping. Here, we found that FKBP12.6 has no effect on either DPc10 binding (Bmax or τwash-in) or vice-versa (Figure 5A, B) and does not quiet the activating effect of DPc10 on the CaSpF (Figure 5E). That is similar to our previous myocyte studies, where FKBP12.6 had very minor effects on Ca2+ sparks.14 Taken together, these results suggest that that DPc10 and FKBP12.6 act through independent mechanisms to modulate RyR2 function.
Relationship between CaM and DPc10 binding to RyR2
In myocytes containing a CPVT-linked RyR2 mutation •-adrenergic stimulation decreases CaM binding at the Z-lines, and this effect is mimicked in healthy myocytes by treatment with DPc10.19 Here, we used methods designed to monitor CaM and DPc10 binding specifically at the RyR2, in myocytes, aiming to understand the structural basis of the inhibition of CaM-RyR2 binding by DPc10. One important finding in the present study is that saturating CaM binding at the RyR2 dramatically reduced F-DPc10 binding and Ca2+ spark activation (Figure 5), presumably by stabilizing the zipped RyR2 state.
Our novel FRET-based method allows direct assessment of CaM-RyR2 binding in the native cardiac myocyte environment (using FKBP12.6-CaM FRET).22 Using this method, we found that unzipping the RyR2 by treatment with saturating [DPc10], reciprocally inhibits CaM binding to RyR2 (Figure 6B, right panel). There are two possible explanations for this reciprocal binding inhibition: (1) DPc10 and CaM compete to bind at overlapping sites (orthosteric mechanism) or (2) the DPc10 and CaM binding sites are separate but coupled in a mutually inhibitory interaction (allosteric mechanism).
To discern between these possibilities we assessed if CaM and DPc10 can coexist on RyR2. In Figure 6C, we show strong FRET between donor-labeled CaM and acceptor-labeled DPc10 at Z-lines, indicating that CaM and DPc10 binding sites in neighboring regions are simultaneously occupied. This conclusion is further supported by FRET measurements using two different donor-acceptor pairs and two different labeling sites on CaM, which indicate distances of 63 ± 1 and 53 ± 1 Å between DPc10 and the N- and C-lobes of CaM, respectively (Figure 6E). Moreover, FRET between FKBP12.6 and DPc10 indicates a distance of 53 ± 3 Å between the probes, which can be compared to the 67 ± 5 Å distance between a donor at position 14 of FKBP12.6 and an acceptor at position 34 of CaM we previously reported.22,28 Taken together, these results strongly support the conclusion that DPc10 and CaM bind at separate sites on RyR2, and these interact through an allosteric mutually inhibitory mechanism.
Our working hypothesis (Figure 8A) which merits further study is as follows. The resting zipped RyR2 does not readily allow DPc10 access to its site (i) and CaM binding at a different site may stabilize this zipped state (iii). We suppose that the RyR2 can transition spontaneously between zipped and the unzipped states (i-ii) but that the low probability at rest causes the slow, but eventual access of DPc10 to its site. This transition may be favored when the channel is open (caffeine+ ryanodine) and also in pathological conditions (e.g. HF). Once the central domain-mimicking DPc10 gains access and binds, it stabilizes the unzipped state (ii) which reciprocally facilitates channel opening and inhibits CaM binding (iv).
Topology of the DPc10 binding site on RyR2
Although our aim here is not a detailed mapping of the DPc10 binding site within the cryo-EM 3D-structure of RyR2, our FRET measurements help narrow down the range of possible locations. The location of FKBP12.6 and CaM on the RyR2 structure are known from cryo-EM structural analysis, and their relative positions agree with our prior FKBP-CaM FRET studies.21,22,27,28 These are represented by the centers of the blue and red spheres in Figure 8B. Our FRET data between DPc10 and both CaM and FKBP12.6 (Figure 6-7) allows us to initially triangulate the likely location of DPc10 in the RyR2 3D structure, that is where the edges of the blue and red spheres intersect. The green arrows in Figure 8B suggest a DPc10 location in the handle domain, between the FKBP and CaM. The clamp domain location previously proposed seems quite far from the most probable location suggested by the FRET results.32 DPc10 is expected to bind the RyR2 within a 150 kDa N-terminal segment,33 containing the first 600 residues that form a hot-spot of pathogenic mutations.34 The high-resolution structure of domain 1-559 has been reported and authoritatively docked into cryo-EM densities forming a vestibule in the cytoplasmic headpiece of RyR (see dashed black circles in Figure 8B).35 A different view in Online Figure X shows that our triangulation puts DPc10 close to, but not exactly at that location. More detailed FRET analysis to triangulate the F-DPc10 marker is expected to more precisely locate the interdomain contact site.
We represent the FKBP12.6, CaM and DPc10 sites all on the same face of the RyR2 tetramer. We previously showed that this is true for the FKBP-CaM FRET pair,21,22 but we also tested whether the potential DPc10 site could be between CaM and FKBP sites on adjacent RyR2 faces. Online Figure IX shows that this possibility is implausible, based on our FRET measurements.
Relevance to heart failure
Until now, methods to monitor local conformational changes occurring in the interacting regulatory domains of RyR have relied on a large fluorescence quencher (used in isolated SR vesicles),11,12 or on FRET between a yellow fluorescent protein inserted into the N-terminal region and a cyan fluorescent protein inserted into the central region of RyR2 (in HEK293 cells).24 In this study, we show how the F-DPc10 wash-in kinetics can be used in the more native environment of permeabilized cardiomyocytes to evaluate domain interaction between the N-terminal and central domains of RyR2. This could serve as a powerful and versatile investigative tool in pre-clinical and clinical studies with respect to the domain unzipping hypothesis. For example, the time course of F-DPc10 wash-in can be monitored in myocytes from failing hearts in which “unzipping” has already occurred,11,20 or to gauge RyR function under pathological conditions (e.g., oxidative stress, phosphorylation state etc) and in the evaluation (or validation) of drug candidates that act to stabilize the RyR2 “zipped” state.11,36
Supplementary Material
Novelty and Significance.
What Is Known?
A synthetic peptide DPc10, which is a part of the central domain (Gly2460-Pro2495) of the RyR2, can destabilize RyR2 function by interfering with domain interaction between central and N-terminal domains (namely, domain unzipping).
Calmodulin (CaM) inhibits RyR2 channel activity at all [Ca2+]i (both at diastolic and elevated [Ca2+]i), indicating that CaM stabilizes the closed state of RyR2. Domain unzipping can also reduce affinity of RyR2 for CaM.
FKBP12.6 is reported to bind RyR2 tightly as an important regulator in RyR2 gating, but no direct effect of DPc10 on FKBP12.6 binding has been seen in SR vesicles.
What New Information Does This Article Contribute?
DPc10 binding affinity and kinetics to the RyR2 in the native myocyte environment using FRET between fluorescent FKBP12.6 (F-FKBP12.6: donor) and fluorescent DPc10 (F-DPc10: acceptor). The Kd for F-DPc10 is ≈ 500 nmol/L and F-DPc10 binding is extremely slow, even at high [F-DPc10], indicating F-DPc10 access to its binding site is sterically hindered in resting RyR2.
F-DPc10 wash-in kinetics provide a sensitive tool for detecting the RyR2 unzipped state in permeabilized myocytes.
Saturating RyR2 with CaM (but not with FKBP12.6) reduced DPc10 binding and Ca2+ spark activation.
Measurements of both CaM-DPc10 FRET and FKBP12.6-DPc10 suggest that CaM and DPc10 binding interact via an allosteric (rather than orthosteric) mechanism.
The location of the DPc10 binding site on the 3D RyR structure was detected using FRET between FKBP-DPc10 and CaM-DPc10 FRET, and is near the handle domain and far from the clamp domain of RyR.
Defects in domain interaction between central and N-terminal domains of the RyR2 can destabilize RyR2 channel gating and increase Ca leak. However, details of DPc10 binding properties (Kd, kon, and koff) to RyR2, and its effect on CaM and FKBP12.6 binding in native cardiac myocytes, are unknown. Here, we show that DPc10 association to its binding site is slow due to steric hindrance in resting RyR2. DPc10 wash-in kinetics were shown to be a useful tool for detecting domain unzipping in the native cardiac myocytes environment. We found that domain unzipping differs from channel opening state, although unzipped state enhances the open probability of the RyR2. CaM binding at RyR2, but not FKBP12.6 binding, stabilizes RyR2 in the zipped state (blocking DPc10 binding and RyR activation), however; once DPc10 gains access and binds, it causes RyR unzipping by inhibiting CaM binding. Moreover, FRET measurements demonstrate that DPc10 and CaM binding sites are distinct, but interact allosterically.
SOURCES OF FUNDING
Supported by NIH R01-HL092097 (DMB, RLC) and P01-HL080101 (DMB) and by Banyu Life Science Foundation International (TO).
Non-standard Abbreviations
- τ
time constant
- τwash-in
wash-in time constant
- τwash-out
wash-out time constant
- AIP
autocamtide-2 related inhibitory peptide
- Bmax
binding maximum
- CaM
calmodulin
- CaSpF
calcium spark frequency
- CPVT
catecholaminergic polymorphic ventricular tachycardia
- EAF
enhanced acceptor fluorescence
- F
fluorescent
- FDHM
full duration at half maximum
- F-FKBP12.6
fluorescent FKBP12.6
- FWHM
full width at half maximum
- FRET
fluorescence resonance energy transfer
- HF
heart failure
- Kd
dissociation constant
- kin
wash-in rate constant, 1/τwash-in
- kout
wash-out rate constant, 1/τwash-out
- koff
dissociation rate constant
- kon
association rate constant
- RyR2
cardiac ryanodine receptor
- SR
sarcoplasmic reticulum
Footnotes
DISCLOSURES None.
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Bers DM. Macromolecular complexes regulating cardiac ryanodine receptor function. J Mol Cell Cardiol. 2004;37:417–429. doi: 10.1016/j.yjmcc.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 2.Yano M, Ono K, Ohkusa T, Suetsugu M, Kohno M, Hisaoka T, Kobayashi S, Hisamatsu Y, Yamamoto T, Kohno M, Noguchi N, Takasawa S, Okamoto H, Matsuzaki M. Altered stoichiometry of FKBP12.6 versus ryanodine receptor as a cause of abnormal Ca2+ leak through ryanodine receptor in heart failure. Circulation. 2000;102:2131–2136. doi: 10.1161/01.cir.102.17.2131. [DOI] [PubMed] [Google Scholar]
- 3.Ai X, Curran JW, Shannon TR, Bers DM, Pogwizd SM. Ca2+/Calmodulin-dependent protein kinase modulates cardiac ryanodine receptor phosphorylation and sarcoplasmic reticulum Ca2+ leak in heart failure. Circulation Research. 2005;97:1314–1322. doi: 10.1161/01.RES.0000194329.41863.89. [DOI] [PubMed] [Google Scholar]
- 4.Wehrens XHT, Lehnart SE, Reiken S, Vest JA, Wronska A, Marks AR. Ryanodine receptor/calcium release channel PKA phosphorylation: A critical mediator of heart failure progression. Proc Natl Acad Sci U S A. 2006;103:511–518. doi: 10.1073/pnas.0510113103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wehrens XHT, Lehnart SE, Huang F, Vest JA, Reiken S, Mohler P, Sun J, Guatimosim S, Song LS, Rosemblit N, D’Armiento JM, Napolitano C, Memmi M, Priori SG, Lederer WJ, Marks AR. FKBP12.6 deficiency and defective calcium release channel (ryanodine receptor) function linked to exercise-induced sudden cardiac death. Cell. 2003;113:829–840. doi: 10.1016/s0092-8674(03)00434-3. [DOI] [PubMed] [Google Scholar]
- 6.Yamamoto T, Ikemoto N. Peptide probe study of the critical regulatory domain of the cardiac ryanodine receptor. Biochem Biophys Res Commun. 2002;291:1102–1108. doi: 10.1006/bbrc.2002.6569. [DOI] [PubMed] [Google Scholar]
- 7.Uchinoumi H, Yano M, Suetomi T, Ono M, Xu XJ, Tateishi H, Oda T, Okuda S, Doi M, Kobayashi S, Yamamoto T, Ikeda Y, Ohkusa T, Ikemoto N, Matsuzaki M. Catecholaminergic polymorphic ventricular tachycardia is caused by mutation-linked defective conformational regulation of the ryanodine receptor. Circ Res. 2010;106:1413–1424. doi: 10.1161/CIRCRESAHA.109.209312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ikemoto N, Yamamoto T. Regulation of calcium release by interdomain interaction within ryanodine receptors. Front Biosci. 2002;7:d671–d683. doi: 10.2741/A803. [DOI] [PubMed] [Google Scholar]
- 9.Ikemoto N. Intra-molecular domain-domain interaction: a key mechanism for calcium channel regulation of ryanodine receptors. In: Wehrens XHT, Marks AR, editors. Ryanodine Receptors: Structure, Function and Dysfunction in Clinical Disease. Springer; New York, NY: 2004. pp. 53–65. [Google Scholar]
- 10.Yamamoto T, El-Hayek R, Ikemoto N. Postulated role of interdomain interaction within the ryanodine receptor in Ca2+ channel regulation. J Bio Chem. 2000;275:11618–11625. doi: 10.1074/jbc.275.16.11618. [DOI] [PubMed] [Google Scholar]
- 11.Oda T, Yano M, Yamamoto T, Tokuhisa T, Okuda S, Doi M, Ohkusa T, Ikeda Y, Kobayashi S, Ikemoto N, Matsuzaki M. Defective regulation of interdomain interactions within the ryanodine receptor plays a key role in the pathogenesis of heart failure. Circulation. 2005;111:3400–3410. doi: 10.1161/CIRCULATIONAHA.104.507921. [DOI] [PubMed] [Google Scholar]
- 12.Tateishi H, Yano M, Mochizuki M, Suetomi T, Ono M, Xu X, Uchinoumi H, Okuda S, Oda T, Kobayashi S, Yamamoto T, Ikeda Y, Ohkusa T, Ikemoto N, Matsuzaki M. Defective domain-domain interactions within the ryanodine receptor as a critical cause of diastolic Ca2+ leak in failing hearts. Cardiovasc Res. 2009;81:536–545. doi: 10.1093/cvr/cvn303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bers DM. Ryanodine receptor S2808 phosphorylation in heart failure smoking gun or red herring. Circ Res. 2012;110:796–799. doi: 10.1161/CIRCRESAHA.112.265579. [DOI] [PubMed] [Google Scholar]
- 14.Guo T, Cornea RL, Huke S, Camors E, Yang Y, Picht E, Fruen BR, Bers DM. Kinetics of FKBP12.6 binding to ryanodine receptors in permeabilized cardiac myocytes and effects on Ca sparks. Circ Res. 2010;106:1743–1752. doi: 10.1161/CIRCRESAHA.110.219816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamaguchi N, Xu L, Pasek DA, Evans KE, Meissner G. Molecular basis of calmodulin binding to cardiac muscle Ca2+ release channel (ryanodine receptor) J Bio Chem. 2003;278:23480–23486. doi: 10.1074/jbc.M301125200. [DOI] [PubMed] [Google Scholar]
- 16.Fruen BR, Bardy JM, Byrem TM, Strasburg GM, Louis CF. Differential Ca2+ sensitivity of skeletal and cardiac muscle ryanodine receptors in the presence of calmodulin. Am J Physiol. 2000;279:C724–C733. doi: 10.1152/ajpcell.2000.279.3.C724. [DOI] [PubMed] [Google Scholar]
- 17.Balshaw DM, Yamaguchi N, Meissner G. Modulation of intracellular calcium-release channels by calmodulin. J Mem Biol. 2002;185:1–8. doi: 10.1007/s00232-001-0111-4. [DOI] [PubMed] [Google Scholar]
- 18.Guo T, Zhang T, Mestril R, Bers DM. Ca2+/Calmodulin-dependent protein kinase II phosphorylation of ryanodine receptor does affect calcium sparks in mouse ventricular myocytes. Circ Res. 2006;99:398–406. doi: 10.1161/01.RES.0000236756.06252.13. [DOI] [PubMed] [Google Scholar]
- 19.Xu XJ, Yano M, Uchinoumi H, Hino A, Suetomi T, Ono M, Tateishi H, Oda T, Okuda S, Doi M, Kobayashi S, Yamamoto T, Ikeda Y, Ikemoto N, Matsuzaki M. Defective calmodulin binding to the cardiac ryanodine receptor plays a key role in CPVT-associated channel dysfunction. Biochem Biophys Res Commun. 2010;394:660–666. doi: 10.1016/j.bbrc.2010.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ono M, Yano M, Hino A, Suetomi T, Xu XJ, Susa T, Uchinoumi H, Tateishi H, Oda T, Okuda S, Doi M, Kobayashi S, Ikemoto N, Matsuzaki M. Dissociation of calmodulin from cardiac ryanodine receptor causes aberrant Ca2+ release in heart failure. Cardiovasc Res. 2010;87:609–617. doi: 10.1093/cvr/cvq108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cornea RL, Nitu FR, Gruber S, Kohler K, Satzer M, Thomas DD, Fruen BR. FRET-based mapping of calmodulin bound to the RyR1 Ca2+ release channel. Proc Natl Acad Sci U S A. 2009;106:6128–6133. doi: 10.1073/pnas.0813010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo T, Fruen BR, Nitu FR, Nguyen TD, Yang Y, Cornea RL, Bers DM. FRET detection of calmodulin binding to the cardiac RyR2 calcium release channel. Biophys J. 2011;101:2170–2177. doi: 10.1016/j.bpj.2011.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Kranias EG, Mignery GA, Bers DM. Protein Kinase A Phosphorylation of the ryanodine receptor does not affect calcium sparks in mouse ventricular myocytes. Circ Res. 2002;90:309–316. doi: 10.1161/hh0302.105660. [DOI] [PubMed] [Google Scholar]
- 24.Liu Z, Wang R, TianX, Zhong X, Gangopadhyay J, Cole R, Ikemoto N, Chen SR, Wagenknecht T. Dynamic, inter-subunit interactions between the N-terminal and central mutation regions of cardiac ryanodine receptor. J Cell Sci. 2010;123:1775–1784. doi: 10.1242/jcs.064071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song Q, Saucerman J, Bossuyt J, Bers DM. Differential integration of Ca2+-calmodulin signal in intact ventricular myocytes at low and high affinity Ca2+-calmodulin targets. J Biol Chem. 2008;283:31531–31540. doi: 10.1074/jbc.M804902200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Y, Chakraborty A, Guo T, Cornea RL, Meissner G, Bers DM. In situ measurement of RyR2-calmodulin binding in permeabilized cardiomyocytes. Biophys J (abstract) 2011;100:413a–414a. [Google Scholar]
- 27.Samsó M, Shen X, Allen PD. Structural characterization of the RyR1–FKBP12 interaction. J Mol Biol. 2006;356:917–927. doi: 10.1016/j.jmb.2005.12.023. [DOI] [PubMed] [Google Scholar]
- 28.Cornea RL, Nitu FR, Samsó M, Thomas DD, Fruen BR. Mapping the ryanodine receptor FK506-binding protein subunit using fluorescence resonance energy transfer. J Bio Chem. 2010;285:19219–19226. doi: 10.1074/jbc.M109.066944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bers DM, Stiffel VM. Ratio of ryanodine to dihydropyridine receptors in cardiac and skeletal muscle and implications for E-C coupling. Am J Physiol. 1993;264:C1587–1593. doi: 10.1152/ajpcell.1993.264.6.C1587. [DOI] [PubMed] [Google Scholar]
- 30.Prestle J, Janssen PM, Janssen AP, Zeitz O, Lehnart SE, Bruce L, Smith GL, Hasenfuss G. Overexpression of FK506-binding protein FKBP12.6 in cardiomyocytes reduces ryanodine receptor–mediated Ca2+ leak from the sarcoplasmic reticulum and increases contractility. Circ Res. 2001;88:188–194. doi: 10.1161/01.res.88.2.188. [DOI] [PubMed] [Google Scholar]
- 31.Xiao J, Tian X, Jones PP, Bolstad J, Kong H, Wang R, Zhang L, Duff HJ, Gillis AM, Fleischer S, Kotlikoff M, Copello JA, Chen SR. Removal of FKBP12.6 does not alter the conductance and activation of the cardiac ryanodine receptor or the susceptibility to stress-induced ventricular arrhythmias. J Bio Chem. 2007;282:34828–34838. doi: 10.1074/jbc.M707423200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang R, Chen W, Cai S, Zhang J, Bolstad J, Wagenknecht T, Liu Z, Chen SR. Localization of an NH2-terminal disease-causing mutation hot spot to the ‘Clamp’ region in the three-dimensional structure of the cardiac ryanodine receptor. J Bio Chem. 2007;282:17785–17793. doi: 10.1074/jbc.M700660200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamamoto T, Ikemoto N. Spectroscopic monitoring of local conformational changes during the intramolecular domain–domain interaction of the ryanodine receptor. Biochemistry. 2002;41:1492–1501. doi: 10.1021/bi015581z. [DOI] [PubMed] [Google Scholar]
- 34.Priori SG, Chen SR. Inherited dysfunction of sarcoplasmic reticulum Ca2+ handling and arrhythmogenesis. Circ Res. 2011;108:871–883. doi: 10.1161/CIRCRESAHA.110.226845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tung C, Lobo PA, Kimlicka L, Petegem FA. The Amino-terminal disease hotspot of ryanodine receptors forms a cytoplasmic vestibule. Nature. 2010;468:585–588. doi: 10.1038/nature09471. [DOI] [PubMed] [Google Scholar]
- 36.Kobayashi S, Yano M, Suetomi T, Ono M, Tateishi H, Mochizuki M, Xu XJ, Uchinoumi H, Okuda S, Yamamoto T, Koseki N, Kyushiki H, Ikemoto N, Matsuzaki M. Dantrolene, a therapeutic agent for malignant hyperthermia, markedly improves the function of failing cardiomyocytes by stabilizing interdomain interactions within the ryanodine receptor. J Am Coll Cardiol. 2009;53:1993–2005. doi: 10.1016/j.jacc.2009.01.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samsó M, Wagenknecht T. Apocalmodulin and Ca2+-calmodulin bind to neighboring locations on the ryanodine receptor. J Biol Chem. 2002;277:1349–1353. doi: 10.1074/jbc.M109196200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.