Abstract
Amyloid fibril formation is associated with misfolding diseases, as well as fulfilling a functional role. The cross-β molecular architecture has been reported in increasing numbers of amyloid-like fibrillar systems. The Waltz algorithm is able to predict ordered self-assembly of amyloidogenic peptides by taking into account the residue type and position. This algorithm has expanded the amyloid sequence space, and in the present study we characterize the structures of amyloid-like fibrils formed by three peptides identified by Waltz that form fibrils but not crystals. The structural challenge is met by combining electron microscopy, linear dichroism, CD and X-ray fibre diffraction. We propose structures that reveal a cross-β conformation with ‘steric-zipper’ features, giving insights into the role for side chains in peptide packing and stability within fibrils. The amenity of these peptides to structural characterization makes them compelling model systems to use for understanding the relationship between sequence, self-assembly, stability and structure of amyloid fibrils.
Keywords: amyloid, amyloidogenic peptide, Waltz algorithm, X-ray fibre diffraction
Abbreviations: CCD, charge-coupled-device; FTIR, Fourier-transform infrared; LD, linear dichroism; PrP, prion protein; TEM, transmission electron microscopy; XRFD, X-ray fibre diffraction
INTRODUCTION
Amyloid is a fibrillar self-assembled non-covalent proteinacious polymer [1]. A great number of proteins have been shown to be able to access a self-assembled β-sheet conformation and it has been suggested that under the right conditions any protein may form amyloid. It has been proposed that the amyloid conformation represents an energetically stable conformation that may be accessed by any polypeptide [2]. Indeed, a diverse range of proteins form amyloid with large variations in protein sequence and amyloidogenic precursor structure. Proteins that form amyloid are typically associated with a degenerative disease; amyloid-β with Alzheimer's disease, transthyretin with familial amyloidotic polyneuropathy, IAPP (islet amyloid polypeptide) with Type 2 diabetes and prion or PrP (prion protein) with TSEs (transmissible spongiform encephalopathies). Potentially other diseases that have previously had unknown underlying pathology are caused by an aggregating peptidic monomer. For example, it has recently been suggested that phenylketonuria may be caused by an accumulation of amyloid-like assemblies of the single amino acid phenylalanine [3]. The ability of a single amino acid to self-assemble in an ordered manner, when compared with the 42-residue protein amyloid-β [4] or the larger ~200 residue PrP [5], is a striking example of the variety found in amyloid systems. This variety is not only fascinating, but promises to have great utility in the application of such systems to bionanomaterial design.
The number of amyloidogenic systems associated with disease has led to numerous investigations into their fibrillar structure [4,6–8], each revealing a common shared architecture named cross-β [9]. The cross-β architecture is defined by β-strands orientated perpendicular to the fibril long axis stacked to form long β-sheets that extend into a fibril. Two or more of these sheets may associate laterally to form the protofilaments of mature amyloid fibrils. These molecular details were first proposed on the basis of the interpretation of X-ray fibre diffraction data from amyloid-like systems [8,10,11]. This architecture has since been found in various other amyloid systems using ssNMR (solid-state NMR) [4,6,7] and also X-ray crystallography [12]. These same advances have further provided structural details that have revealed insights into side-chain interactions that form the basis for the β-sheet association [6,8,12].
Lately, efforts have been made to expand the sequence space of structurally characterized amyloid systems. These sequences may be identified within larger amyloidogenic sequences or be identified by bioinformatics. Identification of amyloidogenic sequences via a bioinformatics approach is not only able to expand the amyloid sequence space, but in the identification algorithms required, highlight the characteristics of a sequence that give it a propensity to assemble [13]. These algorithms are numerous and based on varying principles, but typically identify short peptide sequences, often hexapeptides [14–18]. A systematic study of the peptide with the sequence STVIIE revealed the important positional and chemical characteristic of a hexapeptide by studying the amyloid-forming propensity of variants of the peptide sequence [19]. We previously reported a new algorithm, called Waltz, for identifying short amyloidogenic sequences [18] on the basis of a sequence position scoring matrix. It has successfully identified a small library of amyloidogenic peptide sequences that self-assemble in solution to form ordered amyloid aggregates.
Charge, hydrophobicity and aromaticity have all been implicated in the structural basis for assembly [20]. The present study provides a basis from which to understand the relationship between sequence and the ability of the peptide to form amyloid fibrils and their resulting structures. We report on the structural and biophysical characterization of three of the peptides, identified by Waltz, able to form highly ordered amyloid-like fibrils with the sequences HYFNIF, RVFNIM and VIYKI. Waltz was designed to identify not only amyloid-forming sequences, but also to better distinguish between amyloid sequences and amorphous aggregates [18]. The characterization of these three Waltz peptides further re-enforces the algorithm's ability to do this. This investigation and the analysis of fibrillar systems uses time-resolved XRFD (X-ray fibre diffraction) and analysis of LD (linear dichroism) and CD spectroscopy to identify structural characteristics and produce models representative of the fibrillar semi-crystalline structure. In doing so we reveal insights into the relationship between sequence and structure relevant to understanding amyloid formation in disease and as a basis for designing new self-assembling bionanomaterials.
EXPERIMENTAL
Peptide synthesis and materials
The short peptides were synthesized as freeze-dried powder with N-acetylated and C-amidated capped termini to >95% purity by HPLC (JPT Peptide Technologies). Peptides were incubated in water at 10 mg/ml under quiescent conditions at room temperature (20–25°C) for 1 week unless otherwise stated. Fibres allowed to assemble for longer incubation periods of up to 3 months are referred to as mature. Water was purified by reverse osmosis and filter-sterilized using 0.2 μm membranes (Minisart).
TEM (transmission electron microscopy)
Samples were examined by TEM to confirm fibril formation. Samples were diluted to 0.25–1 mg/ml prior to TEM visualization. For grid preparation, a 4 μl drop of fibril solution was incubated on Formvar/Carbon 400 Mesh Copper grids (Agar Scientific) for 1 min, followed by a 1 min wash with 4 μl of water and negatively stained twice with 4 μl of 2% uranyl acetate for 1 min. Between each stage, excess liquid was removed by blotting with filter paper. Care was taken not to dry out the grid until after the final negative stain step. Grids were visualized using a Hitachi-7100 transmission electron microscope running at 100 kV. Images were taken using an axially mounted (2000 pixels×2000 pixels) Gatan Ultrascan 1000 CCD (charge-coupled-device) camera. The program ImageJ (http://rsbweb.nih.gov/ij/) was used to analyse TEM images [21] and individual fibril morphologies were inspected after applying the in-built fast Fourier-transform band-pass filter.
CD
A Jasco J-715 spectropolarimeter with a peltier temperature control system was used to collect CD spectra. All measurements were collected at 20°C with a sample concentration of 1–2 mg/ml, 180–320 nm with a pitch of 0.1 nm at a scan speed of 50 nm/min, a response time of 4 s, slit widths of 1 nm and standard sensitivity. Control buffer spectra were averaged from triplicate accumulations and subtracted from the averaged triplicate accumulations of samples. Pathlengths were adjusted according to the degree of signal against PMT (photomultiplier tube) high-tension voltage to ensure high data quality and were 0.1 mm using quartz demountable cells (Starna Scientific). To check for contributions arising from LD artefacts due to orientation effects, cuvettes were rotated by 90° in the instrument. Where artefacts arising from orientation of fibrils were found, a combination of tip (Sonics Vibra-Cell VCX500, 20 kHz, 40% amplitude, 2 min) and water bath (Fisher Scientific FB15051, 37 kHz, 1 min) sonication was used to disrupt alignment.
Secondary structure analysis was performed on spectra confirmed to contain only CD signals using the online server Dichroweb [23] with the CDSSTR analysis programme [24] and best available reference set: SP175 190–240 nm [25]. Careful attention was paid to the closeness of fit between secondary structure model predictions and the experimental data where low NRMSD (normalized root mean square deviation) values indicate a high goodness of fit of the secondary structure model prediction and successful analysis [26].
LD
A Jasco J-815 spectropolarimeter modified for LD was used to collect LD spectra. Measurements were taken in a 0.5 mm pathlength at 20°C at 200 μg/ml, 180–320 nm with a pitch of 0.2 nm at a scan speed of 100 nm/min, a response time of 1 s, bandwidth of 1 nm and standard sensitivity. Spectra were collected as an average of three accumulated measurements and processed by subtraction of the background signal from water and zeroing at 300–320 nm. Three channels were monitored; LD, HT[V] (high tension [voltage]) and absorbance. To unambiguously detect LD signals, fibrillar samples were aligned by a Couette flow cell apparatus [27] using rotation speeds of 1500 rev./min, creating fibre alignment parallel with the orientation axis (//). Assembly of the Couette flow cell apparatus was found to align fibres perpendicular to the orientation axis (⊥) and thus comparison spectra were also collected with no Couette flow.
FTIR (Fourier-transform infrared) spectroscopy
Infrared spectra were recorded using a Bruker Tensor 27 infrared spectrophotometer equipped with a Bio-ATR II accessory. Spectra were recorded of dried films at a spectral resolution of 4 cm−1 with 120 accumulations performed per measurement at a wavenumber range of 900–3500 cm−1. Buffer and baseline subtraction from the obtained spectra were made with rescaling in the wave number range of 900–1800 cm−1.
XRFD analysis
Fibril samples were aligned by a variety of methods to produce different textures. Fibrous-textured alignments were created by suspending a 10 μl droplet of 10 mg/ml fibril solution between two wax-tipped 1.2 mm outside diameter, 0.94 mm internal diameter borosilicate capillaries (Harvard apparatus) and placing in a parafilm-sealed Petri dish to air-dry at room temperature. Film-textured alignments were formed by drawing a 10–50 μl droplet of 10 mg/ml fibril solution into a X-ray transmissible 0.7 mm borosilicate capillary (Capillary Tube Suppliers) and sealed at one end to prevent further capillary action. This solution was allowed to dry by evaporation to create a film-texture. These methods are further described in [28]. In order to monitor real-time fibre alignment, a cell similar to that described previously was used [29]. Briefly, the cell comprises a semi-enclosed chamber that can support the wax-tipped capillaries used for alignment, such that this process can be monitored by X-ray fibre diffraction in real-time. Samples for fibre diffraction were tested on a home source Rigaku 007HF Cu Kα [λ=1.5419 Å (1 Å=0.1 nm)] rotating anode generator with VariMax-HF mirrors and Saturn 944+ CCD detector. Additionally, synchrotron data were collected at the Diamond I24 microfocus beamline with a wavelength of 0.9778 Å and an MARCCD detector. Clearer [30] was used to measure signal positions in XRFD patterns. Graphical traces were produced by sampling a 60° radial slice of the XRFD on either meridional or equatorial axes and plotted using Braggs law. Determined diffraction signals were entered into the unit cell determination module within Clearer [30] and possible unit cells were explored by comparing experimental diffraction signal positions with calculated signals and examining for logical indexing schemes.
Modelling
Initial peptide models were built in Insight II (Molecular Dimensions) in an ideal β-strand conformation. Rotamer conformations were assigned using the Dunbrack backbone-dependent rotamer library in PyMOL (http://www.pymol.org), variable architectures were explored and assessed using Molprobity [31], checking for clashes, favourable Φ-Ψ angles and side-chain positions. Crystal lattices were constructed in PyMOL using the unit cell determined by Clearer [30], and these were minimized to remove side-chain clashing and unfavourable conformations. Explicit lattice minimizations were performed using the CHARMM Prot_all_22_forcefield [33] through NAMD [34] as implemented in VMD [35] in a (3 3 3) solvated lattice.
Simulated XRFD and model validation
Using Clearer, fibre-diffraction patterns were simulated from minimized models. Briefly, the central model of a minimized lattice was used to construct a fibre texture from which the intensity and position of reflections were simulated. The diffraction settings were equivalent to the experimental collection parameters (i.e. specimen-to-detector distance, wavelength and detector parameters). The fibre disorder parameters σθ and σΦ were 0.2 and ∞ respectively, and the crystallite size was set to 400 Å3 with a sampling interval of 1 pixel. All other simulation settings were default.
Pattern comparison was initially qualitative and based on visual comparison of simulated XRFD patterns against the experimental pattern. To more accurately identify closeness of fit (RF), the experimental and simulated reflections were tabulated and a quantitative comparison was made on the basis of signal position and relative intensity.
RESULTS
Sequence identification and origin
The three short peptides, identified by Waltz and shown in the present paper, are five or six residues in length and taken from much larger polypeptides with variable native functions, as shown in Table 1. Of the three sequences, the crystal structure of HYFNIF within the context of the full-length native protein structure is available for assessment (PDB code 2KV2). Interestingly the predicted secondary structure by PSIPred [36] and observed secondary structure are different (Supplementary Figure S1 at http://www.biochemj.org/bj/450/bj4500275add.htm). Although HYFNIF was identified by Waltz and predicted to form β-strands, the crystal structure of the native protein reveals this sequence adopts an α-helical structure. RVFNIM and VIYKI are predicted to respectively adopt α-helical and β-strand conformations.
Table 1. The Waltz peptide sequences and their origin with regard to the full-length proteins in which they are found.
The Waltz algorithm predicts the likelihood of an amino acid being found at a particular position of an amyloidogenic sequence. The probability of a residue occurring at a particular position in an amyloidogenic peptide as predicted by Waltz is expressed as a log-odd score [18]. See also Supplementary Figure S1 (at http://www.biochemj.org/bj/450/bj4500275add.htm). eIF, eukaryotic initiation factor.
| Origin | Residue position scoring | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Waltz sequence | UniProt ID | Length | Segment | Name | Function | Organism | 1 | 2 | 3 | 4 | 5 | 6 |
| HYFNIF | P54132 | 1417 | 27–33 | Bloom syndrome protein | DNA helicase | Human | 0 | 0 | 1 | 1 | 2 | 0 |
| RVFNIM | P20042 | 333 | 182–187 | eIF-2 | Translation regulation | Human | −1 | 0 | 1 | 1 | 2 | 0 |
| VIYKI | P07182 | 284 | 183–187 | Chorion protein | Egg shell protein | Drosophila | 0 | 1 | −1 | −2 | −2 | |
In the case of the hexapeptides, the terminal residues are either neutral or slightly unfavoured in the Waltz scoring algorithm, indicating that the core residues are key and may represent a possible core interaction motif. This core sequence is Phe-Asn-Ile at positions 3–5 in the hexapeptides, whereas in the case of the pentapeptide the equivalent positions 3–5 have the sequence Tyr-Lys-Ile. In general the core may be described as having the aromatic pattern A-X-X and the hydrophobicity pattern H-X-H.
Morphology characterization by TEM
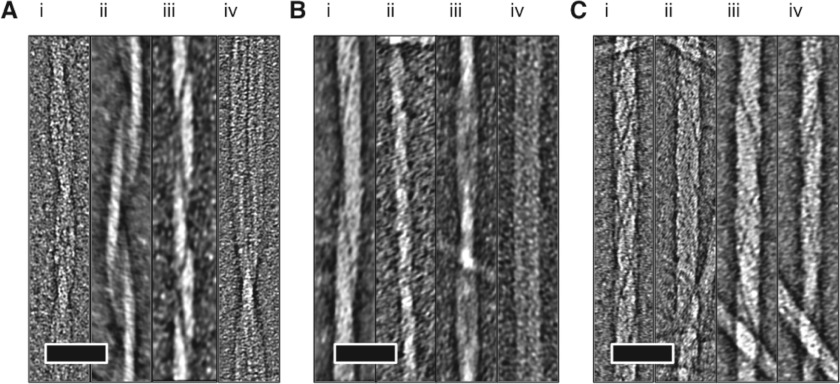
TEM of the Waltz peptides revealed that in water they spontaneously form amyloid-like fibrillar morphologies, with widths of approximately 20 nm (Figure 1). Each peptide fibril exhibits characteristic morphologies, inferring the complex association of individual protofilaments in a hierarchical structure.
Figure 1. The characteristic morphologies of Waltz fibrils.
(A) HYFNIF, (B) RVFNIM and (C) VIYKI as analysed by TEM. Micrographs have been cropped and processed by fast Fourier-transform bandpass filtering in ImageJ [21]. The scale bars represent 50 nm. See also Supplementary Figure S2 (at http://www.biochemj.org/bj/450/bj4500275add.htm).
Fibrils formed by HYFNIF exhibited the greatest range of morphologies, which can broadly be described as having a twisted morphology. The paired filament helices (Figure 1A, i) have a greatest width of 17.04 nm (S.D.±1.06; n=7) and periodicity of 95.16 nm (S.D.±3.15; n=6). Other helical arrangements of ribbons are observed with variable widths and periodicities (Figure 1A, ii and iii). Following long incubation (several months) the fibrils developed into structures that may represent a tubular architecture with widths of 26.03 nm (S.D.±3.29; n=3) (Figure 1A, iv and Supplementary Figure S2 at http://www.biochemj.org/bj/450/bj4500275add.htm).
RVFNIM was more consistently found to form tightly wound ‘ropes’ with a width of 17.61 nm (S.D.±1.23; n=5) with some indication of protofilament structure (Figure 1B, i and ii). Some examples of rope-like protofilaments twisting into a helical arrangement were also observed (Figure 1B, iii) with widths at their widest points of 19.69 nm (S.D.±1.59; n=28). The helical pitch of these morphologies varies dramatically from 147.81 nm (S.D.±21.40; n=11) to indeterminate over a single TEM micrograph (Figure 1B, iv).
VIYKI exhibited the most regular morphology where all observed fibres formed twisted ‘ropes’ (Figure 1C, i–iv) with a width of 21.0 nm (S.D.±1.2; n=6) in a helical twist with a periodicity of 58.1 nm (S.D.±1.7; n=6).
Secondary structure determination by CD and FTIR
The secondary structures of the fibrillar Waltz peptides were investigated using CD and FTIR spectroscopy. Characterization by CD is complicated by both the anisotropic nature and high aromatic content of these systems. A classic β-sheet spectrum typically has a positive and a negative maximum at ~195 and ~216 nm respectively. The CD spectra from the Waltz peptides are more complicated. Each CD spectrum exhibits a strong dependence on sample orientation, with signal intensities that are far greater than normal (Supplementary Figure S3 at http://www.biochemj.org/bj/450/bj4500275add.htm) and signal positions that are not as expected for typical β-sheet peptides. The fibril structure and thus potential for the shear alignment of these systems may give rise to LD artefacts in the CD measurements. To obtain CD spectra with minimal contribution from artefactual LD signals, the Waltz fibrils were sonicated in an attempt to abolish effects from alignment (Supplementary Figure S3) as reported elsewhere [22]. TEM confirmed that, following sonication, fibrils remained present in the solution, but sample orientation CD signal dependence was no longer observed, revealing the true CD signals representative of the Waltz peptides. Dichroweb analysis revealed predominantly β-sheet and other non-helical structures (Supplementary Table S1 at http://www.biochemj.org/bj/450/bj4500275add.htm). We presume the non-helical conformations represent peptides in a random-coil conformation that have been liberated from fibres, but the remaining predominant β-strand structures constitute the Waltz fibrils.
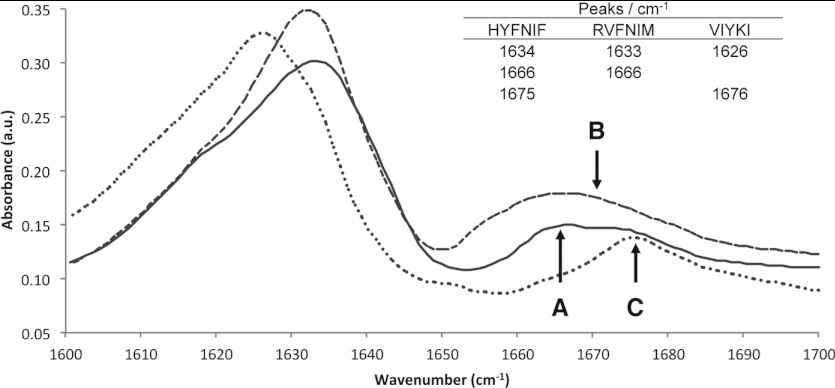
Since the high aromatic content and anisotropy complicates CD analysis, we corroborated these results with FTIR of mature Waltz fibrils. The expected absorption bands for β-sheet structure with two maxima at 1626–1633 and 1666–1676 cm−1 were observed (α-helices exhibit bands at ~1654 cm−1 [37]) for all three peptide assemblies (Figure 2). FTIR has been used to distinguish between parallel and antiparallel β-sheets by the presence of the longer wavenumber band at 1695 cm−1 [38], but the reliability of this interpretation is still a matter of debate [37] and so here we interpret the FTIR only in terms of the presence of β-sheet conformation.
Figure 2. The secondary structure of the Waltz fibrils.
The amide I FTIR region of dried films of mature assembled (A) HYFNIF, (B) RVFNIM and (C) VIYKI, each indicating the presence of a β-sheet conformation. See also Supplementary Figure S3 (at http://www.biochemj.org/bj/450/bj4500275add.htm).
Chromophore orientation determination by LD
Biophysical analyses identified the β-sheet-rich structure of the Waltz peptide fibrils. The presence of LD artefacts in the CD spectra suggests that a regular and ordered arrangement of structural elements is apparent in the assemblies. The alignment of the Waltz peptide fibrils in the initial CD experiments was the result of sample loading. This cannot be controlled and so we chose to measure the LD signals using a micro-volume Couette flow cell apparatus [39] as this provides a reproducible alignment methodology to gain more information regarding the orientation of structural elements and chromophores within the amyloid fibrils.
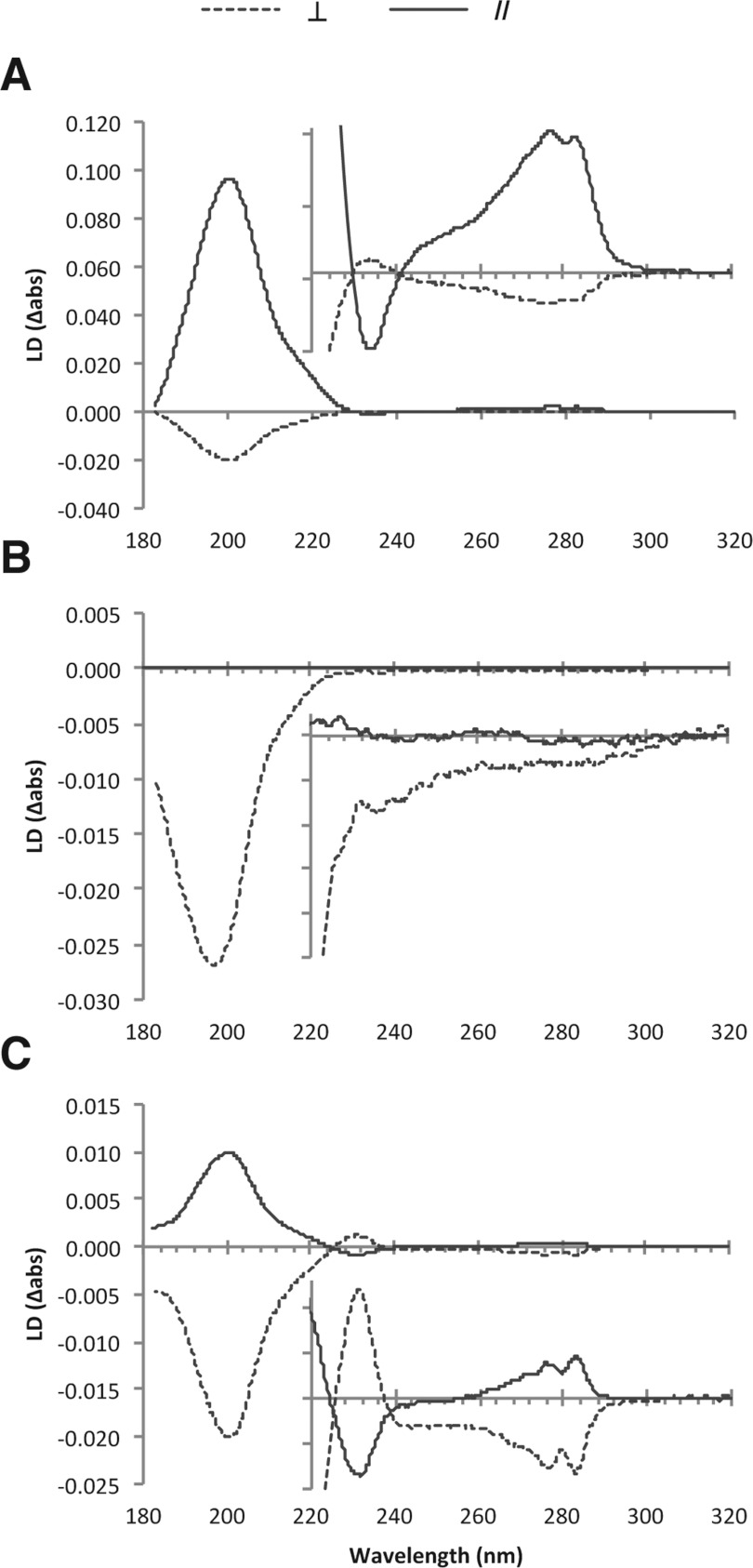
LD on fibrillar systems, in particular amyloid, has been reported previously [22,27,40,41]. Transition moments in chromophores that are regularly orientated produce positive and negative LD signals depending on their orientation. The orientation of chromophores relative to the fibre axis can thus be determined from the magnitude and sign of their LD signals [42]. The LD spectra for the Waltz fibrils were collected when aligned parallel with (//) and perpendicular to (⊥) the orientation axis (Figure 3), where in our instrument the orientation axis is defined as horizontal. Our intention had been to observe orientation under Couette flow; however, the signals shown in Figure 3 indicate that the loading of the sample provided sufficient shear force to induce perpendicular fibril orientation. All three peptides show a large negative signal at ~200 nm arising from the π-π* transition of the β-sheet peptide backbone. The sign of the β-sheet π-π* transition is thus consistent with its polarization being parallel with the fibre long axis. Since this transition occurs perpendicular to the β-strand direction [42], the data are consistent with β-strands arranged perpendicular to the fibre long axis and are consistent with the cross-β architecture, as observed for other self-assembled systems [22,40].
Figure 3. The LD spectra arising from the Waltz fibrils.
Alignment of fibrils was induced by shear force perpendicular (⊥; broken lines) and Couette flow parallel (//; continuous lines) to the orientation axis of (A) HYFNIF, (B) RVFNIM and (C) VIYKI. Peaks are tabulated in Supplementary Table S2 (at http://www.biochemj.org/bj/450/bj4500275add.htm).
Couette flow created significantly more orientation for HYFNIF fibrils, not enough orientation for RVFNIM fibrils to overrule the loading-induced orientation and some orientation for VIYKI fibrils. For HYFNIF and VIYKI fibrils, the signals were also observed in the region expected to arise from aromatic LD spectra (Figure 3). RVFNIM fibrils gave no evidence of orientation of its aromatic chromophores, although this is likely to be due to the fact that phenylalanine has a low absorption coefficient. By way of contrast, HYFNIF and VIYKI have clearly resolved aromatic tyrosine LD signals, the La (230 nm) and Lb (275 nm) transitions, indicating a regular ordered arrangement of these aromatic residues in these structures. The splitting of the Lb tyrosine transition into two components is an excitonic coupling effect indicating that the tyrosine residues in these systems are closely associated and possibly involved in π-π stacking interactions [43]. This phenomenon has been previously noted in LD spectra from fibrils formed by the heptapeptide GNNQQNY [40]. Thus the LD data indicate that the tyrosine long axes for both HYFNIF and VIYKI fibrils are orientated greater than 54.7° from the fibre axis, whereas the tyrosine short axes are orientated less than 54.7° and may adopt a stacked geometry. Knowing the β-sheet content and orientation with the likelihood of aromatic stacking interactions we sought to gather more structural details from XRFD.
XRFD from Waltz peptide fibrils
Fibrils of Waltz assemblies were readily aligned, producing fibrous-textured alignments, by methods previously described [28]. These alignments exhibit birefringence by cross-polarizing light microscopy, indicating a para-crystalline order of the assemblies (Supplementary Figure S4 at http://www.biochemj.org/bj/450/bj4500275add.htm). Each system exhibited the major meridional and equatorial features associated with the cross-β architecture commonly observed for amyloid-like assemblies [9]. These results are thus consistent with the spectroscopic observations and the deduction of the cross-β arrangement within these peptide assembly systems. The reflections arising from the repetitive interatomic separations within the Waltz fibrils differ between the systems, indicating differences in the packing arrangements of the different peptides.
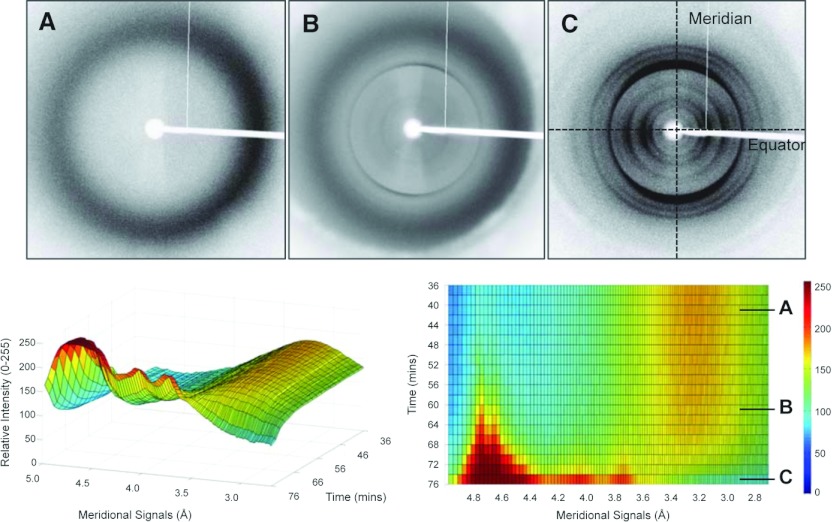
We also monitored XRFD in real-time over the course of fibre alignment. To our knowledge, we report for the first time the XRFD exhibited by an amyloid-like system over the course of fibre alignment in the hydrated, semi-hydrated and dried aligned state as shown in Figure 4. Importantly, we find that the reflection positions arising from the amyloid architecture are present, and the diffuse scattering from water (~3.5 Å) is still observable. This supports the view that the structures of the fibres are the same in the solution and dried state. The same phenomenon was confirmed for each Waltz system individually (Supplementary Figures S4D and S4E). We also explored the effect of texture on the observed XRFD patterns. Samples of fibres were aligned to produce film-textured alignments where all fibre axes are aligned parallel with the film plane. We note that the film-textured alignments also report the same diffraction signals as the fibrous alignments (Supplementary Figure S5 at http://www.biochemj.org/bj/450/bj4500275add.htm), but believe that the film texture may abolish contribution from fibril packing. In the case of RVFNIM, additional equatorial information is observed in the film-textured alignment and so this pattern was used in subsequent analysis.
Figure 4. Real-time XRFD of VIYKI over the course of alignment.
In the (A) hydrated, (B) semi-hydrated and (C) dried and aligned state. The corresponding graphical representation shows that the meridional signals arising from the cross-β structure are present before the complete loss of water and thus are present in the normal hydrated state. All fibre axes are vertical. See also Supplementary Figures S4(D) and S4(E) (at http://www.biochemj.org/bj/450/bj4500275add.htm).
The relatively large number of reflections exhibited by these systems is indicative of a para-crystalline order greater than that typically observed for amyloid-like systems [9]. The quality of the patterns thus presents an opportunity to determine a unit cell on the basis of the reflection positions (Supplementary Table S3 at http://www.biochemj.org/bj/450/bj4500275add.htm) and model the molecular structure within the Waltz fibrillar assemblies. All three peptides share a strong meridional reflection, the distance of which varies between 4.66 and 4.76 Å (Supplementary Table S3), arising from the separation between hydrogen-bonded β-strands along the fibre axis. Higher-order reflections of the principal meridional reflections are present, but the precise position of the meridional reflections is dependent on β-strand separation and relative displacement of these along the fibre axis. Knowing the fibre axis repeat, the equatorial reflections were indexed and predicted unit cell dimensions were determined using Clearer [30]. The determined unit cell dimensions for each system (Supplementary Table S4 at http://www.biochemj.org/bj/450/bj4500275add.htm) are defined as: a, the β-strand chain length; b, the β-sheet spacing; and c, the hydrogen bonding spacing along the fibre axis.
The strongest equatorial reflections pertaining to the distance between β-sheets for the peptides HYFNIF, RVFNIM and VIYKI are 12.4, 10.8 and 9.21 Å respectively. This information allowed for the basic arrangement of the peptides within the repeating cells to be constructed. From X-ray crystallography analysis of amyloidogenic short peptides, a range of cross-β architectures have been proposed and grouped into eight classes [12]. The arrangement of β-strands within β-sheets can be antiparallel, β-sheet relative orientation (face-to-face or face-to-back) and β-sheet direction (up or down). This information was used as the basis for using molecular modelling to explore the possible arrangements of the Waltz peptides within their respective repeating cells (Supplementary Online Data at http://www.biochemj.org/bj/450/bj4500275add.htm).
Modelling and assessment by simulated XRFD
Diffraction patterns were simulated from models systematically constructed (Supplementary Figure S6 at http://www.biochemj.org/bj/450/bj4500275add.htm) using the descriptions of parallel amyloid structural classes [12] and quantitatively compared with the experimental diffraction patterns (Supplementary Table S5 at http://www.biochemj.org/bj/450/bj4500275add.htm). The model structures shown in Figure 5 are those that were explored and found to best fit the diffraction data. A comparison between the simulated and experimental diffraction patterns confirming the validity of the constructed models is shown in Figure 6.
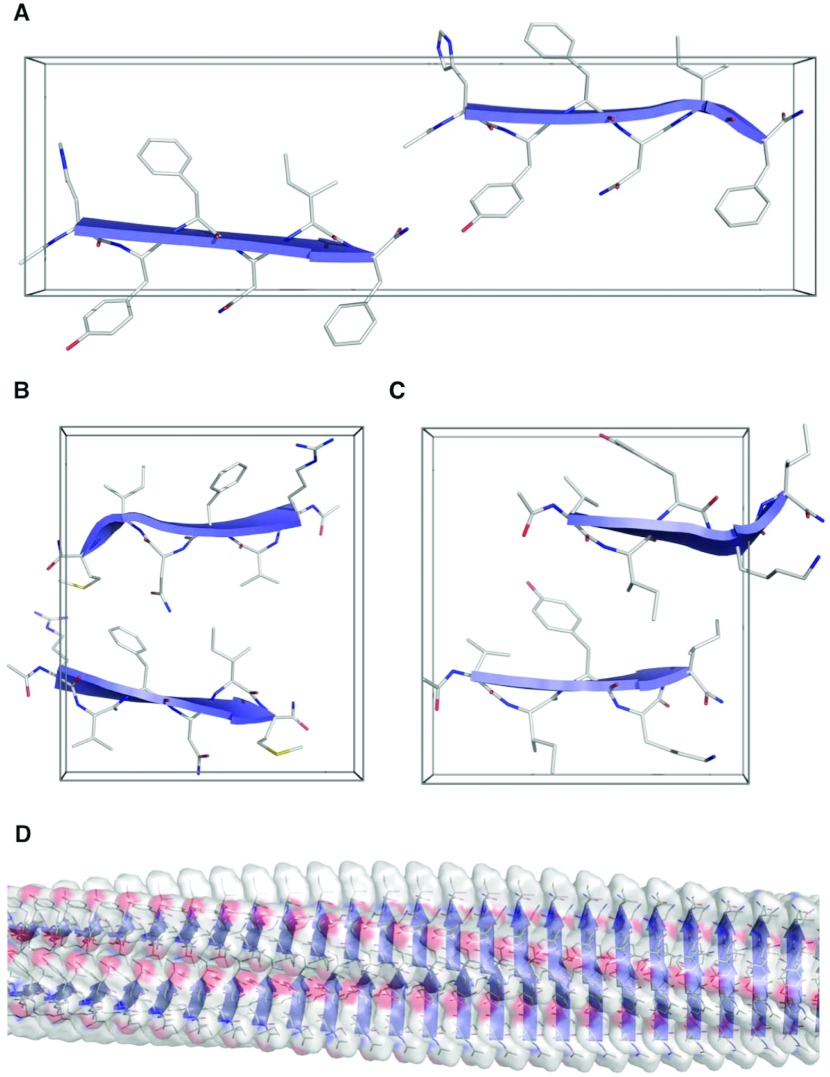
Figure 5. Representative models of the Waltz peptides.
The peptides are shown modelled into the unit cells determined from XRFD: (A) HYFNIF, (B) RVFNIM and (C) VIYKI. The visualization (D) shows the possible protofilament context of VIYKI. See also Supplementary Figure S6 and the Supplementary Online Data (at http://www.biochemj.org/bj/450/bj4500275add.htm).
Figure 6. Experimental XRFD patterns compared with patterns simulated from modelled structures.
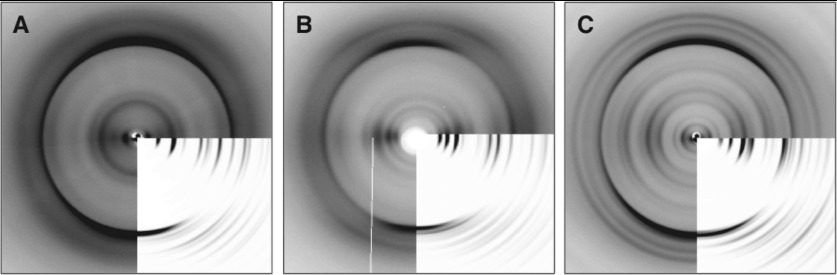
XRFD patterns are shown for alignments of (A) HYFNIF, (B) RVFNIM and (C) VIYKI. Fibre axes are vertical and insets for each show the simulated diffraction pattern for comparison with the experimental data. See also Supplementary Tables S3–S5 (at http://www.biochemj.org/bj/450/bj4500275add.htm). RVFNIM is compared with a film-textured alignment, see Supplementary Figure S5 (at http://www.biochemj.org/bj/450/bj4500275add.htm).
Figure 5 shows the most representative models for each of the Waltz systems. Of the models constructed, the majority of simulated patterns compared well with experimental fibre diffraction data, and so we postulate that the experimental fibre diffraction patterns may result from a mixture of architectures. However, the structures shown in Figure 5 represent the predominant representative architectures. The modelled β-sheet separations closely correspond to the major equatorial reflections (Supplementary Table S3) and the β-strand separation along the fibre axis varies between 4.70 and 4.78 Å (Supplementary Table S4). The low-resolution reflections found on the HYFNIF and VIYKI patterns were not easily reproduced, but we attribute this to our simulations modelling the structures as a continuous lattice rather than discrete protofilaments. HYFNIF is found to adopt an up-up architecture where two sheets are displaced by b/2, where the β-sheets are parallel with respect to one another. RVFNIM and VIYKI adopt more classical steric zippers in classes IV and II respectively; the former has parallel β-sheets in a face-to-back and up-down arrangement, whereas the latter β-sheets are face-to-back in an up-up arrangement. The meridional reflections of RVFNIM and VIYKI were better reproduced with β-strands displaced along the fibre axis by c/2.
DISCUSSION
The present study confirms further the Waltz algorithm's ability to predict ordered aggregation, and we have characterized two hexapeptides and one pentapeptide uniquely identified during the iterative sequence exploration that developed Waltz [18]. The sequences are found to have varying secondary structure propensities, but in their native structures adopt a conformation dependent on their surrounding amino acid sequence and structural environment. When removed from their native sequences, the Waltz peptides form a β-strand conformation that adopts a cross-β architecture, but with differing lateral packing arrangements. Previous work by Johansson and co-workers suggested that amyloidogenic sequences were often ‘promiscuous’ and able to adopt both α-helical and β-strand conformations depending on their context and environment [44]. The results of the present study re-enforce this and the concept that the flanking amino acid sequences in amyloidogenic portions of proteins protect against aggregation and self-assembly by forcing the adoption of particular secondary and tertiary structures [45].
On the nanoscopic scale these systems are observed to adopt characteristic and discrete fibril morphologies that may be based on the underlying differences in molecular packing, but are ultimately typical for amyloid fibrils [46]. The exact relationship between these two levels of structure remains unclear. To model the molecular packing of these peptides, the investigations reported in the present study have required careful consideration of the use of the biophysical and structural techniques employed. It was found that CD data is highly dependent on the fibrillar nature of the assemblies, but we highlight methods, as reported elsewhere [22,27], for the successful interpretation of this data. The identification of LD artefacts in CD data has been reported previously [47] and the results of the present study clearly reiterate that LD artefacts can occur in CD experiments, the identification of which have broad methodological implications for using CD to study anisotropic systems. Clearly care should be taken in analyses of these sorts of data, but the previously described phenomena can be usefully rationalized and used in LD experiments to reveal information about chromophore orientation.
Particular care was taken to ensure that the hydrated state of these systems was the same as the dried, through the use of a semi-enclosed chamber to monitor XRFD in real-time over the course of fibril alignment. The structures reported in the present study are found to be unaffected by drying as reported previously [48].
Using XRFD and Clearer, unit cells were determined for the Waltz peptides in the fibrillar state and the architectures that best represent the fibril structure of the Waltz peptides were determined. These peptides do not crystallize in the conditions tested by us (results not shown) and we have focussed on the fibrillar structure due to concerns of the comparability of crystal structures with the absolute structure of self-assembled fibres [40,49]. Despite this, the proposed crystalline class models of amyloid systems provide a useful framework within which to structurally explore the possible architectures of short amyloidogenic sequences. It is probable that the Waltz fibrils contain a mixture of these architectures where the difference between simulated patterns of different class models is slight. It must be considered that more structural information may be obtained from crystal structures, but they represent a single global minimum, which may not be representative of the local minima of the many polymorphs present in real amyloid fibrils.
Although likely to be polymorphic, we have judged quantitatively the most representative predominant polymorphs that constitute the Waltz peptide assemblies. Structurally, RVFNIM and VIYKI are found to adopt arrangements with displacement of β-sheets along the fibre axis (c/2) with respect to one another. This has been observed for crystallized short amyloidogenic peptides [12], but, at the time of writing, the results from the present study are the first example of this being demonstrated through experimental and simulated XRFD. The arrangements adopted are variable between the Waltz peptides, which is perhaps surprising given the position sequence similarity of these peptides, but is a reflection of the adaptability of this conformation. Interestingly, aromatic amino acids appear to provide a possible driving force for assembly in some amyloidogenic peptides [20,40] and others have suggested an important contribution of hydrophobicity [50]. Examination of the peptide arrangements reveals that, although hydrophobicity may drive elongation of β-sheets, it is likely that charge and aromaticity modulate the possible arrangements and architecture within the final fibrillar structure via sheet–sheet interactions and lateral association of protofilaments.
In the future, studies of these systems have the potential to make valuable and unique contributions to the model systems currently describing the structural space of amyloid assemblies. The characterization of these systems not only further confirms the ability of the Waltz algorithms to predict ordered amyloid aggregation, but also introduces an opportunity to further understand this conformation while presenting new highly ordered and well-characterized systems for nanotechnological applications.
Online data
AUTHOR CONTRIBUTION
Kyle Morris conducted the experimental work, performed analysis and wrote the paper. Alison Rodger provided equipment and expertise and contributed to data analysis. Matthew Hicks contributed to LD data collection and analysis. Maya Debulpaep collected FTIR data. Joost Schymkowitz and Frederic Rousseau contributed materials and wrote the Waltz algorithm on which the paper is based. They also advised on the interpretation and the paper. Louise Serpell managed the project, analysed the results and wrote the paper.
ACKNOWLEDGEMENTS
XRFD data collected at the Diamond I24 microfocus beamline was done so with the expert help of Dr Gwyndaf Evans, Dr Danny Axford and Dr Robin Owen. We thank Dr Julian Thorpe for essential electron microscopy support, Dr Peter Varnai for valuable advice on model minimization and Youssra Al-Hilaly for insightful discussions. The apparatus used to generate the real-time XRFD data was based on a prototype design kindly lent to us by Professor Pawel Sikorski, to whom we are indebted.
FUNDING
This work was supported by the Biotechnology and Biological Sciences Research Council funded Synthetic components network awarded to L.C.S. and K.L.M. L.C.S. is supported by Alzheimer's Research UK. The Switch Laboratory was supported by grants from the Flanders Institute for Biotechnology (VIB), The University of Leuven, the Funds for Scientific Research Flanders (FWO), the Flanders Institute for Science and Technology (IWT) and the Federal Office for Scientific Affairs, Belgium IUAP P7.
References
- 1.Morris K., Serpell L. From natural to designer self-assembling biopolymers, the structural characterisation of fibrous proteins & peptides using fibre diffraction. Chem. Soc. Rev. 2010;39:3445–3453. doi: 10.1039/b919453n. [DOI] [PubMed] [Google Scholar]
- 2.Dobson C. M. The structural basis of protein folding and its links with human disease. Philos. Trans. R. Soc. London Ser. B. 2001;356:133–145. doi: 10.1098/rstb.2000.0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adler-Abramovich L., Vaks L., Carny O., Trudler D., Magno A., Caflisch A., Frenkel D., Gazit E. Phenylalanine assembly into toxic fibrils suggests amyloid etiology in phenylketonuria. Nat. Chem. Biol. 2012;8:701–706. doi: 10.1038/nchembio.1002. [DOI] [PubMed] [Google Scholar]
- 4.Luhrs T., Ritter C., Adrian M., Riek-Loher D., Bohrmann B., Dobeli H., Schubert D., Riek R. 3D structure of Alzheimer's amyloid-β(1–42) fibrils. Proc. Natl. Acad. Sci. U.S.A. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wille H., Bian W., McDonald M., Kendall A., Colby D. W., Bloch L., Ollesch J., Borovinskiy A. L., Cohen F. E., Prusiner S. B., Stubbs G. Natural and synthetic prion structure from X-ray fiber diffraction. Proc. Natl. Acad. Sci. U.S.A. 2009;106:16990–16995. doi: 10.1073/pnas.0909006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petkova A. T., Ishii Y., Balbach J. J., Antzutkin O. N., Leapman R. D., Delaglio F., Tycko R. A structural model for Alzheimer's β-amyloid fibrils based on experimental constraints from solid state NMR. Proc. Natl. Acad. Sci. U.S.A. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Madine J., Copland A., Serpell L. C., Middleton D. A. Cross-β spine architecture of fibrils formed by the amyloidogenic segment NFGSVQFV of medin from solid-state NMR and X-ray fiber diffraction measurements. Biochemistry. 2009;48:3089–3099. doi: 10.1021/bi802164e. [DOI] [PubMed] [Google Scholar]
- 8.Makin O. S., Atkins E., Sikorski P., Johansson J., Serpell L. C. Molecular basis for amyloid fibril formation and stability. Proc. Natl. Acad. Sci. U.S.A. 2005;102:315–320. doi: 10.1073/pnas.0406847102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jahn T. R., Makin O. S., Morris K. L., Marshall K. E., Tian P., Sikorski P., Serpell L. C. The common architecture of cross-β amyloid. J. Mol. Biol. 2010;395:717–727. doi: 10.1016/j.jmb.2009.09.039. [DOI] [PubMed] [Google Scholar]
- 10.Geddes A. J., Parker K. D., Atkins E. D., Beighton E. “Cross-β” conformation in proteins. J. Mol. Biol. 1968;32:343–358. doi: 10.1016/0022-2836(68)90014-4. [DOI] [PubMed] [Google Scholar]
- 11.Kirschner D. A., Abraham C., Selkoe D. J. X-ray diffraction from intraneuronal paired helical filaments and extraneuronal amyloid fibers in Alzheimer disease indicates cross-β conformation. Proc. Natl. Acad. Sci. U.S.A. 1986;83:503–507. doi: 10.1073/pnas.83.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sawaya M. R., Sambashivan S., Nelson R., Ivanova M. I., Sievers S. A., Apostol M. I., Thompson M. J., Balbirnie M., Wiltzius J. J., McFarlane H. T., et al. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 13.Rousseau F., Schymkowitz J., Serrano L. Protein aggregation and amyloidosis: confusion of the kinds? Curr. Opin. Struct. Biol. 2006;16:118–126. doi: 10.1016/j.sbi.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 14.Thompson M. J., Sievers S. A., Karanicolas J., Ivanova M. I., Baker D., Eisenberg D. The 3D profile method for identifying fibril-forming segments of proteins. Proc. Natl. Acad. Sci. U.S.A. 2006;103:4074–4078. doi: 10.1073/pnas.0511295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zibaee S., Makin O. S., Goedert M., Serpell L. C. A simple algorithm locates β-strands in the amyloid fibril core of α-synuclein, Aβ, and tau using the amino acid sequence alone. Protein Sci. 2007;16:906–918. doi: 10.1110/ps.062624507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tartaglia G. G., Pawar A. P., Campioni S., Dobson C. M., Chiti F., Vendruscolo M. Prediction of aggregation-prone regions in structured proteins. J. Mol. Biol. 2008;380:425–436. doi: 10.1016/j.jmb.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez-Escamilla A.-M., Rousseau F., Schymkowitz J., Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- 18.Maurer-Stroh S., Debulpaep M., Kuemmerer N., de la Paz M. L., Martins I. C., Reumers J., Morris K. L., Copland A., Serpell L., Serrano L., et al. Exploring the sequence determinants of amyloid structure using position-specific scoring matrices. Nat. Methods. 2010;7:237–242. doi: 10.1038/nmeth.1432. [DOI] [PubMed] [Google Scholar]
- 19.de la Paz M., Serrano L. Sequence deteminants of amyloid fibril formation. Proc. Natl. Acad. Sci. U.S.A. 2004;101:87–92. doi: 10.1073/pnas.2634884100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marshall K. E., Morris K. L., Charlton D., O’Reilly N., Lewis L., Walden H., Serpell L. C. Hydrophobic, aromatic, and electrostatic interactions play a central role in amyloid fibril formation and stability. Biochemistry. 2011;50:2061–2071. doi: 10.1021/bi101936c. [DOI] [PubMed] [Google Scholar]
- 21.Abramoff A. M., Magelhaes P. J., Ram S. J. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- 22.Andersen C. B., Hicks M. R., Vetri V., Vandahl B., Rahbek-Nielsen H., Thogersen H., Thogersen I. B., Enghild J. J., Serpell L. C., Rischel C., Otzen D. E. Glucagon fibril polymorphism reflects differences in protofilament backbone structure. J. Mol. Biol. 2010;397:932–946. doi: 10.1016/j.jmb.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 23.Whitmore L., Wallace B. A. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32:668–673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Compton L. A., Johnson W. C. Analysis of protein circular-dichroism spectra for secondary structure using a simple matrix multiplication. Anal. Biochem. 1986;155:155–167. doi: 10.1016/0003-2697(86)90241-1. [DOI] [PubMed] [Google Scholar]
- 25.Lees J. G., Miles A. J., Wien F., Wallace B. A. A reference database for circular dichroism spectroscopy covering fold and secondary structure space. Bioinformatics. 2006;22:1955–1962. doi: 10.1093/bioinformatics/btl327. [DOI] [PubMed] [Google Scholar]
- 26.Mao D., Wachter E., Wallace B. A. Folding of the mitochondrial proton adenosine-triphosphatase proteolipid channel in phospholipid-vesicles. Biochemistry. 1982;21:4960–4968. doi: 10.1021/bi00263a020. [DOI] [PubMed] [Google Scholar]
- 27.Dafforn T. R., Rajendra J., Halsall D. J., Serpell L. C., Rodger A. Protein fiber linear dichroism for structure determination and kinetics in a low-volume, low-wavelength couette flow cell. Biophys. J. 2004;86:404–410. doi: 10.1016/S0006-3495(04)74116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morris K. L., Serpell L. C. X-ray fibre diffraction studies of amyloid fibrils. In: Sigurdsson E. M., Calero M., Gasset M., editors. Amyloid Proteins: Methods and Protocols. Second Edition. Springer; 2012. pp. 121–135. [DOI] [PubMed] [Google Scholar]
- 29.McDonald M., Kendalla A., Tanaka M., Weissman J. S., Stubbs G. Enclosed chambers for humidity control and sample containment in fiber diffraction. J. Appl. Crystallogr. 2008;41:206–209. [Google Scholar]
- 30.Makin O. S., Sikorski P., Serpell L. C. CLEARER: a new tool for the analysis of X-ray fibre diffraction patterns and diffraction simulation from atomic structural models. J. Appl. Crystallogr. 2007;40:966–972. [Google Scholar]
- 31.Wang J. M., Cieplak P., Kollman P. A. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 2000;21:1049–1074. [Google Scholar]
- 32. Reference deleted.
- 33.Brooks B. R., Brooks C. L., III, Mackerell A. D., Jr, Nilsson L., Petrella R. J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., et al. CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kale L., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graphics Model. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 36.Jones D. T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 37.Barth A., Zscherp C. What vibrations tell us about proteins. Q. Rev. Biophys. 2002;35:369–430. doi: 10.1017/s0033583502003815. [DOI] [PubMed] [Google Scholar]
- 38.Miyazawa T., Blout E. R. Infrared spectra of polypeptides in various conformations–amide I and II bands. J. Am. Chem. Soc. 1961;83:712–719. [Google Scholar]
- 39.Rodger A., Marrington R., Geeves M. A., Hicks M., de Alwis L., Halsall D. J., Dafforn T. R. Looking at long molecules in solution: what happens when they are subjected to Couette flow? Phys. Chem. Chem. Phys. 2006;8:3161–3171. doi: 10.1039/b604810m. [DOI] [PubMed] [Google Scholar]
- 40.Marshall K. E., Hicks M. R., Williams T. L., Hoffmann S. V., Rodger A., Dafforn T. R., Serpell L. C. Characterizing the assembly of the Sup35 yeast prion fragment, GNNQQNY: structural changes accompany a fiber-to-crystal switch. Biophys. J. 2010;98:330–338. doi: 10.1016/j.bpj.2009.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adachi R., Yamaguchi K.-i., Yagi H., Sakurai K., Naiki H., Goto Y. Flow-induced alignment of amyloid protofilaments revealed by linear dichroism. J. Biol. Chem. 2007;282:8978–8983. doi: 10.1074/jbc.M611738200. [DOI] [PubMed] [Google Scholar]
- 42.Hicks M. R., Kowalski J., Rodger A. LD spectroscopy of natural and synthetic biomaterials. Chem. Soc. Rev. 2010;39:3380–3393. doi: 10.1039/b912917k. [DOI] [PubMed] [Google Scholar]
- 43.Rodger A., Norden B. Circular Chroism & Linear Dichroism. Oxford: Oxford University Press; 1997. [Google Scholar]
- 44.Kallberg Y., Gustafsson M., Persson B., Thyberg J., Johansson J. Prediction of amyloid fibril-forming proteins. J. Biol. Chem. 2001;276:12945–12950. doi: 10.1074/jbc.M010402200. [DOI] [PubMed] [Google Scholar]
- 45.Reumers J., Maurer-Stroh S., Schymkowitz J., Rousseau F. Protein sequences encode safeguards against aggregation. Hum. Mutat. 2009;30:431–437. doi: 10.1002/humu.20905. [DOI] [PubMed] [Google Scholar]
- 46.Goldsbury C. S., Cooper G. J. S., Goldie K. N., Muller S. A., Saafi E. L., Gruijters W. T. M., Misur M. P. Polymorphic fibrillar assembly of human amylin. J. Struct. Biol. 1997;119:17–27. doi: 10.1006/jsbi.1997.3858. [DOI] [PubMed] [Google Scholar]
- 47.Norden B. Linear and circular dichroism of polymeric pseudoisocyanine. J. Phys. Chem. 1977;81:151–159. [Google Scholar]
- 48.Squires A. M., Devlin G. L., Gras S. L., Tickler A. K., MacPhee C. E., Dobson C. M. X-ray scattering study of the effect of hydration on the cross-β structure of amyloid fibrils. J. Am. Chem. Soc. 2006;128:11738–11739. doi: 10.1021/ja063751v. [DOI] [PubMed] [Google Scholar]
- 49.Lewandowski J. R., van der Wel P. C. A., Rigney M., Grigorieff N., Griffin R. G. Structural complexity of a composite amyloid fibril. J. Am. Chem. Soc. 2011;133:14686–14698. doi: 10.1021/ja203736z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tracz S. M., Abedini A., Driscoll M., Raleigh D. P. Role of aromatic interactions in amyloid formation by peptides derived from human amylin. Biochemistry. 2004;43:15901–15908. doi: 10.1021/bi048812l. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.