Abstract
The integral membrane protein complex between phospholamban (PLN) and sarcoplasmic reticulum Ca2+-ATPase (SERCA) regulates cardiac contractility. In the unphosphorylated form, PLN binds SERCA and inhibits Ca2+ flux. Upon phosphorylation of PLN at Ser16, the inhibitory effect is reversed. Although structural details on both proteins are emerging from X-ray crystallography, cryo-electron microscopy, and NMR studies, the molecular mechanisms of their interactions and regulatory process are still lacking. It has been speculated that SERCA regulation depends on PLN structural transitions (order to disorder, i.e., folding/unfolding). Here, we investigated PLN conformational changes upon chemical unfolding by a combination of electron paramagnetic resonance and NMR spectroscopies, revealing that the conformational transitions involve mostly the cytoplasmic regions, with two concomitant phenomena: (1) membrane binding and folding of the amphipathic domain Ia and (2) folding/unfolding of the juxtamembrane domain Ib of PLN. Analysis of phosphorylated and unphosphorylated PLN with two phosphomimetic mutants of PLN (S16E and S16D) shows that the population of an unfolded state in domains Ia and Ib (T′ state) is linearly correlated to the extent of SERCA inhibition measured by activity assays. Inhibition of SERCA is carried out by the folded ground state (T state) of the protein (PLN), while the relief of inhibition involves promotion of PLN to excited conformational states (Ser16 phosphorylated PLN). We propose that PLN population shifts (folding/unfolding) are a key regulatory mechanism for SERCA.
Keywords: phospholamban, excited states, NMR, Ca-ATPase, membrane proteins
Introduction
Phospholamban (PLN) is a 52-residue integral membrane protein embedded in the sarcoplasmic reticulum (SR) membrane of cardiac muscle1 that reversibly inhibits the SR Ca2+-ATPase (SERCA), which regulates calcium flux and muscle contractility.1–4
Although several X-ray crystallography and cryo-electron microscopy studies focused on SERCA have revealed important details on calcium translocation,5–9 structural data on the SERCA/PLN complex10–13 are sparse and do not offer mechanistic details on the regulatory function of PLN.
In the SR membrane, PLN exists as a homopentamer in equilibrium with monomers that bind SERCA.1,2,4,14 Both the monomer and the pentamer adopt an L-shaped topology, with a cytoplasmic α-helix (domain Ia; residues 1–16) connected to a membrane-spanning α-helix (residues 23–52) by a short loop (residues 17–22).15–17 The membrane-spanning helix has two dynamically distinct domains: domain Ib (residues 23–30) that is positioned at the hydrophilic portion of the lipid bilayer and domain II (residues 31–52) that exists in the hydrophobic interior.15,18,19
Previously, using NMR and electron paramagnetic resonance (EPR) spectroscopies, we detected a conformational equilibrium between two major states: an ordered T state and a disordered R state.20,21 According to EPR, the R-state population is ~16% (T state, ~84%) in the absence of SERCA22 and becomes slightly more populated in the presence of the enzyme.21 We then speculated that the regulation of SERCA by PLN occurs via conformational transitions between the T and R states. This hypothesis was supported by our studies on the structural effects of PLN phosphorylation at Ser1623,24 and by NMR and functional studies of PLN reconstituted in different lipid mimetics.25–27 In fact, phosphorylation of PLN at Ser16 by protein kinase A (PKA) relieves SERCA inhibition, allowing for more efficient muscle relaxation.1–3,28 We found that, structurally and dynamically, phosphorylation causes an order-to-disorder transition, with an unwinding of domain Ia at residues 14–16 around the phosphorylation site.16,21,23,24 As a result, the phosphoryl transfer shifts the PLN conformational equilibrium, increasing the R-state population.21 The latter can be exploited for the design of mutants to control SERCA function.29 It is also possible to rationalize the effects of phosphomimetic mutations (S16E)29 used in gene therapy.30 Although we established the foundations for a rational design of mutants and showed that the excited states of PLN are important for SERCA inhibition,29 we did not provide a quantitative interpretation of the conformational transitions of PLN between the ground and excited states or elucidate the structural and dynamic features of the excited conformations.
Here, we used EPR and NMR spectroscopies in conjunction with biological activity assays to define the nature of PLN excited states and, at the same time, to establish a quantitative correlation between the population of the excited state and the extent of SERCA inhibition. We show that the conformational equilibrium of PLN is more complex than previously anticipated. We present a new model for the PLN conformational pre-equilibrium and show a linear correlation between the loss of function (LOF) character of PLN phosphomimetic mutants and their excited-state populations.
Results
A four-state model of PLN conformational equilibrium
Based on our previous studies, we hypothesized that PLN undergoes conformational interconversion between two states: a motionally restricted T state with the cytoplasmic domain absorbed on the membrane surface and a more dynamic R state with domain Ia detached from the membrane and mostly unfolded.20,21,29 To test this hypothesis, we monitored the chemical unfolding of PLN upon addition of guanidine hydrochloride (Gdn HCl) using both EPR in 4:1 DOPC:DOPE (1,2-dioleoyl-sn-glycero-3-phosphocholine:1,2-dioleoyl-sn-glycero-3-phosphoethanolamine) lipid bilayers and NMR in dodecylphosphocholine (DPC) micelles. We utilized a synthetic (for EPR) or recombinant (for NMR) monomeric variant of PLN (AFA-PLN) that has the same inhibitory power and similar structural topology as wild-type PLN.18,31
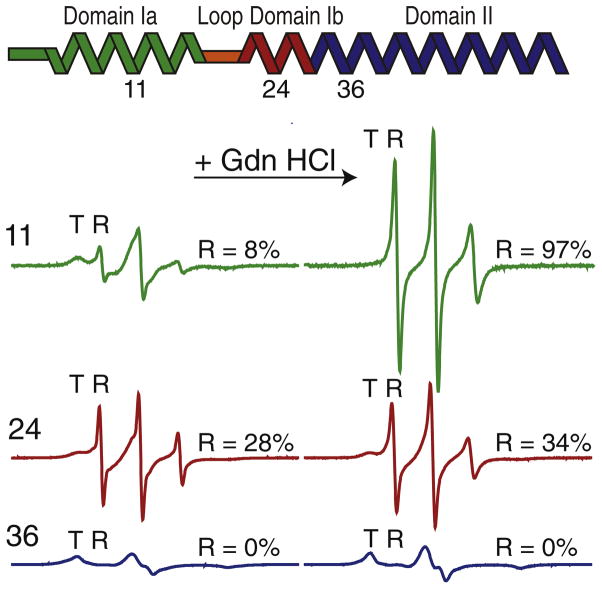
EPR spectra of TOAC (2,2,6,6-tetramethyl-piperidine-1-oxyl-4-amino-4-carboxylic acid) spin label engineered at position 11 revealed the presence of two resolved peaks: one broad resonance that was assigned to the T state and a sharper peak assigned to the R state (Fig. 1).22 The R state is very dynamic, with an order parameter threefold lower than the corresponding T state.21 To follow the chemical unfolding of PLN, we carried out EPR experiments by titrating Gdn HCl into the PLN samples and monitoring the spectral changes of TOAC at positions 11 (domain Ia), 24 (domain Ib), and 36 (domain II). In the absence of chemical denaturant, the populations of the R state were 8% and 28% for positions 11 and 24, respectively; while only the T state was detected at position 36 (Fig. 1). The addition of Gdn HCl caused the disappearance of the TOAC 11 T-state component and a concomitant increase in the peak intensity of the R-state component. This demonstrates a shift in the PLN conformational equilibrium toward the R state for domain Ia. In contrast, the TOAC spectrum for 36 remained unperturbed upon addition of the denaturant and showed that, while domain Ia unfolded completely, the membrane-embedded domain II was still helical in the presence of chemical denaturant. At position 24 (domain Ib), the addition of the Gdn HCl increased the R-state population from 28% to 34%.
Fig. 1.
Chemical unfolding of PLN in lipid vesicles. Continuous-wave EPR spectra of TOAC spin label engineered at positions 11 (domain Ia), 24 (domain Ib), and 36 (domain II). Left: EPR spectra in the absence of Gdn HCl. Right: EPR spectra in the presence of 6 M Gdn HCl (right).
To analyze the conformational transitions at the atomic level, we monitored the NMR chemical shifts for both the methyl and the amide groups of PLN upon addition of Gdn HCl using two-dimensional heteronuclear single quantum coherence (HSQC) spectra. As a reference for the unfolded R state of PLN, we used a peptide corresponding to the cytoplasmic residues of PLN (PLN1–20). In aqueous solutions, this peptide is soluble and unfolded, while in the presence of DPC (or lipid vesicles), it is helical and associated with the micelle surface (Supplementary Fig. 1).32–35
Titration of Gdn HCl into a PLN sample in DPC micelles caused gradual changes to the chemical shifts for all of the resonances (methyls and amides) in domain Ia, loop, and domain Ib (Figs. 2 and 3). Domain II remained mostly unperturbed with the exception of the C-terminal residues, which is in agreement with the EPR spectra for position 36. The chemical shifts for the PLN methyl groups follow trajectories that are close to linear trajectories (Fig. 3) and at a high concentration of denaturant, overlay with those of PLN1–20 without DPC (unfolded R state) (Fig. 4a).
Fig. 2.
Chemical shift perturbations of AFA-PLN upon unfolding with Gdn HCl. (a) 15N amide and (b) 13C methyl chemical shift changes as a function of residue. Colors reflect the different structural domains of PLN: domain Ia (green), loop (orange), domain Ib (red), and domain II (blue). The asterisk indicates an overlapped resonance as a result of Gdn HCl unfolding.
Fig. 3.
Chemical unfolding of AFA-PLN and pS16-AFA-PLN. Representative spectra of methyl [1H,13C] and amide [1H,15N] HSQC experiments for residues in domain Ia (Val4, Leu7, and Ile12) and domain Ib (Gln26) upon Gdn HCl titration for AFA-PLN (black) and pS16-AFA-PLN (red). AFA-PLN and pS16-AFA-PLN peaks correspond to Gdn HCl concentrations of 0 M, 0.067 M, 0.178 M, 0.48 M, 0.89 M, 1.81 M, and 2.79 M. The direction of the arrows represents the increase in Gdn HCl concentration.
Fig. 4.
Methyl and amide fingerprint spectra of PLN1–20 and unfolded AFA-PLN. (a) Overlay of [1H,13C] HSQC spectra of AFA-PLN in the presence of 2.79 M Gdn HCl (black) and PLN1–20 in the absence of DPC (purple). (b) Overlay of the [1H,15N] HSQC spectra for the species reported in (a). With the exception of the terminal residues, the chemical shifts of the resonances of the unfolded domain Ia of AFA-PLN and PLN1–20 (in the absence of DPC) are very similar, showing that the PLN1–20 free peptide is a good mimic of the R state.
The deviations from linearity, however, are more apparent from the amide chemical shifts. In particular, amide chemical shifts of domain Ia show at least three different states (see triangulation of chemical shifts for I12), while more linear behavior is observed for resonances located in domain Ib (Fig. 3 and Supplementary Figs. 3 and 4). At high Gdn HCl concentrations, amide chemical shifts in domain Ia overlap with the R state (Fig. 4b). These results demonstrate that, while EPR detects an apparent two-state equilibrium, the conformational states visited by PLN during chemical unfolding are at least three as revealed by NMR spectroscopy. The latter supports the three-state folding model of amphipathic helices proposed by Ladokhin and White and White and Wimley.36,37 For PLN, the unfolding process highlights three major conformational ensembles: (1) a helical T state, (2) a partially unfolded membrane-associated R′ state, and (3) a totally unfolded membrane-dissociated R state. Note that while the chemical shifts of the methyl groups were sensitive to environmental they only show an apparent two-state equilibrium like the EPR measurements. In contrast, the amide groups are more sensitive to secondary structural changes and therefore are able to detect the presence of the R′ state more clearly.
Interestingly, the plots of the chemical shift changes as a function of the concentration of Gdn HCl show that residues in domains Ia and Ib have different melting profiles (Fig. 5). Specifically, unfolding curves for residues in domain Ia have a more cooperative unfolding behavior than residues in domain Ib [larger Hill coefficient; 2.2±0.2 versus 1.6±0.1 (average±standard error)] (Supplementary Fig. 5 and Supplementary Table 1). Thus, a lower concentration of Gdn HCl is required to induce unfolding for residues in domain Ib. This behavior is typical of less compact states that are likely to interconvert non-cooperatively.38,39
Fig. 5.
Unfolding of domains Ia and Ib upon titration of Gdn HCl. Plots of the 15N chemical shifts as a function of Gdn HCl concentration for Leu7 and Ala11 in domain Ia (left) and Gln26 and Asn27 in domain Ib (right). The unfolding of domain Ia is more cooperative than the unfolding of domain Ib.
Phosphorylation of PLN at Ser16 causes a complete LOF. This state is responsible for calcium reuptake into the SR membrane and regulates the diastolic phase of the cardiac cycle. Guided by the EPR data,21 we originally interpreted phosphorylation of PLN at Ser16 as a shift of the conformational equilibrium toward the R state.24 To test this, we compared the chemical shifts of both methyl and amide groups with the unfolding experiments carried out in Gdn HCl. While the methyl group chemical shifts of pS16-AFA-PLN are in line with the trajectories toward the unfolded R state, supporting an apparent two-state equilibrium, the amide group chemical shifts do not overlap with those from the Gdn HCl titration. This distinct state (T′ state), which is present in most of the resonances of domain Ia, is partially unfolded as revealed by the 13C chemical shift index (CSI) (Fig. 6)23,24 but is primarily membrane attached.23,27 Upon titration with Gdn HCl, the T′ state of phosphorylated PLN is perturbed toward the unfolded and membrane-detached R state (Fig. 4 and Supplementary Figs. 3 and 4). Note that for most of the resonances the unfolded states of both phosphorylated and unphosphorylated PLN overlap completely. However, slight differences in chemical shifts around the phosphorylation site were observed and were probably due to the charged phosphate group. Also, pS16-AFA-PLN unfolds at significantly lower Gdn HCl concentration than AFA-PLN (average k values of 0.7±0.1 and 1.6±0.1) (Supplementary Fig. 6), consistent with the partially unfolded nature of the T′ state.
Fig. 6.
13C CSI for AFA-PLN and pS16-AFA-PLN. The CSI is calculated as 13Cα – 13Cβ – (13Cα,RC – 13Cβ,RC), where RC stands for the random-coil chemical shifts reported by Zhang et al.40 Consecutive positive CSI values indicate helix propensity.
Based on the above results, we can conclude that the conformational interconversions of PLN include at least four major states for domain Ia and two states for domain Ib. The folding/unfolding equilibrium of domain Ia is dictated by its interactions with the lipids and is regulated by phosphorylation (Fig. 7). The T state can proceed to the R state either through the unfolded and membrane-associated R′ state or through the T′ state. Domain Ib transitions between two states, folded and unfolded, independent of domain Ia.
Fig. 7.
Model of the conformational equilibrium of PLN. (Top) The T state of domain Ia undergoes unfolding to the R state via an intermediate T′ state or R′ state. (Bottom) Two-state conformational equilibrium for domain Ib.
Pseudo-phosphorylated mutants of PLN and conformational equilibrium
To further test the presence of the excited T′ state, we monitored the effects of phosphomimetic mutations of PLN (S16D and S16E) by monitoring the chemical shifts of both methyl and amide groups. As with the unfolding studies, the methyl group chemical shifts follow an apparent linear trajectory toward the unfolded R state (Fig. 8). In contrast, the amide chemical shifts of the mutants do not fall along the unfolding trajectory for pS16-AFA-PLN or AFA-PLN, rather, they reside in intermediate positions between the resonances of pS16-AFA-PLN and of AFA-PLN. Therefore, it is possible to conclude that pseudo-phosphorylation and phosphorylation promote a gradual shift in population toward the T′ state, consistent with previous reports that pS16 phosphorylation leads to partial unfolding.26
Fig. 8.

Methyl and amide HSQC spectra for Ser16 phosphorylated AFA-PLN and the pseudo-phosphorylated variants. [1H,13C] and [1H,15N] HSQC spectra for AFA-PLN (black), S16E-AFA-PLN (blue), S16D-AFA-PLN (gray), and pS16-AFA-PLN (red). The continuous-line arrows point to the AFA-PLN resonance in the presence of Gdn HCl, while the broken-line arrow visually traces the transition from AFA-PLN (T state) to pS16-AFA-PLN (T′ state).
Quantitative correlation between the population of excited state and SERCA inhibition
To investigate a relationship between the population of the excited states and the loss of inhibitory power of PLN, we correlated structure (chemical shifts), dynamics [HX-nuclear Overhauser effect (NOE)] (Supplementary Fig. 9), and water accessibility [solvent exposed amides-CLEAN chemical exchange (SEA-CLEANEX)] (Supplementary Fig. 10) with the extent of PLN inhibition on SERCA’s calcium affinity (inhibition; ΔpKCa) induced by the different mutants. Consistent with previous results,29 we found the following overall order of increasing inhibition: AFA-PLN → S16E-AFA-PLN→ S16D-AFA-PLN → pS16-AFA-PLN (Table 1). In order to link the structure to the function, we adopted the approach proposed by Li et al.,41 correlating the average chemical shift change (corresponding to T to T′) for each mutant with the respective SERCA inhibitory potency. The resonances of AFA-PLN were defined to be 100% T state, while those from pS16-AFA-PLN were set to 100% T′ state. The structure–function correlation plot is reported in Fig. 9a. The linearity of this plot (correlation coefficient=0.98) indicates a direct correlation between the increase in the population of the T′ state and the loss of inhibition. Similar correlations with the inhibitory potency are obtained for both fast dynamics (ps-ns) and solvent accessibility (Fig. 9b and c). This is in agreement with the partially unfolded character of the T′ state, which makes it more dynamic and solvent accessible than the T state and allows it to exchange directly with the completely unfolded R state (Fig. 7). These data suggest that the promotion of PLN from the ground T state to the excited T′ state (and likely R state) results in LOF.
Table 1.
SERCA inhibition for PLN mutants
| PLN variant | ΔpKCa |
|---|---|
| AFA-PLN | 0.20±0.02 |
| S16E-AFA-PLN | 0.14±0.04 |
| S16D-AFA-PLN | 0.08±0.04 |
| pS16-AFA-PLN | 0.02±0.02 |
pKCa values were determined from fits to the Hill equation (see Materials and Methods). Inhibition was measured from the difference in pKCa in the absence and in the presence of a 10-fold excess of PLN (ΔpKCa). Standard errors were calculated from the average of three measurements.
Fig. 9.
Structure–dynamics function relationships for AFA-PLN, Ser16 phosphorylated AFA-PLN, and pseudo-phosphorylated mutants. (a) Correlation between combined 15N-HSQC chemical shift change (normalized Δδ), averaged [1H,15N] NOE in domain Ia and the loop (b), and normalized solvent exchange from SEA-CLEANEX experiments (c) with SERCA inhibition from ATPase assays (ΔpKCa) for AFA-PLN (black), S16E-AFA-PLN (blue), S16D-AFA-PLN (gray), and pS16-AFA-PLN (red). See Materials and Methods for calculation of ΔpKCa, normalized Δδ, 〈[1H,15N] NOE〉, and normalized solvent exchange.
Discussion
Based on our previous investigations,16 we hypothesized that the conformational equilibrium of PLN is central to SERCA regulation. However, in our original papers, both EPR and NMR studies revealed the presence of an apparent two-state equilibrium (T and R states).20,21,42 Here, we provide evidence that PLN has pre-existing equilibria between several conformational states, exemplified by the folding/unfolding of both domain Ia and Ib. For domain Ia methyl groups, the chemical shift trajectories for chemical unfolding follow an apparent two-state equilibrium in agreement with EPR data. In contrast, the trajectories for amide group chemical shifts showed the presence of four conformational states, which is in agreement with the folding model of amphipathic helices.36,37 On the other hand, for domain Ib, the folding/unfolding process involves a simpler two-state model, as we also found using Carr–Purcell–Meiboom–Gill relaxation dispersion methods.42 This new, more complex model agrees with recent data from Hughes et al. and Abu-Baker and Lorigan,25,26 who proposed an active role of lipids in modulating PLN structural dynamics and SERCA inhibition. At the same time, it explains previous mutagenesis studies, which identified a number of sites in domain Ia of PLN that lead to relief of SERCA inhibition.43 Specifically, residues populating the hydrophobic face of the amphipathic domain Ia (V4, L7, and I12) are among these hot spots,43 which are inserted into the lipid bilayer.17 Shortening the side chain of these residues would likely attenuate the interactions of domain Ia with the membrane environment, supporting the hypothesis that lipid bilayers influence the regulatory process in the context of membrane detachment and helix unfolding. Interestingly, attachment of a lipid anchor to the N-terminus completely shifts PLN to the T state, with Ser16 phosphorylation of this lipidated PLN failing to relieve SERCA inhibition.21,33 This result also strongly supports the structure–function relationship reported in Fig. 9. It is important to stress that the relative population of states can potentially be biased by the choice of membrane mimic. For example, in DPC micelles, the T and T′ states are primarily detected. However, solid-state NMR experiments in dimyristoylphosphatidylcholine lipid bilayers found domain Ia to be predominantly unfolded.44 This suggests an important role of lipid charge, head groups, and possibly bilayer curvature in influencing the conformational equilibria and extent of aggregation of PLN.
Tuning PLN conformational equilibria leads to the control of SERCA activity. Recently, we showed that to gain complete control over PLN inhibition it is necessary to map the transition to the excited states.29 Our new data show that pseudo-phosphorylation and phosphorylation of PLN increase the population of T′ state. Since the T′ state exchanges more readily with the fully unfolded R state (Fig. 7), it is likely that the R state also increases in population. The correlation between membrane detachment/unfolding and SERCA inhibition (Fig. 9) suggests that these states need to be considered in the design of new, more efficient LOF PLN mutants. To this extent, the S16D mutation is more similar in structure and dynamics to pS16-AFA-PLN than is the S16E mutant. This is supported by SERCA inhibition assays that show a greater LOF character for S16D-AFA-PLN than for S16E-AFA-PLN and would make the S16D mutant a better candidate for positive inotropic therapy.45
An important corollary of this study is the intrinsic flexibility and versatility of the cytoplasmic domain of PLN that when detached from the membrane surface behaves as an intrinsically disordered domain that can adopt several conformations.46,47 In the heart muscle, PLN interacts with several different partners to elicit its regulatory function. Specifically, PLN associates with the lipid bilayer, SERCA, PKA, Ca2+/calmodulin-dependent protein kinase II, A-kinase anchoring protein 18δ, protein phosphatase 1, HS-1-associated protein X-1, and itself in the form of a pentamer state. How can this protein with such a simple structure and topology bind these diverse biomolecules? We believe that the answer is encoded in its flexibility and malleability (i.e., dynamics) that enable PLN to undergo conformational changes and mold into different binding sites.
Finally, it is worth noting that PLN can be divided into two main regions: an inhibitory transmembrane segment and a cytoplasmic regulatory region. The latter is reminiscent of the autoinhibitory elements found in Vav and light–oxygen–voltage proteins, where an amphipathic helical switch is activated upon phosphorylation or light.41,48 In the case of Vav and light–oxygen–voltage, this regulatory element is covalently attached to the proteins with the amphipathic helix undergoing folding/unfolding transitions upon interaction with the protein surface. For SERCA, the reversible regulatory element is provided by PLN that is modulated by the folding/unfolding transitions via the lipid bilayer and phosphorylation. Given the high level of similarity between PLN and other single-pass membrane protein subunits such as the FYXD family, it is likely that the mechanism described in this paper is a general one, also found in the regulation of other ion pumps.
Materials and Methods
Purification of PLN protein
Recombinant AFA-PLN, S16D-AFA-PLN, and S16E-AFA-PLN were grown and purified as described by Buck et al.49 PLN1–20 was expressed in Escherichia coli and was purified as described by Masterson et al.50 To obtain full phosphorylation at Ser16, we dissolved lyophilized AFA-PLN protein at a concentration of 0.3 mg/ml in 20 mM Mops buffer (pH 7.0) containing 1% (w/v) n-octyl-β-D-glucopyranoside, ATP (1 mM), and MgCl2 (1 mM).23 Phosphorylation was accomplished by the addition of purified recombinant PKA51,52 at a 1:2000 PKA:AFA-PLN molar ratio and was incubated for ~12 h at 30 °C. After purification by reversed-phase HPLC, the pS16-AFA-PLN was lyophilized and stored at −20 °C.
Assay for SERCA activity
For SERCA activity measurements, AFA-PLN, pS16-AFA-PLN, S16E-AFA-PLN, and S16D-AFA-PLN were reconstituted into 4:1 DOPC:DOPE lipid bilayers. SERCA was purified in octaethylene glycol monododecyl ether (C12E8)5 and was added to the lipid vesicle suspension containing 700:1 lipid:SERCA and 10:1 PLN: SERCA molar ratios. Subsequently, the samples were incubated with Bio-Beads (30:1 Bio-Beads:C12E8 by weight) (Bio-Rad Laboratories) to remove excess detergent. A coupled NADH assay was used to measure SERCA activity (hydrolysis of ATP) at 37 °C as a function of calcium concentration.53 The rate of enzyme activity was measured as a decrease of NADH absorption at 340 nm using a SpectraMax plate reader (Molecular Devices). Data were fit using the Hill equation to extract the maximum activity (Vmax), Hill coefficient (n), and calcium concentration needed to achieve half maximal activity (pKCa).54
EPR spectroscopy
For EPR experiments, AFA-PLN with the TOAC spin label substituted at 11, 24, or 36 was synthesized and reconstituted into lipid vesicles containing DOPC: DOPE (4:1, 200 lipids per PLN).54 EPR spectra were acquired using a Bruker ELEXSYS 500 spectrometer with the SHQ cavity. Samples (20 μl in a 0.6-mm-i.d. quartz capillary) were maintained at 4 °C using the Bruker temperature controller with a quartz Dewar insert. The field modulation frequency was 100 kHz, with a peak-to-peak amplitude of 2 G. The microwave power was 7.9 mW, producing moderate saturation (so that signal intensity was at least 50% of maximum) without a significant effect on the spectral lineshape. EPR spectra were simulated as a sum of one or two components, each component produced by a population having a static random distribution of membrane orientations and undergoing rotational diffusion in a restricting potential (defined by order parameter S) with a single rotational correlation time.21 Experimental spectra were fit by summing simulated components, yielding mole fractions.
NMR spectroscopy
To prepare NMR samples of the AFA-PLN variants, we dissolved lyophilized protein in NMR sample buffer containing 20 mM NaHPO4, 120 mM NaCl, 0.01% NaN3, 5% D2O, 6 M Gdn HCl, 300 mM deuterated DPC, and pH 6.0 uncorrected for isotopic effect. Gdn HCl was subsequently dialyzed against 1 l of NMR sample buffer. This procedure gave more homogenous spectral lines and avoided protein aggregation. All of the NMR experiments were performed on Varian VNMRS and Inova spectrometers operating at a 1H frequency of 600 MHz and at a temperature of 37 °C. Data were processed using NMRPipe55 and were viewed and analyzed in Sparky.56 Protein amide fingerprints were analyzed using gradient-enhanced [1H,15N] HSQC experiments.57 The data were acquired using 1544 complex points in the direct 1H dimension and 50–80 increments in the indirect 15N dimension. Gradient-enhanced constant-time [1H,13C] HSQC experiments were utilized to measure methyl group chemical shifts. For PLN1–20 and each AFA-PLN mutant, 8 scans with 2048 complex points and a spectral width of 12,000 Hz in the direct 1H dimension and 64 points with a spectral width of 3500 Hz in the indirect 13C dimension were acquired. After Fourier transformation, the data were zero filled to a final matrix size of 4096×2048.
Heteronuclear [1H 15N] steady-state NOE measurements were acquired using the pulse sequence of Farrow et al.58 Two-dimensional spectra with and without a 3-s 1H saturation period were acquired to obtain the ratio between peak intensities in the saturated (Isat) unsaturated (Iunsat) spectra:
| (1) |
The uncertainty of the measurement was estimated for each residue from the relationship:58
| (2) |
where σsat and σunsat are the baseline noise in the saturated and unsaturated spectra, and σHNNOE is the estimated error for the heteronuclear NOE (HN NOE) values.
To measure the solvent exchange for amide protons, we performed [1H,15N] SEA-CLEANEX experiments.59 This pulse sequence utilizes a spin-echo filter to select for water magnetization followed by a mixing period to allow for exchange between protein and solvent.60 To eliminate NOE contributions, we implemented the CLEANEX-PM mixing scheme.61 Four different mixing times (30, 60, 120, and 200 ms) were used to follow the buildup of peaks corresponding to solvent exchanging amide protons. A total of 64–96 scans were recorded with 2048 complex points in the direct 1H dimension and 40 increments in the indirect 15N dimension. Spectral widths were 12,000 Hz and 1200 Hz in the direct and indirect dimensions, respectively. A shifted sine-bell window function was applied to all data sets, which were zero filled to 4096 in both dimensions prior to Fourier transformation.
Gdn-HCl-induced denaturation was done by titrating Gdn HCl from a 6 M stock solution to AFA-PLN at pH 6.0. Six additions were made to a final Gdn HCl concentration of 2.8 M, and [1H,15N] HSQC and [1H,13C] HSQC spectra were acquired after each addition as described above. For the analysis of unfolding, spectra were referenced to 10 mM 2,2-dimethyl-2-silapentane-5-sulfonate, and 15N peak shifts were plotted as a function of Gdn HCl concentration. The data were fit using the Hill equation:
where n is the Hill coefficient, and k is the unfolding half point. Hill coefficients for residues in domain Ib of PLN (22–28) and domain Ia (3–16) were compared using a t-test. Residue 9 was excluded, since it could not be assigned; residues 2, 29, and 30 were excluded, since those fit poorly to the Hill equation. k values for domain Ia and Ib of AFA-PLN and pS16-AFA-PLN (residues 3–16 and 22–28) were compared with a t-test.
Structure, dynamics, function relationship
Chemical shift changes for the different mutants were calculated according to the expression:
| (6) |
where Δδcombined is the difference in chemical shift relative to AFA-PLN. Several resonances within the HSQC spectra (Supplementary Fig. 2) that had a gradual change in chemical shifts from AFA-PLN → S16E-AFA-PLN→ S16D-AFA-PLN → pS16-AFA-PLN (Leu7, Thr8, Ala11, Ile12, Ala15, Ile18, Glu19, Gln22, Gln23, Arg25, Gln26, Asn27, Leu28, Gln29, and Asn30) were used in the analysis. Each resonance shift was calculated using the following formula:
| (7) |
where ω is the combined chemical shift, and p is the relative peak position for each species. In the formula, the AFA-PLN chemical shifts corresponded to 0, and pS16-AFA-PLN to 1. The HN NOE value was calculated as an average of HN NOE values for all residues in domain Ia, loop, and domain Ib (residues 2–31). To determine the normalized solvent exchange, we measured peak intensities from [1H,15N] SEA-CLEANEX spectra using the software Sparky.55 The intensities were corrected for the differences in sensitivity for residues in a [1H–15N] HSQC spectrum and were then normalized among the different PLN species (AFA-PLN, pS16-AFA-PLN, S16D-AFA-PLN, and S16E-AFA-PLN) by assuming Glu2 to have the same solvent exchange for all mutants. The averaged normalized solvent exchange value included residues in domain Ia, loop, and domain Ib (2–30). Residues 15–17 were excluded from the analysis due to their proximity to the phosphorylated/mutated Ser16, and residues 5, 9, and 25 were removed due to spectral overlap.
Supplementary Material
Acknowledgments
This work was supported by grants to GV (GM64742) and DDT (GM27906) and a pre-doctoral fellowship to MG from the American Heart Association (10PRE3860050). We thank Larry Masterson, Kim Ha, Raffaello Verardi and Tao Yu for technical assistance and helpful discussions.
Abbreviations used
- PLN
phospholamban
- SERCA
sarcoplasmic reticulum Ca2+-ATPase
- EPR
electron paramagnetic resonance
- SR
sarcoplasmic reticulum
- LOF
loss of function
- Gdn HCl
guanidine hydrochloride
- DOPC
DOPE, 1,2-dioleoyl-sn-glycero-3-phosphocholine:1,2-dioleoyl-sn-glycero-3-phosphoethanolamine
- DPC
dodecylphosphocholine
- TOAC
2,2,6,6-tetramethyl-piperidine-1-oxyl-4-amino-4-carboxylic acid
- HSQC
heteronuclear single quantum coherence
- NOE
nuclear Overhauser effect
- SEA-CLEANEX
solvent exposed amides-CLEAN chemical exchange
- PKA
protein kinase A
- CSI
chemical shift index
- HNNOE
heteronuclear NOE
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j. jmb.2011.03.015
References
- 1.Simmerman HK, Collins JH, Theibert JL, Wegener AD, Jones LR. Sequence analysis of phospholamban. Identification of phosphorylation sites and two major structural domains. J Biol Chem. 1986;261:13333–13341. [PubMed] [Google Scholar]
- 2.Reddy LG, Jones LR, Thomas DD. Depolymerization of phospholamban in the presence of calcium pump: a fluorescence energy transfer study. Biochemistry. 1999;38:3954–3962. doi: 10.1021/bi981795d. [DOI] [PubMed] [Google Scholar]
- 3.Tada M, Kadoma M. Regulation of the Ca2+ pump ATPase by cAMP-dependent phosphorylation of phospholamban. BioEssays. 1989;10:157–163. doi: 10.1002/bies.950100505. [DOI] [PubMed] [Google Scholar]
- 4.MacLennan DH, Kranias EG. Phospholamban: a crucial regulator of cardiac contractility. Nat Rev, Mol Cell Biol. 2003;4:566–577. doi: 10.1038/nrm1151. [DOI] [PubMed] [Google Scholar]
- 5.Stokes DL, Green NM. Three-dimensional crystals of CaATPase from sarcoplasmic reticulum. Symmetry and molecular packing. Biophys J. 1990;57:1–14. doi: 10.1016/S0006-3495(90)82501-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toyoshima C, Nakasako M, Nomura H, Ogawa H. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 Å resolution. Nature. 2000;405:647–655. doi: 10.1038/35015017. [DOI] [PubMed] [Google Scholar]
- 7.Toyoshima C, Nomura H. Structural changes in the calcium pump accompanying the dissociation of calcium. Nature. 2002;418:605–611. doi: 10.1038/nature00944. [DOI] [PubMed] [Google Scholar]
- 8.Sorensen TL, Moller JV, Nissen P. Phosphoryl transfer and calcium ion occlusion in the calcium pump. Science. 2004;304:1672–1675. doi: 10.1126/science.1099366. [DOI] [PubMed] [Google Scholar]
- 9.Olesen C, Sorensen TL, Nielsen RC, Moller JV, Nissen P. Dephosphorylation of the calcium pump coupled to counterion occlusion. Science. 2004;306:2251–2255. doi: 10.1126/science.1106289. [DOI] [PubMed] [Google Scholar]
- 10.Stokes DL, Pomfret AJ, Rice WJ, Glaves JP, Young HS. Interactions between Ca2+-ATPase and the pentameric form of phospholamban in two-dimensional co-crystals. Biophys J. 2006;90:4213–4223. doi: 10.1529/biophysj.105.079640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young HS, Jones LR, Stokes DL. Locating phospholamban in co-crystals with Ca2+-ATPase by cryoelectron microscopy. Biophys J. 2001;81:884–894. doi: 10.1016/S0006-3495(01)75748-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seidel K, Andronesi OC, Krebs J, Griesinger C, Young HS, Becker S, Baldus M. Structural characterization of Ca2+-ATPase-bound phospholamban in lipid bilayers by solid-state nuclear magnetic resonance (NMR) spectroscopy. Biochemistry. 2008;47:4369–4376. doi: 10.1021/bi7024194. [DOI] [PubMed] [Google Scholar]
- 13.Schmitt JP, Ahmad F, Lorenz K, Hein L, Schulz S, Asahi M, et al. Alterations of phospholamban function can exhibit cardiotoxic effects independent of excessive sarcoplasmic reticulum Ca2+-ATPase inhibition. Circulation. 2009;119:436–444. doi: 10.1161/CIRCULATIONAHA.108.783506. [DOI] [PubMed] [Google Scholar]
- 14.Cornea RL, Jones LR, Autry JM, Thomas DD. Mutation and phosphorylation change the oligomeric structure of phospholamban in lipid bilayers. Biochemistry. 1997;36:2960–2967. doi: 10.1021/bi961955q. [DOI] [PubMed] [Google Scholar]
- 15.Zamoon J, Mascioni A, Thomas DD, Veglia G. NMR solution structure and topological orientation of monomeric phospholamban in dode-cylphosphocholine micelles. Biophys J. 2003;85:2589–2598. doi: 10.1016/s0006-3495(03)74681-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Traaseth NJ, Ha KN, Verardi R, Shi L, Buffy JJ, Masterson LR, Veglia G. Structural and dynamic basis of phospholamban and sarcolipin inhibition of Ca2+-ATPase. Biochemistry. 2008;47:3–13. doi: 10.1021/bi701668v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Traaseth NJ, Shi L, Verardi R, Mullen DG, Barany G, Veglia G. Structure and topology of monomeric phospholamban in lipid membranes determined by a hybrid solution and solid-state NMR approach. Proc Natl Acad Sci USA. 2009;106:10165–10170. doi: 10.1073/pnas.0904290106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mascioni A, Karim C, Zamoon J, Thomas DD, Veglia G. Solid-state NMR and rigid body molecular dynamics to determine domain orientations of monomeric phospholamban. J Am Chem Soc. 2002;124:9392–9393. doi: 10.1021/ja026507m. [DOI] [PubMed] [Google Scholar]
- 19.Metcalfe EE, Zamoon J, Thomas DD, Veglia G. H/15N heteronuclear NMR spectroscopy shows four dynamic domains for phospholamban reconstituted in dodecylphosphocholine micelles. Biophys J. 2004;87:1205–1214. doi: 10.1529/biophysj.103.038844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zamoon J, Nitu F, Karim C, Thomas DD, Veglia G. Mapping the interaction surface of a membrane protein: unveiling the conformational switch of phospholamban in calcium pump regulation. Proc Natl Acad Sci USA. 2005;102:4747–4752. doi: 10.1073/pnas.0406039102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karim CB, Zhang Z, Howard EC, Torgersen KD, Thomas DD. Phosphorylation-dependent conformational switch in spin-labeled phospholamban bound to SERCA. J Mol Biol. 2006;358:1032–1040. doi: 10.1016/j.jmb.2006.02.051. [DOI] [PubMed] [Google Scholar]
- 22.Nesmelov YE, Karim CB, Song L, Fajer PG, Thomas DD. Rotational dynamics of phospholamban determined by multifrequency electron paramagnetic resonance. Biophys J. 2007;93:2805–2812. doi: 10.1529/biophysj.107.108910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Metcalfe EE, Traaseth NJ, Veglia G. Serine 16 phosphorylation induces an order-to-disorder transition in monomeric phospholamban. Biochemistry. 2005;44:4386–4396. doi: 10.1021/bi047571e. [DOI] [PubMed] [Google Scholar]
- 24.Traaseth NJ, Thomas DD, Veglia G. Effects of Ser16 phosphorylation on the allosteric transitions of phospholamban/Ca2+-ATPase complex. J Mol Biol. 2006;358:1041–1050. doi: 10.1016/j.jmb.2006.02.047. [DOI] [PubMed] [Google Scholar]
- 25.Hughes E, Clayton JC, Middleton DA. Cytoplasmic residues of phospholamban interact with membrane surfaces in the presence of SERCA: a new role for phospholipids in the regulation of cardiac calcium cycling? Biochim Biophys Acta. 2009;1788:559–566. doi: 10.1016/j.bbamem.2008.10.029. [DOI] [PubMed] [Google Scholar]
- 26.Abu-Baker S, Lorigan GA. Phospholamban and its phosphorylated form interact differently with lipid bilayers: a 31P, 2H, and 13C solid-state NMR spectroscopic study. Biochemistry. 2006;45:13312–13322. doi: 10.1021/bi0614028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chu S, Abu-Baker S, Lu J, Lorigan GA. N Solid-state NMR spectroscopic studies on phospholamban at its phosphorylated form at ser-16 in aligned phospholipid bilayers. Biochim Biophys Acta. 2010;1798:312–317. doi: 10.1016/j.bbamem.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wegener AD, Simmerman HK, Lindemann JP, Jones LR. Phospholamban phosphorylation in intact ventricles. Phosphorylation of serine 16 and threonine 17 in response to β-adrenergic stimulation. J Biol Chem. 1989;264:11468–11474. [PubMed] [Google Scholar]
- 29.Ha KN, Traaseth NJ, Verardi R, Zamoon J, Cembran A, Karim CB, et al. Controlling the inhibition of the sarcoplasmic Ca2+-ATPase by tuning phospholamban structural dynamics. J Biol Chem. 2007;282:37205–37214. doi: 10.1074/jbc.M704056200. [DOI] [PubMed] [Google Scholar]
- 30.Hoshijima M, Ikeda Y, Iwanaga Y, Minamisawa S, Date MO, Gu Y, et al. Chronic suppression of heart-failure progression by a pseudo-phosphorylated mutant of phospholamban via in vivo cardiac rAAV gene delivery. Nat Med. 2002;8:864–871. doi: 10.1038/nm739. [DOI] [PubMed] [Google Scholar]
- 31.Karim CB, Marquardt CG, Stamm JD, Barany G, Thomas DD. Synthetic null-cysteine phospholamban analogue and the corresponding transmembrane domain inhibit the Ca-ATPase. Biochemistry. 2000;39:10892–10897. doi: 10.1021/bi0003543. [DOI] [PubMed] [Google Scholar]
- 32.Clayton JC, Hughes E, Middleton DA. Spectroscopic studies of phospholamban variants in phospholipid bilayers. Biochem Soc Trans. 2005;33:913–915. doi: 10.1042/BST20050913. [DOI] [PubMed] [Google Scholar]
- 33.Lockwood NA, Tu RS, Zhang Z, Tirrell MV, Thomas DD, Karim CB. Structure and function of integral membrane protein domains resolved by peptide-amphiphiles: application to phospholamban. Biopolymers. 2003;69:283–292. doi: 10.1002/bip.10365. [DOI] [PubMed] [Google Scholar]
- 34.Hubbard JA, MacLachlan LK, Meenan E, Salter CJ, Reid DG, Lahouratate P, et al. Conformation of the cytoplasmic domain of phospholamban by NMR and CD. Mol Membr Biol. 1994;11:263–269. doi: 10.3109/09687689409160436. [DOI] [PubMed] [Google Scholar]
- 35.Huggins JP, England PJ. Evidence for a phosphorylation-induced conformational change in phospholamban from the effects of three proteases. FEBS Lett. 1987;217:32–36. doi: 10.1016/0014-5793(87)81236-x. [DOI] [PubMed] [Google Scholar]
- 36.Ladokhin AS, White SH. Folding of amphipathic α-helices on membranes: energetics of helix formation by melittin. J Mol Biol. 1999;285:1363–1369. doi: 10.1006/jmbi..1998.2346. [DOI] [PubMed] [Google Scholar]
- 37.White SH, Wimley WC. Membrane protein folding and stability: physical principles. Annu Rev Biophys Biomol Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 38.Fersht A. Structure and Mechanism in Protein Science. W. H. Freeman & Co; New York, NY: 1999. [Google Scholar]
- 39.D’Onofrio M, Ragona L, Fessas D, Signorelli M, Ugolini R, Pedo M, et al. NMR unfolding studies on a liver bile acid binding protein reveal a global two-state unfolding and localized singular behaviors. Arch Biochem Biophys. 2009;481:21–29. doi: 10.1016/j.abb.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 40.Zhang H, Neal S, Wishart DS. RefDB: a database of uniformly referenced protein chemical shifts. J Biomol NMR. 2003;25:173–195. doi: 10.1023/a:1022836027055. [DOI] [PubMed] [Google Scholar]
- 41.Li P, Martins IR, Amarasinghe GK, Rosen MK. Internal dynamics control activation and activity of the autoinhibited Vav DH domain. Nat Struct Mol Biol. 2008;15:613–618. doi: 10.1038/nsmb.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Traaseth NJ, Veglia G. Probing excited states and activation energy for the integral membrane protein phospholamban by NMR CPMG relaxation dispersion experiments. Biochim Biophys Acta. 2010;1798:77–81. doi: 10.1016/j.bbamem.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.MacLennan DH, Kimura Y, Toyofuku T. Sites of regulatory interaction between calcium ATPases and phospholamban. Ann N Y Acad Sci. 1998;853:31–42. doi: 10.1111/j.1749-6632.1998.tb08254.x. [DOI] [PubMed] [Google Scholar]
- 44.Andronesi OC, Becker S, Seidel K, Heise H, Young HS, Baldus M. Determination of membrane protein structure and dynamics by magic-angle-spinning solid-state NMR spectroscopy. J Am Chem Soc. 2005;127:12965–12974. doi: 10.1021/ja0530164. [DOI] [PubMed] [Google Scholar]
- 45.Stevenson LW. Clinical use of inotropic therapy for heart failure: looking backward or forward? Part II: chronic inotropic therapy. Circulation. 2003;108:492–497. doi: 10.1161/01.CIR.0000078349.43742.8A. [DOI] [PubMed] [Google Scholar]
- 46.Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mittag T, Kay LE, Forman-Kay JD. Protein dynamics and conformational disorder in molecular recognition. J Mol Recognit. 2009;23:105–116. doi: 10.1002/jmr.961. [DOI] [PubMed] [Google Scholar]
- 48.Yao X, Rosen MK, Gardner KH. Estimation of the available free energy in a LOV2-Jα photoswitch. Nat Chem Biol. 2008;4:491–497. doi: 10.1038/nchembio.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buck B, Zamoon J, Kirby TL, DeSilva TM, Karim C, Thomas D, Veglia G. Over-expression, purification, and characterization of recombinant Ca-ATPase regulators for high-resolution solution and solid-state NMR studies. Protein Expr Purif. 2003;30:253–261. doi: 10.1016/s1046-5928(03)00127-x. [DOI] [PubMed] [Google Scholar]
- 50.Masterson LR, Bortone N, Yu T, Ha KN, Gaffarogullari EC, Nguyen O, Veglia G. Expression and purification of isotopically labeled peptide inhibitors and substrates of cAMP-dependant protein kinase A for NMR analysis. Protein Expression Purif. 2009;64:231–236. doi: 10.1016/j.pep.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yonemoto WM, McGlone ML, Slice LW, Taylor SS. Prokaryotic expression of catalytic subunit of adenosine cyclic monophosphate-dependent protein kinase. Methods Enzymol. 1991;200:581–596. doi: 10.1016/0076-6879(91)00173-t. [DOI] [PubMed] [Google Scholar]
- 52.Masterson LR, Mascioni A, Traaseth NJ, Taylor SS, Veglia G. Allosteric cooperativity in protein kinase A. Proc Natl Acad Sci USA. 2008;105:506–511. doi: 10.1073/pnas.0709214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reddy LG, Cornea RL, Winters DL, McKenna E, Thomas DD. Defining the molecular components of calcium transport regulation in a reconstituted membrane system. Biochemistry. 2003;42:4585–4592. doi: 10.1021/bi026995a. [DOI] [PubMed] [Google Scholar]
- 54.Karim CB, Kirby TL, Zhang Z, Nesmelov Y, Thomas DD. Phospholamban structural dynamics in lipid bilayers probed by a spin label rigidly coupled to the peptide backbone. Proc Natl Acad Sci USA. 2004;101:14437–14442. doi: 10.1073/pnas.0402801101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 56.Goddard TD, Kneller DG. SPARKY 3. 1999. [Google Scholar]
- 57.Kay LE, Keifer E, Saarinen T. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J Am Chem Soc. 1992;114:10663–10665. [Google Scholar]
- 58.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, et al. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 59.Lin D, Sze KH, Cui Y, Zhu G. Clean SEA-HSQC: a method to map solvent exposed amides in large non-deuterated proteins with gradient-enhanced HSQC. J Biomol NMR. 2002;23:317–322. doi: 10.1023/a:1020225206644. [DOI] [PubMed] [Google Scholar]
- 60.Mori S, Berg JM, van Zijl PC. Separation of intramolecular NOE and exchange peaks in water exchange spectroscopy using spin-echo filters. J Biomol NMR. 1996;7:77–82. doi: 10.1007/BF00190459. [DOI] [PubMed] [Google Scholar]
- 61.Hwang TL, van Zijl PC, Mori S. Accurate quantitation of water–amide proton exchange rates using the phase-modulated CLEAN chemical EXchange (CLEANEX-PM) approach with a Fast-HSQC (FHSQC) detection scheme. J Biomol NMR. 1998;11:221–226. doi: 10.1023/a:1008276004875. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.