Abstract
With multi-drug resistant cases of tuberculosis increasing globally, better antibiotic drugs and novel drug-targets are becoming an urgent need. Traditional β-lactam antibiotics that disrupt the D,D-transpeptidases are not effective against mycobacteria, in part because mycobacteria rely mostly on β-lactam insensitive L,D-transpeptidases for biosynthesis and maintenance of their peptidoglycan layer. This reliance plays a major role in drug-resistance and persistence of Mycobacterium tuberculosis (Mtb) infections. The crystal structure at 1.7 Å resolution of the Mtb L,D-transpeptidase LdtMt2 containing a bound peptidoglycan fragment, reported here, provides information about catalytic site organization as well as substrate recognition by the enzyme. Based on our structural, kinetic, and calorimetric data, we propose a catalytic mechanism for LdtMt2 in which both acyl-acceptor and acyl-donor substrates reach the catalytic site from the same, rather than different, entrances. Together, this information provides vital insights for the development of novel drugs targeting this validated yet unexploited enzyme.
Introduction
Multidrug-resistant (MDR) and extensively drug-resistant (XDR) strains of Mycobacterium tuberculosis (Mtb) are emerging at alarming rates (Fauci, 2008). A possible reason for this is poor patient compliance with existing treatments, which require multiple drugs to be taken daily for at least 6 months (ATS, 1997) to eliminate the resilient ~1% of bacilli, commonly known as persisters (Jindani et al., 2003; Lewis, 2007). Any strategy aimed at shortening duration of tuberculosis therapy must be effective in eliminating the persisters. Mtb persists in the lungs of infected individuals in a slow growing, non-replicating dormant state in which the bacillus is resistant to the host immune response and is transiently tolerant of antibiotics (Betts et al., 2002; Bishai, 2000). Mtb persisters extracted from lung lesions show morphological changes similar to those observed in bacilli that are subject to nutrient starvation or anaerobic stress (Betts et al., 2002). These changes involve, among other physiologic alterations, the remodeling of the Mtb peptidoglycan layer (Lavollay et al., 2008), a vital layer consisting of an elaborate network of peptidoglycan chains with cross-linked peptide stems.
Formation of most common type of cross-link, the (D,D) 4→3 linkage (Fig. 1A: top), is catalyzed by the D,D-transpeptidase activity of enzymes commonly referred to as penicillin binding proteins. These enzymes catalyze the transfer of the peptide bond between the fourth residue (D chiral-center) and the fifth residue of a pentapeptide donor stem to a side chain amide group (also a D chiral-center) at the third residue of an adjacent acceptor stem (usually a di-amino acid). A second type of cross-link, the (L,D) 3→3 linkage (Fig. 1A: bottom), is catalyzed by L,D-transpeptidases such as the one studied here. These enzymes transfer the peptide bond between the third residue (L chiral-center) of a tetrapeptide donor stem to the side chain amide group of the third residue (D chiral-center) of an adjacent acceptor-stem. In both types of transpeptidases, the catalysis proceeds by a two-step mechanism: acylation of the enzyme by the penultimate peptide of the donor-stem with the release of the stem C-terminal residue, followed by de-acylation of this acyl-enzyme intermediate by an acceptor stem.
Figure 1.
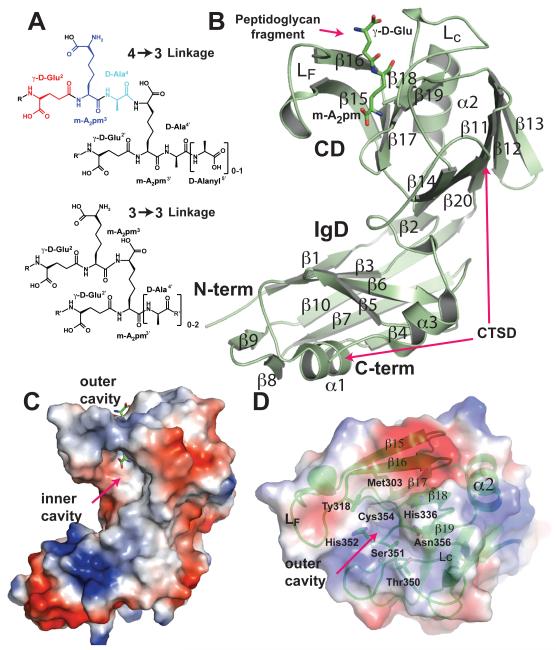
Peptidoglycan linkages and structure of LdtMt2. (A) 4→3 and 3→3 linkages. The C-terminal residues from the γ-D-Glu of donor and acceptor peptidoglycan stems are depicted in different colors, γ-D-Glu (red), m-A2pm (blue), and D-Ala (cyan). Residue numbers of the acceptor stem are primed. (B) Cartoon representation of the overall structure of ex-LdtMt2 with elements of secondary structure labeled. Two pink arrows mark the beginning and end of the CTSD. The peptidoglycan fragment bound to the active site is also shown in a stick representation. (C and D) Views of the solvent accessible surface of ex-LdtMt2 colored by electrostatic surface-potential (from blue positive to red negative). (C) side view (same orientation as part (B). The stick representation of peptidoglycan-fragment bound to the active site is shown. Oxygen atoms are colored in red, nitrogen atoms in blue, sulfur atoms in yellow and carbon atoms in green. (D) top view of the outer cavity (rotated 90° around a horizontal axis from B). The surface was rendered transparent and the peptidoglycan fragment removed to show residues and secondary structure elements lining the outer cavity.
The 3→3 linkages were first identified In 1974 by Wietzerbin et al. in the peptidoglycan of mycobacteria (Wietzerbin et al., 1974), but their significance and biosynthetic pathway remained unknown until recently, when their predominance in Mtb was demonstrated and LdtMt1 (or MT0125, the product of gene Rv0116c) was identified as an L,D-transpeptidase that generates 3→3 linkages (Lavollay et al., 2008). Frequently associated with virulence and β-lactam resistance, predominance of 3→3 linkage has been observed in spontaneous mutation of E. faecium (Mainardi et al., 2005), in other mycobacteria M. abscessus (Lavollay et al., 2011), and in the spore-forming bacteria Clostridium difficile (Peltier et al., 2011).
Recently, another 3→3 L,D-transpeptidase, LdtMt2 (MT2594, the product of gene Rv2518c), was characterized in Mtb (Gupta et al., 2010). An Mtb strain lacking LdtMt2 loses virulence and has attenuated growth during the chronic phase of the disease (Gupta et al., 2010). In addition, this strain is more susceptible to the therapeutic combination of amoxicillin and clavulanic acid, suggesting that the (3→3) L,D-transpeptidase activity is a major contributor to β-lactam resistance. It was also shown that a vaccine utilizing an M. bovis BCG strain that overexpresses L,D-transpeptidases has enhanced protective efficacy against Mtb persisters (Nolan and Lamichhane, 2011), suggesting that the higher degree of 3→3 linkages resulting from the overexpression of the enzyme better represents the peptidoglycan layer of Mtb persisters.
Despite the obvious significance of L,D-transpeptidases in Mtb growth and virulence, no structural or binding information exists for the five members of this class of Mtb enzymes identified to date (Gupta et al., 2010). Here, we present the characterization of the extramembrane portion of LdtMt2 (ex-LdtMt2), including the determination of its structure at 1.7-Å resolution. Ex-LdtMt2 is composed of two domains: an N-terminal immunoglobulin-like domain and a C-terminal catalytic ErfK/YbiS/YhnG domain (Pfam accession number PF03734). Based on this structure, comparative modeling of the identified Mtb homologues indicates that the N-terminal domain fold and the enzyme’s overall conformation distinguish this group from other structurally characterized ErfK/YbiS/YhnG-domain-containing proteins such as Bacillus subtilis ykuD (Bielnicki et al., 2006) and Enterococcus faecium L,D-transpeptidase Ldtfm (Biarrotte-Sorin et al., 2006).
The catalytic site of LdtMt2, located at the entrance of a narrow tunnel connecting two cavities that open to the solvent, is similar to that observed in Ldtfm (Biarrotte-Sorin et al., 2006). Unexpectedly, the crystal structure shows the presence of a fragment of the peptidoglycan of the expression host (E. coli) in the catalytic site of the enzyme. Until now, no related L,D-transpeptidase has been structurally characterized with a bound peptidoglycan fragment. LdtMt2 recognizes the peptidoglycan (acyl-donor-like) using residues located in both cavities, including the catalytic residues Cys354 and His336. The observed mode of binding of this short peptidoglycan fragment led us to propose a catalytic mechanism for these transpeptidases that differs from that previously posited (Biarrotte-Sorin et al., 2006).
Kinetic measurements show that LdtMt2 has residual hydrolase-activity towards certain β-lactams. Isothermal titration calorimetry experiments show that the carbapenems (Bonfiglio et al., 2002), imipenem and meropenen, bind only the catalytically active enzyme and they do so with apparent submicromolar affinities. The structural, biochemical, and thermodynamic data presented here provide vital information about the interactions of the enzyme with its substrates and inhibitors that will help advancing this unexploited enzyme into a target for the design of novel compounds for the treatment of Mtb infections.
Results
Ex-LdtMt2 crystal structure
The extramembrane portion of the L,D-transpeptidase from M. tuberculosis, LdtMt2 (ex-LdtMt2, residues 120 to 408), was cloned and expressed in E. coli. A crystal of a seleno-methionine (SeMet) derivative of ex-LdtMt2 was used to determine the structure using Multiple Anomalous Dispersion (MAD) methods. The structure was refined to a final Rwork of 0.19 and an Rfree of 0.23 using diffraction data collected from a di-μ-Iodo-bis[ethylenediamine]-di-Pt[II] (PIP) derivative crystal (Fig. 1B and Table 1; see also Fig S1), which had the best diffraction among all the crystals tested (1.7 Å resolution). Crystals of ex-LdtMt2 belong to the orthorhombic space group I212121 and contain two monomers (A and B) in the asymmetric unit. These two molecules do not correspond to a physiological dimer, since ex-LdtMt2 behaves as a monomer in solution as determined by size exclusion chromatography (data not shown). In addition, areas buried by the two possible dimers in the crystal asymmetric unit, 810 Å2 and 1360 Å2, are not consistent with a dimer stable in solution (Krissinel, 2011).
Table 1.
Data collection statistics of the PIP complex used to refine the structure and of the SeMet crystal used for phase determination. See also Table S1 for statistics of other crystal forms.
| Crystal | PIP (Refinement) | SeMet (phase determination) | ||
|---|---|---|---|---|
| Space group | I212121 | |||
| Wavelength | CuKα | 0.96785 Å | 0.97907 Å | 0.97939 Å |
| Cell dimensions | ||||
| a, b, c (Å) | 119.1, 120.8, 122.8 | 118.3, 121.1, 122.5 | ||
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Resolution range (Last shell) (Å) |
85.7-1.89 (1.89-1.72) | 50.0-2.78(2.83-2.78) | 50.0-2.45(2.49-2.45) | 50.0-2.54 (2.58-2.54) |
| Rsym | 0.051 (0.310) | 0.099 (0.683) | 0.095 (0.478) | 0.087 (0.615) |
| I/σ(I) | 29.2 (2.5) | 18.1 (1.7) | 22.6 (2.9) | 20.9 (2.0) |
| Completeness (%) |
99.6 (99.6) | 97.6 (83.0) | 97.8 (85.4) | 96.9 (80.5) |
| Redundancy | 3.5 (2.3) | 9.4 (6.6) | 9.4 (6.6) | 9.4 (6.0) |
| Refinement | ||||
| Resolution (Å) | 50 – 1.72(1.765-172) | |||
| No. reflections | 84134 (287) | |||
| Rwork/Rfree | 0.19/0.23 (0.3/0.32) | |||
| No. atoms | ||||
| Protein | 4075 | |||
| Ligands/ions | |111 | |||
| Peptoglycans | 44 | |||
| PIP adducts | |67 | |||
| Waters | 471 | |||
| B-factors (Å2) | ||||
| Protein | 36.9 | |||
| Ligands/ions | 67.1 | |||
| Waters | 49.7 | |||
| Rms deviations | ||||
| Bond lengths (Å) | 0.009 | |||
| Bond angles (°) | 1.297 | |||
| Ramachandran | ||||
| % Most favorable region |
98.5 | |||
| % allowed regions |
1.5 | |||
| % outliers | 0 | |||
A portion of the N-terminal region of ex-LdtMt2, spanning residues 122 to 132, is observed only in monomer B with low occupancy (50%) and disconnected from the bulk of the fold. Aminoacids from 150 to 407 are observed in both monomers of the crystal asymmetric unit with an rms deviation of 0.31 Å between the aligned 258 Cα-atoms of monomers A and B. Each monomer consists of two globular domains: an N-terminal domain (residues 150 to 250) folded as an anti-parallel β-barrel resembling an immunoglobulin domain (IgD) (Amzel and Poljak, 1979), and a C-terminal catalytic domain (CD, residues 254 to 407), consisting of a β-sandwich with two mixed β-sheets characteristic of the ErfK/YbiS/YhnG-fold (Bielnicki et al., 2006). A short linker (residues 251 to 253) joins the two domains (Fig. 1B). A small C-terminal sub-domain (CTSD; residues 379 to 407) extends the ErfK/YbiS/YhnG-fold. In this sub-domain, Trp394 and two Trp residues of the C-terminal helix α3 (398 and 401) make a zipper-like interaction with the IgD N-terminal domain that fixes the relative orientation of the two domains.
The N-terminal domain has characteristics not commonly found in Ig-domains (Amzel and Poljak, 1979): the interstrands loops are unusually long, including a three-turn helix (α1) in the loop connecting strands β3 and β4.
In the refined structure, PIP was found to have reacted with solvent exposed Nδ atoms of histidine residues and with sulfur atoms of methionine residues (His214 in monomer A, and Met153, Met157 Met237 and His347 in both monomers), to form five (four in monomer B) iodo-Pt(II) ethylenediamine adducts (Fig. S1) (O’Halloran et al., 1987).
Active site
A two-step enzymatic mechanism resembling that of D,D-transpeptidases was proposed for the ErfK/YbiS/YhnG-fold family of transpeptidases, except that, the catalytic serine of D,D-transpeptidases is replaced by an absolutely conserved cysteine residue (Mainardi et al., 2007). A highly conserved sequence motif of this group of L,D-transpeptidases, HXX14-17[S/T]HGChN (where ‘h’ stands for a hydrophobic residue, Fig. 2) contains three residues (in bold letters) analogous to the catalytic triad of cysteine-proteases: a cysteine, a histidine, and a third residue that accept an H-bond from the histidine stabilizing the competent imidazole tautomer (Bielnicki et al., 2006; Dodson and Wlodawer, 1998).
Figure 2.
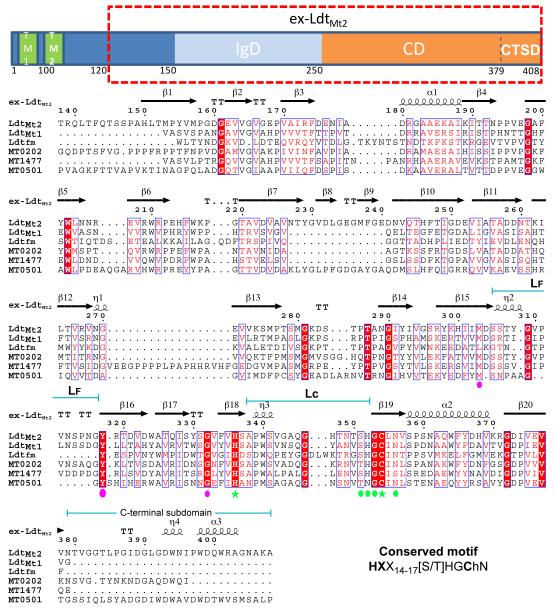
Sequence alignment of LdtMt2, the L,D-transpeptidase of E. faecium (1ZAT) and other Mtb LdtMt2 homologues. Top: domain distribution of LdtMt2. The ex-LdtMt2 protein construct used in this work is indicated by a dashed red box. Bottom: sequence alignment among amino acids 138 to 408 of LdtMt2, LdtMT1, Ldtfm and other LdtMt2 homologues in Mtb. The observed secondary structure is indicated above the corresponding residues. Identical residues are white with red background, regions scoring higher than 70 % using a Risler’s similarity scoring-matrix (Risler et al., 1988) are boxed with red letters. Residues involved in binding and catalysis are labeled: the conserved motif is indicated with green marks and the rest with pink marks. Stars label main catalytic residues and ovals label substrate recognition residues. The figure was made using the web server ESPript (http://espript.ibcp.fr/ESPript/ESPript/).
In ex-LdtMt2, those catalytic residues, His336, Ser337, and Cys354, reside under a flap formed by a long insert (residues 299 to 323) that is not part of the canonical ErfK/YbiS/YhnG-fold (Bielnicki et al., 2006). This insert, strand β15, loop LF and strand β16, folds over the β-sheet formed by strands β17, β18 and β19 (residues 324 to 357). The closed flap creates two large cavities (vestibules) connected by a narrow tunnel. These cavities are open to solvent, one at the end of the molecule, and the other at the interface between the CD and IgD (Fig. 1C, outer and inner cavities, respectively). Strand β16 (residues 318 to 323), the C-terminal end of β18 (residues 336 and 337), the loop Lc (residues 338 to 352) and the strand β19 (residues 353 to 357) line the outer cavity (Fig. 1D). The residues of the conserved motif (Fig. 2) are readily accessible through the outer cavity, with the catalytic residues located deep within it. With the exceptions of His336 and Ser337 (both in β18), the conserved motifs extend through the end of loop Lc and strand β19. Ser351, His352, and Gly353 are located in loop Lc while Cys354 and Asn356 in strand β19 (Fig. 1D). Other highly conserved residues in similar sequences of gram-positive bacteria, Tyr318 and Gly332 in LdtMt2 (Fig. 2), are located at the inner cavity. Tyr318 and Met303 line the internal side of the flap and, together with Cys354, form the walls of the narrow tunnel. His336 and His352 flank the tunnel at the outer cavity (Fig. 1D). The carbonyl group of Ser337 accepts a H-bond from the Nδ of His336 (2.8 Å). This H-bond stabilizes the tautomer of His336 protonated at Nδ.
Comparison of LdtMt2 with Mtb paralogs
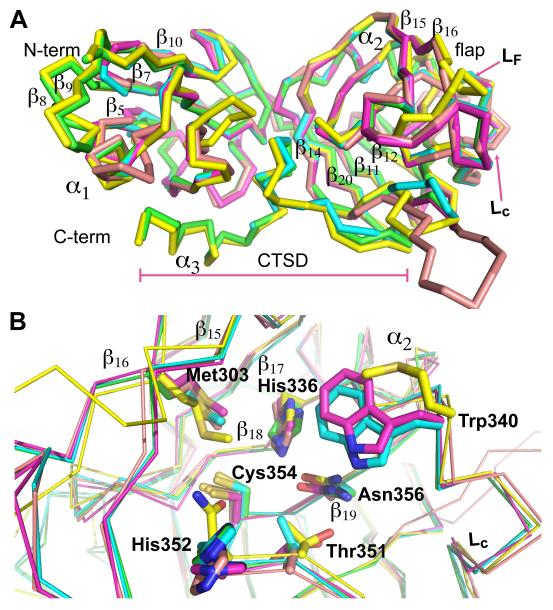
The presence of five genes in Mtb that code for proteins with potential L,D-transpeptidase activity has been reported (Gupta et al., 2010). Of these proteins, only LdtMt1 (Lavollay et al., 2008) and LdtMt2 (Gupta et al., 2010) have been shown to be functional L,D-transpeptidases. The multiple alignment of LdtMt2 with the other Mtb proteins shows pairwise sequence identities range from 33 % to 45 % (Fig. 2), suggesting a high structural similarity of these proteins to ex-LdtMt2. Comparative models of these proteins were built using LdtMt2 as a template (Fig. 3). The overall fold of the IgD is conserved among these proteins. Three homologues, MT0202 (also know as Rv0192, 45% identity with LdtMt2), LdtMt1 (36 %) and MT1477 (Rv1433, 33 %), have a short loop connecting strands β7 and β10 and lack strands β8 and β9 of the ex-LdtMt2 IgD (Fig. 2 and Fig. 3A). The bulk of the CD has little variation among homologues; MT0501 (Rv0483, 34 %) has the shortest loop LF among the five proteins and MT1477 shows an eighteen-residue insertion after β12. The C-terminal sub-domains of these homologues, however, show significant differences with LdtMt2. The only exception is MT0501, which contains a CTSD of the same size and features as LdtMt2. MT1477 and LdtMt1 completely lack this sub-domain and MT0202 has a significantly shortened version. Most of the motif residues (Fig. 2) are conserved in these proteins, with the exception of the replacement of His 352 (LdtMt2 numbering; Fig. 2) by an asparagine in MT0501. In the neighborhood of the active site a methionine in MT0501 replaces the highly conserved Trp340 of LdtMt2 (Fig. 2 and Fig. 3B).
Figure 3.
Comparative Modeling of Mtb homologues. (A) Overlay of Cα-trace of the modeled structures of the Mtb homologues with the crystal structure of ex-LdtMt2 (green). (B) Overlay of the modeled catalytic sites. Residues participating in the interactions with the substrate are displayed; carbons atoms are colored with the same color as the corresponding Cα-trace: Green, LdtMt2; yellow, MT1477; pink, MT0501; magenta, LdtMt1; and cyan, MT0202. Non-carbon atoms are colored as Fig. 1.
Peptidoglycan fragment
Electron density not belonging to protein residues was present in the catalytic site of the SeMet and the PIP derivative crystals. This density, longer and more branched than either a succinic acid or a PEG molecule from the crystallization media, was interpreted as belonging to a di-peptide fragment of a muropeptide stem, probably a fragment of the E. coli peptidoglycan bound from the lysis supernatant and carried through the purification process. Ex-LdtMt2 probably recognizes common features between its physiological substrate and a muropeptide of E. coli. The peptidoglycan of Mtb contains muropeptides composed of an N-acetylglucosamine joined by a β (1-4) glycosidic bond to an N-acetylmuramic acid decorated with 4 or 5 aminoacid-long peptide stems (NAcGlc-β[1,4]-MurNAc-L-Ala1-γ-D-GLu2-m-A2pm-NH23-D-Ala4 -[D-Alanyl5]) (Barreteau et al., 2008). The electron density, 14 Å in length, broadens at three places that correspond to carboxylates of the peptidoglycan (Fig. 4 A, see also Fig. S2). The broadest section, at the center of the catalytic site, is connected by narrow zig-zag densities to two broad densities located at the tunnel entrances across from the active site tunnel. Based on the separation and size of these features, a γ-D-Glu-m-A2pm dipeptide was built into this electron density (Fig. 4A), with the terminal peptide L chiral-center placed near the catalytic His336 and the D chiral-center in the inner cavity of the tunnel, hydrogen-bonded by Tyr318 and main-chain carboxyl of Gly332. The SH group of Cys354 is 4.6 Å from the carboxylate group of the dipeptide A2pm moiety (Fig. 4A). This carboxylate makes H-bonds with the Nε of His336 and the Nδ1 of Asn356, both residues of the conserved motif (Fig. 2). The γ-D-Glu-m-A2pm dipeptide may correspond to the second and third peptide moieties of a peptidoglycan stem.
Figure 4.
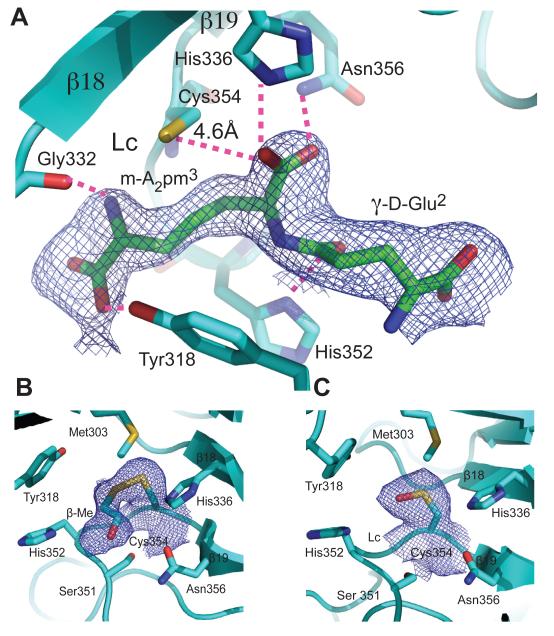
Electron densities at the catalytic site of LdtMt2. (A) Final refinement 2DFo - mFc map of the co-crystallized peptidoglycan fragment contoured at 0.5 σ (see below) around the γ-D-Glu-m-A2pm fragment at the active site of ex-LdtMt2. The fitted fragment, binding residues and the catalytic cysteine are shown. Hydrogen bond interactions with conserved residues are shown with pink dashed lines. As the two binding sites in the non-crystallographic dimer occur side-by-side, overlapping density for two equivalent peptidoglycan fragments was observed. The density was interpreted as two equivalent ligands related by the non-crystallographic two fold and refined as alternative orientations with occupancy of 0.5. (See also Fig. S2 for a stereo view of an omit map). (B-C) Modifications of Cys354 observed in native forms. (B) β-mercapto-ethanol covalently bound to Cys354 (2.0 Å bond distance). The rms deviation between this structure (native form) and the refined (PIP derivative) is 0.176 Å for 520 Cα-atoms of the asymmetric unit. Final refinement 2DFo – mFc electron density map contoured at 1 sigma around Cys354. (C) Oxidized Cys354 (suphenic acid form). Atoms are colored as Fig. 1.
Chemical modification of Cys354 in the absence of ligand
During the structural analysis, several of the structures of different native crystal forms showed modifications of Cys354 by buffer components or molecular oxygen (Fig. 4B and C; Table S1). When β-mercaptoethanol (β-Me) is present, a disulfide bridge is formed between the Sγ of Cys354 and β-Me (Fig. 4B). The rest of the molecule remains unperturbed (rms 0.17 Å for 524 Cα’s of the crystal asymmetric unit). In protein crystals without endogenous ligand (obtained by purifying the protein under denaturing conditions; see Supplemental Experimental Procedures), Cys354 is in its sulfenic acid form (Fig. 4C). The rest of the structure, including the active site does not show significant changes (rms 0.23 Å for 524 Cα’s of the crystal asymmetric unit).
Binding of carbapenems to wild type and C354A LdtMt2
Two classes of antibiotics inhibit polymerization of the cell wall peptidoglycan: β-lactams and glycopeptides. Carbapenems are a subfamily of β-lactam antibiotics that irreversibly inhibit classical D,D-transpeptidases (Bonfiglio et al., 2002). Three carbapenems -- imipenem (Fig. 5A), meropenem (Fig. 5B), and ertapenem -- have been shown to also be active against L,D-transpeptidases. Adducts of these carbapenems to Ldtfm (Mainardi et al., 2007), LdtMt1 (Lavollay et al., 2008) and LdtMt2 (Gupta et al., 2010) were identified using mass spectrometry.
Figure 5.
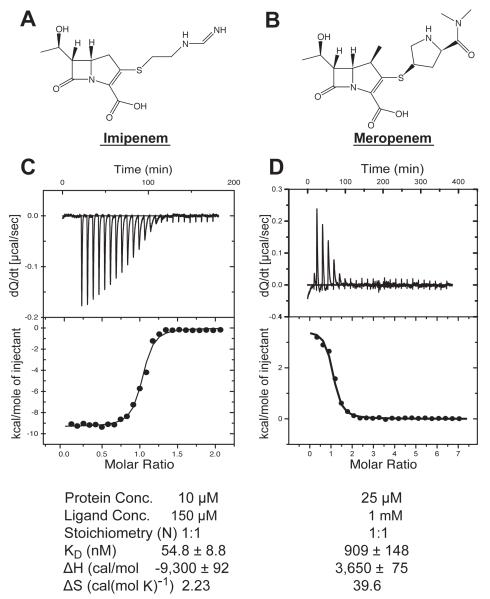
Isothermal titration calorimetry experiments. The chemical structures of (A) Imipenem ((5R,6S)-6-[(1R)-1-hydroxymethyl]-3-({2-[iminomethyl)amino]ethyl}thio)-7-oxo-1 azabicyclo[3.2.0]hept-2-ene-2-carboxylic acid), and (B) meropenem (3-[5-(dimethylcarbamoyl) pyrrolidin-2-yl] sulfanyl-6-(1-hydroxyethyl)-4-methyl-7-oxo-1-azabicyclo[3.2.0]-hept-2-ene-2-carboxylic acid) are shown. Titrations of (C) imipenem and (D) meropenem binding to LdtMt2 are shown in the upper portion of the panels. The lower portion of the panel displays plots of the total heat released as a function of the molar ratio between each carbapenem and LdtMt2. The solid line represents the non-linear least square fit of the data using a single-site binding model. Thermodynamic parameters of binding to ex-LdtMt2 --KD, ΔH and ΔS-- are tabulated at the bottom of the figure.
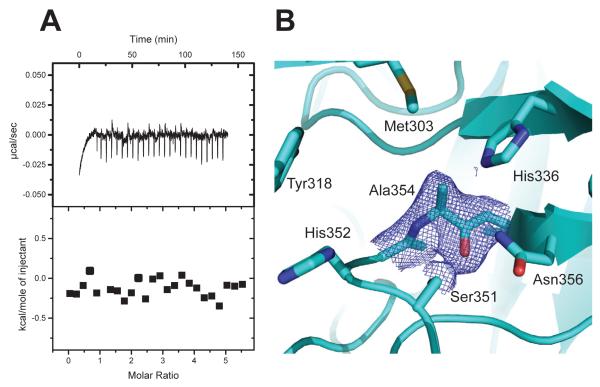
To characterize direct binding of imipenem and meropenem to LdtMt2 in the absence of enzymatic activity, we performed isothermal titration calorimetry using a catalytically inactive mutant C354A. Surprisingly, no heat of complex formation was observed with the mutant protein with either imipenem (Fig. 6A) or meropenem (data not shown) at ligand concentrations up to the mM range. In contrast, ITC experiments with the wild-type ex-LdtMt2 show strong binding of both carbapenems to the enzyme (Fig. 5C, D).
Figure 6.
C354A mutant. (A) ITC of Imipenem binding to the C354A mutant. Upper panel: Titration of the C354 mutant with imipenem. Lower panel: plot of the total heat released as a function of total ligand concentration for the titration shown in the upper panel. No heat of binding is detected, only heat of dilution. (B) Refined electron density of the C354A mutant around the mutated residue. Final refinement 2DFo – mFc map contoured at 1 σ around Ala354 of the mutant. Carbons atoms are colored in cyan, the rest as Fig. 1.
Lack of binding by the C354A mutant is not due to a large structural change: the crystal structure of the mutant (Fig. 6B, Table S1) shows no significant changes with respect to the wild-type (rms deviation of 0.26 Å for 516 Cα-atoms of the asymmetric unit). The absence of a thiol group at the residue 354 slightly widens the narrowest section of the tunnel, producing only minor disturbances in other binding site residues not enough change to explain the lack of binding (Fig 6B).
Imipenem has approximately 17 times higher affinity for the enzyme than meropenem (Fig. 5 C, D). While binding of imipenem to LdtMt2 is exothermic, the same reaction with meropenem is endothermic (Fig. 5C, D). The meropenem positive ΔS of binding, a consequence of its large hydrophobic surface area, partially compensates for the unfavorable enthalpic contribution.
In addition to meropenem and imipenem, a potent β-lactamase inhibitor, clavulanic-acid, was tested as ligand of ex-LdtMt2. ITC experiments did not detect binding by this compound to wild-type ex-LdtMt2 even at concentrations in the mM range (data not shown). The β-Me disulfide-adduct (Fig. 4B) observed when protein samples are treated with β-Me interferes with imipenem and meropenem binding: the KD of these compounds increases an order of magnitude (data not shown).
β – Lactamase activity of ex- LdtMt2
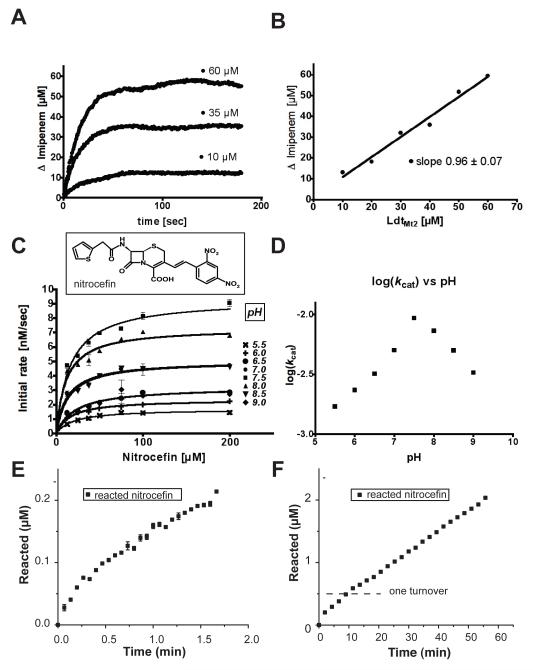
The kinetics of opening the lactam-ring of imipenem by ex-LdtMt2 displays the expected irreversible inhibitor characteristics (Fig. 7A, B). The enzymatic reaction stops after one turnover. The ratio of the total amount of imipenem rings opened to the amount of enzyme added is close to one (0.96 ± 0.07, Fig. 7B).
Figure 7.
Enzymatic activity of LdtMt2. (A) and (B) Irreversible inhibition by iminopenem. (A) three selected concentrations of enzyme (10, 35 and 60 μM) reacting with 100 μM of imipenem. (B) Plot of the concentration of imipenem reacted versus the concentration of enzyme used (10, 20, 35, 40, 50 and 60 μM). (C) Initial-rate experiments of the ex-LdtMt2 β-lactamase activity using nitrocefin as substrate (structural formula show in the Figure). Solid lines are the non-linear fit of the initial rate to the Michaelis-Menten equation. (Note that pH 7 and 8.5, and 6.5 and 9 overlap.) (D) Plot of the log of kcat for nitrocefin as a function of pH. See also Table S2. (E-F) Progress of the reaction of nitrocefin (100 μM) and LdtMt2 (500 nM). (E) Initial phase of the reaction (controlled by binding and acylation rates). (F) Later phase of the reaction (controlled by de-acylation rate). The amount of product (open ring) is monitored by its absorption at 486 nM.
To further characterize the enzyme, the β-lactamase activity of ex-LdtMt2 was measured using the chromogenic substrate nitrocefin as a reporter (O’Callaghan et al., 1972). A low β-lactamase activity was observed, with a Km of 16 μM and a kcat of 0.01 sec−1 (Fig. 7C; see also Table S2). The pH profile of the reaction shows a maximum catalytic rate at pH 7.8 (Fig. 7D, Table S3). The enzymatic reaction shows the biphasic kinetics observed before in β-lactamases (Fig. 7E and F).
An initial burst of activity (Fig. 7E) is followed by a linear slower rate (kde-acylation = 33.4 nM/min; Fig. 7F) characteristic of a bifurcated kinetic path consistent with the formation of an alternative, more labile, acyl-enzyme intermediate species (Monks and Waley, 1988).
Two pieces of evidence reduce the possibility that contamination with β-lactamases from the bacterial expression-host is the source of the weak activity observed. First, the C354A mutant of ex-LdtMt2 expressed and purified the same way as the wild-type shows no detectable β-lactamase activity (data not shown). Second, no inhibition of ex-LdtMt2 β-lactamase activity was detected with clavulanic acid (data not shown), that at the concentrations tested (ten of mMs) should have inhibited most knwon bacterial β-lactamases.
Discussion
The degree of cross-linking and proportion of each type of linkage in the peptidoglycan layer vary among different bacterial species as well as during different growth phases. For example, the degree of cross-linking of the E. coli peptidoglycan is approximately 50%, with 4→3 linkages accounting for 95% (90% in stationary phase) of the linkages (Vollmer and Holtje, 2004). In contrast, the degree of cross-linking of the Mycobacterium spp peptidoglycan is as high as 80 %, with 3→3 linkages comprising 70% (80% in stationary phase) of total linkages (Matsuhashi, 1966). This high frequency of 3→3 linkages in mycobacteria contrasts sharply with the low level commonly observed in most other bacteria. For example, in E. coli and E. faecium, 3→3 linkages represent only a relatively small fraction of the total --5 % during growth phases and 10 % in stationary phase (Tuomanen and Cozens, 1987).
During the stationary phase of growth, formation of new 4→3 linkages is impaired because the mature peptidoglycan layer has few pentapeptide stems, the required substrates of D,D-transpeptidases, and the de novo synthesis of precursors is significantly reduced (Goffin and Ghuysen, 2002). In contrast, L,D-transpeptidases could still cross-link the available tetrapeptide donors.
Expression of the Mtb L,D transpeptidase LdtMt1 is upregulated in the transcriptome of laboratory models of persisters (Betts et al., 2002; Keren et al., 2011) and shows a seventeen-fold increase during the activation of the dormancy regulon of Mtb (Lavollay et al., 2008). Interestingly, in all growth phases of Mtb, expression of LdtMt2 is ten fold higher than that of LdtMt1 (Gupta et al., 2010). These data underscore the importance of L,D-transpeptidase activity as a chemotherapeutic target.
Structural comparison with other L,D-transpeptidases
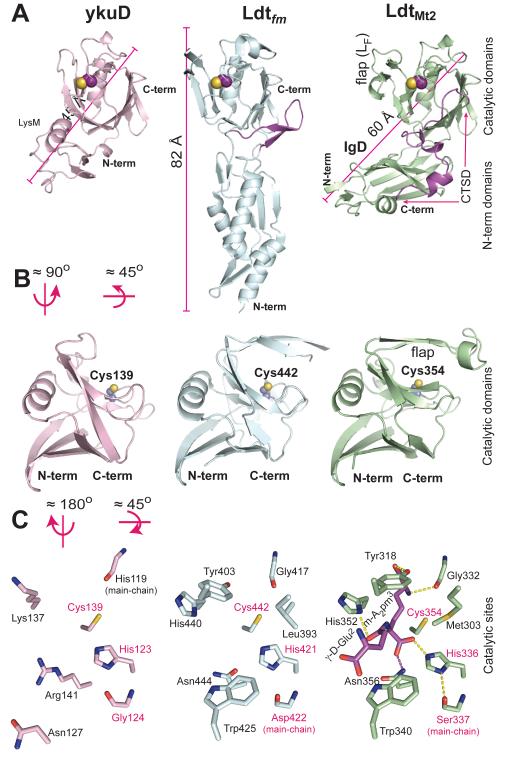
The structure determined here shows that the bulk of the ex-LdtMt2 CD (251-379 aa) has a fold similar to those of the C-terminal domain of B. subtilis ykuD (Bielnicki et al., 2006) and the catalytic domain of the L,D-transpeptidase of E. faecium Ldtfm (Biarrotte-Sorin et al., 2006). LdtMt2 shows 23 % sequence identity with Ldtfm, mostly concentrated in the CD. Despite having similar two-domain architectures and catalytic domains, the three enzymes differ significantly in their N-terminal domains (Fig. 8). YkuD shows a small N-terminal domain with a fold, LysM, that is associated with cell wall degradation (Bateman and Bycroft, 2000). The Ldtfm stalk domain is a 60 Å long cylinder formed by a four α-helix bundle, with each helix of the bundle connected by a small two-strand antiparallel β-sheet (Fig. 8A). The relative orientation of the two domains of Ldtfm is maintained by a long loop that follows the third α-helix of the stalk domain resulting in an 82 Å long cylindrical molecule (Fig. 8A). In contrast, the orientation of the two domains of ex-LdtMt2 is maintained by direct interaction of residues of the CTSD with residues of the IgD, resulting in a maximum dimension of only 60 Å (Fig. 8A).
Figure 8.
Structural comparison among L,D transpeptidases from B. subtilus, E. faecium and M. tuberculosis. (A) Side by side ribbon diagrams of the crystal structures of the L,D-transpeptidases: ykuD (PDB id. 1Y7M) in pink (rms deviation of 0.97 Å between 104 Cα-atoms aligned with ex-LdtMt2), Ldtfm (PDB id. 1ZAT) in cyan (1.0 Å rms deviation between 116 Cα-atoms aligned) and LdtMt2 in green. In the E faecium and Mtb case, the regions that apparently fix the domains orientation are colored in dark green. The catalytic domains of the three enzymes were used to align the structures. (B) Comparison of the catalytic domains. The catalytic cysteine residue is depicted in a CPK representation with carbon atoms purple and sulphur atom yellow. (C) Catalytic site residues of the three enzymes. View them from the outer cavity; the catalytic site residues are shown as side-chains or main-chains according to their expected function. The catalytic site triad is labeled with pink letters. The γ-glutamyl-meso-diaminopymelic acid (γ-D-Glu-m-A2pm) is shown only for ldtMt2 in a stick representation with magenta colored carbon atoms. Non-carbon atoms are colored as Fig. 1. H-bond interactions of the peptidoglycan fragment with residues of the catalytic site are shown with dashed lines.
The solvent accessibility of the catalytic site is very different in these three bacterial enzymes: ykuD shows a completely open site, whereas LdtMt2 and Ldtfm show a site cover by the β-hairpin flap (Fig. 8B). The β-hairpin flap of LdtMt2 is similar to that of Ldtfm, although it has a seven residues longer loop LF that narrows the inner cavity of LdtMt2. The catalytic site of ykuD shows the greatest divergence of the three sites (Fig. 8C). Although, these three enzymes preserve a similar arrangement of the catalytic triad residues (Fig. 8C). Most of LdtMt2 and Ldtfm catalytic site residues are conserved with only a few changes that (nevertheless) maintain the overall shape of the site (Fig. 8C).
Mtb LdtMt2 homologues
Comparative modeling of LdtMt2 homologues in Mtb predicts localized variations in their IgD and CD that may perturb the IgD-CD interaction. Loop modifications at the interface between the IgD and CD, and the loss of CTSD interaction with the IgD (Fig. 3A), could change each enzyme’s overall shape and flexibility. It is worth noting that these homologues are membrane-associated proteins and have different levels of expression during different phases of growth (Gupta et al., 2010). Their different shape and flexibility could reflect the fact that these enzymes are specialized to target different peptidoglycan layer constituents, act at different levels in the three-dimensional peptidoglycan layer (Vollmer and Holtje, 2004), or form a particular cross-linking pattern by acting only on stems that present a spatial arrangement that is optimum for a given enzyme. These multiple enzymes may provide a mechanism for modulating the three-dimensional structure of the peptidoglycan layer during different phases of growth.
Binding of Imipenem to LdtMt2
The shape of the binding isotherm indicates the presence of equilibrium between bound and unbound species (Fig. 5C). Kinetics experiments using fluorescence and absorption spectrophotometric-techniques to follow the inactivation by imipenem of two closely related enzymes, Ldtfm (Triboulet et al., 2011) and LdtMt1 (Dubee et al., 2012), showed the presence of an enzyme-inhibitor complex before the irreversible acylation. Such complex is consistent with the apparent tight binding of imipenem to LdtMt2 we observed using ITC (KD = 54.8 ± 8.8 nM). The observation of a tight complex enzyme-inhibitor previous to enzyme irreversible acylation, the one-to-one relation between inactivated enzyme and consumed inhibitor (Fig. 7A, B), and the lack of binding of the inhibitor to the catalytically inactive C354A mutant suggest that the formation of a reversible tetrahedral intermediate (see below) is required for a strong binding of imipenem to LdtMt2.
LdtMt2 catalytic mechanism
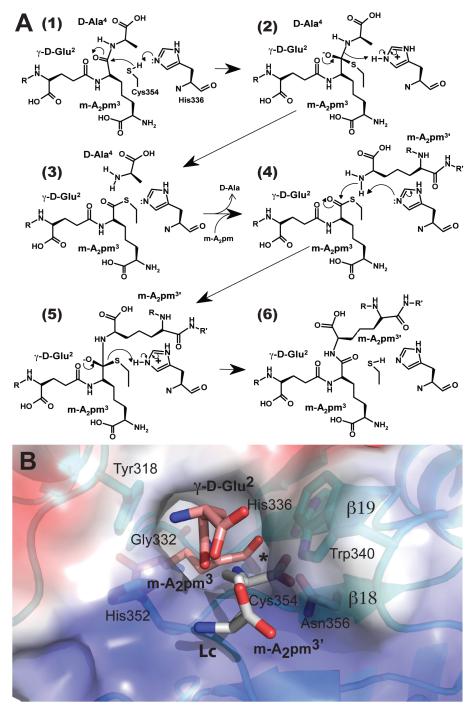
Biarrotte-Sorin et al. (2006) proposed that in this kind of L,D-transpeptidases the two paths to the catalytic cysteine (via the inner or the outer cavity; Fig. 1C), are used one for the acyl-donor and the other for the acyl-acceptor substrates. However in this mechanism, the enzyme would need to go through multiple conformational changes of the flap to accommodate entrance of substrates and release of products using the catalytic site tunnel. The mode of binding of the peptidoglycan fragment found in the structure of LdtMt2 suggests a much simpler mechanism, in which the acyl-donor peptidoglycan stem binds to both cavities through the tunnel (Fig. 1C and Fig. 4) and remains bound during most of the catalytic cycle, reducing the need for conformational changes during the catalytic cycle of the enzyme. We built models of the peptidoglycan-stem substrates bound to the active site of the enzyme based on the structure of LdtMt2 and use it to propose atomic details of the catalytic mechanism of LdtMt2 (Fig. 9). In this model, the catalytic site tunnel and features at either side of the tunnel tightly bind the stem di-aminoacid (A2pm3). At one end of the tunnel, within the inner cavity, Tyr318 and the carbonyl group of Gly332 recognize the D chiral-center of the di-aminoacid side chain, making hydrogen bonds to its carboxylate and amide group. The tunnel recognizes the aliphatic portion of the side chain. At the other end, within the outer cavity, the L chiral-center is surrounded by an “anion hole” formed by the backbone NH-groups of residues 352 to 354 at the C-terminus of loop Lc. In this position, the acyl-group of the m-A2pm3 L chiral-center is within reach of Cys354. These features are consistent with a nucleophilic attack by this cysteine on the acyl-carbon.
Figure 9.
Proposed mechanism of the L,D-transpeptidation reaction and structural model of the acyl-acceptor and donor-peptidoglycan stems bound to the outer cavity of ex-LdtMt2. (A) Steps 1 – 6 of the mechanism of (3,3) L,D-transpeptidation by LdtMt2. (B) The figure represents step 4 of the mechanism in part A, after the cleaved D-Ala4 has left and a thioester intermediate is formed between acyl-donor (carbon atoms colored pink) and the enzyme (cyan). Non-carbon atoms are colored as Fig. 1. Only the last two residues of the donor stem and the m-A2pm (light gray) of the acceptor stem are shown. The peptidoglycan stem of the acyl-acceptor binds to the site with its A2pm3′ side chain (D chiral-center) near the thioester bond (marked with an *). (B) Solvent accessible surface at the outer cavity. The surface is colored by electrostatic surface-potential (from blue positive to red negative). The two substrates of the bi-substrate reaction are shown.
In the crystal structure of ex-LdtMt2, the Sγ atom of Cys354 points toward the inside entrance of the catalytic site, away from the putative acyl-group. Identical cysteine conformations (χ ≅ −73°) have been consistently observed in all the other crystal structures of ErfK/YbiS/YhnG-domain proteins (Biarrotte-Sorin et al., 2006; Bielnicki et al., 2006). Therefore, one can expect that earlier in the catalytic cycle the cysteine may adopt another conformation (χ ≅ 58°) that points toward the carbonyl carbon of the scissile bond. In this conformation the SH group would be briefly at 2.9 Å from the Nε atom of His336, allowing deprotonation of the Cys354 SH-group for the nucleophilic attack at the peptide carbonyl group. The imidazolium cation of the protonated His336 may then be the donor for the protonation of the departing D-Ala amide. His336 and Asn356 make hydrogen bonds to this CO-group, providing further recognition of the donor-stem L chiral-center (Fig. 4A). The pH dependence of the reaction rate shows an increase of β-lactamase activity compatible with the ionization of a group with a pKa of around 7.8 (Fig. 6D, see also Table S3). Participation of the Cys354-thiolate/His336-Imidazolium ion pair in the concerted acid-base catalysis of the acyl-transfer is compatible with this pH optimum.
The rest of the interactions between the LdtMt2 and the bound peptidoglycan fragment involve the side chain of Asn356, which coordinates the main-chain NH-group and the terminal carboxylate of D-Ala4 (D-Ala4 binding site). Trp340 provides steric constraints to the stem terminus that may explain the preference of the enzyme for tetrapeptide stems (Lavollay et al., 2008). Interestingly, in one of the Mtb homologues of LdtMt2, MT0501, a less bulky methionine residue replaces Trp340 (Fig. 3B), suggesting that this enzyme can accommodate pentapeptides, perhaps broadening its specificity from D-Ala to non-canonical D-aminoacids observed by Cava et al. (2011). Towards the N-terminus of the peptidoglycan stem, the conserved His352, provides a hydrogen bond to groups of the iso-glutamyl side-chain.
In the observed tautomer, forced by the hydrogen bond to the carbonyl of Ser337, His336 has its Nε ready to accept a H+ from Cys354 (Fig. 9A step 1). The cysteine thiolate formed by this H+ abstraction attacks the acyl-carbon and forms a tetrahedral intermediate stabilized by the “anion hole” and by the just protonated His336 (Fig. 9A step 2). After the intermediate thioester formation and protonation by His336, D-Ala4 is released (Fig. 9A step 3). Then another peptide stem can enter the catalytic site, also through the external vestibule, and bind to active site residues with the side-chain amide of the A2pm3′ residue (also a D chiral-center and isomorphic to D-Ala4), (Fig. 9B). His336 is in position to activate the amine group of the acceptor stem D chiral-center (Fig 9A step 4). Nucleophilic attack by this amine group, forms the new peptide bond that cross-links the two stems with release and protonation of the cysteine thiolate by His336 (Fig. 9 step 5-6). It is clear that the last step in the transpeptidation reaction requires that donor and acceptor peptidoglycan stems both be at the catalytic site for the reaction to occur (Fig. 9B). The wide and shallow entrance of the outer cavity has the required characteristics to accommodate both peptidoglycans stems (Fig. 9B).
Higher order cross-linked stems are also observed in Mtb, though with low frequency (3 %) (Lavollay et al., 2008), suggesting that already linked peptidoglycan stems are not precluded from participating in new cross-links. Linked stems can dock in the vestibule with their unreacted D chiral-center reaching the catalytic residues without any conformational change in the enzyme. The use of the unreacted L chiral-center of an already crosslinked stem as acyl-donor imposes more steric constrains and most likely involves the opening of the flap. After formation of the new cross-link, release of the higher-order linked peptide stems will require opening the flap again.
Concluding Remarks
The crystal structure of the complex of a Mtb L,D-transpeptidase with a peptidoglycan reported here provides a three-dimensional map of the extensive interactions between the peptidoglycan fragment and LdtMt2, involving residues that are conserved in all members of this Mtb family of L,D-transpeptidases. Knowing this map is an important step toward a rational design of high affinity drugs engineered to inhibit L,D-transpeptidase activity.
Experimental Procedures
Protein preparation, purification and crystallization
A truncated construct of MT2594 (Rv2518c) spanning residues 120-408 (ex-LdtMt2) was expressed in E. Coli BL21(DE3) strains and in a methionine auxotroph strain supplemented with SeMet. Proteins were separated from the lysates using (Ni)-affinity chromatography (Ni-Histrap GE HealthCare Inc.). Size exclusion chromatography was performed using a sepharose column (Sepharose 12 GE HealthCare Inc.). Native and SeMet-derivatized protein samples were used in the crystallization trials. Diffraction quality crystals of all forms used in this study were obtained in 48 hrs at 20 °C with hanging drops vapor diffusion methods. Drops of 1μl of protein (20 mg/ml) and 1μl of reservoir solution were equilibrated against a reservoir containing 0.1 M HEPES (pH 7.5), 1 M succinic acid and 1% (w/v) PEG MME 2000, except for SeMet crystals in where HEPES at pH 7.0 was used. For heavy atom derivates, crystals were and soaked in 10 μl of reservoir solution containing the heavy atom compounds at the desire concentration. In the case of PIP (di-μ-iodo-bis(ethylenediamine)-di-Pt(II)), ex-LdtMt2 crystals were soaked with 100 μM for 48 hrs. (See Supplemental Experimental procedures for additional details)
Data Collection, Structure Determination and Refinement
Diffraction data of the PIP derivative, mutant, and β-Me disulfide-adduct and oxidized forms of the native were collected using an in house CuKα X-ray source (FRE+ super bright ™ generator with a Saturn 944+ CCD Detector; Rigaku USA, Inc.). The SeMet-derivative crystal used in the phase determination was collected on beamline X4A of the National Synchrotron Light Source of the Brookhaven National Laboratory (Table 1). X-ray diffraction experiments were carried out under cryogenic conditions after transferring the crystal into crystallization solution containing 10% v/v glycerol. Intensities were indexed and integrated using HKL2000 (HKL Research Inc). Data were scaled with the CCP4 program suite (CCP4, 1994). Phases were determined using the program autoSHARP (Vonrhein et al., 2007). Refinement was carried out using REFMAC5 (Murshudov et al., 2011) and rebuilding was done with Coot (Emsley et al., 2004).
Calorimetric Studies
Isothermal Titration Calorimetry (ITC) experiments were performed using a high-precision VP-ITC titration calorimeter system (Microcal Inc.). The buffer of the protein samples was replaced by 20 mM HEPES 300 mM NaCl pH 7.5 using a desalting column (HiTrap G-25, GE HealthCare Inc.). Inhibitors and substrates used in these experiments were dissolved in the same buffer. ITC experiments using PBS and PIPES showed similar ΔH values, indicating that no H+s were released or bound during the reaction. Titration experiments consisted in the addition of 25 injections of 10 μl of concentrated inhibitor into the protein solution contained in the cell (1.38 ml) with 300 to 450 sec equilibration between injections. Data were analyzed with Origin 5.0 software (Microcal Software, Inc., Northampton, MA) with a single site model. Imipenem monohydrate and meropenem were purchased from SIGMA Inc., and ertapenem was purchased from Merck Sharpe and Dhome-Chibret.
Enzyme Kinetics
Initial rate measurements were used to determine the steady state kinetic parameters of the β-lactamase activity against nitrocefin —3-(2,4-Dinitrostyryl)-(6R, 7R)-7-(2-thienylacetamido)-ceph-3-em-4-carboxylic Acid, EMD Inc., (Fig. 7D)— a chromogenic β-lactam. The β-Lactamase activity was measured by following the absorbance of the product produced by opening the lactam-ring at 486 nm using a molar extinction coefficient of 20500 M−1cm−1 (O’Callaghan et al., 1972) in metal free buffer of 20 mM HEPES at pH 7.5 and 300 mM NaCl. Nitrocefin was added at concentrations ranging from 12.5 to 200 μM. The kinetic parameters, Km and kcat were fitted by a nonlinear least-squares fit of Michaelis-Menten equation the to the initial rate measurements.
Enzyme-acylation by imipenem was measured by quantifying the amount of reacted imipenem at a given time, following the formation of the open penem-ring product by monitoring the absorbance at 299 nm. The decrease in absorbance values was converted into concentration units using the difference between molar extinction coefficients of the open and closed penem-ring of -7100 M−1cm−1 (Triboulet et al., 2011).
Supplementary Material
The structure of Mtb LdtMt2 shows the binding of a peptidoglycan fragment.
The structure of the complex maps the interaction between acyl-donor and enzyme.
Ldts show a conserved C-terminal ykuD domain with a divergent N-terminal domain.
Based on the structure of this complex a reaction mechanism is proposed.
Acknowledgements
Use of beam line X4A and X4C at Brookhaven National Laboratoty NSLS and the technical support of John Schwanof and Randy Abramowitz are acknowledged. Use of the Advanced Photon Source at Argonne National Laboratory during early stages of this crystal structure determination was supported by the U. S. Department of Energy, Office of Science, and Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. Eli Lilly & Company, who operates the facility, provided the use of the SGX Collaborative Access Team (SGX-CAT) beamline at Sector 31 of the Advanced Photon Source. This work was supported by NIH grant 1R56AI087749 and DP2OD008459 to GL.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Amzel LM, Poljak RJ. Three-dimensional structure of immunoglobulins. Annu Rev Biochem. 1979;48:961–997. doi: 10.1146/annurev.bi.48.070179.004525. [DOI] [PubMed] [Google Scholar]
- ATS Diagnosis and treatment of disease caused by nontuberculous mycobacteria. This official statement of the American Thoracic Society was approved by the Board of Directors, March 1997. Medical Section of the American Lung Association. Am J Respir Crit Care Med. 1997;156:S1–25. doi: 10.1164/ajrccm.156.2.atsstatement. [DOI] [PubMed] [Google Scholar]
- Barreteau H, Kovac A, Boniface A, Sova M, Gobec S, Blanot D. Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol Rev. 2008;32:168–207. doi: 10.1111/j.1574-6976.2008.00104.x. [DOI] [PubMed] [Google Scholar]
- Bateman A, Bycroft M. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD) J Mol Biol. 2000;299:1113–1119. doi: 10.1006/jmbi.2000.3778. [DOI] [PubMed] [Google Scholar]
- Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Molecular Microbiology. 2002;43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- Biarrotte-Sorin S, Hugonnet JE, Delfosse V, Mainardi JL, Gutmann L, Arthur M, Mayer C. Crystal structure of a novel beta-lactam-insensitive peptidoglycan transpeptidase. J Mol Biol. 2006;359:533–538. doi: 10.1016/j.jmb.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Bielnicki J, Devedjiev Y, Derewenda U, Dauter Z, Joachimiak A, Derewenda ZS. B. subtilis ykuD protein at 2.0 A resolution: insights into the structure and function of a novel, ubiquitous family of bacterial enzymes. Proteins. 2006;62:144–151. doi: 10.1002/prot.20702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishai W. Lipid lunch for persistent pathogen. Nature. 2000;406:683–685. doi: 10.1038/35021159. [DOI] [PubMed] [Google Scholar]
- Bonfiglio G, Russo G, Nicoletti G. Recent developments in carbapenems. Expert Opin Investig Drugs. 2002;11:529–544. doi: 10.1517/13543784.11.4.529. [DOI] [PubMed] [Google Scholar]
- Cava F, de Pedro MA, Lam H, Davis BM, Waldor MK. Distinct pathways for modification of the bacterial cell wall by non-canonical D-amino acids. EMBO J. 2011;30:3442–3453. doi: 10.1038/emboj.2011.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CCP4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Dodson G, Wlodawer A. Catalytic triads and their relatives. Trends in Biochemical Sciences. 1998;23:347–352. doi: 10.1016/s0968-0004(98)01254-7. [DOI] [PubMed] [Google Scholar]
- Dubee V, Triboulet S, Mainardi JL, Etheve-Quelquejeu M, Gutmann L, Marie A, Dubost L, Hugonnet JE, Arthur M. Inactivation of Mycobacterium tuberculosis L,D-Transpeptidase LdtMt1 by Carbapenems and Cephalosporins. Antimicrob Agents Chemother. 2012;56:4189–4195. doi: 10.1128/AAC.00665-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K, Lohkamp B, Scott WG. Coot: model-building tools for molecular graphics. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fauci AS. Multidrug-resistant and extensively drug-resistant tuberculosis: the National Institute of Allergy and Infectious Diseases Research agenda and recommendations for priority research. J Infect Dis. 2008;197:1493–1498. doi: 10.1086/587904. [DOI] [PubMed] [Google Scholar]
- Goffin C, Ghuysen JM. Biochemistry and comparative genomics of SxxK superfamily acyltransferases offer a clue to the mycobacterial paradox: presence of penicillin-susceptible target proteins versus lack of efficiency of penicillin as therapeutic agent. Microbiol Mol Biol Rev. 2002;66:702–738. doi: 10.1128/MMBR.66.4.702-738.2002. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Lavollay M, Mainardi JL, Arthur M, Bishai WR, Lamichhane G. The Mycobacterium tuberculosis protein LdtMt2 is a nonclassical transpeptidase required for virulence and resistance to amoxicillin. Nat Med. 2010;16:466–469. doi: 10.1038/nm.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugonnet JE. Efficient combination of clavulanate and beta-lactam antibiotics against extensively drug-resistant M. tuberculosis. Med Sci (Paris) 2009;25:661–663. doi: 10.1051/medsci/2009258-9661. [DOI] [PubMed] [Google Scholar]
- Hugonnet JE, Blanchard JS. Irreversible inhibition of the Mycobacterium tuberculosis beta-lactamase by clavulanate. Biochemistry. 2007;46:11998–12004. doi: 10.1021/bi701506h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jindani A, Dore CJ, Mitchison DA. Bactericidal and sterilizing activities of antituberculosis drugs during the first 14 days. Am J Respir Crit Care Med. 2003;167:1348–1354. doi: 10.1164/rccm.200210-1125OC. [DOI] [PubMed] [Google Scholar]
- Keren I, Minami S, Rubin E, Lewis K. Characterization and Transcriptome Analysis of Mycobacterium tuberculosis Persisters. MBio. 2011;2:00100–00111. doi: 10.1128/mBio.00100-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E. Macromolecular complexes in crystals and solutions. Acta Crystallogr D Biol Crystallogr. 2011;67:376–385. doi: 10.1107/S0907444911007232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavollay M, Arthur M, Fourgeaud M, Dubost L, Marie A, Veziris N, Blanot D, Gutmann L, Mainardi JL. The peptidoglycan of stationary-phase Mycobacterium tuberculosis predominantly contains cross-links generated by L,D-transpeptidation. J Bacteriol. 2008;190:4360–4366. doi: 10.1128/JB.00239-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavollay M, Fourgeaud M, Herrmann JL, Dubost L, Marie A, Gutmann L, Arthur M, Mainardi JL. The peptidoglycan of Mycobacterium abscessus is predominantly cross-linked by L,D-transpeptidases. J Bacteriol. 2011;193:778–782. doi: 10.1128/JB.00606-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K. Persister cells Persister cells, dormancy and infectious disease. Annu Rev Microbiol. 2007;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- Mainardi JL, Fourgeaud M, Hugonnet JE, Dubost L, Brouard JP, Ouazzani J, Rice LB, Gutmann L, Arthur M. A novel peptidoglycan cross-linking enzyme for a beta-lactam-resistant transpeptidation pathway. J Biol Chem. 2005;280:38146–38152. doi: 10.1074/jbc.M507384200. [DOI] [PubMed] [Google Scholar]
- Mainardi JL, Hugonnet JE, Rusconi F, Fourgeaud M, Dubost L, Moumi AN, Delfosse V, Mayer C, Gutmann L, Rice LB, Arthur M. Unexpected inhibition of peptidoglycan LD-transpeptidase from Enterococcus faecium by the beta-lactam imipenem. J Biol Chem. 2007;282:30414–30422. doi: 10.1074/jbc.M704286200. [DOI] [PubMed] [Google Scholar]
- Matsuhashi M. Biosynthesis in the bacterial cell wall. Tanpakushitsu Kakusan Koso. 1966;11:875–886. [PubMed] [Google Scholar]
- Monks J, Waley SG. Imipenem as substrate and inhibitor of beta-lactamases. Biochem J. 1988;253:323–328. doi: 10.1042/bj2530323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan ST, Lamichhane G. Protective efficacy of BCG overexpressing an L,D-transpeptidase against M. tuberculosis infection. PLoS One. 2011;5:e13773. doi: 10.1371/journal.pone.0013773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Callaghan CH, Morris A, Kirby SM, Shingler AH. Novel method for detection of beta-lactamases by using a chromogenic cephalosporin substrate. Antimicrob Agents Chemother. 1972;1:283–288. doi: 10.1128/aac.1.4.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Halloran TV, Lippard SJ, Richmond TJ, Klug A. Multiple heavy-atom reagents for macromolecular X-ray structure determination. Application to the nucleosome core particle. J Mol Biol. 1987;194:705–712. doi: 10.1016/0022-2836(87)90248-8. [DOI] [PubMed] [Google Scholar]
- Peltier J, Courtin P, El Meouche I, Lemee L, Chapot-Chartier MP, Pons JL. Clostridium difficile has an original peptidoglycan structure with high level of N-acetylglucosamine deacetylation and mainly 3-3 cross-links. J Biol Chem. 2011;17 doi: 10.1074/jbc.M111.259150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risler JL, Delorme MO, Delacroix H, Henaut A. Amino acid substitutions in structurally related proteins. A pattern recognition approach. Determination of a new and efficient scoring matrix. J Mol Biol. 1988;204:1019–1029. doi: 10.1016/0022-2836(88)90058-7. [DOI] [PubMed] [Google Scholar]
- Triboulet S, Arthur M, Mainardi JL, Veckerle C, Dubee V, Nguekam-Nouri A, Gutmann L, Rice LB, Hugonnet JE. Inactivation kinetics of a new target of β-lactam antibiotics. J Biol Chem. 2011;2011:4. doi: 10.1074/jbc.M111.239988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuomanen E, Cozens R. Changes in peptidoglycan composition and penicillin-binding proteins in slowly growing Escherichia coli. J Bacteriol. 1987;169:5308–5310. doi: 10.1128/jb.169.11.5308-5310.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W, Holtje J-V. The Architecture of the Murein (Peptidoglycan) in Gram-Negative Bacteria: Vertical Scaffold or Horizontal Layer(s)? J. Bacteriol. 2004;186:5978–5987. doi: 10.1128/JB.186.18.5978-5987.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- Wietzerbin J, Das BC, Petit JF, Lederer E, Leyh-Bouille M, Ghuysen JM. Occurrence of D-alanyl-(D)-meso-diaminopimelic acid and meso-diaminopimelyl-meso-diaminopimelic acid interpeptide linkages in the peptidoglycan of Mycobacteria. Biochemistry. 1974;13:3471–3476. doi: 10.1021/bi00714a008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.