Abstract
The HIV-1 capsid protein consists of two independently folded domains connected by a flexible peptide linker (residues 146–150), the function of which remains to be defined. To investigate the role of this region in virus replication, we made alanine or leucine substitutions in each linker residue and two flanking residues. Three classes of mutants were identified: (i) S146A and T148A behave like wild type (WT); (ii) Y145A, I150A, and L151A are noninfectious, assemble unstable cores with aberrant morphology, and synthesize almost no viral DNA; and (iii) P147L and S149A display a poorly infectious, attenuated phenotype. Infectivity of P147L and S149A is rescued specifically by pseudotyping with vesicular stomatitis virus envelope glycoprotein. Moreover, despite having unstable cores, these mutants assemble WT-like structures and synthesize viral DNA, although less efficiently than WT. Collectively, these findings demonstrate that the linker region is essential for proper assembly and stability of cores and efficient replication.
Keywords: HIV-1 capsid protein, HIV-1 assembly, HIV-1 cores, VSV-G pseudotyping, interdomain linker, in vitro assembly, TRIM5 proteins, host restriction, virus disassembly, reverse transcription
Introduction
The capsid protein (CA) of human immunodeficiency virus type 1 (HIV-1) has a major role in virus assembly and early postentry events and is derived from the multidomain Gag polyprotein precursor (Pr55gag) (Freed, 1998; Vogt, 1997). During or shortly after budding of immature particles, Gag is cleaved by the viral protease to generate the individual mature viral proteins including (from the N- to C-terminus) matrix, CA, nucleocapsid (NC), and p6, as well as two spacer peptides (SP1 and SP2) (Henderson et al., 1992); reviewed in (Adamson and Freed, 2007; Ganser-Pornillos et al., 2008; Swanstrom and Wills, 1997). Once Gag is cleaved, dramatic structural rearrangements of the proteins occur (virus “maturation”), resulting in the formation of mature, infectious virions containing electron-dense conical cores (de Marco et al., 2010; Gross et al., 2000; von Schwedler et al., 1998; Wiegers et al., 1998); reviewed in Adamson and Freed, 2007; Briggs and Kräusslich, 2011; Ganser-Pornillos et al., 2008).
The HIV-1 core is a fullerene cone composed of approximately 250 hexamers and 12 pentamers that cap both ends of the cone (Ganser et al., 1999; Li et al., 2000; Pornillos et al., 2011). The N-terminal domain (NTD) of CA forms the hexameric lattice and each hexamer is linked to six others by interactions with C-terminal CA dimers (Briggs et al., 2006; Ganser et al., 1999; Ganser-Pornillos et al., 2007; Ganser-Pornillos et al., 2004; Huseby et al., 2005; Lanman et al., 2003; Lanman and Prevelige, 2005; Li et al., 2000; Pornillos et al., 2009). The interior of the core is a ribonucleoprotein complex containing NC, reverse transcriptase (RT), integrase (IN), Vpr, Nef, two copies of genomic RNA, and tRNALys3 (Thomas and Gorelick, 2008; Vogt, 1997).
Following entry of the virus into the cell, the core disassembles, i.e., the CA shell is removed and the ribonucleoprotein complex is released, resulting in the formation of reverse transcription complexes (RTCs) (Fassati and Goff, 2001). The time at which uncoating is initiated is still not clear (reviewed in Arhel, 2010; Levin et al., 2010) and has been reported to be: (i) soon after viral entry (Fassati and Goff, 2001; Grewe et al., 1990; Hulme et al., 2011); (ii) several hours postentry (McDonald et al., 2002; Sayah et al., 2004; Shi and Aiken, 2006; Stremlau et al., 2006); or (iii) just prior to nuclear import of preintegration complexes (Arhel et al., 2007). Both proper assembly and disassembly of viral cores as well as optimal core stability are essential for reverse transcription and virus replication (Bowzard et al., 2001; Fitzon et al., 2000; Forshey et al., 2002; Reicin et al., 1996; Tang et al., 2001; Tang et al., 2003b; von Schwedler et al., 1998; von Schwedler et al., 2003).
Interestingly, monkey cells express host restriction proteins TRIM5α (Hatziioannou et al., 2004; Keckesova et al., 2004; Stremlau et al., 2004; Yap et al., 2004) or TRIMCyp, a fusion protein in which the C-terminal SPRY domain of TRIM5α has been replaced by cyclophilin A (CypA) (Newman et al., 2008; Nisole et al., 2004; Sayah et al., 2004; Virgen et al., 2008); both TRIM5 proteins target intact HIV-1 CA. This results in premature uncoating of the viral core, degradation of CA, abrogation of reverse transcription, and loss of infectivity (Black and Aiken, 2010; Rold and Aiken, 2008; Sebastian and Luban, 2005; Stremlau et al., 2006; Wu et al., 2006; Zhao et al., 2011). The mechanism by which TRIM5 functions relies on pattern recognition (Ganser-Pornillos et al., 2011; Pertel et al., 2011) and the ability of TRIM5 to promote innate immune signaling (Pertel et al., 2011).
Early structural studies indicated that HIV-1 CA consists of two independently folded domains, an NTD (residues 1–145) and a C-terminal domain (CTD) (residues 151–231), which are connected by a short, flexible linker (146–150) (Berthet-Colominas et al., 1999; Gamble et al., 1997; Gitti et al., 1996; Momany et al., 1996) that becomes more structured upon CA multimerization (Lanman et al., 2003). It has been proposed that linker flexibility may allow movement of the CTD relative to the NTD (Pornillos et al., 2011; Worthylake et al., 1999) to ensure that both domains can have similar interactions in the CA hexamer and pentamer (Pornillos et al., 2011). Despite advances in our knowledge of CA structure and the considerable sequence conservation of the linker residues, only limited information on their biological and molecular properties has been available thus far. In the case of murine leukemia virus (MLV), several linker mutations inserted into the CA domain of Gag (i.e., extending the length of the interdomain linker) led to a defect in virus assembly and release as well as the appearance of virions with aberrant morphology (Arvidson et al., 2003).
Recently, the phenotypes of mutations in two residues in the HIV-1 region of interest have been described. One of these residues is Y145, which in a number of structural studies has been assigned to helix 7 in the NTD (Gamble et al., 1996; Gitti et al., 1996; Momany et al., 1996; Pornillos et al., 2009). Surprisingly, a new high resolution NMR structure of HIV-1 CA consisting of residues 144–231 has shown that Y145 is crucial for formation of the CTD dimer interface (Byeon et al., 2009) (also see below). To assess the biological significance of this finding, two Y145 mutants (replacement with A or F) were constructed and were found to exhibit a severely compromised phenotype, including loss of infectivity (Byeon et al., 2009). Other work on CA residues S108, S149, and S178, which have the potential to be phosphorylated, showed that an S149A mutant has low infectivity (Brun et al., 2008; Wacharapornin et al., 2007) and is only partially defective in other assays (Brun et al., 2008).
In the present work, we set out to investigate the biological function of all of the residues between the NTD and CTD, i.e., S146-I150, as well as the flanking residues Y145 and L151 (Fig. 1). Alanine substitution mutations were made for each of the residues with the exception of P147, which was changed to leucine. Two mutants (S146A and T148A) behave like WT, whereas three other mutants (Y145A, I150A, and L151A) are noninfectious and exhibit severe defects in core assembly and stability and viral DNA synthesis in infected cells. Two mutants (P147L and S149A), while poorly infectious, have a novel, attenuated phenotype. Thus, despite the instability of P147L and S149A cores, these mutants retain an ability to undergo reverse transcription (albeit less efficiently than WT) and can assemble conical cores that resemble those of WT virions. These results clearly demonstrate that the interdomain linker region has a critical role in facilitating proper core assembly and stability, which in turn ultimately impact the ability of the virus to undergo efficient replication.
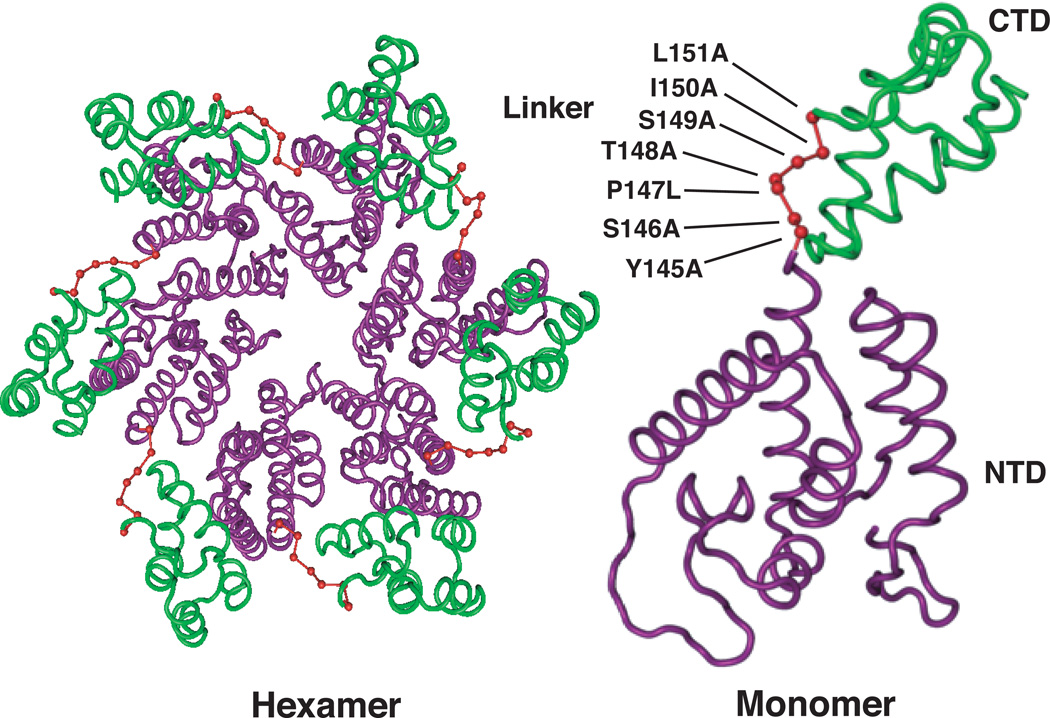
Fig. 1.
HIV-1 CA hexamer and monomer illustrating the positions of residues in the interdomain linker region. Residues in this region are shown in red. In both the monomer and hexamer structures, the NTD and CTD are highlighted in purple and green, respectively. The positions of the five linker residues and the two flanking residues that were mutated to Ala or Leu are shown in the monomer diagram. Structures were derived from PDB ID: 3GV2, using Cn3D 4.3 macromolecular structure viewer (NCBI) from Pornillos et al. (Pornillos et al., 2009).
Results
Single amino acid substitutions in the interdomain linker region of HIV-1 CA
The aim of this study was to examine the function of residues 146–150 in HIV-1 replication and in particular, to determine whether they are important for proper virus assembly. At the outset, we also decided to include residues Y145 and L151, which flank the interdomain linker region. Single alanine substitutions were made in all residues except P147, which was changed to leucine (Fig. 1A), since a P147A mutant was unable to produce virus particles (i.e., RT activity in the supernatant fluid of transfected cells was not detectable). The mutations were introduced into the WT HIV-1 pNL4-3 clone (Adachi et al., 1986) or the env− derivative pNL4-3KFS (Freed et al., 1992; Freed and Martin, 1995). A diagram of the CA structure highlighting the positions of the residues of interest in the hexamer and in the monomer are shown in Fig. 1.
Virus production and infectivity of mutants
To determine whether the CA mutants are able to produce virus particles, the supernatant fluids of transfected cells were assayed for RT activity (Freed et al., 1995). As shown in Table 1, column 2, all of the mutants produced significant amounts of particles and even the lowest producer, L151A, exhibited 50% of WT activity. When infectivity of env+ virus was measured, it was observed that mutants S146A and T148A had significant levels of infectivity, similar to those of the WT, whereas P147L and S149A had only 4–5% of WT infectivity (column 3). In contrast, infectivity was at background level for Y145A, I150A, and L151A.
Table 1.
Virus production and infectivity.
| Virus | Virus Production (%)a |
Infectivity (%)b | |
|---|---|---|---|
| HIV-1 env+ | VSV-G Pseudotyped HIV-1c |
||
| WT | 100 | 100 | 100 |
| Y145A | 63 ± 13d | ≤0.5 | ≤10 |
| S146A | 81 ± 8 | 61 ± 11 | 105 ± 18 |
| P147L | 65 ± 19 | 4 ± 2 | 86 ± 32 |
| T148A | 77 ± 14 | 86 ± 11 | 110 ± 18 |
| S149A | 65 ± 24 | 5 ± 1 | 102 ± 32 |
| I150A | 59 ± 12 | ≤0.5 | ≤10 |
| L151A | 50 ± 11 | ≤0.5 | ≤10 |
Virus production was assayed as RT activity in the supernatant of transfected cells. Values were converted to percentage of WT level for each of three or more independent transfections and were averaged.
Infectivity was measured in the single-round LuSIV assay (Roos et al., 2000). WT infectivity was set at 100%.
HIV-1 env − virions were pseudotyped with VSV-G envelope protein, as described in Materials and methods.
Numbers to the right of the “±” represent the standard deviation.
There are instances in which retroviral infectivity can be rescued or enhanced if env− particles are pseudotyped with a heterologous envelope protein, such as the vesicular stomatitis virus envelope glycoprotein (VSV-G) (Aiken, 1997; Brun et al., 2008; Emi et al., 1991; Jorgenson et al., 2009; Khan et al., 2003; Naldini et al., 1996; Yee et al., 1994). Although it is known that VSV-G pseudotyped particles enter via pH-dependent endocytosis (Matlin et al., 1982) rather than by direct fusion at the plasma membrane, the underlying mechanism for rescue is still not understood. Nevertheless, this assay can give information on subtle differences between mutants.
As might be expected, the infectivity of the completely noninfectious mutants, i.e., Y145A, I150A, and L151A, could not be rescued by VSV-G pseudotyping (Table 1, column 4). The env+ mutants that were infectious, i.e., S146A and T148A, had WT levels of infectivity when env− virions were pseudotyped with VSV-G. Surprisingly, despite the poor infectivity of P147L and S149A bearing HIV-1 Env, when pseudotyped with VSV-G, these mutants displayed essentially WT levels of infectivity (86% and 102%, respectively).
To demonstrate that rescue of P147L and S149A infectivity by VSV-G can also occur in a different target cell type, the experiment was repeated using the TZM-bl assay for infectivity (Table 2) (Derdeyn et al., 2000; Platt et al., 1998; Wei et al., 2002). This assay utilizes a HeLa-derived indicator cell line that supports VSV-G-mediated infection, but not as efficiently as observed in T cells (E.O. Freed, unpublished observation); a positive read-out is therefore unlikely to be simply the result of “saturating” the infectivity assay. In this case, we found that P147L infectivity was partially rescued (~44%), whereas S149A infectivity was rescued almost completely (~80%) (Table 2). This suggests that at least with respect to this parameter, P147L may be somewhat more defective than S149A. Importantly, rescue was highly specific for pseudotyping with VSV-G (Table 2). For example, MLV Env was unable to rescue the infectivity of P147L and S149A, despite the high level of infectivity of ”WT” HIV-1 env− virions pseudotyped with MLV Env in TZM-bl cells (data not shown). Collectively, the VSV-G infectivity data suggest that the phenotypes exhibited by P147L and S149A differ from those of the three noninfectious mutants.
Table 2.
Rescue of infectivity of pseudotyped WT and mutant env− virions is Env-specific.a
| Virus | HIV-1 (pIIINL4env) |
A-MLVenv | VSV-G (pHCMV-Genv) |
|---|---|---|---|
| WT | 100 | 100 | 100 |
| P147L | 10 ± 6b | 12 ± 8 | 44 ± 15 |
| S149A | 14 ± 10 | 9 ± 5 | 80 ± 15 |
Infectivity was measured in a single-cycle assay using TZM-bl indicator cells (Derdeyn et al., 2000; Platt et al., 1998; Wei et al., 2002). WT infectivity was set at 100%. Three independent experiments were performed. The data shown are from a representative experiment, in which each sample was assayed in triplicate.
Numbers to the right of the “±” represent the standard deviation.
Viral protein composition in cell and viral lysates
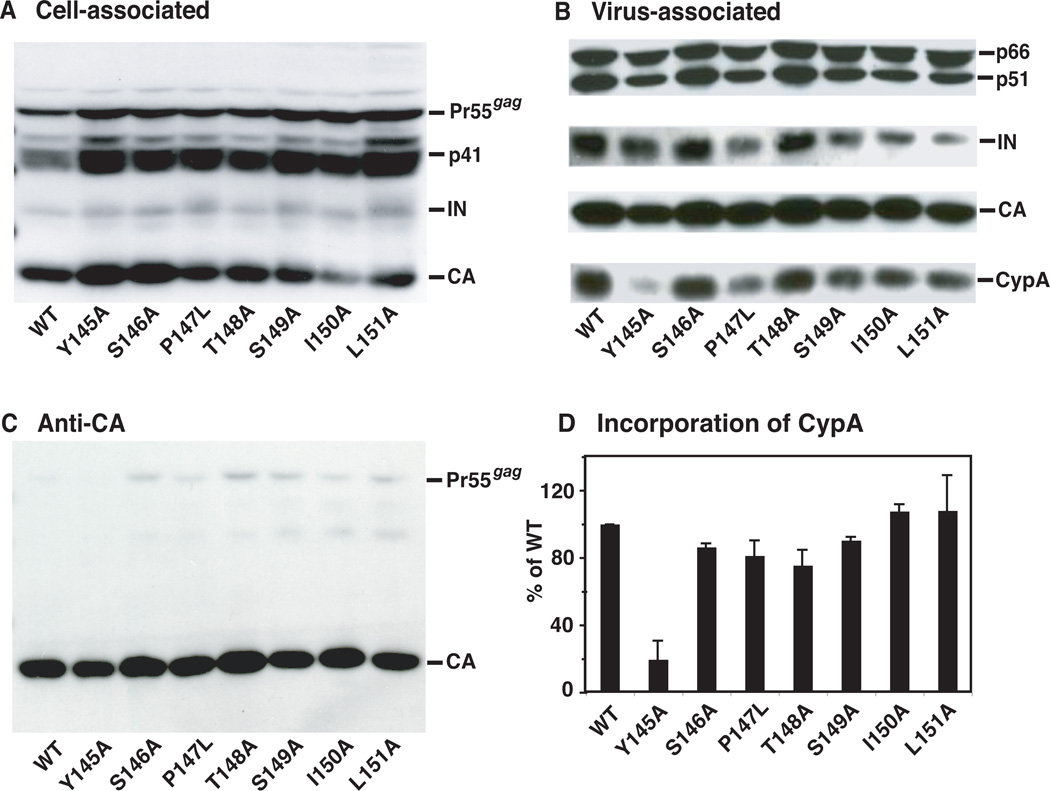
It was also of interest to determine the viral protein composition in cell and viral lysates. Cell lysates were prepared as described in Materials and methods and were subjected to Western blot analysis with HIV-1 neutralizing serum, which reacted strongly with Pr55gag, Gag cleavage product p41, IN, and CA (Fig. 2A). To examine the protein composition of WT and mutant virions, viral lysates were analyzed with antisera specific for RT, IN, CA, and CypA. Strong bands representing p66/p51 (RT), IN, CA, and CypA (Fig. 2B) were visualized after probing by Western blot, whereas Pr55gag bands, detected with anti-CA antibodies, were very weak (Fig. 2C). This indicates that the mutants do not have major processing defects.
Fig. 2.
Analysis of WT and mutant viral proteins present in cell lysates and in virions. HeLa cells were transfected with WT or mutant plasmid DNAs. Proteins present in cell and virion lysates were separated by SDS-PAGE in 10% gels and were detected by Western blot analysis. (A) Cell-associated viral proteins probed with HIV-1 neutralizing serum. Note that analysis of cell-associated proteins suggests that I150A might have a processing defect, but importantly, this defect was not detected in an anti-CA blot of the viral lysate (see (C), below). (B) Virion-associated proteins probed sequentially with a mixture of anti-CypA and anti-IN, anti-RT, and anti-CA antibodies. (C) Entire Western blot analyzed with anti-CA antibodies. The CA bands are the same as those shown in (B). (D) Relative incorporation levels of CypA into WT and mutant virions. The CypA and IN band intensities were quantified as described in Materials and methods. The data were normalized for each sample by calculating the ratio of the level of CypA to the level of IN and then multiplying by 100. The WT ratio was set at 100%.
Note that in this figure and in Figs. 5, 7, and 8, error bars indicate the standard deviation from at least two or three independent experiments.
To determine the relative amount of CypA in virions, the band intensities of CypA to IN were quantified; the ratio of CypA to IN was determined and multiplied by 100 (Fig. 2D). IN was chosen for normalization, since the virion-associated IN is unlikely to be affected by the mutations in CA (Tang et al., 2003b). The WT value was set at 100%. All of the mutants except for Y145A had approximately WT levels of CypA. However, the Y145A mutant had ~5-fold less CypA than WT, suggesting that this mutant is even more defective than the other noninfectious mutants.
Transmission electron microscopy (TEM)
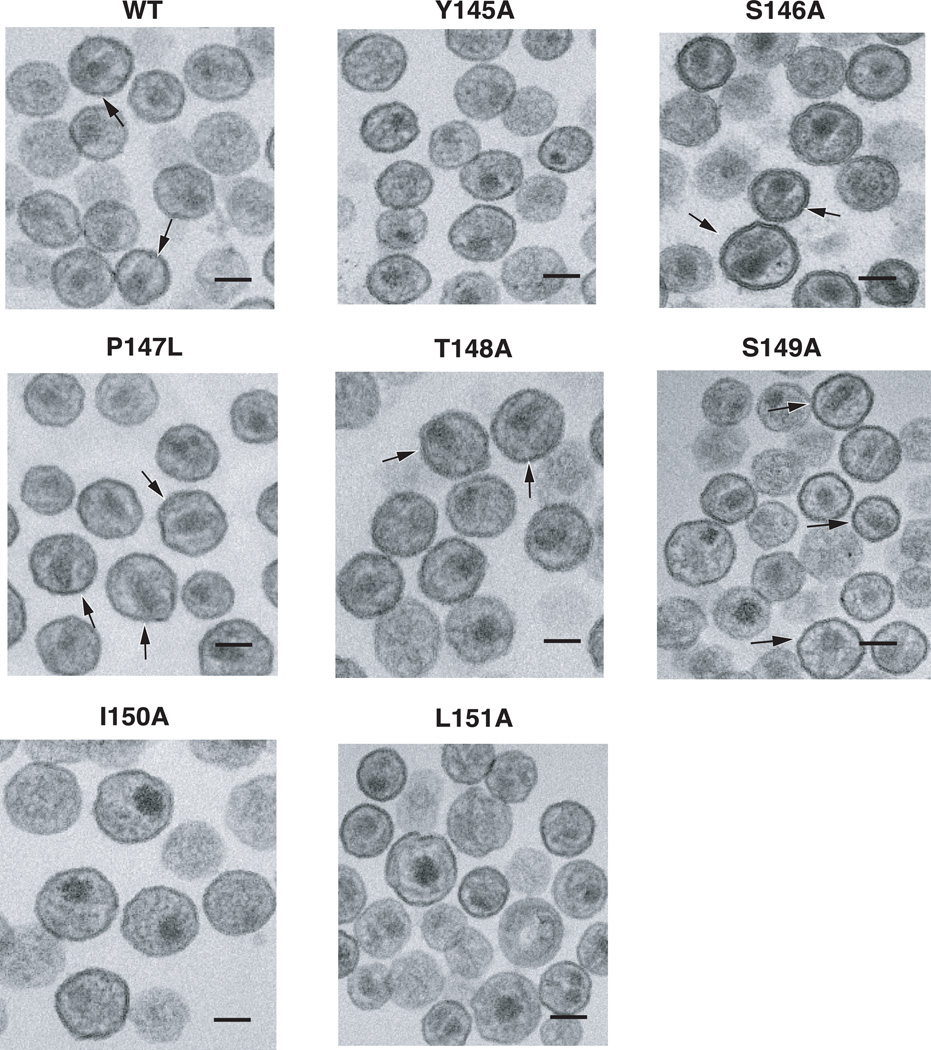
The ability to form conical cores is a stringent test for proper assembly of HIV-1 particles. To investigate the architecture of mutant cores, WT or mutant virions were imaged by TEM (Fig. 3). In addition to conical cores (arrows), centric and acentric cores (cores that are proximal to the viral membrane) were also observed. Infectious HIV-1 is expected to have a significant percentage of conical cores in the virion population. By analyzing the core structures in a large number of particles (ranging from ~100 – ~400), we determined that for WT and infectious mutants S146A and T148A, the percentage of conical cores was between 40 and 45% (Table 3). Conical cores were detected in P147L and S149A virus populations at a frequency approximately half that of the WT. However, as is shown below, these cores display some biochemical properties that differ from those of authentic WT cores. The noninfectious mutants lacked conical cores, and instead had approximately the same number of centric and acentric cores, as also observed in earlier studies (Auerbach et al., 2006; Dorfman et al., 1994; Fitzon et al., 2000; Forshey et al., 2002; Reicin et al., 1996; Scholz et al., 2005; Tang et al., 2007; Tang et al., 2001; von Schwedler et al., 1998; von Schwedler et al., 2003).
Fig. 3.
TEM analysis of WT and mutant virions present in extracellular fluids of transfected HeLa cells. Env− virions were used for this experiment. Mutants are identified above each image. The arrows point to representative virus particles in the field with conical cores. The scale bars are 100 nm.
Table 3.
Analysis of HIV-1 WT and mutant virions by TEM.
| Virus | No. of Particles |
Particle size (nm) |
No. of particles whose cores are: | ||
|---|---|---|---|---|---|
| Cone-Shaped | Centric | Acentric | |||
| WT | 252 | 126 ± 13a | 115 (46)b | 93 (27) | 44 (17) |
| Y145A | 126 | 152 ± 28 | 0 | 51 (40) | 75 (60) |
| S146A | 388 | 134 ± 12 | 156 (40) | 137 (35) | 95 (25) |
| P147L | 138 | 136 ± 25 | 33 (24) | 58 (42) | 47 (34) |
| T148A | 234 | 131 ± 14 | 103 (44) | 74 (32) | 57 (24) |
| S149A | 250 | 141 ± 21 | 65 (26) | 101 (40) | 84 (34) |
| I150A | 108 | 146 ± 19 | 0 | 42 (39) | 66 (61) |
| L151A | 170 | 146 ± 22 | 0 | 92 (55) | 78 (45) |
Numbers to the right of "+/−" represent the standard deviation.
Numbers in parentheses are percentages of the total number of particles.
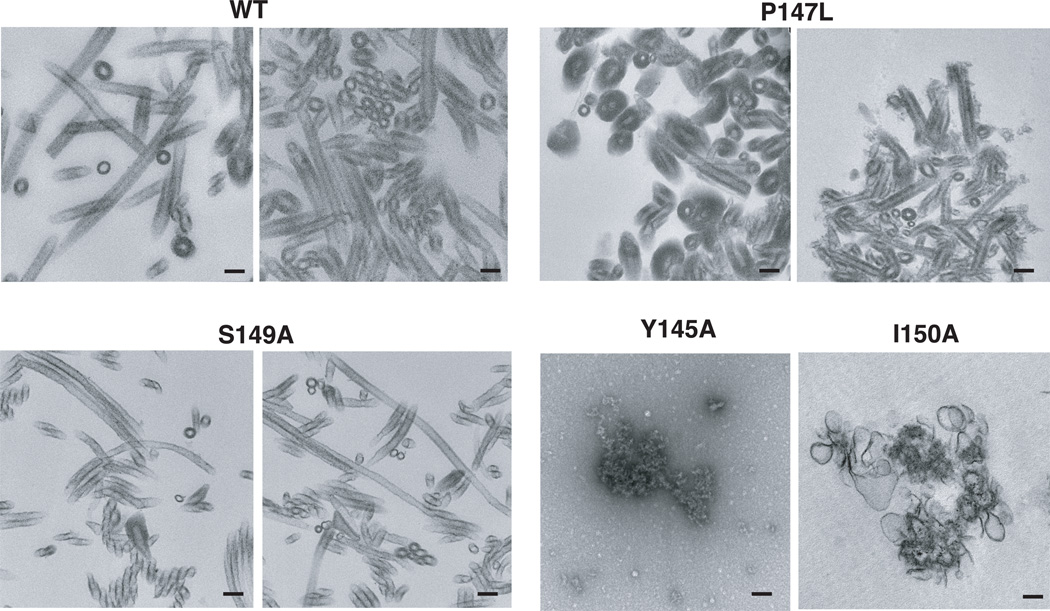
Another approach to evaluate the assembly competency of Gag mutants is to perform in vitro assembly reactions with purified viral proteins (Campbell and Rein, 1999; Campbell and Vogt, 1995; Ehrlich et al., 1992; von Schwedler et al., 1998). Here, we purified the CA proteins of WT, P147L, and S149A, as well as the Y145A and I150A CA proteins, which were chosen to represent the class of noninfectious mutants that form only aberrant cores (Fig. 3). The in vitro assemblies were analyzed by TEM (Fig. 4). As is typical for WT CA, long tubular structures were formed (Ehrlich et al., 1992). The P147L and S149A assemblies had structures that were similar to WT. In some reactions, subtle differences were observed between the WT and these mutants (e.g., formation of somewhat shorter tubes by P147L and elongated narrow tubes by S149A). What is particularly striking, however, is that in contrast to these CA proteins, neither Y145A nor I150A CA formed any recognizable structures.
Fig. 4.
Analysis of in vitro assemblies of WT and mutant CA proteins by TEM. Purified CA proteins were assembled and then processed for TEM, as described in Materials and methods. Mutants are identified above each image (Y145A, I150A) or pair of images (WT, P147L, S149A). The scale bars are 100 nm.
Taken together, the EM data demonstrate that while the noninfectious mutants are severely defective with respect to virus assembly both in vivo and in vitro, mutants P147L and S149A exhibit an attenuated assembly phenotype.
Synthesis of viral DNA in infected cells and in detergent-treated virions
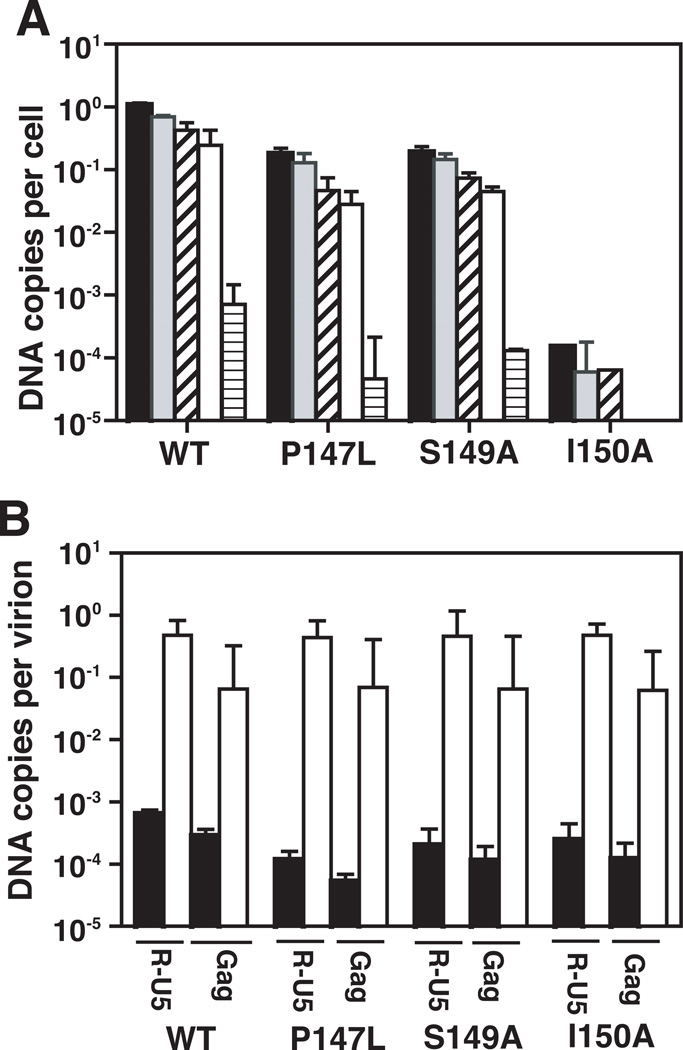
Several studies have shown that a defect in conical core assembly is associated with an inability to synthesize viral DNA in infected cells (Brun et al., 2008; Fitzon et al., 2000; Reicin et al., 1996; Rulli et al., 2006; Tang et al., 2001). It was therefore of interest to determine the efficiencies of viral DNA synthesis in infections with WT or each of the P147L, S149A, and I150A mutants. Quantitative PCR (qPCR) was used to detect the major products of reverse transcription: R-U5, (−) strong-stop DNA; U3–U5, minus-strand transfer DNA; Gag, late minus-strand DNA; R-5’UTR, plus-strand transfer DNA; and full-junction (FJ), 2-long terminal repeat (2-LTR) circles, which are a marker for entry of viral DNA into the nucleus (Fig. 5A).
Fig. 5.
Analysis of DNA synthesized in WT- and mutant-infected cells and in ERT assays with detergent-treated particles. The mutants used for the assays were P147L, S149A, and I150A. Viral DNA from infected cells and from detergent-treated virions was quantified as described in Materials and Methods. (A) Analysis of viral DNA in infected cells. The data are plotted in bar graphs as DNA copies per cell for the following viral DNA products synthesized by RT: R-U5 (−) strong-stop DNA, closed; U3–U5, minus-strand transfer DNA, gray; Gag, late minus-strand DNA, slanted stripe; R-5’UTR, plus-strand transfer DNA, open; FJ 2-LTR circles, horizontal stripe. The amounts of DNA were normalized for the number of cells recovered and for the number of viruses used to infect the cells. (B) Viral DNA synthesis in ERT assays. The data are expressed as DNA copies per virion. Values for the R-U5 and Gag DNA products are plotted in bar graphs. Amount of DNA products present in virions (i.e., intravirion DNA) prior to the ERT assay, closed; amount of DNA products after the ERT assay, open.
The data showed that P147L and S149A synthesized ~10-fold less of each viral DNA product compared to WT. The results for the two mutants were essentially the same, although it appears that S149A made more 2-LTR circles than P147L. With I150A, the amount of DNA products synthesized was ~104 lower than WT and the two late products were not detectable. This suggests that the inability to synthesize viral DNA in I150A-infected cells is likely due to the complete absence of normal cores in the virion population (Fig. 3; Table 3). Collectively, the results demonstrate a dramatic contrast between the levels of viral DNA detected in cells infected with P147L and S149A and the noninfectious I150A.
We also examined DNA synthesis in detergent-treated virions by using the endogenous RT (ERT) assay. The amounts of early (R-U5) and late minus-strand (Gag) products were measured using qPCR (Fig. 5B). Samples were taken before the initiation of the ERT assay (pre-ERT) and 24 h later. In this case, following incubation for 24 h, WT and all of the mutant samples showed the same levels of viral DNA synthesis for both early and late products of reverse transcription. This finding is consistent with previous studies demonstrating that the absence of conical cores does not block reverse transcription in the ERT assay (Brun et al., 2008; Kaplan et al., 1994; Tang et al., 2001).
Assay of CA function in vivo
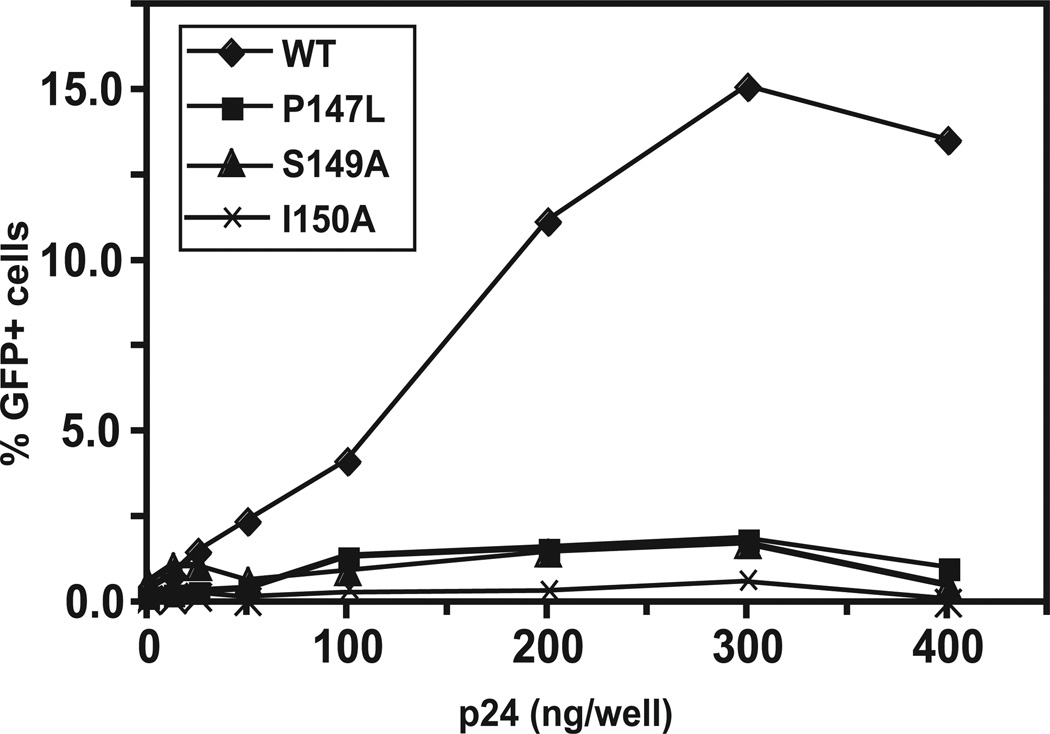
To learn more about the ability of the CA mutants to function in the infected cell, we performed an abrogation assay to test whether the CA mutants could saturate the post-entry defect in owl monkey kidney (OMK) cells conferred by TRIMCyp, a potent host restriction factor that interacts with HIV-1 CA and blocks virus infection. In our experiments, OMK cells were infected with WT and mutants P147L, S149A, and I150A. After brief (4 to 6 h) incubation, the infected cells were challenged with an HIV-1-green fluorescent protein (GFP) reporter virus. If the CA protein of the initial virus is functional, then TRIMCyp will be saturated and the reporter virus should be able to replicate. As a consequence, a significant number of the infected cells should register as GFP+ (Fig. 6). In fact, this is what we observed when WT was used as the initial virus. If mutant CA proteins are not able to interact with TRIMCyp, we should obtain the opposite result. In this case, almost no infectivity was observed when the mutants were tested. In some experiments, there was a very small difference between the activity of P147L and S149A compared with the activity of I150A, but in other experiments, no difference was detected.
Fig. 6.
TRIMCyp abrogation assay. An ELISA assay was used for measuring CA concentrations in the virus preparations (Tang and Hewlett, 2010). The ability of WT and three mutants (P147L, S149A, and I150A) to interact with TRIMCyp expressed in OMK cells was determined as described in Materials and methods. The percentage of GFP+ cells was plotted against CA concentration (ng/well). Three independent experiments were performed. The data shown are from a representative experiment, in which each sample was assayed in triplicate. The error bars represent the standard deviation.
These results show that the three mutants were all essentially inactive in the TRIMCyp abrogation assay. Moreover, the data further indicate that differences between the two poorly infectious mutants (i.e., P147L and S149A) and the noninfectious mutants that were observed in other assays (see above) do not affect the assay for CA-TRIMCyp interaction.
Determination of viral core stability
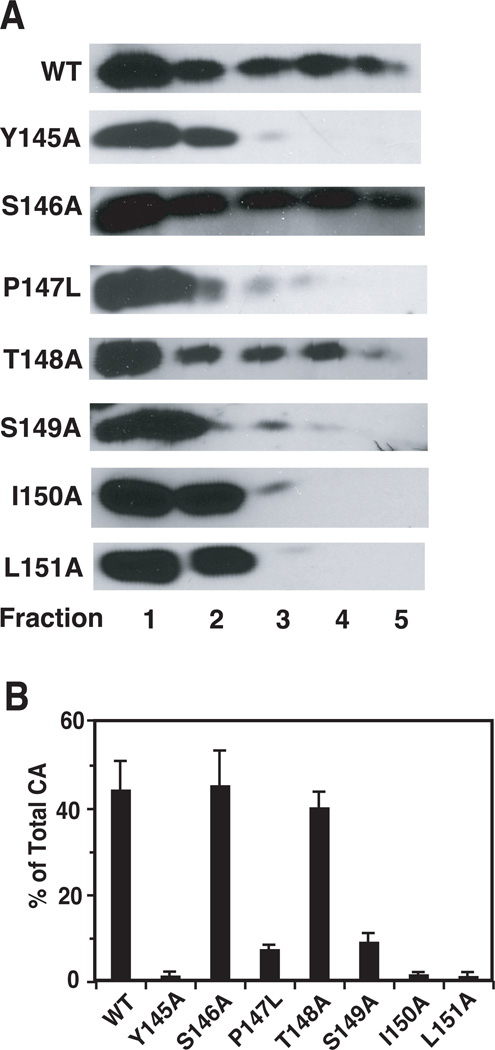
In view of the fact that TRIM5 proteins interact most efficiently with CA present in an intact core, i.e., assembled CA (Black and Aiken, 2010; Ganser-Pornillos et al., 2011; Kar et al., 2008; Langelier et al., 2008; Sebastian and Luban, 2005; Stremlau et al., 2006; Zhao et al., 2011), it seemed possible that the results of the TRIMCyp assay might reflect common defects in the core stability of the mutants. Optimal core stability is an important determinant of core function; virions with hyperstable or unstable cores are not infectious (Bowzard et al., 2001; Forshey et al., 2005; Forshey et al., 2002; Tang et al., 2003b; von Schwedler et al., 2003). To address this issue, we determined the amounts of CA and RT associated with core structures. WT and mutant virions were treated with 0.2% (vol/vol) Triton X-100 prior to sedimentation in sucrose gradients. If detergent concentrations greater than 0.2% (even as low as 0.3%) were used, only low amounts of CA protein could be detected for all of the mutants with the exception of S146A and T148A, which have a WT phenotype (Table 1). This suggested that in the case of the other five mutants, viral cores are detergent-sensitive and unstable.
For analysis of CA content, detergent-treated particles were sedimented in sucrose step gradients; cores were found in fractions 3, 4, and 5 (Tang et al., 2003b). Fractions were probed by Western blot (Fig. 7A) and the ratio of the amounts of CA in fractions 3 to 5 to total CA was calculated and multiplied by 100 (Fig. 7B). WT, S146A, and T148A had ~40% of total CA associated with the core, whereas Y145A, I150A, and L151A had barely detectable amounts. For P147L and S149A, between 3 and 5% of total CA was recovered in the core fractions.
Fig. 7.
Retention of CA in core fractions of WT and mutant virions. Virions treated with 0.2% Triton X-100 (vol/vol) were subjected to sedimentation in sucrose step gradients. Aliquots of each fraction were subjected to Western blot analysis by using anti-CA serum. (A) HIV-1 CA bands are shown for WT and mutant samples. (B) The band intensities of CA in each fraction were determined as described in Materials and methods. The values for fractions 3, 4, and 5 (detergent-resistant core fractions) were combined and are shown in bar graphs as the percentage of total CA protein recovered from the gradient.
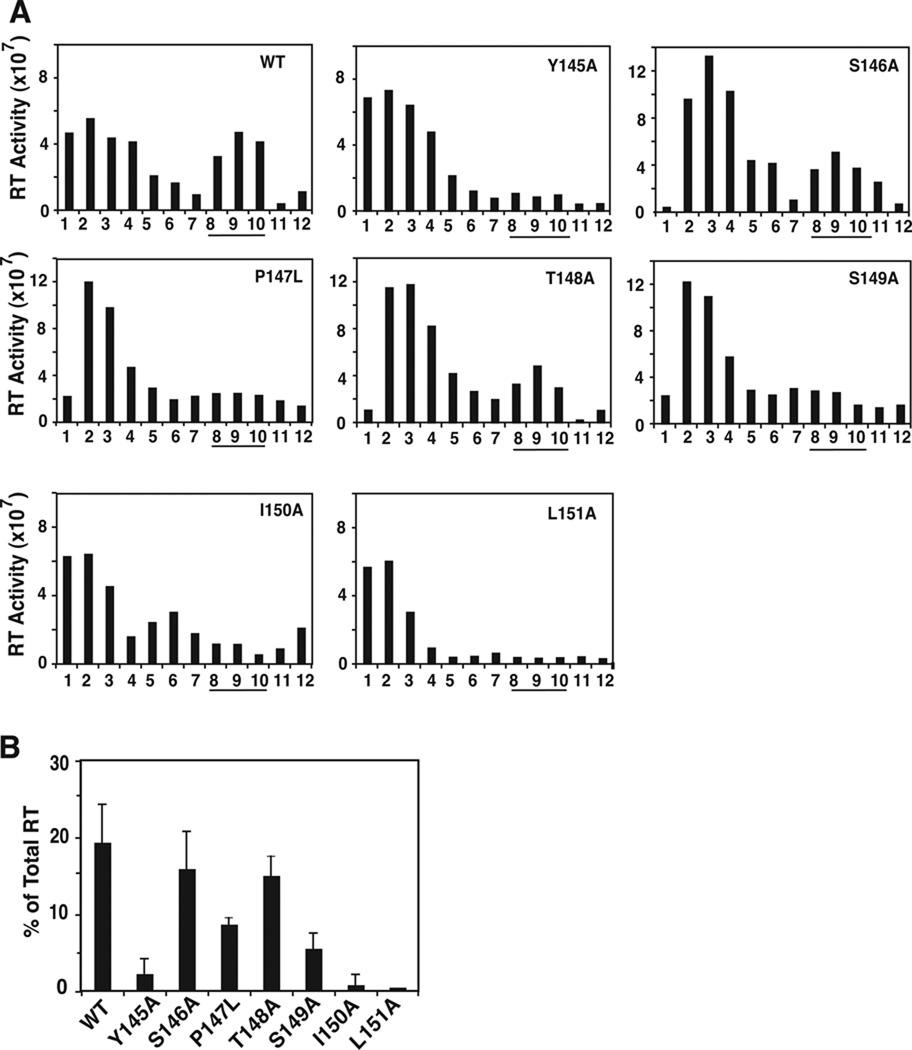
RT content of virions was measured by sedimenting detergent-treated virions in 20 to 70% (wt/wt) linear sucrose gradients followed by assay of RT polymerase activity (Freed et al., 1995) (Fig. 8A). Viral cores, which sediment in sucrose gradients with a density of 1.24–1.28 g/ml, were found in fractions 8 to 10. WT, S146A, and T148A each had between 15 and 20% of total RT in core fractions, whereas the noninfectious mutants had ≤1% RT in cores (Fig. 8B). Mutants P147L and S149A showed broad peaks of activity, in contrast to the sharp peak seen for WT (Fig. 8A). The percentage of RT associated with the cores of these mutants was between 5 and 10% of total RT activity (Fig. 8B).
Fig. 8.
Retention of HIV-1 RT in WT and mutant viral cores. Virions were treated with 0.2% Triton X-100 (vol/vol) and were sedimented through 20% to 70% (wt/wt) linear sucrose density gradients (Tang et al., 2003b). Aliquots of each fraction were assayed for RT activity. (A) RT activity of WT and mutant fractions plotted in a bar graph. (B) RT activity in core fractions (fractions 8, 9, and 10, underlined) was quantified. The data are plotted as the percentage of total RT activity in the gradient. The value for percent RT activity in L151A core fractions was at the detection limit and was therefore subtracted from the values for all of the other samples.
These results show that mutants P147L and S149A retain minimal amounts of CA and somewhat higher levels of RT in core fractions. This suggests that the cores of these mutants are also unstable, although not to the same extent as the cores of the noninfectious mutants. Thus, there appear to be small, but reproducible differences between the core stability of P147L and S149A compared with the core stability of the noninfectious mutants. Apparently, these small differences do not affect the outcome of the TRIMCyp assay, although as seen above, other properties of P147L and S149A are clearly distinguishable from the corresponding properties of the noninfectious mutants in a variety of assays including infectivity (Table 1).
Discussion
In the present study, we sought to investigate the biological role of residues between the two domains of HIV-1 CA (Y145-L151) (Fig. 1) and in particular, to assess their importance for virus assembly and replication. Our approach was to make point mutations in residues encompassing the entire region, i.e., alanine substitutions in all residues except P147, which was changed to leucine. The results clearly demonstrate that the CA interdomain linker is crucial for facilitating proper core stability and architecture.
Analysis of mutant phenotypes shows that the mutants can be divided into three classes: (i) infectious virions (S146A, T148A); (ii) completely noninfectious virions with severe defects in viral morphology and function (Y145A, I150A, L151A); and (iii) virions with very low infectivity, having an attenuated phenotype (P147L, S149A) (Table 1). Interestingly, the infectivity data for some of the mutants can be correlated with what is known about CA structure in this region. Recently, a study combining high resolution NMR analysis of recombinant HIV-1 CA containing residues 144–231 with cryo-EM data for tubular assemblies of this protein revealed the importance of residues Y145, T148, and L151 for CTD dimer formation (Byeon et al., 2009). Residue Y145 is crucial for formation of this interface and for biological function, since Y145A or Y145F mutants are not infectious and have defects in core stability and assembly (Byeon et al., 2009), consistent with our Y145A data (Table 1; Figs. 2–4; Figs. 7 and 8) (see below). Moreover, of all the mutants studied here, Y145A is the only one that exhibits a CypA deficiency (Fig. 2D), presumably as a result of global changes in CA folding. A similar defect was also observed with two severely defective N-terminal CA mutants (W23A and F40A) (Tang et al., 2003b). A significant reduction in CypA incorporation would be expected to be associated with mutated residues in the NTD, although our data do not rule out such an effect when mutation of a residue near the NTD also results in global disruption of CA structure. The apparent discrepancy concerning the localization of Y145 is likely due to differences in the techniques and protein constructs used for the structure determinations. An additional possibility, although highly speculative, is that CA might adopt different conformations that place Y145 in either the NTD or CTD, depending on the particular stage in the virus life cycle.
Residues L151 and T148 (in order of importance) also exhibit intermolecular interactions that are involved in CTD dimer formation (Byeon et al., 2009) (I.L. Byeon, personal communication). Thus, the lack of infectivity of L151A is consistent with the structural data (Table 1). Most likely, retention of the WT phenotype in the case of T148A (Table 1) is due to the conservative change from threonine to alanine. The S146A mutation may not impact infectivity because this residue is exposed on the surface of the protein (A.M. Gronenborn, personal communication).
As mentioned above, a major conclusion that emerges from our study is that residues Y145-L151, with the possible exception of S146, are critical for proper core formation. Indeed, point mutations that lead to abrogation or severe reduction of infectivity are often associated with formation of virion cores that are highly unstable: retention of CA in core fractions is negligible (Y145A, I150A, L151A) or only slightly higher than background (P147L, S149A), whereas ~40% of total CA in the sample is associated with WT, S146A, and T148A cores under our conditions (Fig. 7). Core instability of S149A was also observed in studies on HIV-1 CA residues that are potential substrates for phosphorylation (Brun et al., 2008; Wacharapornin et al., 2007). Reduction of RT activity associated with unstable mutant cores is less severe than depletion of CA in the case of P147L and S149A (Fig. 8). Thus, removal of the mutant CA shells might occur shortly after entry by premature uncoating (Stremlau et al., 2006); reviewed in (Arhel, 2010; Levin et al., 2010), whereas RT might be less labile due to its location in the interior of the virus. In accord with the CA sedimentation data, we also find that P147L, S149A, and I150A are unable to saturate TRIMCyp in the assay for abrogation of host restriction (Fig. 6), which gives a positive readout only if the cores have optimal stability (Forshey et al., 2005).
A striking result of this work is the novel finding that despite the poor replication capacity of P147L and S149A, infectivity can be rescued in an efficient and specific manner by pseudotyping env− virions with VSV-G (Tables 1 and 2). Similar results were obtained for S149A and S178A in one of the studies of CA phosphorylation (Brun et al., 2008). Additionally, these authors showed that the pseudotyped virions undergo more efficient viral DNA synthesis. In contrast, VSV-G pseudotyping does not rescue the infectivity of the three noninfectious mutants in our study (Table 1).
It is generally thought that HIV-1 entry into cells depends on interaction with cellular receptors followed by fusion at the plasma membrane (Hunter, 1997). However, there is also evidence supporting the possibility that entry occurs by an endosomal pathway (Miyauchi et al., 2009). With respect to pseudotyping with VSV-G, it is known that VSV-G-mediated virus entry occurs via pH-dependent endocytosis (Matlin et al., 1982), but the reason why this mode of entry can result in rescue of infectivity is not known. We speculate that endocytosis might allow delivery of the cores to the nuclear pore without premature uncoating and a requirement to traverse cytoplasmic microfilament/microtubule networks (Arhel et al., 2007; Bukrinskaya et al., 1998; McDonald et al., 2002), thereby bypassing the core instability defect. A somewhat similar suggestion was made in an earlier study on VSV-G pseudotyping of nef-defective HIV-1 virions (Aiken, 1997). Alternatively, some other aspect of VSV-G-mediated entry, e.g., faster fusion kinetics with VSV-G than with HIV-1 Env (Hulme et al., 2011; Iordanskiy et al., 2006), might result in bypass of the infectivity defect conferred by the P147L and P149A mutations. Collectively, these considerations lead us to suggest that the two mutants might be useful tools for studies on the viral entry pathway.
The P147L and S149A assembly properties and core architecture are also of great interest. Thus, despite the instability of P147L and S149A cores (Fig. 7), a significant number of mutant virions contain conical cores that are indistinguishable from WT structures in images generated by TEM (Fig. 3; Table 3). Moreover, in contrast to the Y145A (Fig. 4) (Byeon et al., 2009), Y145F (Byeon et al., 2009), and I150A (Fig. 4) CA proteins, P147L and S149A CA form tubular assemblies that are very similar to the long tubes assembled by WT CA, although some subtle differences can be detected by TEM (Fig. 4). Experiments exploiting higher resolution EM techniques are being initiated in an effort to identify potential ultrastructural differences between WT and mutant core structures.
It is now established that in addition to other defects, virions having unstable or hyperstable cores are unable to undergo or complete reverse transcription in infected cells (Aiken, 2006; Bowzard et al., 2001; Forshey et al., 2002; Tang et al., 2001; Tang et al., 2003b; von Schwedler et al., 2003). Accordingly, we find that the noninfectious mutant I150A makes ~104 less viral DNA products than WT and no late products (Fig. 5A). However, in agreement with the EM analysis, synthesis of viral DNA products by the P147L and S149A mutants is reduced by only 10-fold compared with WT and all of the expected DNA products are detected (Fig. 5A).
Taken together, our results raise the following question: “How can we reconcile our finding that in two assays affected by core stability, P147L and S149A give negative results (Figs. 6 and 7), whereas in other assays reflecting core structure, assembly, and reverse transcription activity (Figs. 3, 4, and 5; Fig. 8), these mutants exhibit an attenuated phenotype?” It would appear that differences between the WT and mutant CA protein structures, which in turn dictate core structure and biological activity (reviewed in (Adamson and Freed, 2007; Vogt, 1997), are subtle and may not be sufficient to generate a positive readout in some assays. This is most likely the case in the TRIMCyp abrogation assay (Fig. 6). Data from a recent structural study support the idea that the hexagonal scaffold of TRIM5α proteins is assembled using the symmetry and spacing of the CA lattice as a template (“pattern recognition”) (Ganser-Pornillos et al., 2011). Thus, even subtle defects in CA structure and especially defects that lead to core instability (Forshey et al., 2005) could prevent significant binding of TRIM5α proteins to CA. Additionally, the low recovery of CA from isolated mutant cores is not entirely unexpected, since sedimentation at high speeds, even for relatively short times, is likely to lead to disruption of cores that are unstable. In contrast, EM and PCR analysis of DNA synthesis involve more gentle treatment of virions, thereby generating a modulated response in these assays.
These findings have important implications for understanding the molecular nature of HIV-1 assembly, since they underscore the unusual plasticity of CA, which despite the rigorous structural requirements that govern assembly and integrity of viral cores, permits some expression of biological activity even under less than optimal circumstances (Tang et al., 2007). Additional studies on the structure of the P147L and S149A CA proteins should be invaluable for elucidating the ultrastructure of HIV-1 conical cores and its relation to CA function.
In summary, we have shown for the first time that residues between the NTD and CTD of HIV-1 CA (Y145-L151) have a crucial role in virus assembly and formation of conical cores. Mutations in three of these residues (Y145A, I150A, and L151A) lead to a total loss of infectivity, defects in core morphology and stability, as well as virtually complete abrogation of viral DNA synthesis. However, two mutants (P147L and S149A), while poorly infectious, exhibit an attenuated phenotype, including rescue of infectivity by pseudotyping with VSV-G, modest ability to undergo reverse transcription, and assembly of cores with seemingly normal architecture. These findings provide new insights into the biological function of a region in HIV-1 CA that has only recently become the subject of intense interest and represents a potential target for anti-HIV therapy.
Materials and methods
Cell culture, transfection, and RT assay
OMK cells (a generous gift from Christopher Aiken, Vanderbilt University Medical Center, Nashville, TN), HeLa cells, and 293T cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (Hyclone), 2 mM glutamine, penicillin (50 IU/ml), and streptomycin (50 µg/ml). Three cell lines were obtained from the AIDS Research and Reagent Program, Division of AIDS, NIAID, NIH: (i) LuSIV cells (from Jason W. Roos and Janice E. Clements, catalog no. 5460) (Roos et al., 2000), which were grown in RPMI 1640 medium with 300 µg/ml of hygromycin B (Invitrogen, Carlsbad, CA) and the same supplements as described above; TZM-bl cells (from John C. Kappes, Xiaoyun Wu, and Tranzyme Inc., catalog no. 8129) (Derdeyn et al., 2000; Platt et al., 1998; Wei et al., 2002), which were grown in the same medium used for HeLa cells (see above); and (iii) HeLa CD4+ cells (HeLa CD4 Clone 1022 from Bruce Chesebro, catalog no. 1109), which were maintained as recommended (Chesebro et al., 1990; Chesebro and Wehrly, 1988; Chesebro et al., 1991).
To produce HIV-1 virions, HeLa cells were transfected in 100-mm dishes with 10 µg of WT or mutant proviral DNA, using Lipofectamine™ 2000 (Invitrogen), according to the manufacturer’s instructions. Supernatant fluids were collected 36 to 48 h after transfection. Cellular debris was removed by low-speed centrifugation and subsequent passage of the clarified fluids through 0.45-µm-pore-size syringe filters. The fluids were adjusted to pH 7.3 with HEPES buffer (final concentration, 10 mM) and were divided into several tubes prior to storage at −80 °C. Particle production was determined by an exogenous RT assay (Freed et al., 1995). For ERT assays, virus was prepared by transfecting 293T cells with mutant or wild-type pNL4-3-based proviral plasmids using the (Ca)2PO4 method (Julias et al., 2001). In this case, exogenous RT assays were performed as described by Gorelick et al. (Gorelick et al., 1990).
Single-cycle infectivity assays
Viral infectivity was measured using a single-cycle assay. Equivalent amounts of virus particles, as determined by RT assay, were used to infect LuSIV cells (Roos et al., 2000). Luciferase activity was measured using the Luciferase Assay System kit from Promega (Madison, WI), according to the manufacturer’s instructions. Some single-cycle assays were performed by using TZM-bl indicator cells (Derdeyn et al., 2000; Platt et al., 1998; Wei et al., 2002). The Env vectors used for pseudotyping HIV-1 env− virions were as follows. The VSV-G expression vector pHCMV-Genv (Yee et al., 1994) was a generous gift from Jane Burns (University of California at San Diego School of Medicine, La Jolla, CA). The HIV-1 Env expression vector pIIINL4env has been described previously (Murakami and Freed, 2000). The plasmid pSV-A-MLVenv (Landau et al., 1991), which encodes amphotropic (A) MLV Env, was obtained from Dan Littman and Nathaniel Landau through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
Plasmids and site-specific mutagenesis
The plasmids used to make site-specific mutations in the CA linker coding region were as follows: (i) HIV-1 pNL4-3 (GenBank accession no. AF324493) (Adachi et al., 1986); and (ii) env− subclone, pNL4-3KFS, which has a frameshift mutation at a KpnI site in the env coding region (Freed et al., 1992; Freed and Martin, 1995). PCR products were generated by nested PCR with appropriate primers. The products were digested with BamHI and SphI, so that the BamHI-SphI fragment in pNL4-3 could be replaced by the corresponding fragment containing the desired mutation. The same restriction sites were used to transfer mutations into the env− pNL4-3KFS proviral vector. In each case, the mutation was confirmed by sequencing performed by AGCT, Inc. Plasmid DNAs were prepared with a QIAFilter Maxi Kit (QIAGEN Inc.-USA, Valencia, CA). The HIV-1 GFP reporter construct in which nef was replaced by the coding region for GFP was a generous gift from Christopher Aiken (He et al., 1997; Shi and Aiken, 2006).
Western blot analysis of WT and mutant viral proteins
Cell and viral lysates were prepared and fractionated in 10% SDS-polyacrylamide gels as described previously (Freed and Martin, 1994; Huang et al., 1995). A chemiluminescence kit (SuperSignal West Dura Extended Duration Substrate), anti-human horseradish peroxidase (HRP), and anti-rabbit HRP were purchased from Pierce (division of Thermo Fisher Scientific, Inc.). HIV-1 neutralizing serum (catalog no. 1983 and 1984) and anti-CA sera (HIV-1SF2 p24 antiserum, catalog no. 4250) were obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. Rabbit anti-CypA was purchased from Biomol International (Plymouth Meeting, PA). Rabbit anti-HIV IN and RT sera are described in an earlier publication (Klutch et al., 1998). Band intensities were quantified by first scanning the film and then converting the image into a jpg file that could be analyzed by using NIH Image (http://rsbweb.nih.gov/nih-image/about.html).
Preparation of purified CA proteins expressed in E. coli
WT and mutant (Y145A, P147L, S149A, and I150A) CA proteins were expressed in E. coli Rosetta (DE3) pLysS (Novagen) with a C-terminal His tag, using a modification of the protocol kindly provided by Michael Summers, University of Maryland Baltimore County, Baltimore, MD (Tang et al., 2003a). The final eluate from the cobalt affinity column (TALON Superflow Metal Affinity Resin, Clonetech, Palo Alto, CA) was dialyzed against 50 mM sodium phosphate buffer, pH 7.2, and 5 mM 2-mercaptoethanol.
Transmission Electron Microscopy
HeLa cells transfected with WT and mutant virions were fixed with 2% glutaraldehyde in 0.1 M sodium cacodylate and examined by TEM as described (Freed and Martin, 1994; Tang et al., 2001). For in vitro assembly, purified CA protein (60 to 100 µg) was incubated in 1 M NaCl at 4 °C overnight. Reactions were analyzed by TEM as described above for virus-infected cells.
Assay of viral DNA in infected cells and in detergent-treated virions (ERT)
Virus produced by transfecting 293T cells was used to infect HeLa CD4+ cells. At 24 h postinfection, total cellular DNA was extracted (Julias et al., 2001). Primers and probes used for quantitation of viral and cellular nucleic acids have been described (Buckman et al., 2003; Thomas et al., 2006). Briefly, viral sequence targets of the primers and probes used for quantitation of viral DNA included (−) SSDNA (R-U5), minus-strand transfer product (U3–U5), late minus-strand synthesis product (gag), plus-strand transfer product (R-5’UTR), FJ (i.e., 2-LTR circles), gag for genomic RNA, and CCR5 to determine cell equivalents present in extracts from infected HeLa CD4+ cells. Quantities of viral DNA were normalized for cell recovery and exogenous RT activity.
For ERT reactions, virus produced from transfections was sequentially treated with DNase I (Sigma-Aldrich, St. Louis, MO) and then subtilisin A (Sigma-Aldrich) (Thomas et al., 2008); ERT assays were performed as described (Thomas et al., 2008). Viral DNA and RNA species were quantified using the primers and probes mentioned above. The copies of viral DNA were normalized to the number of virions present in the corresponding pre-ERT sample (calculated by assuming 2 copies of genomic RNA per virion).
CA (p24) ELISA Assay
Supernatant fluids from cells transfected by WT and mutant viruses were assayed for p24 by a Europium nanoparticle immunoassay (ENIA), as described previously (Tang and Hewlett, 2010). Briefly, each well of the 96-well microtiter plate was coated with 60 µl of monoclonal anti-HIV-1 p24 antibody (final concentration, 2 µg/ml) (catalog no. C65690M, Meridian Life Science, Inc., Saco, ME) and was incubated overnight at 4 °C. After washing five times with buffer containing PBS and 0.05% Tween 20 (PBST) (each new step was always preceded by washing with PBST), the plate was blocked by adding 350 µl of blocking buffer (Starting Block™ Blocking Buffer, Thermo Fisher Scientific, Inc.) to each well. One hundred µl each of p24 standards (ranging from 0.05 ng/ml to 1 ng/ml) and experimental samples were added and were then incubated for 1 h at 37 °C. Note that the samples were prepared by diluting viral supernatant fluids 5-fold in RT buffer containing 0.05% Nonidet P-40 (Freed et al., 1995) and subsequently making serial dilutions in PBST containing 1% bovine serum albumin (BSA). Addition of 100 µl per well of the detection antibody, which is biotinylated polyclonal anti-HIV-1 p24 antibody (PerkinElmer, Waltham, MA), was followed by incubation for 1 h at 37 °C. Finally, 107 europium nanoparticles conjugated with streptavidin in PBST containing 1%BSA were added and the plates were incubated for 30 min at 37 °C. Fluorescence was measured with a Victor fluorometer (PerkinElmer) as described (Tang and Hewlett, 2010). The concentration of HIV-1 p24 in the supernatants was determined based on the standard curve obtained in the experiments.
Abrogation of host restriction assay
Assays to determine the ability of WT and mutant viral CA to restrict infection in OMK cells expressing TRIMCyp, a fusion of TRIM5α and CypA (Nisole et al., 2004; Sayah et al., 2004), were performed essentially as reported previously (Forshey et al., 2005; Shi and Aiken, 2006). All virus samples were assayed for CA (p24) content so that the same concentrations of CA could be used for each sample. Serial dilutions of WT or mutant virus pseudotyped with VSV-G were used to infect OMK cells and were incubated for 4 to 6 h at 37 °C. HIV-1-GFP reporter virus pseudotyped with VSV-G was then added to the infected cells and incubated for 24 h at 37 °C. The reporter virus was subjected to preliminary titration on OMK cells and an amount giving less than 1% of total cells registered as GFP+ was chosen for the assay. In the final step, the cells were treated with trypsin and were fixed in a freshly prepared solution of 4% paraformaldehyde in PBS. GFP expression was analyzed with a Becton-Dickinson FACScan flow cytometer.
Isolation of viral cores
For analysis of CA and RT retention in viral cores, supernatant fluids collected from transfected HeLa cells were sedimented at 100,000 × g. The pellets were resuspended in PBS and Triton X-100 (final concentration 0.2% (vol/vol)) was added. Samples were then subjected to sucrose gradient centrifugation without prior incubation. For CA, step gradients were used, since the CA yield for all samples was higher and more reproducible after centrifugation for 2 h, compared with the amounts detected following overnight centrifugation (required for linear gradients). Five fractions were collected from the top of the tube and the amount of CA in each fraction was measured by Western blot analysis using anti-CA serum. Fractions 1 and 2 represent the soluble and detergent-soluble fractions, respectively; fractions 3, 4, and 5 represent detergent-resistant core fractions (Tang et al., 2003b).
To analyze RT retention in viral cores, detergent-treated virions (see above) were sedimented through 20% to 70% (wt/wt) linear sucrose gradients as described (Tang et al., 2003b). (Assay of enzymatic activity is more sensitive than Western blotting and in this case, sufficient amounts of activity were detected after overnight centrifugation.) Twelve 0.4-ml fractions were collected from the top of the gradient and were assayed for exogenous RT activity. The density of the fractions was determined as described in an earlier report (Tang et al., 2003b). Cores were found in fractions with a density of 1.24 to 1.28 g/ml (Fig. 8A, fractions 8–10).
Highlights.
-
>
Mutational analysis of residues 145–151 (linker region) in HIV-1 capsid protein.
-
>
Y145A, I150A, L151A noninfectious, severe defects in core morphology and stability.
-
>
P147L, S149A poorly infectious, infectivity rescued by VSV-G, attenuated phenotype.
-
>
Latter mutants make some viral DNA, assemble normal looking, but unstable cores.
-
>
Linker residues essential for proper assembly and stability of HIV-1 cores.
Acknowledgments
We thank Christopher Aiken for his generous gift of OMK cells and the HIV-1-GFP reporter construct as well as for helpful advice regarding the TRIMCyp assay, Michael Summers for kindly providing his protocol for bacterial expression and purification of HIV-1 CA, Jane Burns for the VSV-G expression vector (pHCMV-Genv), and the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH for antisera, cells, and plasmids, as detailed in the text. We are also indebted to Angela M. Gronenborn and In-Ja L. Byeon for valuable discussion. This work was supported in part by the Intramural Research Program at the National Institutes of Health (Eunice Kennedy Shriver National Institute of Child Health and Human Development [J.J., S.D., K.H., and J.G.L.] and the National Cancer Institute, Center for Cancer Research [S.A. and E.O.F.]) and in part by the Office of Science and Health Coordination, Food and Drug Administration, National Heart, Lung, and Blood Institute, National Institutes of Health (S.T. and I.H.). This project has also been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E with SAIC-Frederick, Inc. (F.S., K.N., J.A.T., and R.J.G.). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson CS, Freed EO. Human immunodeficiency virus type 1 assembly, release, and maturation. Adv. Pharmacol. 2007;55:347–387. doi: 10.1016/S1054-3589(07)55010-6. [DOI] [PubMed] [Google Scholar]
- Aiken C. Pseudotyping human immunodeficiency virus type 1 (HIV-1) by the glycoprotein of vesicular stomatitis virus targets HIV-1 entry to an endocytic pathway and suppresses both the requirement for Nef and the sensitivity to cyclosporin A. J. Virol. 1997;71:5871–5877. doi: 10.1128/jvi.71.8.5871-5877.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiken C. Viral and cellular factors that regulate HIV-1 uncoating. Curr. Opin. HIV AIDS. 2006;1:194–199. doi: 10.1097/01.COH.0000221591.11294.c1. [DOI] [PubMed] [Google Scholar]
- Arhel N. Revisiting HIV-1 uncoating. Retrovirology. 2010;7:96. doi: 10.1186/1742-4690-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arhel NJ, Souquere-Besse S, Munier S, Souque P, Guadagnini S, Rutherford S, Prevost MC, Allen TD, Charneau P. HIV-1 DNA Flap formation promotes uncoating of the pre-integration complex at the nuclear pore. EMBO J. 2007;26:3025–3037. doi: 10.1038/sj.emboj.7601740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvidson B, Seeds J, Webb M, Finlay L, Barklis E. Analysis of the retrovirus capsid interdomain linker region. Virology. 2003;308:166–177. doi: 10.1016/s0042-6822(02)00142-3. [DOI] [PubMed] [Google Scholar]
- Auerbach MR, Brown KR, Kaplan A, de Las Nueces D, Singh IR. A small loop in the capsid protein of Moloney murine leukemia virus controls assembly of spherical cores. J. Virol. 2006;80:2884–2893. doi: 10.1128/JVI.80.6.2884-2893.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthet-Colominas C, Monaco S, Novelli A, Sibaï G, Mallet F, Cusack S. Head-to-tail dimers and interdomain flexibility revealed by the crystal structure of HIV-1 capsid protein (p24) complexed with a monoclonal antibody Fab. EMBO J. 1999;18:1124–1136. doi: 10.1093/emboj/18.5.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black LR, Aiken C. TRIM5α disrupts the structure of assembled HIV-1 capsid complexes in vitro. J. Virol. 2010;84:6564–6569. doi: 10.1128/JVI.00210-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowzard JB, Wills JW, Craven RC. Second-site suppressors of Rous sarcoma virus CA mutations: evidence for interdomain interactions. J. Virol. 2001;75:6850–6856. doi: 10.1128/JVI.75.15.6850-6856.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs JAG, Grünewald K, Glass B, Förster F, Kräusslich HG, Fuller SD. The mechanism of HIV-1 core assembly: Insights from three-dimensional reconstructions of authentic virions. Structure. 2006;14:15–20. doi: 10.1016/j.str.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Briggs JAG, Kräusslich HG. The molecular architecture of HIV. J. Mol. Biol. 2011;410:491–500. doi: 10.1016/j.jmb.2011.04.021. [DOI] [PubMed] [Google Scholar]
- Brun S, Solignat M, Gay B, Bernard E, Chaloin L, Fenard D, Devaux C, Chazal N, Briant L. VSV-G pseudotyping rescues HIV-1 CA mutations that impair core assembly or stability. Retrovirology. 2008;5:57. doi: 10.1186/1742-4690-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckman JS, Bosche WJ, Gorelick RJ. Human immunodeficiency virus type 1 nucleocapsid Zn2+ fingers are required for efficient reverse transcription, initial integration processes, and protection of newly synthesized viral DNA. J. Virol. 2003;77:1469–1480. doi: 10.1128/JVI.77.2.1469-1480.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukrinskaya A, Brichacek B, Mann A, Stevenson M. Establishment of a functional human immunodeficiency virus type 1 (HIV-1) reverse transcription complex involves the cytoskeleton. J. Exp. Med. 1998;188:2113–2125. doi: 10.1084/jem.188.11.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byeon IJL, Meng X, Jung J, Zhao G, Yang R, Ahn J, Shi J, Concel J, Aiken C, Zhang P, Gronenborn AM. Structural convergence between Cryo-EM and NMR reveals intersubunit interactions critical for HIV-1 capsid function. Cell. 2009;139:780–790. doi: 10.1016/j.cell.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S, Rein A. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J. Virol. 1999;73:2270–2279. doi: 10.1128/jvi.73.3.2270-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S, Vogt VM. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 1995;69:6487–6497. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesebro B, Buller R, Portis J, Wehrly K. Failure of human immunodeficiency virus entry and infection in CD4-positive human brain and skin cells. J. Virol. 1990;64:215–221. doi: 10.1128/jvi.64.1.215-221.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesebro B, Wehrly K. Development of a sensitive quantitative focal assay for human immunodeficiency virus infectivity. J. Virol. 1988;62:3779–3788. doi: 10.1128/jvi.62.10.3779-3788.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesebro B, Wehrly K, Metcalf J, Griffin DE. Use of a new CD4-positive HeLa cell clone for direct quantitation of infectious human immunodeficiency virus from blood cells of AIDS patients. J. Infect. Dis. 1991;163:64–70. doi: 10.1093/infdis/163.1.64. [DOI] [PubMed] [Google Scholar]
- de Marco A, Müller B, Glass B, Riches JD, Kräusslich HG, Briggs JA. Structural analysis of HIV-1 maturation using cryo-electron tomography. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001215. e1001215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derdeyn CA, Decker JM, Sfakianos JN, Wu X, O'Brien WA, Ratner L, Kappes JC, Shaw GM, Hunter E. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 2000;74:8358–8367. doi: 10.1128/jvi.74.18.8358-8367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman T, Bukovsky A, Öhagen Å, Höglund S, Göttlinger HG. Functional domains of the capsid protein of human immunodeficiency virus type 1. J. Virol. 1994;68:8180–8187. doi: 10.1128/jvi.68.12.8180-8187.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich LS, Agresta BE, Carter CA. Assembly of recombinant human immunodeficiency virus type 1 capsid protein in vitro. J. Virol. 1992;66:4874–4883. doi: 10.1128/jvi.66.8.4874-4883.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emi N, Friedmann T, Yee JK. Pseudotype formation of murine leukemia virus with the G protein of vesicular stomatitis virus. J. Virol. 1991;65:1202–1207. doi: 10.1128/jvi.65.3.1202-1207.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassati A, Goff SP. Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J. Virol. 2001;75:3626–3635. doi: 10.1128/JVI.75.8.3626-3635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzon T, Leschonsky B, Bieler K, Paulus C, Schröder J, Wolf H, Wagner R. Proline residues in the HIV-1 NH2-terminal capsid domain: structure determinants for proper core assembly and subsequent steps of early replication. Virology. 2000;268:294–307. doi: 10.1006/viro.1999.0178. [DOI] [PubMed] [Google Scholar]
- Forshey BM, Shi J, Aiken C. Structural requirements for recognition of the human immunodeficiency virus type 1 core during host restriction in owl monkey cells. J. Virol. 2005;79:869–875. doi: 10.1128/JVI.79.2.869-875.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forshey BM, von Schwedler U, Sundquist WI, Aiken C. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J. Virol. 2002;76:5667–5677. doi: 10.1128/JVI.76.11.5667-5677.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed EO. HIV-1 Gag proteins: diverse functions in the virus life cycle. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- Freed EO, Delwart EL, Buchschacher GL, Jr, Panganiban AT. A mutation in the human immunodeficiency virus type 1 transmembrane glycoprotein gp41 dominantly interferes with fusion and infectivity. Proc. Natl. Acad. Sci. USA. 1992;89:70–74. doi: 10.1073/pnas.89.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed EO, Englund G, Martin MA. Role of the basic domain of human immunodeficiency virus type 1 matrix in macrophage infection. J. Virol. 1995;69:3949–3954. doi: 10.1128/jvi.69.6.3949-3954.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed EO, Martin MA. Evidence for a functional interaction between the V1/V2 and C4 domains of human immunodeficiency virus type 1 envelope glycoprotein gp120. J. Virol. 1994;68:2503–2512. doi: 10.1128/jvi.68.4.2503-2512.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed EO, Martin MA. Virion incorporation of envelope glycoproteins with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix. J. Virol. 1995;69:1984–1989. doi: 10.1128/jvi.69.3.1984-1989.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble TR, Vajdos FF, Yoo S, Worthylake DK, Houseweart M, Sundquist WI, Hill CP. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- Gamble TR, Yoo S, Vajdos FF, von Schwedler UK, Worthylake DK, Wang H, McCutcheon JP, Sundquist WI, Hill CP. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- Ganser-Pornillos BK, Chandrasekaran V, Pornillos O, Sodroski JG, Sundquist WI, Yeager M. Hexagonal assembly of a restricting TRIM5α protein. Proc. Natl. Acad. Sci. U.S.A. 2011;108:534–539. doi: 10.1073/pnas.1013426108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganser-Pornillos BK, Cheng A, Yeager M. Structure of full-length HIV-1 CA: a model for the mature capsid lattice. Cell. 2007;131:70–79. doi: 10.1016/j.cell.2007.08.018. [DOI] [PubMed] [Google Scholar]
- Ganser-Pornillos BK, von Schwedler UK, Stray KM, Aiken C, Sundquist WI. Assembly properties of the human immunodeficiency virus type 1 CA protein. J. Virol. 2004;78:2545–2552. doi: 10.1128/JVI.78.5.2545-2552.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganser-Pornillos BK, Yeager M, Sundquist WI. The structural biology of HIV assembly. Curr. Opin. Struct. Biol. 2008;18:203–217. doi: 10.1016/j.sbi.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitti RK, Lee BM, Walker J, Summers MF, Yoo S, Sundquist WI. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- Gorelick RJ, Nigida SM, Jr, Bess JW, Jr, Arthur LO, Henderson LE, Rein A. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J. Virol. 1990;64:3207–3211. doi: 10.1128/jvi.64.7.3207-3211.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewe C, Beck A, Gelderblom HR. HIV: early virus-cell interactions. J. Acquir. Immune Defic. Syndr. 1990;3:965–974. [PubMed] [Google Scholar]
- Gross I, Hohenberg H, Wilk T, Wiegers K, Grättinger M, Müller B, Fuller S, Kräusslich HG. A conformational switch controlling HIV-1 morphogenesis. EMBO J. 2000;19:103–113. doi: 10.1093/emboj/19.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatziioannou T, Perez-Caballero D, Yang A, Cowan S, Bieniasz PD. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5α. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10774–10779. doi: 10.1073/pnas.0402361101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Chen Y, Farzan M, Choe H, Ohagen A, Gartner S, Busciglio J, Yang X, Hofmann W, Newman W, Mackay CR, Sodroski J, Gabuzda D. CCR3 and CCR5 are co-receptors for HIV-1 infection of microglia. Nature. 1997;385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- Henderson LE, Bowers MA, Sowder RC, II, Serabyn SA, Johnson DG, Bess JW, Jr, Arthur LO, Bryant DK, Fenselau C. Gag proteins of the highly replicative MN strain of human immunodeficiency virus type 1: posttranslational modifications, proteolytic processings, and complete amino acid sequences. J. Virol. 1992;66:1856–1865. doi: 10.1128/jvi.66.4.1856-1865.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Orenstein JM, Martin MA, Freed EO. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulme AE, Perez O, Hope TJ. Complementary assays reveal a relationship between HIV-1 uncoating and reverse transcription. Proc. Natl. Acad. Sci. U.S.A. 2011;108:9975–9980. doi: 10.1073/pnas.1014522108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter E. Viral entry and receptors. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor, N. Y.: Cold Spring Harbor Laboratory Press; 1997. pp. 71–119. [PubMed] [Google Scholar]
- Huseby D, Barklis RL, Alfadhli A, Barklis E. Assembly of human immunodeficiency virus precursor Gag proteins. J. Biol. Chem. 2005;280:17664–17670. doi: 10.1074/jbc.M412325200. [DOI] [PubMed] [Google Scholar]
- Iordanskiy S, Berro R, Altieri M, Kashanchi F, Bukrinsky M. Intracytoplasmic maturation of the human immunodeficiency virus type 1 reverse transcription complexes determines their capacity to integrate into chromatin. Retrovirology. 2006;3:4. doi: 10.1186/1742-4690-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgenson RL, Vogt VM, Johnson MC. Foreign glycoproteins can be actively recruited to virus assembly sites during pseudotyping. J. Virol. 2009;83:4060–4067. doi: 10.1128/JVI.02425-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julias JG, Ferris AL, Boyer PL, Hughes SH. Replication of phenotypically mixed human immunodeficiency virus type 1 virions containing catalytically active and catalytically inactive reverse transcriptase. J. Virol. 2001;75:6537–6546. doi: 10.1128/JVI.75.14.6537-6546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan AH, Krogstad P, Kempf DJ, Norbeck DW, Swanstrom R. Human immunodeficiency virus type 1 virions composed of unprocessed Gag and Gag-Pol precursors are capable of reverse transcribing viral genomic RNA. Antimicrob. Agents Chemother. 1994;38:2929–2933. doi: 10.1128/aac.38.12.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar AK, Diaz-Griffero F, Li Y, Li X, Sodroski J. Biochemical and biophysical characterization of a chimeric TRIM21-TRIM5α protein. J. Virol. 2008;82:11669–11681. doi: 10.1128/JVI.01559-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keckesova Z, Ylinen LM, Towers GJ. The human and African green monkey TRIM5α genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10780–10785. doi: 10.1073/pnas.0402474101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M, Garcia-Barrio M, Powell MD. Treatment of human immunodeficiency virus type 1 virions deleted of cyclophilin A by natural endogenous reverse transcription restores infectivity. J. Virol. 2003;77:4431–4434. doi: 10.1128/JVI.77.7.4431-4434.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klutch M, Woerner AM, Marcus-Sekura CJ, Levin JG. Generation of HIV-1/HIV-2 cross-reactive peptide antisera by small sequence changes in HIV-1 reverse transcriptase and integrase immunizing peptides. J. Biomed. Sci. 1998;5:192–202. doi: 10.1007/BF02253469. [DOI] [PubMed] [Google Scholar]
- Landau NR, Page KA, Littman DR. Pseudotyping with human T-cell leukemia virus type I broadens the human immunodeficiency virus host range. J. Virol. 1991;65:162–169. doi: 10.1128/jvi.65.1.162-169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier CR, Sandrin V, Eckert DM, Christensen DE, Chandrasekaran V, Alam SL, Aiken C, Olsen JC, Kar AK, Sodroski JG, Sundquist WI. Biochemical characterization of a recombinant TRIM5α protein that restricts human immunodeficiency virus type 1 replication. J. Virol. 2008;82:11682–11694. doi: 10.1128/JVI.01562-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanman J, Lam TT, Barnes S, Sakalian M, Emmett MR, Marshall AG, Prevelige PE., Jr Identification of novel interactions in HIV-1 capsid protein assembly by high-resolution mass spectrometry. J. Mol. Biol. 2003;325:759–772. doi: 10.1016/s0022-2836(02)01245-7. [DOI] [PubMed] [Google Scholar]
- Lanman J, Prevelige PE., Jr Kinetic and mass spectrometry-based investigation of human immunodeficiency virus type 1 assembly and maturation. Adv. Virus Res. 2005;64:285–309. doi: 10.1016/S0065-3527(05)64009-2. [DOI] [PubMed] [Google Scholar]
- Levin JG, Mitra M, Mascarenhas A, Musier-Forsyth K. Role of HIV-1 nucleocapsid protein in HIV-1 reverse transcription. RNA Biol. 2010;7:754–774. doi: 10.4161/rna.7.6.14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Hill CP, Sundquist WI, Finch JT. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature. 2000;407:409–413. doi: 10.1038/35030177. [DOI] [PubMed] [Google Scholar]
- Matlin KS, Reggio H, Helenius A, Simons K. Pathway of vesicular stomatitis virus entry leading to infection. J. Mol. Biol. 1982;156:609–631. doi: 10.1016/0022-2836(82)90269-8. [DOI] [PubMed] [Google Scholar]
- McDonald D, Vodicka MA, Lucero G, Svitkina TM, Borisy GG, Emerman M, Hope TJ. Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 2002;159:441–452. doi: 10.1083/jcb.200203150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyauchi K, Kim Y, Latinovic O, Morozov V, Melikyan GB. HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell. 2009;137:433–444. doi: 10.1016/j.cell.2009.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momany C, Kovari LC, Prongay AJ, Keller W, Gitti RK, Lee BM, Gorbalenya AE, Tong L, McClure J, Ehrlich LS, Summers MF, Carter C, Rossmann MG. Crystal structure of dimeric HIV-1 capsid protein. Nat. Struct. Biol. 1996;3:763–770. doi: 10.1038/nsb0996-763. [DOI] [PubMed] [Google Scholar]
- Murakami T, Freed EO. The long cytopasmic tail of gp41 is required in a cell type-dependent manner for HIV-1 envelope glycoprotein incorporation into virions. Proc. Natl. Acad. Sci. U.S. A. 2000;97:343–348. doi: 10.1073/pnas.97.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- Newman RM, Hall L, Kirmaier A, Pozzi LA, Pery E, Farzan M, O'Neil SP, Johnson W. Evolution of a TRIM5-CypA splice isoform in old world monkeys. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000003. e1000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisole S, Lynch C, Stoye JP, Yap MW. A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc. Natl. Acad. Sci. U.S.A. 2004;101:13324–13328. doi: 10.1073/pnas.0404640101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertel T, Hausmann S, Morger D, Züger S, Guerra J, Lascano J, Reinhard C, Santoni FA, Uchil PD, Chatel L, Bisiaux A, Albert ML, Strambio-De-Castillia C, Mothes W, Pizzato M, Grütter MG, Luban J. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 2011;472:361–365. doi: 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pornillos O, Ganser-Pornillos BK, Kelly BN, Hua Y, Whitby FG, Stout CD, Sundquist WI, Hill CP, Yeager M. X-ray structures of the hexameric building block of the HIV capsid. Cell. 2009;137:1282–1292. doi: 10.1016/j.cell.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pornillos O, Ganser-Pornillos BK, Yeager M. Atomic-level modelling of the HIV capsid. Nature. 2011;469:424–427. doi: 10.1038/nature09640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reicin AS, Ohagen A, Yin L, Hoglund S, Goff SP. The role of Gag in human immunodeficiency virus type 1 virion morphogenesis and early steps of the viral life cycle. J. Virol. 1996;70:8645–8652. doi: 10.1128/jvi.70.12.8645-8652.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rold CJ, Aiken C. Proteasomal degradation of TRIM5α during retrovirus restriction. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000074. e1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos JW, Maughan MF, Liao Z, Hildreth JEK, Clements JE. LuSIV cells: a reporter cell line for the detection and quantitation of a single cycle of HIV and SIV replication. Virology. 2000;273:307–315. doi: 10.1006/viro.2000.0431. [DOI] [PubMed] [Google Scholar]
- Rulli SJ, Jr, Muriaux D, Nagashima K, Mirro J, Oshima M, Baumann JG, Rein A. Mutant murine leukemia virus Gag proteins lacking proline at the N-terminus of the capsid domain block infectivity in virions containing wild-type Gag. Virology. 2006;347:364–371. doi: 10.1016/j.virol.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Sayah DM, Sokolskaja E, Berthoux L, Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430:569–573. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- Scholz I, Arvidson B, Huseby D, Barklis E. Virus particle core defects caused by mutations in the human immunodeficiency virus capsid N-terminal domain. J. Virol. 2005;79:1470–1479. doi: 10.1128/JVI.79.3.1470-1479.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian S, Luban J. TRIM5α selectively binds a restriction-sensitive retroviral capsid. Retrovirology. 2005;2:40. doi: 10.1186/1742-4690-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Aiken C. Saturation of TRIM5α-mediated restriction of HIV-1 infection depends on the stability of the incoming viral capsid. Virology. 2006;350:493–500. doi: 10.1016/j.virol.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:847–852. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, Sodroski J. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5α restriction factor. Proc. Natl. Acad. Sci. U.S.A. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanstrom R, Wills JW. Synthesis, assembly, and processing of viral proteins. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor, N. Y.: Cold Spring Harbor Laboratory Press; 1997. pp. 263–334. [PubMed] [Google Scholar]
- Tang C, Loeliger E, Kinde I, Kyere S, Mayo K, Barklis E, Sun Y, Huang M, Summers MF. Antiviral inhibition of the HIV-1 capsid protein. J. Mol. Biol. 2003a;327:1013–1020. doi: 10.1016/s0022-2836(03)00289-4. [DOI] [PubMed] [Google Scholar]
- Tang S, Ablan S, Dueck M, Ayala-López W, Soto B, Caplan M, Nagashima K, Hewlett IK, Freed EO, Levin JG. A second-site suppressor significantly improves the defective phenotype imposed by mutation of an aromatic residue in the N-terminal domain of the HIV-1 capsid protein. Virology. 2007;359:105–115. doi: 10.1016/j.virol.2006.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Hewlett I. Nanoparticle-based immunoassays for sensitive and early detection of HIV-1 capsid (p24) antigen. J. Infect. Dis. 2010;201(Suppl. 1):S59–S64. doi: 10.1086/650386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Murakami T, Agresta BE, Campbell S, Freed EO, Levin JG. Human immunodeficiency virus type 1 N-terminal capsid mutants that exhibit aberrant core morphology and are blocked in initiation of reverse transcription in infected cells. J. Virol. 2001;75:9357–9366. doi: 10.1128/JVI.75.19.9357-9366.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Murakami T, Cheng N, Steven AC, Freed EO, Levin JG. Human immunodeficiency virus type 1 N-terminal capsid mutants containing cores with abnormally high levels of capsid protein and virtually no reverse transcriptase. J. Virol. 2003b;77:12592–12602. doi: 10.1128/JVI.77.23.12592-12602.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JA, Bosche WJ, Shatzer TL, Johnson DG, Gorelick RJ. Mutations in human immunodeficiency virus type 1 nucleocapsid protein zinc fingers cause premature reverse transcription. J. Virol. 2008;82:9318–9328. doi: 10.1128/JVI.00583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JA, Gagliardi TD, Alvord WG, Lubomirski M, Bosche WJ, Gorelick RJ. Human immunodeficiency virus type 1 nucleocapsid zinc-finger mutations cause defects in reverse transcription and integration. Virology. 2006;353:41–51. doi: 10.1016/j.virol.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Thomas JA, Gorelick RJ. Nucleocapsid protein function in early infection processes. Virus Res. 2008;134:39–63. doi: 10.1016/j.virusres.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgen CA, Kratovac Z, Bieniasz PD, Hatziioannou T. Independent genesis of chimeric TRIM5-cyclophilin proteins in two primate species. Proc. Natl. Acad. Sci. U.S.A. 2008;105:3563–3568. doi: 10.1073/pnas.0709258105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt VM. Retroviral virions and genomes. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor, N. Y.: Cold Spring Harbor Laboratory Press; 1997. pp. 27–69. [PubMed] [Google Scholar]
- von Schwedler U, Stemmler TL, Klishko VY, Li S, Albertine KH, Davis DR, Sundquist WI. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 1998;17:1555–1568. doi: 10.1093/emboj/17.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schwedler UK, Stray KM, Garrus JE, Sundquist WI. Functional surfaces of the human immunodeciency virus type 1 capsid protein. J. Virol. 2003;77:5439–5450. doi: 10.1128/JVI.77.9.5439-5450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacharapornin P, Lauhakirti D, Auewarakul P. The effect of capsid mutations on HIV-1 uncoating. Virology. 2007;358:48–54. doi: 10.1016/j.virol.2006.08.031. [DOI] [PubMed] [Google Scholar]
- Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 2002;46:1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegers K, Rutter G, Kottler H, Tessmer U, Hohenberg H, Kräusslich HG. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J. Virol. 1998;72:2846–2854. doi: 10.1128/jvi.72.4.2846-2854.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthylake DK, Wang H, Yoo S, Sundquist WI, Hill CP. Structures of the HIV-1 capsid protein dimerization domain at 2.6 A resolution. Acta Crystallogr D Biol Crystallogr. 1999;55:85–92. doi: 10.1107/S0907444998007689. [DOI] [PubMed] [Google Scholar]
- Wu X, Anderson JL, Campbell EM, Joseph AM, Hope TJ. Proteasome inhibitors uncouple rhesus TRIM5α restriction of HIV-1 reverse transcription and infection. Proc. Natl. Acad. Sci. U.S.A. 2006;103:7465–7470. doi: 10.1073/pnas.0510483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap MW, Nisole S, Lynch C, Stoye JP. Trim5α protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10786–10791. doi: 10.1073/pnas.0402876101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee JK, Friedmann T, Burns JC. Generation of high-titer pseudotyped retroviral vectors with very broad host range. Methods Cell Biol. 1994;43(Pt. A):99–112. doi: 10.1016/s0091-679x(08)60600-7. [DOI] [PubMed] [Google Scholar]
- Zhao G, Ke D, Vu T, Ahn J, Shah VB, Yang R, Aiken C, Charlton LM, Gronenborn AM, Zhang P. Rhesus TRIM5α disrupts the HIV-1 capsid at the inter-hexamer interfaces. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002009. e1002009. [DOI] [PMC free article] [PubMed] [Google Scholar]