Abstract
This paper is the first report of the impact on the bacterial community in the midgut of the Asian corn borer (Ostrinia furnacalis) by the chitinase from the transgenic Trichoderma strain. In this study, we detected a change of the bacterial community in the midgut of the fourth instar larvae by using a culture-independent method. Results suggested that Proteobacteria and Firmicutes were the most highly represented phyla, being present in all the midgut bacterial communities. The observed species richness was simple, ranging from four to five of all the 16S rRNA clone libraries. When using Trichoderma fermentation liquids as additives, the percentages of the dominant flora in the total bacterial community in larval midgut changed significantly. The community of the genus Ochrobactrum in the midgut decreased significantly when the larvae were fed with the fermentation liquids of the transgenic Trichoderma strain Mc4. However, the Enterococcus community increased and then occupied the vacated niche of the Ochrobactrum members. Furthermore, the Shannon–Wiener (H) and the Simpson (1-D) indexes of the larval midgut bacterial library treated by feeding fermentation liquids of the transgenic Trichoderma strain Mc4 was the lowest compared with the culture medium, fermentation liquids of the wild type strain T30, and the sterile artificial diet. The Enterococcus sp. strain was isolated and characterized from the healthy larvae midgut of the Asian corn borer. An infection study of the Asian corn borer larvae using Enterococcus sp. ACB-1 revealed that a correlation existed between the increased Enterococcus community in the larval midgut and larval mortality. These results demonstrated that the transgenic Trichoderma strain could affect the composition of the midgut bacterial community. The change of the midgut bacterial community might be viewed as one of the factors resulting in the increased mortality of the Asian corn borer larvae.
Introduction
The Asian corn borer [Ostrinia furnacalis (Guenée)] is a Lepidopteran insect distributed in East and Southeast Asian countries, including countries such as China, Japan, Korea, Thailand, the Philippines, Indonesia, Malaysia, as well as some islands in the Pacific Ocean [1]. The species remains the most significant economic insect pest for corn. Our previous study revealed that an overexpression of the Metarhizium anisopliae chit42 gene with a chitin-binding domain (chBD) in Trichoderma koningii increases the biocontrol activity, particularly the virulence against the Asian corn borer larvae [2]. Transgenic Trichoderma could also hinder the growth and development of the larvae. We speculated that the bacterial community of the alimentary tract might be one of the important factors for insect health. Studies on bacterial communities focusing on the Lepidopteran midgut are limited [3], [4], [5], [6]. To date, no reports are available on the change of the bacterial diversity in the midgut of the Asian corn borer larvae induced by the transgenic Trichoderma strain overexpressing entomopathogenic genes.
Insect digestive system is a complex ecosystem harboring a large number of microorganisms. These microorganisms constitute communities. Microflora in the digestive system is an important factor in the nutritional, immune, and defense functions under normal circumstances [7], [8]. On one hand, the nutritional physiology of the insects is closely linked with their gut microorganisms [9]. The community structures and metabolic activities of microorganisms are affected by the gut microenvironment of the insect. On the other hand, intestinal microorganisms can also affect the physiological activity of the insect. Studies on the microbial diversity of the insect midgut are limited and primarily focus on termites [10], aphids [11] and honeybees [12].
Microbial communities are dynamic. Feeding, digestion, and excretion can facilitate changes in composition and structure [4]. These changes can be attributed to a variety of factors, including nutrient availability, physical aspects of the environment, and proximity to other organisms [13], [14], [15]. Chitinases from Trichoderma and Metarhizium have been proven useful in biocontrol activities against a wide range of plant fungal pathogens [16] and insects [17]. Therefore, chitinase from both sources seemed to be an adverse factor facilitating changes of the gut bacterial composition and structure.
Our previous study revealed that an overexpression of the Metarhizium anisopliae chit42 gene with a chitin-binding domain in Trichoderma koningii displayed an increased biocontrol activity, particularly in the virulence against the Asian corn borer larvae. However, little is known about the anti-insect mechanism of the transgenic Trichoderma strain in terms of a midgut microbial ecology. In this study, we used a culture-independent method to detect the change in whole bacterial community of the fourth instar Asian corn borer larvae in response to the fermentation liquid from the transgenic Trichoderma strain overexpressing the chit42 gene with a chBD.
Materials and Methods
Microbial Strains
The Escherichia coli host strain DH5 α was grown in an LB medium. Wild type (WT) T. koningii T30 was provided by the Biocontrol Laboratory of Plant Diseases, Shenyang Agricultural University, China. This species has been used for the biocontrol of soil-borne diseases in a variety of vegetable crops. The chit42 gene was cloned from M. anisopliae. The chBD from bitter melons (Momordica charantia) was fused with the chit42 gene in the C-terminal to constitute the hybrid chitinase gene. The hybrid chitinase gene was controlled under the TrpC promoter and TrpC terminator to constitute an overexpression cassette. The overexpression cassette was transformed into the WT Trichoderma strain T. koningii T30 by using an Agrobacterium-mediated transformation technique. The Trichoderma transformants harboring the hybrid chitinase gene overexpression cassette were selected and labeled as T. koningii Mc4. T. koningii Mc4 had been proven to have an enhanced insecticidal activity against the Asian corn borer larvae and antifungal capability toward Fusarium verticillioides and Rhizoctonia solani [2].
Preparation of Fermentation Liquids from Trichoderma Strain
Mycelia of the WT strain Trichoderma T30 and transgenic Trichoderma strain Mc4 each were isolated and transferred into a 100 mL fermentation medium (MgSO4.7H2O 0.6 g/L, FeSO4.7H2O 0.1 g/L, NH4NO3 3.0 g/L, KH2PO4 2.0 g/L, colloidal chitin 10% v/v, powdered chitin 2% w/w), inoculated at 28°C for 180 rpm in a shaking table at constant temperature until the highest chitinase activity was achieved. The highest chitinase activity values of T30 and Mc4 were 12.4 U/mL and 46.8 U/mL, respectively. One unit of chitinase activity is defined as the amount of enzyme required to release 1 µmol of N-acetyl-ß-glucosamine per hour. Fermentation liquids were prepared by collecting the supernatant after removing insoluble substances as well as the debris of the mycelium by centrifugation at 4°C, 12000 g for 30 min. The artificial diets were mixed with the fermentation liquids of T30 and Mc4 (artificial diet/fermentation liquids = 1∶1.2 w/v), whereas the artificial diets mixed with the supernatant of the control fermentation medium was used as the control. The Asian corn borer larvae reared on the artificial diets without any additives were used as the control.
Chitinase produced by Trichoderma in the fermentation medium could degrade chitin into N-acetyl-D-(+)-glucosamine (NAG). This derivative could more easily be used as a carbon source compared with chitin. A 3, 5-dinitrosalicylic acid method was used to determine the NAG content in the fermentation liquids. The chitinase fermentation liquids were treated by boiling for 5 min to inactivate the enzyme content. The effect of enzyme-inactivated fermentation liquids on the midgut bacterial community of fourth instar larvae was analyzed against the control treatment, in which larvae were reared on the sterile artificial diets.
Insect Rearing and Treatments
Second instar larvae of the Asian corn borer were obtained from the Institute of Plant Protection, Chinese Academy of Agricultural Sciences, and were reared on the sterile artificial diet with for 14 h: 10 h (light: dark) at 25°C under 40% to 60% relative humidity until the fourth instar.
Strong and healthy larvae with the similar sizes were selected and starved for 24 h before being fed with the artificial diets under different treatments. The selected larvae were divided into six groups. Each group contained 20 larvae, among which 10 were kept for further feeding on sterile artificial diets. Another 10 were selected for comparison of the midgut bacterial libraries between different treatments. The treatments included larvae reared on the sterile artificial diets as well as artificial diets supplemented with the supernatant of the control fermentation liquids, artificial diets supplemented with the supernatant of the fermentation liquids of Trichoderma WT strain T30, artificial diets supplemented with the supernatant of the fermentation liquids of the transgenic Trichoderma strain Mc4, artificial diets supplemented with the supernatant of the enzyme-inactivated fermentation liquids of T30, and artificial diets supplemented with the supernatant of the enzyme-inactivated fermentation liquids of Mc4. All feeding treatments were completed for up to three days. The experiments were repeated thrice.
Midgut Separation and DNA Extraction
Fourth instar larvae were placed in sterilized culture dishes and starved for 6 h to reduce the artificial diets content. The larvae were surface-sterilized and dissected with anatomic scissors and forceps. The midguts were removed and stored at −80°C prior to DNA isolation. Total microbial DNA was extracted from the midgut by using the protocol mentioned in Broderick et al. [3]. DNA was quantified and adjusted to 10 ng/µL [18] using Quant-iT™ PicoGreen® dsDNA Reagent and Kits (Invitrogen). A 2× PicoGreen working reagent was prepared by adding 5 µL of a 200-fold dilution into 995 µL of 1× TE buffer. Thereafter, 10 µL of the DNA extracts were added into 990 µL of 1× TE buffer to constitute the DNA sample. Subsequently, 1 mL of the DNA sample was added into 1 mL of the 2× PicoGreen working reagent and reacted for 2 min at room temperature and protected from light. The sample fluorescence was measured using a spectrofluorometer. The DNA standard curve was prepared using a lambda DNA.
Construction of the Midgut16S rRNA Gene Library
The midgut 16S rRNA gene libraries of the fourth instar larvae after treatment were constructed. Total DNA of the samples was separately PCR-amplified by using the primers 27F and 1492R [19]. PCR reactions were performed in a 25 µL solution containing 1 ng/µL DNA, 1× PCR reaction buffer (TaKaRa, Japan), 0.5 mM additional MgCl2 (TaKaRa, Japan), 0.2 µM of each primer (Sangon Biotech, Shanghai, China), 0.4 mM of each dNTP (TaKaRa, Japan), and 1 U of rTaq DNA polymerase (TaKaRa, Japan). PCR was performed in vapor (Eppendorf, Germany) under appropriate conditions: starting at 94°C for 5 min, followed by 30 cycles at 94°C for 30 s, 55°C for 30 s, 72°C for 1 min and 30 s, and 72°C for 10 min. The purified PCR products were cloned into a pMD19-T simple vector system (TaKaRa, Japan) according to the manufacturer’s instructions. The ligation products were transformed into the competent E. coli DH5α cells. The clones were screened using an LB medium containing ampicillin (100 µg/L) with isopropyl-β-D-thiogalactopyranoside and 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside. At least 150 white clones were selected randomly for each clone library.
Phylogenetic Analysis of the Midgut16S rDNA Gene Library
The PCR products amplified from each clone were digested by two Fast-Digest restriction enzymes (HinfI and AfaI, 37°C for 30 min). The digestion reactions (30 µL of the total volume) contained 15 µL of the PCR products, 3 µL of 10× Fast-Digest buffer (Fermentas), 1 µL of the Fast-Digest enzyme (HinfI or AfaI, Fermentas), and 11 µL of dH2O. The restriction enzyme analysis of the positive clones in the 16S rDNA library was conducted through an agarose gel electrophoresis (2%). All the positive clones were classified according to the patterns of the restriction fragments digested with the restriction enzymes in agarose gel. The clones that exhibited the same patterns of restriction fragments after digestion were classified into the same operational taxonomic units (OTUs). The sequences were initially analyzed using SeqMan and then rapidly aligned using the Greengenes Web application (http://greengenes.lbl.gov). Chimeras were verified against Ribosomal Database Project II and then removed from the group for further analysis.
An estimate of the library was employed to assess whether the constructed midgut 16S rDNA gene libraries were sufficiently large by using the formula
where C is the percent coverage, n is the number of sequence appearing once in each library, and N is the total number of clones in each library [20].
SChao1 was calculated for each clone library to assess whether the libraries were sufficiently large to yield a stable estimate of the phylotype richness using a Web-based program (http://www.aslo.org/lomethods/free/2004/0114a.html). PAST software was used to calculate the Shannon–Wiener (H) and the Simpson (1-D) indexes for each of the libraries constructed [21].
The sequences from each OTU were submitted to GenBank for BLAST analyses [22]. Most of the associated sequences were downloaded in a FASTA format and then aligned using the program MEGA 4.0. The aligned tests were converted to the MEGA format before the phylogenetic trees were constructed using the neighbor-joining method with 1000 bootstrap replicates [23].
Analysis of the Microflora Distribution
All the positive clones in the library were PCR-amplified using the universal primers in the pMD19-T simple vector. The PCR products were digested. Clones belonging to the same OTU were counted. The percent of microflora distribution was obtained using the formula
where A is the percentage of microflora distribution in each clone library, b is the total clone numbers belonging to the same OTU in each library, and N is the total number of clones in each library.
Bacterial Isolation from the Midgut of the Asian Corn Borer Larvae
Enterococcus sp. was isolated from the midgut of the healthy Asian corn borer larvae. The cutting instruments, including anatomic scissors and forceps, were disinfected by immersion in 95% ethanol. Larvae surface were sterilized before dissecting the midguts using anatomic scissors and forceps. The digestive tract was shredded and deposited in an Eppendorf tube containing 200 µL of sterile saline solution. The decimally diluted digestive tract solutions were mixed with the Enterococcus Agar (Qingdao Hope Biol-Technology Co., Ltd, China) to isolate and culture the Enterococcus sp. from different species. The plates were sealed with parafilm and then incubated at 37°C for at least 24 h.
Phylogenic Analysis of Enterococcus sp. Isolated from the Larval Midgut
Enterococcus sp. was cultured in the Enterococcus broth. The genomic DNA of Enterococcus sp. was extracted according to the protocols by using a Wizard® Genomic DNA Purification Kit (Promega, USA). The 16S rRNA gene sequence of Enterococcus sp. was PCR-amplified using the primers 27F and 1492R. PCR reactions were performed in a system volume of 25 µL system according to the standard protocols under approximate conditions. The PCR products were purified and then ligated with the pMD19T simple vector. Positive clones were obtained by transforming the ligation products into E. coli DH 5α competent cells. The 16S rRNA sequences of Enterococcus sp. was submitted to GenBank for BLAST analyses. Most associated sequences were downloaded in a FASTA format and then aligned using the program MEGA 4.0. The aligned tests were converted to the MEGA format before phylogenetic trees were constructed using the neighbor-joining method with 1000 bootstrap replicates.
Infection of the Asian Corn Borer Larvae Using Enterococcus sp. Under Germfree Conditions
The Asian corn borer larvae were reared on the artificial diets until fourth instar. Larvae with similar dimensions and sizes were selected and starved for 24 h before conducting the infection study. A disinfection test on the body surface of the larvae was performed. The infection study with the isolated Enterococcus sp. was performed on the healthy larvae, which were kept at 25°C and then fed with the sterile artificial diets under germfree conditions. Dilutions (10−3, 10−5, and 10−7) of an overnight pure culture of Enterococcus sp. were harvested and suspended in physiological saline. These dilutions of Enterococcus sp. were mixed thoroughly with the sterile artificial diets. For each experiment, 20 larvae were used. All the experiments were conducted in triplicates with three biological repetitions. The control diets were supplemented with physiological saline and sterile distilled water.
The infectivity of the isolated Enterococcus sp. (dilution of 10−5) was also examined by injecting into the body cavity of the Asian corn borer larvae. A tuberculin syringe with fine intradermal needle was used to inject the body cavity of the larvae with 0.02 mL of the Enterococcus sp. suspension according to the methods described by Lysenko. The control diets were supplemented with physiological saline and sterile distilled water. For each experiment, 20 larvae were used. The experiments were repeated thrice.
Nucleotide Sequence Accession Number
The 16S rRNA gene sequences from this study were deposited in GenBank under accession numbers JX679623 to JX679640. The 16S rRNA gene sequence of the isolated Enterococcus sp. in this study was deposited in GenBank under accession number KC243406.
Results
Construction of the Clone Libraries
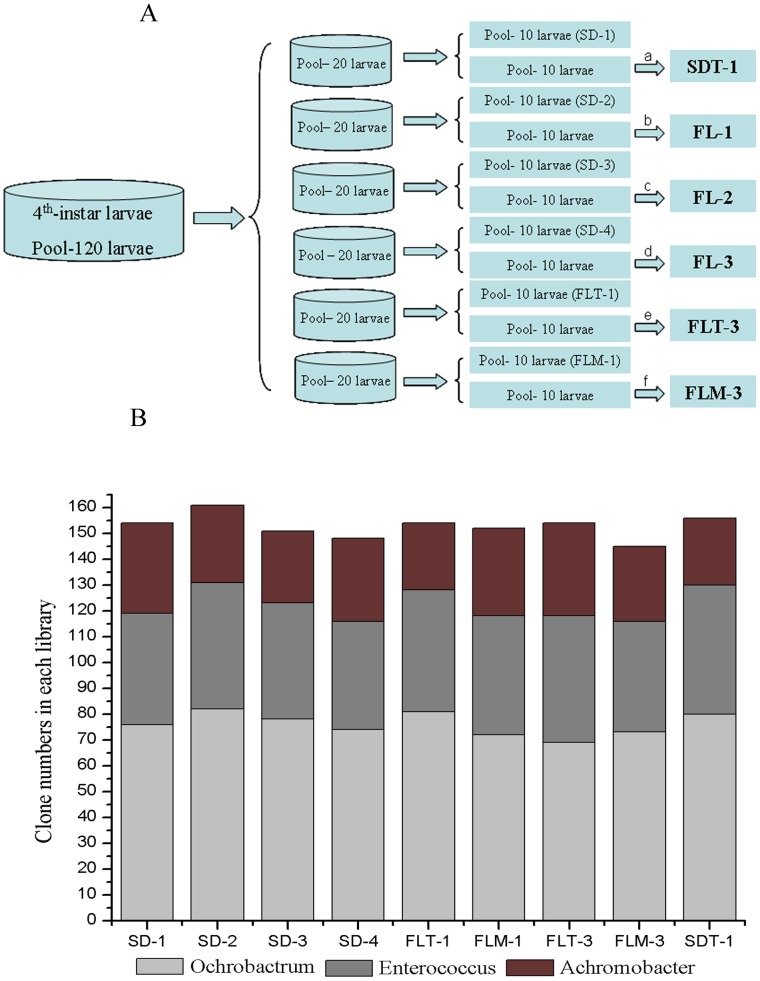
A total of 12 midgut bacterial libraries were obtained (Figure 1A) and called: SD-1 (midgut bacterial library of fourth instar larvae reared on the sterile artificial diets in the first day of feeding), SD-2 (midgut bacterial library of fourth instar larvae reared on the sterile artificial diets in the first day of feeding), SD-3 (midgut bacterial library of fourth instar larvae reared on the sterile artificial diets in the first day of feeding), SD-4 (midgut bacterial library of fourth instar larvae reared on the sterile artificial diets in the first day of feeding), FLT-1 (midgut bacterial library of fourth instar larvae reared on the sterile artificial diets in the first day of feeding), FLM-1 (midgut bacterial library of fourth instar larvae reared on the sterile artificial diets in the first day of feeding), SDT-1 (midgut bacterial library of fourth instar larvae reared on the sterile artificial diets for 3 d), FL-1 (midgut bacterial library of fourth instar larvae reared on the sterile artificial diets supplemented with the supernatant of the control fermentation liquids for 3 d), FL-2 (midgut bacterial library of fourth instar larvae reared on the sterile artificial diets supplemented with the supernatant of T30 fermentation liquids for 3 d), FL-3 (midgut bacterial library of fourth instar larvae reared on the sterile artificial diets supplemented with the supernatant of Mc4 fermentation liquids for 3 d), FLT-3 (midgut bacterial library of fourth instar larvae reared on the sterile artificial diets supplemented with the supernatant of the enzyme-inactivated T30 fermentation liquids for 3 d), and FLM-3 (midgut bacterial library of fourth instar larvae reared on the sterile artificial diets supplemented with the supernatant of the enzyme-inactivated Mc4 fermentation liquids for 3 d).
Figure 1. Construction and analysis of bacterial 16S rRNA clone libraries of larval midgut.
(A) Construction strategy of larval midgut bacterial libraries. SD-1 (midgut bacterial library of fourth instar larvae reared on the sterile artificial diets in the first day of feeding), SD-2 (midgut bacterial library of fourth instar larvae reared on the sterile artificial diets in the first day of feeding), SD-3 (midgut bacterial library of fourth instar larvae reared on the sterile artificial diets in the first day of feeding), SD-4 (midgut bacterial library of fourth instar larvae reared on the sterile artificial diets in the first day of feeding), FLT-1 (midgut bacterial library of fourth instar larvae reared on the sterile artificial diets in the first day of feeding), and FLM-1 (midgut bacterial library of fourth instar larvae reared on the sterile artificial diets in the first day of feeding), SDT-1 (midgut bacterial library of fourth instar larvae reared on the sterile artificial diets for 3 d), FL-1 (midgut bacterial library of fourth instar larvae reared on the sterile artificial diets supplemented with the supernatant of the control fermentation liquids for 3 d), FL-2 (midgut bacterial library of fourth instar larvae reared on the sterile artificial diets supplemented with the supernatant of T30 fermentation liquids for 3 d), FL-3 (midgut bacterial library of fourth instar larvae reared on the sterile artificial diets supplemented with the supernatant of Mc4 fermentation liquids for 3 d), FLT-3 (midgut bacterial library of fourth instar larvae reared on the sterile artificial diets supplemented with the supernatant of the enzyme-inactivated T30 fermentation liquids for 3 d), and FLM-3 (midgut bacterial library of fourth instar larvae reared on the sterile artificial diets supplemented with the supernatant of the enzyme-inactivated Mc4 fermentation liquids for 3 d); a, fourth instar larvae reared on sterile artificial diets for 3 d; b, fourth instar larvae reared on sterile artificial diets supplemented with supernatant of control fermentation liquids for 3 d; c, fourth instar larvae reared on sterile artificial diets supplemented with supernatant of T30 fermentation liquids for 3 d; d, fourth instar larvae reared on sterile artificial diets supplemented with supernatant of Mc4 fermentation liquids for 3 d; e, fourth instar larvae reared on sterile artificial diets supplemented with supernatant of enzyme-inactivated T30 fermentation liquids for 3 d; f, fourth instar larvae reared on sterile artificial diets supplemented with supernatant of enzyme-inactivated Mc4 fermentation liquids for 3 d. (B) Comparison of dominant flora in the larval midgut between different clone libraries.
The bacterial communities were similar regardless of the larvae were collected from the first day of feeding on the sterile artificial diets (clone libraries of SD-1, SD-2, SD-3, SD-4, FLT-B-1, and FLM-B-1) or after feeding for 3 d on the same diets (clone library of SDT-1), or feeding on the artificial diets supplemented with fermentation medium without inoculation (clone library of FL-1). No obvious differences were observed when larvae were fed with the sterile artificial diets supplemented with the supernatant of enzyme-inactivated fermentation liquids for 3 d (clone library of FLT-B-3 and FLM-B-3), as compared with the larvae reared on the artificial diets for 3 d (Figure 1B). By feeding larvae with the artificial diets supplemented with fermentation liquids of T30 (clone library of FL-2) and Mc4 (clone library of FL-3), significant changes were observed in the proportion of each species accounting for the bacterial community. Therefore, the bacterial clone libraries of SDT-1, FL-1, FL-2, and FL-3 were further analyzed.
Statistical Analysis of the Bacterial 16S rRNA Gene Libraries
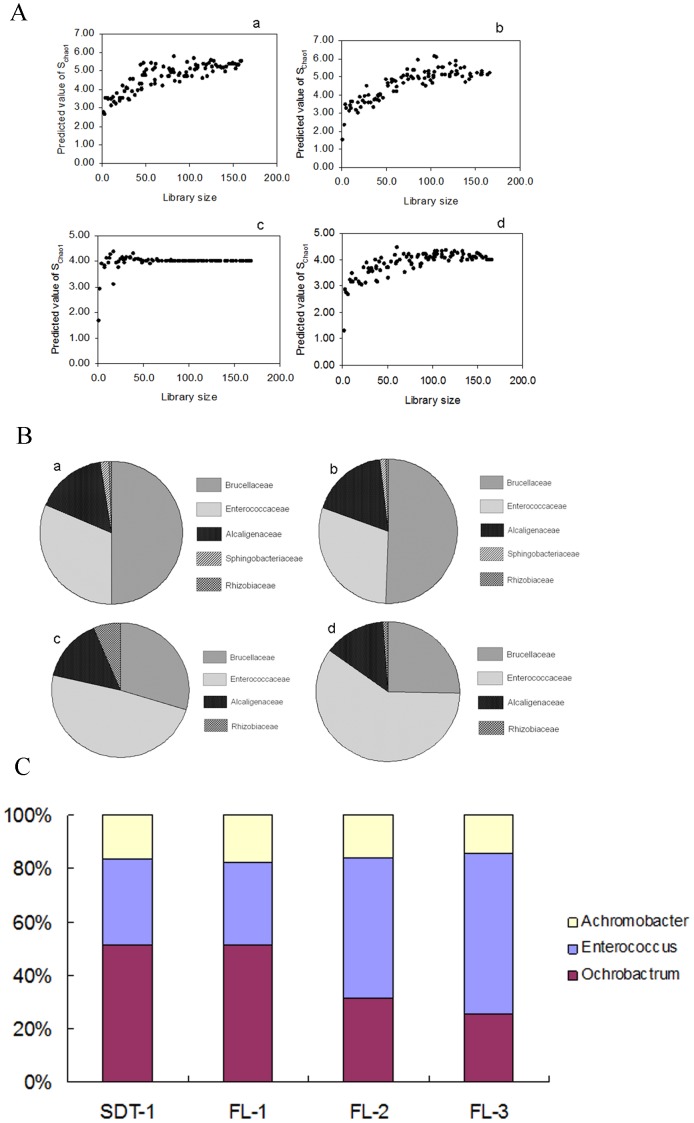
Four bacterial 16S rRNA gene libraries (SDT-1, FL-1, FL-2, and FL-3) were statistically analyzed ( Table 1 ). A total of 160 clones of the SDT-1 library were divided into five OTUs, 158 clones of the FL-1 library were divided into five OTUs, 169 clones of the FL-2 library were divided into four OTUs, and 166 clones of the FL-3 library were divided into four OTUs. The coverage of the four clone libraries ranged from 98.7% to 99.4%, indicating that these 16S rRNA gene sequences represented the majority of the bacterial community in larval midgut of the Asian corn borer. The Shannon–Wiener and Simpson indexes indicated that the diversity of bacteria in FL-3 was the lowest among the four clone libraries.
Table 1. Characteristics of 16S rRNA gene libraries constructed from midgut bacterial communities in fourth instar larvae of the Asian corn borer.
| Library name | Diets | No. ofsequences | No. of OTUSobserved | Good’s coverage | Shannon H | Simpson 1-D |
| SDT-1 | Sterile artificial diets | 160 | 5 | 99.4% | 1.117(1.002–1.204) | 0.6287(0.5779–0.6646) |
| FL-1 | Control artificial diets | 158 | 5 | 98.7% | 1.097(0.9829–1.181) | 0.6219(0.5669–0.6583) |
| FL-2 | T30-treated artificial diets | 169 | 4 | 99.4% | 1.170(1.065–1.247) | 0.6451(0.5953–0.6833) |
| FL-3 | Mc4-treated artificial diets | 166 | 4 | 99.4% | 0.9831(0.8611–1.704) | 0.5610(0.4927–0.6131) |
For the four bacterial 16S rRNA gene clone libraries constructed, Schao1 was calculated for each library and then plotted to the library size (Figure 2A). The Schao1 of the four libraries all reached an asymptote, indicating that the libraries were sufficiently large to yield a stable estimate of the phylotype richness.
Figure 2. Distribution of midgut bacterial communities between clone libraries of SDT-1, FL-1, FL-2, and FL-3.
(A) Predicted numbers of phylotypes based on the SChao1 of the 16S rRNA clone libraries. a, clone library of SDT-1; b, clone library of FL-1; c, clone library of FL-2; d, clone library of FL-3. X axis, Bacterial clone numbers in each library; Y axis, Predicted value of Schao1. (B) The bacterial composition in the larval midgut of fourth instar at the family level. a, clone library of SDT-1; b, clone library of FL-1; c, clone library of FL-2; d, clone library of FL-3. (C) Comparison of genera Achromobacter, Enterococcus, and Ochrobactrum distribution in the larval midguts between clone libraries of SDT-1, FL-1, FL-2, and FL-3.
Phylogenetic Analysis of the Midgut Bacterial Communities of the Fourth Instar Larvae
The composition of the bacterial communities was analyzed based on the bacterial 16S rRNA gene sequences obtained from the midgut of the fourth instar larvae reared on sterile artificial diets.
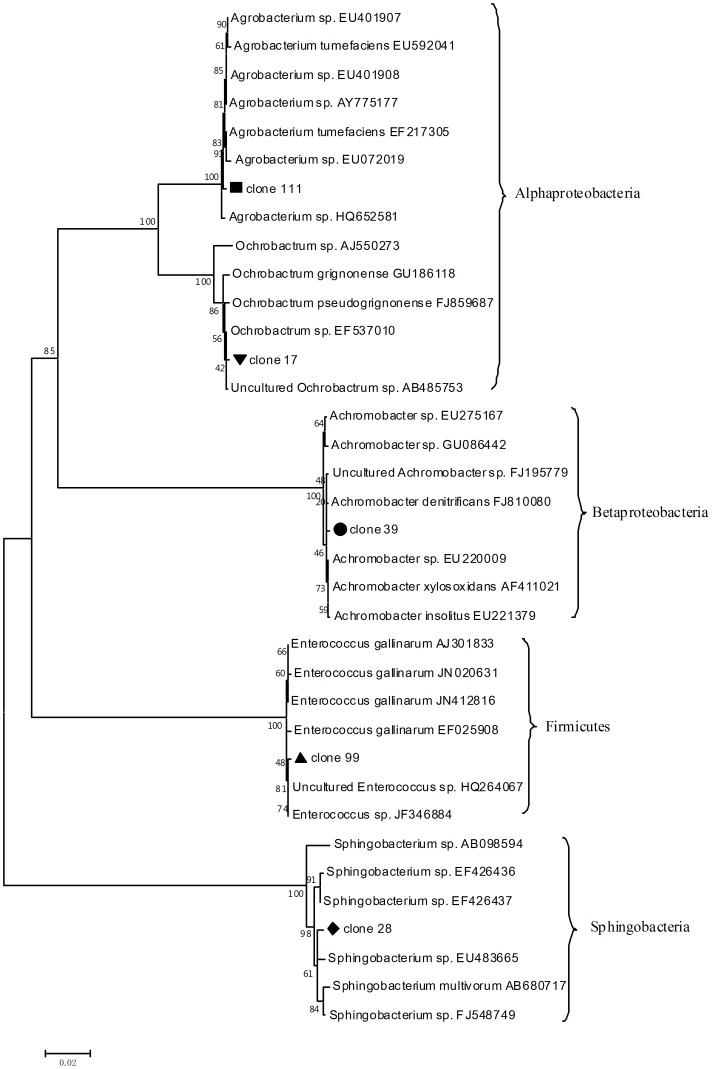
At the family level, the dominant families in SDT-1 were Brucellaceae (50.0%), Enterococcaceae (31.2%), and Alcaligenaceae (16.3%) (Figure 2B). The remaining sequences were affiliated with Sphingobacteriaceae (1.9%) and Rhizobiaceae (0.6%) (Figure 2B). All the sequences were clustered into four classes: Alphaproteobacteria (50.6%), Betaproteobacteria (16.3%), Firmicutes (31.2%), and Sphingobacteria (1.9%) (Figure 3).
Figure 3. Phylogenetic neighbor-joining trees of representative OTUs in the midgut bacterial clone libraries.
The numbers on the branches are bootstrap values obtained from 1000 bootstrap replicates.
BLAST searches were performed against the nr-database of the National Center for Biotechnology Information. Table 2 summarizes the BLAST results of the representative OTUs in the clone libraries.
Table 2. Partial 16S rDNA sequences isolated in this study.
| Closet known relative | Phylogenetically related class | Sequence length (bp) | Identity (%) | Accession no.* |
| Uncultured Ochrobactrum sp. | Alphaproteobacteria | 1446 | 99 | AB485753 |
| Enterococcus sp. GYPB01 | Firmicutes | 1524 | 99 | JF346884 |
| Achromobacter denitrificans | Betaproteobacteria | 1495 | 99 | FJ810080 |
| Sphingobacterium sp. GF2B | Sphingobacteria | 1494 | 99 | FJ548749 |
| Agrobacterium sp. CIP 106281 | Alphaproteobacteria | 1449 | 99 | EU401908 |
The accession number of the closest relative is indicated between brackets.
Effects of the Transgenic Trichoderma Strain Fermentation Liquids on the Larval Midgut Bacterial Communities of the Asian Corn Borer
Among all treatments to the larval midgut, Ochrobactrum, Enterococcus, and Achromobacter were found to be the dominant bacterial microflora. However, significant changes in dominant bacterial communities were observed after feeding on artificial diets supplemented with the fermentation liquids of the transgenic Trichoderma strain (Figure 2C).
In SDT-1, Ochrobactrum and Enterococcus accounted for 50.0% and 31.2% of the 16S rRNA gene sequences, respectively. In FL-1, Ochrobactrum and Enterococcus accounted for 50.6% and 29.7% of the 16S rRNA gene sequences, respectively. In FL-2, however, these numbers shifted to 29.6% and 49.1%, respectively. A similar trend was observed in FL-3, in which the Ochrobactrum and Enterococcus members shifted to 25.3% and 59.6% of the 16S rRNA gene sequences, respectively. Additionally, no significant change was detected in the numbers of genus Achromobacter. In SDT-1, FL-1, FL-2, and FL-3, Achromobacter accounted for 16.3%, 17.7%, 14.8%, and 13.9% of the clones in each library, respectively.
Bacterial Isolation and Phylogenetic Analysis
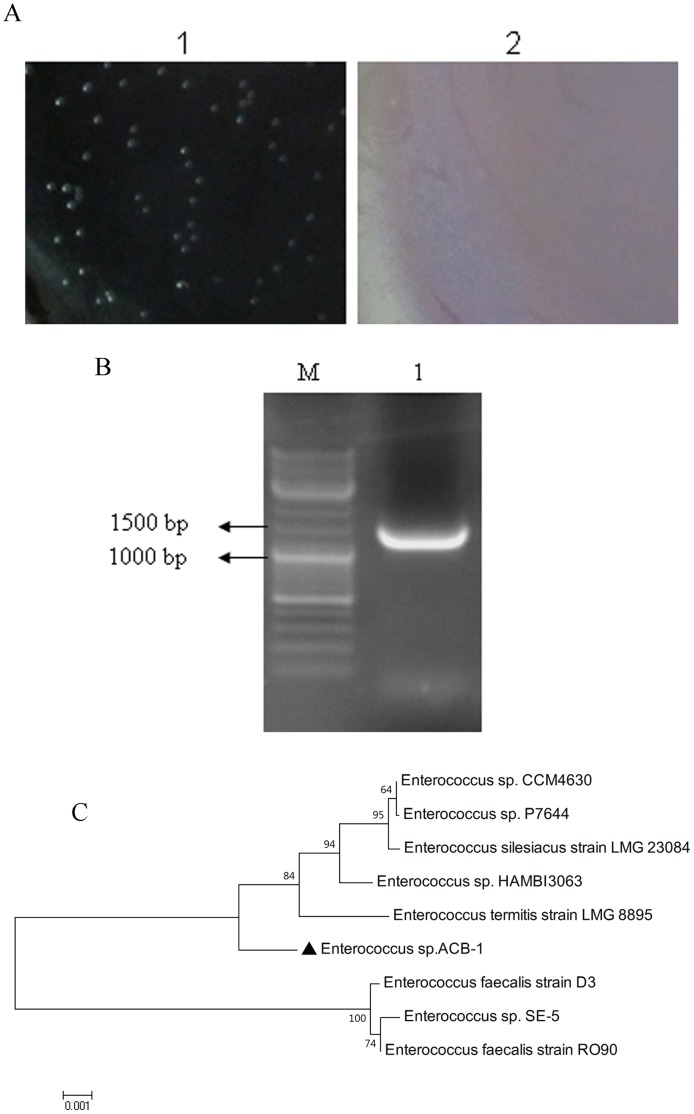
Enterococcus was isolated from midguts of the larvae reared on the sterile artificial diets. The isolated Enterococcus strain could hydrolyze the aesculin content in the medium and then react with an ion to generate a brownish black substance. The colony appeared grayish-white, opaque, round, low-arched, wet, and smooth. The colony exhibited a good growth at 45°C under pH 9.6, and can resist 60°C for 30 min [25].
The 16Sr DNA sequence was cloned using the primers 27f and 1492r (Figure 4A). The results of the phylogenetic analysis are indicated in Figure 4B.
Figure 4. Isolation and characterization of the isolated Enterococcus sp. ACB-1 from healthy midgut of the Asian corn borer larvae.
(A) Colony appearance of the Enterococcus sp. ACB-1 growing on Enterococcus agar. 1, Enterococcus agar with the Enterococcus sp. ACB-1 grown on it; 2, Enterococcus agar without the Enterococcus sp. ACB-1 grown on it. (B) 16S rDNA fragment amplification of the Enterococcus sp. ACB-1. (C) Phylogenetic neighbor-joining trees of the Enterococcus sp. ACB-1 (▴). The numbers on the branches are bootstrap values obtained from 1000 bootstrap replicates.
Infection of the Larvae Rearing on the Sterile Artificial Diets
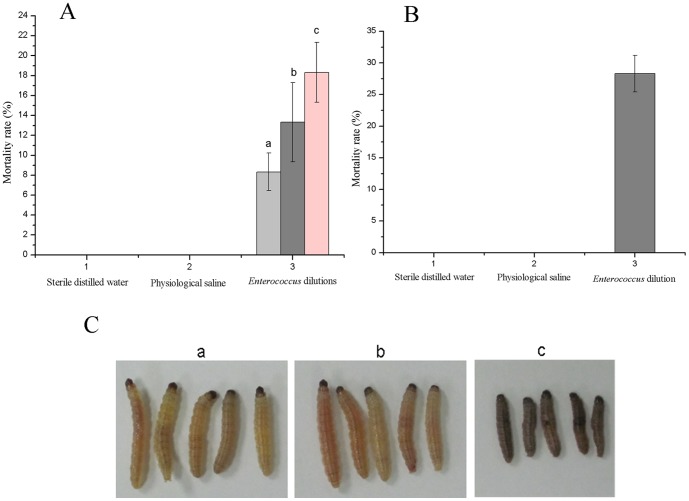
The Enterococcus strain was pathogenic for the Asian corn borer larvae. The body of the larvae appeared black after the infection study (Figure 5C). The infectious symptom of the Enterococcus strain by injection could be observed after an incubation period of 1 d. After an incubation period of 2 d, the larvae began to die. Mortality reached 28.3% after incubation for 5 d (Figure 5B). The infection study by feeding was analyzed using three dilution degrees of a pure of Enterococcus strain culture suspended in physiological saline. A higher Enterococcus concentration resulted in a shorter time for the infectious symptom to appear. Larvae infection by injection was more severe than that by feeding. This result was in accordance with the previous report by Lysenko [24]. When using a dilution of 10−5 pure Enterococcus strain culture by feeding, mortality only reached 13.3% after incubation for 5 d (Figure 5A). No mortality was observed in the physiological saline control and sterile distilled water control in both injection and feeding treatment within 5 d.
Figure 5. Pathogenic studies of the Asian corn borer larvae by the Enterococcus sp. ACB-1.
(A) Mortality of the Asian corn borer larvae after feeding strain of the Enterococcus sp. ACB-1. a, Enterococcus sp. ACB-1 suspension with dilution of 10−3; b, Enterococcus sp. ACB-1 suspension with dilution of 10−5; c, Enterococcus sp. ACB-1 suspension with dilution of 10−7. (B) Mortality of the Asian corn borer larvae after injecting strain of the Enterococcus sp. ACB-1. (C) Larvae appearance after incubation for 5 d by injection strain of the Enterococcus sp. ACB-1 with dilution of 10−5. a, Sterile distilled water; b, Physiological saline; c, Enterococcus sp. ACB-1 suspension with dilution of 10−5.
Discussion
Studies on isolating and characterizing the Lepidopteran intestinal bacterial community are limited. The midgut bacterial community of the gypsy moth larvae was reported to be relatively simple [3]. Cabbage white butterfly larvae were shown to contain a simple bacterial community with a species richness ranging from six to 15 [4]. Phylotypes belonging to five genera were identified from the midguts of the laboratory population of the Helicoverpa armigera larvae [5]. In this study, we surveyed the changes in the bacterial communities in midgut of fourth instar of the Asian corn borer larvae after feeding on artificial diets supplemented with the supernatant of the fermentation liquids of the transgenic strain to understand the association between the bacterial community imbalance in the midgut and the mortality of the corn borer larvae. Results demonstrated that the observed species richness was five, a number that was relatively simple compared with other gut environments such as in the hindguts of isopteran termites [26]. No report is available on the relationship between the complexity of the bacterial community of an insect and robustness to external factors. However, experimental evidence suggests that species diversity in small grassland plots enhances invasion resistance [27]. A study on the species diversity and invasion resistance in a marine ecosystem also revealed that increased species richness significantly decreased invasion success. That is, a more diverse an ecosystem is more resistant to exotic species [28]. Based on these studies, we infer that the community of the Lepidopteran midgut might be easily damaged and consequently lose its ecological buffering capability.
The intestinal dominant bacterial flora depends on different insect species. Ochrobactrum, Enterococcus, and Achromobacter were identified as the dominant genera, which belonged to the phyla Proteobacteria and Firmicutes, respectively. These results are consistent with the previously reported bacterial communities in other Lepidopteran midguts, such as the Lymantria dispar, H. armigera, Bombyx mori, Schistocerca gregaria, [29] and those in the midgut of the natural aphid populations [30]. Although the taxonomic status of the midgut bacterial communities was consistent at the phylum level in the different insects discussed above, significant differences in taxonomic status were observed at the class level. Except for the genus Enterococcus, genera Ochrobactrum and Achromobacter were not detected in other bacterial communities of the Lepidoptera insects’ midguts, such as Lymantria dispar, Pieris rapae, and H. armigera. Similarly, genera Enterobacter, Pantoea, and Acinetobacter detected in the midguts of other Lepidopteran insects were not found in midgut of the Asian corn borer. The bacteria belonging to Sphingobacteria only accounted for less than 2% of the bacterial community in the Asian corn borer midgut, although they were dominant in midgut of the isopteran termites [31], [32], [33]. Proteobacteria has been detected in many insect species, which implies the presence of common Proteobacteria in gut environment of the insect.
We found that the Trichoderma fermentation liquids altered the relative proportions of the different midgut flora components. Ochrobactrum and Enterococcus were detected as the dominant members in all bacterial libraries. In FL-2 and FL-3, the proportion of Ochrobactrum was reduced by 50% in the community compared with SDT-1 and FL-1. Enterococcus, however, was increased by more than 50%. A variety of factors could facilitate the changes of the microbial community composition. The reason why an improved chitinase activity could facilitate a dynamic change in the midgut microbial community is remains unclear. Chitin, a component of the exoskeleton and gut lining of insects, is a metabolic target for pest control agents. Both the chitinases produced by Trichoderma and Metarhizium could degrade chitin. Ingested chitinase can damage the peritrophic membrane, potentially facilitating the changes in intestinal bacterial community structure. In addition, chitin degradation in the midgut and peritrophic membrane might result in the midgut cell electrolyte leakage, possibly providing a new source of nutrition for the midgut bacterial community. When the larvae were treated with chitinase fermentation liquids, the physiological activity of the insect was affected because of chitin content degradation on the peritrophic membrane structure of the larvae. Moreover, intestinal microorganisms were also closely related to the physiological activity of the insect. Based on our results, the Enterococcus community increased when the larvae were fed with artificial diets supplemented with the chitinase fermentation liquids. The typical ecological niche for the Enterococcus species is the intestine of humans and other animals. Enterococci, which were firmly established as major nosocomial pathogens, have also been related to human diseases. A large number of enterococci were also found in the intestinal tract of silkworms, the presence of which is not harmful under normal conditions. However, silkworms could be infectious when the silkworm the larvae are suffering from poor physiological activity. Enterococcus mundtii was shown to be associated with the flacherie disease of the silkworm larvae reared on artificial diets. Silkworm larvae infection analysis with E. mundtii was conducted by using pure E. mundtii culture in a nutrient broth sprinkled directly on the artificial diet. The larvae were reared under germfree conditions. The results indicated that the germfree larvae displayed typical bacterial infection symptoms after a few days. The number of days varied according to the quality and concentration of the E. mundtii inoculum. The infection could be achieved at low E. mundtii concentrations added to the diet. The infection could also enter the body cavity after multiplying in the larval gut [34]. Lysenko used two kinds of methods to prove the pathogenicity of the Streptocossus strains on healthy silkworm larvae through feeding and injection [24]. Silkworm larvae mortality reached 100% after injecting the Streptocossus strains into the larval cavity. In our study, we conducted the infection study of the Asian corn borer larvae using Enterococcus sp. isolated from midgut of the healthy the Asian corn borer larvae. Results indicated that an increase of the Enterococcus community in the larval midgut could indeed increase the mortality of the larvae. We remain unaware of why injecting the Enterococcus strain into the body cavity of the Asian corn borer larvae could result in larval mortality. We speculated that the Enterococcus strain could enter into the larval midgut after multiplying in the body cavity. Based on the results, the increased Enterococcus community in the larval midgut could be an important factor resulting in the death of the Asian corn borer larvae.
Bacterial diversity changes caused by gastro-intestinal diseases have been reported in humans. Bacteroides spp., Clostridium coccoides, and Clostridium leptum are considered to be the dominant populations in the human fecal microflora [35], [36]. The results indicated that patients who suffered Crohn’s disease had a reduced diversity of the bacterial phylum Firmicutes [37]. Furthermore, the numbers of lactobacilli, Clostridium coccoides, and Clostridium leptum decreased in fecal microflora, whereas the number of enterobacteria increased when the patients were suffering inflammatory bowel diseases. The white plague disease (WPD) is a prevalent coral disease that mainly affects massive coral species. Shifts in bacterial communities were observed when the coral species were infected with WPD. A significant increase in Alphaproteobacteria was also observed in WPD-affected coral species. A concomitant decrease in the Beta- and Gammaproteobacteria was observed [38]. The number of Enterococcus clones increased, followed by a reduction of the Ochrobactrum clones. Enterococcus species were present in the larvae of silkworm [39] and gypsy moth [3] regardless of diets. These results were in accordance with our findings, in which Enterococcus species were present in all the larvae fed with different artificial diets. Other dominant bacteria types such as the Enterobacter species in lepidopteran midguts were not detected in this study.
Exploring the association between the Asian corn borer larvae midgut microbial community imbalance and larval mortality will enrich our understanding of the mechanism induced by chitinase from the transgenic Trichoderma strain against corn borers. Further analysis of this midgut ecosystem, including the dynamic changes in the midgut microbial composition using the metagenomic method, will provide more useful markers for the biocontrol of insect pests.
Acknowledgments
We thank Dr. Wang Zhenying for providing us many batches of cultured Asian corn borer larvae.
Funding Statement
The research was supported by China Agriculture Research System (CARS-02), the “948” project of the Ministry of Agriculture of China (2011-G4), the key project from Shanghai Municipal Science and Technology Commission (09391910900), the National Department Public Benefit Research Foundation of China (No. 200903052). The ministry of education research fund for the doctoral specialty (No. 20090073110048) and the “863” national projects (2011AA10A205). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Nafus DW, Schreiner I (1991) Review of the biology and control of the Asian corn borer, Ostrinia furnacalis (Lep: Pyralidae). Trop Pest Manag 37: 41–56. [Google Scholar]
- 2. Li YY, Fu KH, Gao SG, Wu Q, Fan LL, et al. (2013) Increased virulence of transgenic Trichoderma koningi strains to the Asian corn borer larvae by overexpressing heterologous chit42 gene with chitin-binding domains. Journal of Environmental Science and Health, Part B 48: 1–8 (In press).. [DOI] [PubMed] [Google Scholar]
- 3. Broderick NA, Raffa KF, Goodman RM, Handelsman J (2004) Census of the bacterial community of the gypsy moth larval midgut by using culturing and culture-independent methods. Appl Environ Microbiol 70: 293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Robinson CJ, Schloss P, Ramos Y, Raffa K, Handelsman J (2010) Robustness of the bacterial community in the cabbage white butterfly larval midgut. Microb Ecol 59: 199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xiang H, Wei GF, Jia S, Huang J, Miao XX, et al. (2006) Microbial communities in the larval midgut of laboratory and field populations of cotton bollworm (Helicoverpa armigera). Can J Microbiol 52: 1085–1092. [DOI] [PubMed] [Google Scholar]
- 6. Pinto-Tomas AA, Sittenfeld A, Uribe-Lorio L, Chavarria F, Mora M, et al. (2011) Comparison of midgut bacterial diversity in tropical caterpillars (lepidoptera: saturniidae) fed on different diets. Environ Entomol 40: 1111–1122. [DOI] [PubMed] [Google Scholar]
- 7. Kelly D, Conway S, Aminov R (2005) Commensal gut bacteria: mechanisms of immune modulation. Trends Immunol 26: 326–333. [DOI] [PubMed] [Google Scholar]
- 8. O’Neill SL, Giordano R, Colbert AM, Karr TL, Robertson HM (1992) 16S rRNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proc Natl Acad Sci U S A 89: 2699–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nardi JB, Mackie RI, Dawson JO (2002) Could microbial symbionts of arthropod guts contribute significantly to nitrogen fixation in terrestrial ecosystems? Journal of Insect Physiology 48: 751–763. [DOI] [PubMed] [Google Scholar]
- 10. Breznak JA (1982) Intestinal microbiota of termites and other xylophagous insects. Annu Rev Microbiol 36: 323–343. [DOI] [PubMed] [Google Scholar]
- 11. Chen DQ, Purcell AH (1997) Occurrence and transmission of facultative endosymbionts in aphids. Curr Microbiol 34: 220–225. [DOI] [PubMed] [Google Scholar]
- 12. Gilliam M (1997) Identification and roles of non-pathogenic microflora associated with honey bees. FEMS Microbiology Letters 155: 1–10. [Google Scholar]
- 13. Butler JL, Williams MA, Bottomley PJ, Myrold DD (2003) Microbial community dynamics associated with rhizosphere carbon flow. Appl Environ Microbiol 69: 6793–6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carrero-Colon M, Nakatsu CH, Konopka A (2006) Effect of nutrient periodicity on microbial community dynamics. Appl Environ Microbiol 72: 3175–3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kiorboe T, Tang K, Grossart HP, Ploug H (2003) Dynamics of microbial communities on marine snow aggregates: colonization, growth, detachment, and grazing mortality of attached bacteria. Appl Environ Microbiol 69: 3036–3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Limon MC, Chacon MR, Mejias R, Delgado-Jarana J, Rincon AM, et al. (2004) Increased antifungal and chitinase specific activities of Trichoderma harzianum CECT 2413 by addition of a cellulose binding domain. Appl Microbiol Biotechnol 64: 675–685. [DOI] [PubMed] [Google Scholar]
- 17. Bogo MR, Rota CA, Pinto H Jr, Ocampos M, Correa CT, et al. (1998) A chitinase encoding gene (chit1 gene) from the entomopathogen Metarhizium anisopliae: isolation and characterization of genomic and full-length cDNA. Curr Microbiol 37: 221–225. [DOI] [PubMed] [Google Scholar]
- 18. Burgmann H, Pesaro M, Widmer F, Zeyer J (2001) A strategy for optimizing quality and quantity of DNA extracted from soil. J Microbiol Methods 45: 7–20. [DOI] [PubMed] [Google Scholar]
- 19. Lane DJ (1991) 16S/23S rRNA sequencing. In: E Stackebrandt, M Goodfellow, eds. Nucleic Acid Techniques in Bacterial Systematics, John Wiley and Sons, New York. 5–175 55–58. [Google Scholar]
- 20. Ravenschlag K, Sahm K, Pernthaler J, Amann R (1999) High bacterial diversity in permanently cold marine sediments. Appl Environ Microbiol 65: 3982–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammer O, Harper DAT, Ryan PD (2001) PAST: paleontological statistics software package for education and data analysis. Palaeontologia electronica 4(1): 9. http://palaeo-electronica.org/2001_1/past/issue1_01.htm, Accessed 2013 Jan 4.
- 22. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425. [DOI] [PubMed] [Google Scholar]
- 24. LYSENKO O (1958) ‘Streptococcus bombycis’, its Taxonomy and Pathogenicity for Silkworm Caterpillars. Journal of General Microbiology 18: 774–781. [DOI] [PubMed] [Google Scholar]
- 25. Schleifer KH, Kilpper-Bälz R (1984) Transfer of Streptococcus faecalis and Streptococcus faecium to the Genus Enterococcus nom. rev. as Enterococcus faecalis comb. nov. and Enterococcus faecium comb. nov. International Journal of Systematic Bacteriology 34: 31–34. [Google Scholar]
- 26. Ohkuma M, Kudo T (1996) Phylogenetic diversity of the intestinal bacterial community in the termite Reticulitermes speratus . Appl Environ Microbiol 62: 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kennedy TA, Naeem S, Howe KM, Knops JMH, Tilman D, et al. (2002) Biodiversity as a barrier to ecological invasion. Nature 417: 636–638. [DOI] [PubMed] [Google Scholar]
- 28. Stachowicz JJ, Whitlatch RB, Osman RW (1999) Species Diversity and Invasion Resistance in a Marine Ecosystem. Science 286: 1577–1579. [DOI] [PubMed] [Google Scholar]
- 29. Dillon R, Charnley K (2002) Mutualism between the desert locust Schistocerca gregaria and its gut microbiota. Research in Microbiology 153: 503–509. [DOI] [PubMed] [Google Scholar]
- 30. Haynes S, Darby AC, Daniell TJ, Webster G, Van Veen FJ, et al. (2003) Diversity of bacteria associated with natural aphid populations. Appl Environ Microbiol 69: 7216–7223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohkuma M, Hongoh Y, Kudo T (2006) Diversity and Molecular Analyses of Yet-Uncultivated Microorganisms Intestinal Microorganisms of Termites and Other Invertebrates. In: König H, Varma A, editors: Springer Berlin Heidelberg. 303–317.
- 32. Noda S, Iida T, Kitade O, Nakajima H, Kudo T, et al. (2005) Endosymbiotic Bacteroidales bacteria of the flagellated protist Pseudotrichonympha grassii in the gut of the termite Coptotermes formosanus . Appl Environ Microbiol 71: 8811–8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shinzato N, Muramatsu M, Matsui T, Watanabe Y (2007) Phylogenetic analysis of the gut bacterial microflora of the fungus-growing termite Odontotermes formosanus . Biosci Biotechnol Biochem 71: 906–915. [DOI] [PubMed] [Google Scholar]
- 34. Cappellozza S, Saviane A, Tettamanti G, Squadrin M, Vendramin E, et al. (2011) Identification of Enterococcus mundtii as a pathogenic agent involved in the “flacherie” disease in Bombyx mori L. larvae reared on artificial diet. Journal of Invertebrate Pathology 106: 386–393. [DOI] [PubMed] [Google Scholar]
- 35. Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, et al. (2005) Diversity of the human intestinal microbial flora. Science 308: 1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hayashi H, Takahashi R, Nishi T, Sakamoto M, Benno Y (2005) Molecular analysis of jejunal, ileal, caecal and recto-sigmoidal human colonic microbiota using 16S rRNA gene libraries and terminal restriction fragment length polymorphism. J Med Microbiol 54: 1093–1101. [DOI] [PubMed] [Google Scholar]
- 37. Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, et al. (2006) Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 55: 205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cardenas A, Rodriguez RL, Pizarro V, Cadavid LF, Arevalo-Ferro C (2012) Shifts in bacterial communities of two Caribbean reef-building coral species affected by white plague disease. ISME J 6: 502–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xiang H, Li M, Zhao Y, Zhao L, Zhang Y, et al. (2007) Bacterial community in midguts of the silkworm larvae estimated by PCR/DGGE and 16S rDNA gene library analysis. Acta Entomologica Sinica 50: 222–233. [Google Scholar]