Abstract
Purpose
DNA repair deficiencies have been postulated to play a role in the development and progression of cardiovascular disease (CVD). The hypothesis is that DNA damage accumulating with age may induce cell death, which promotes formation of unstable plaques. Defects in DNA repair mechanisms may therefore increase the risk of CVD events. We examined whether the joints effect of common genetic variants in 5 DNA repair pathways may influence the risk of CVD events.
Methods
The PLINK set-based test was used to examine the association to myocardial infarction (MI) of the DNA repair pathway in GWAS data of 866 subjects of the GENetic DEterminants of Restenosis (GENDER) study and 5,244 subjects of the PROspective Study of Pravastatin in the Elderly at Risk (PROSPER) study. We included the main DNA repair pathways (base excision repair, nucleotide excision repair, mismatch repair, homologous recombination and non-homologous end-joining (NHEJ)) in the analysis.
Results
The NHEJ pathway was associated with the occurrence of MI in both GENDER (P = 0.0083) and PROSPER (P = 0.014). This association was mainly driven by genetic variation in the MRE11A gene (PGENDER = 0.0001 and PPROSPER = 0.002). The homologous recombination pathway was associated with MI in GENDER only (P = 0.011), for the other pathways no associations were observed.
Conclusion
This is the first study analyzing the joint effect of common genetic variation in DNA repair pathways and the risk of CVD events, demonstrating an association between the NHEJ pathway and MI in 2 different cohorts.
Introduction
Cardiovascular disease (CVD) is caused by interplay of environmental factors and multiple predisposing genes. DNA damage, caused by for instance oxidative stress and cigarette smoking, has been recognized as a significant contributor to the pathogenesis of CVD. [1], [2] Mechanistically, in cells where the DNA damage is beyond repair apoptosis is induced. [3] The effect of this damage induced cell death is dependent on the cell type. Death of endothelial cells is implicated in plaque erosion and subsequent vessel thrombosis. [4] Vascular smooth muscle cell (VSMC) death has been associated with thinning of the fibrous cap and increasing the risk of plaque rupture. [5], [6] To further complicate matters, apoptosis is not the only response of cells to DNA damage, also cellular senescence has been described. [7], [8] Cellular senescence is a state in which cells remain in cell cycle arrest and in which they have lost their optimal function. With respect to the pathogenesis of atherosclerosis, senescence of for instance vascular endothelial cells can result in a provasoconstrictor and a proinflammatory phenotype. [7] So besides cell death, DNA damage could also increases the risk of CVD by inducing cell senescence.
Adequate DNA repair is crucial for survival of an organism, as the DNA is continuously exposed to various types of external factors, like mutagenic chemicals and radiation, and endogenously generated triggers like reactive oxygen species (ROS) and DNA replication errors, all capable of inducing DNA damage. Human cells possess several innate DNA repair processes to protect against the harmful consequences of DNA damage. [9] Single-strand DNA damage can be repaired by excision repair and mismatch repair pathways that use the undamaged strand as template during the repair process. For the repair of double-strand breaks other repair mechanisms like non-homologous end-joining (NHEJ) or homologous recombination are required. [10].
Evidence of the relation between genomic integrity and cardiovascular disease in the ageing population has been growing over the last years. Up to now, this evidence consists in various forms, ranging from cellular biology studies in vascular endothelial cells [11], [12], vascular smooth muscle cells [13], [14] and macropahges [15], histological examination of human atherosclerotic plaques [14], [16] and animal studies in telomerase deficient mice [17] and DNA repair defective mice. [8], [18], [19] In contrast, only limited studies have focused on single nucleotide polymorphisms (SNPs) in genes related to DNA repair processes and CVD events, although some associations have been reported. The only SNP with some consistent results is the Arg399Gln (rs25487) SNP in the XRCC1 base excision repair (BER) gene which was reported to be associated with stroke [20] and coronary atherosclerosis [21]. Other genes from the excision repair pathway such as OGG1, XRCC3, ERCC2 (XPD) and ERCC5, were found to moderately associated as a combined score to the risk of large artery atherosclerotic stroke in the smoking subset of a Chinese population [22]. The authors suggested that the individual vulnerability to smoking-induced oxidative stress was influenced by carriers of these SNPS.
In genome-wide association studies (GWAS) investigating the genetic background of CVD, no association was found with SNPs in genes related to DNA repair processes. [23], [24] However, considering the multifactorial nature of the condition, it is possible that by a joint effect, genetic variants with small individual effect sizes, could contribute to disease risk and are undetected in a GWAS. [25], [26] The goal of the current study was to examine whether common genetic variants in DNA repair genes are related to the risk of CVD events by using a gene set analysis of the 5 main DNA repair pathways in two large representative CVD populations.
Methods
GENDER Study Population
The design of the GENetic DEterminants of Restenosis (GENDER) study has been described previously. [27] In brief, GENDER included 3,104 consecutive unrelated symptomatic patients treated successfully by PCI for angina. The study protocol conforms to the Declaration of Helsinki and was approved by the ethics committees of each participating institution. Written informed consent was obtained from each participant before the PCI procedure. Experienced operators, using a radial or femoral approach, performed standard angioplasty and stent placement. During the study, no drug-eluting stents were used. Blood samples were collected at the index procedure for DNA isolation. During a follow-up period of 9 months, the endpoint clinical restenosis, defined as renewed symptoms requiring target vessel revascularization (TVR) either by repeated PCI or CABG, by death from cardiac causes or myocardial infarction (MI) not attributable to another coronary event than the target vessel, was recorded. Furthermore, of each patient the occurrence of MI or stroke prior to inclusion into the study, as well as during the follow up period, was recorded. For this study the combination of prevalent and incident MI or stroke was analyzed.
PROSPER Study Population
The design and population of the PROSPective study for the Elderly at Risk (PROSPER) has been described previously. [28] PROSPER is a prospective multicenter randomized placebo-controlled trial to assess whether treatment with pravastatin diminishes the risk of major vascular events in elderly individuals. Between December 1997 and May 1999, subjects were screened and enrolled in Scotland (Glasgow), Ireland (Cork), and The Netherlands (Leiden). Men and women aged 70–82 years were recruited if they had pre-existing vascular disease or increased risk of such disease because of smoking, hypertension, or diabetes. A total number of 5,804 subjects were randomly assigned to pravastatin or placebo. In this study several cardiovascular endpoints were evaluated during a mean follow-up of 3.2 years; the primary endpoint consisted of a composite of fatal/non-fatal MI or fatal/non-fatal stroke. Secondary and tertiary endpoints included stroke and MI separately, all-cause mortality and death due to a vascular cause. [29] The institutional ethics review boards of all centers approved the protocol, and all participants gave written informed consent. The protocol was consistent with the Declaration of Helsinki. For the current study we combined the incident events,myocardial infarction and stroke, that occurred during the follow-up period with the prevalent events that occurred before inclusion into PROSPER, to obtain a lifetime risk for these events.
Genotyping
In GENDER, a GWAS was performed in 325 cases of restenosis and 630 controls matched by gender, age, and some confounding clinical variables for restenosis in the GENDER study such as total occlusion, diabetes, current smoking and residual stenosis. [30] Genotyping was performed using the Illumina Human 610-Quad Beadchips following manufacturer’s instructions. After stringent quality control, bad performing samples (call rate <99%) and assays (call rate <95%, minor allele frequency <1% and deviation from Hardy-Weinberg equilibrium) were excluded from further analysis. The final dataset consisted of 866 individuals (295 cases, 571 controls) and 556,099 SNPs.
In PROSPER, a GWAS was performed using Illumina Human 660-Quad Beadchips following manufacturer’s instructions. After stringent quality control, bad performing samples and assays were excluded from further analysis. Genotypic data was available in 5,244 subjects and a total of 557,192 SNPs [31].
Both datasets were imputed using MaCH software [32] up to ∼2.5 million SNPs based on the HapMap Phase I+II CEU release 22 (hg18/build36) reference.
Gene Set Analysis
We analyzed SNPs within a 10-kb window around the genes encoding proteins belonging to the 5 DNA repair pathways described in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database [33], [34]; the BER pathway, the nucleotide excision repair (NER) pathway, the mismatch repair (MMR) pathway, the homologous recombination pathway and the NHEJ pathway. Gene set analyses were performed with the PLINK set-based test v1.07 in a case-control setting. [35] In the first step of this test, a single SNP analysis of all SNPs within the set is performed. Subsequently, a mean SNP statistic is calculated from the single SNP statistics of a maximum amount of SNPs below a certain P-value threshold. For the current study this threshold was set on 0.20 to ensure that all SNPs with minor effect will be analyzed. If SNPs are not independent, i.e. the LD (expressed in R2) is above a certain threshold, the SNP with the lowest P-value in the single SNP analysis is selected. This analysis is repeated with 10,000 simulated SNP sets, in which the phenotype status of the individuals is permuted. An empirical P-value for the SNP set is computed by calculating the number of times the test statistic of the simulated SNP sets exceeds that of the original SNP set. For the set-based analysis of this study, the parameters were set to P-value threshold <0.20, R2 threshold <0.5, and maximum number of SNPs = 99999. The associations were considered significant, after correction for the 5 analyzed pathways, if the P-value <0.01 (0.05/5). For the pathway analysis we only used the genotyped GWAS sets of both studies, since imputed SNPs were obtained based on their LD pattern with the genotyped SNPs and the LD threshold of the set-based analysis corrects for this, making their added value minimal.
Results
Participant characteristics of the two study populations are presented in table 1. The main differences in the baseline characteristics between the two study populations are the mean age of the participants (GENDER; 62.5 years, PROSPER; 75.3 years) and the proportion of women (GENDER; 27%, PROSPER; 52%) Moreover, diabetes and complaints of stable angina pectoris were more frequent in GENDER, whereas a history of hypertension was more frequent in PROSPER. The incidence of MI in GENDER was 45% and in PROSPER 22%.
Table 1. Baseline characteristics and endpoints of the GENDER and PROSPER studies.
| GENDERN = 866 | PROSPERN = 5,244 | |
| Baseline characteristics | ||
| Age (years) | 62.5±10.8 | 75.3±3.4 |
| Male gender (N) | 634 (73) | 2,524 (48) |
| Current smoking (N) | 216 (25) | 1,392 (27) |
| History of diabetes (N) | 177 (20) | 544 (10) |
| History of hypertension (N) | 349 (40) | 3,257 (62) |
| History of angina (N) | 288 (68) | 1,424 (27) |
| History of myocardial infarction (N) | 365 (42) | 708 (14) |
| Total cholesterol (mmol/L) | 4.9±1.0a | 5.7±0.9 |
| Body mass index (kg/m2) | 27.0±3.7 | 26.8±4.2 |
| Statin treatment | 465 (54) | 2,605 (50) |
| Endpoints | ||
| Myocardial infarctionb | 389 (45) | 1145 (22) |
| Strokeb | 49 (6) | 731 (14) |
| Myocardial infarction or strokeb | 416 (48) | 1714 (33) |
| Clinical restenosisc | 295 (34) | NA |
| All cause mortality | 237 (27)d | 548 (11)c |
| Vascular mortalityc | NA | 266 (5) |
Data are presented as mean ± SD or number (%).
Cholesterol levels available in only 177 patients.
Before inclusion or during follow-up period.
During follow-up period.
During follow up of 10 years after inclusion in GENDER.
Of the 5 selected DNA repair pathways, as described by the KEGG pathway database, the NER pathway was the largest one (44 genes), (Table S1), while the NHEJ pathway was the smallest (13 genes). The BER pathway consisted of 35 genes, the MMR pathway of 23 genes and the homologous recombination pathway of 28 genes.
The set-based analysis of the 5 pathway sets in the GENDER population resulted in a significant association of the homologous recombination pathway with MI (P = 0.011) and the combined endpoint of MI or stroke (P = 0.0039) (Table 2). A significant association with the same endpoints was also found for the NHEJ pathway (P = 0.0083 for MI and P = 0.0089 for MI or stroke). No significant association of any of the DNA repair pathways was found with stroke alone.
Table 2. Set-based analysis of DNA repair pathways in the GENDER and PROSPER study populations.
| MIa | Strokea | MI or Strokea | |||||||
| Pathway | Genes | SNPs | SNPs | P | SNPs | P | SNPs | P | |
| GENDERb | N = 389/477 | N = 49/817 | N = 416/450 | ||||||
| Base excision | 35 | 315 | 37 | 0.48 | 36 | 0.47 | 37 | 0.59 | |
| Nucleotide excision | 44 | 401 | 57 | 0.38 | 49 | 0.34 | 60 | 0.38 | |
| Mismatch repair | 23 | 285 | 39 | 0.34 | 40 | 0.42 | 47 | 0.35 | |
| Homologous recombination | 28 | 418 | 64 | 0.011 | 51 | 0.71 | 60 | 0.0039 | |
| Non-homologous end joining | 13 | 137 | 19 | 0.0084 | 14 | 0.43 | 19 | 0.0089 | |
| PROSPERb | N = 1,145/4,099 | N = 731/4,513 | N = 1,714/3,530 | ||||||
| Base excision | 35 | 298 | 43 | 0.049 | 42 | 0.39 | 39 | 0.43 | |
| Nucleotide excision | 44 | 392 | 62 | 0.34 | 57 | 0.47 | 49 | 0.49 | |
| Mismatch repair | 23 | 285 | 39 | 0.43 | 33 | 0.46 | 31 | 0.91 | |
| Homologous recombination | 28 | 426 | 61 | 0.14 | 42 | 0.49 | 51 | 0.24 | |
| Non-homologous end joining | 13 | 144 | 19 | 0.014 | 30 | 0.35 | 22 | 0.11 | |
The SNPs per endpoint indicate the number of independent SNPs that passed the test constrains (P<0.2 and R2<0.5) and were thus jointly analyzed in 10,000 permutations.
Before inclusion or during follow-up period. MI, myocardial infarction.
Cases/controls.
Analysis in the PROSPER dataset resulted in an association of the NHEJ pathway with MI (P = 0.014). A borderline significant association of the BER pathway with MI was also observed (P = 0.049). The other pathways did not show significant associations in PROSPER (Table 2).
To determine by which genes the association of the NHEJ pathway with MI was driven, we examined the SNP set from each gene of this pathway separately. We found that the association was driven by several genes from this pathway. In GENDER the XRCC4 gene demonstrated a borderline significant association (P = 0.055) and in PROSPER the genes PRKDC (P = 0.029) and LIG4 (P = 0.030) were individually associated with MI. The strongest association was found with the MRE11A gene in both GENDER (P = 0.0001) and PROSPER (P = 0.0017), driven by 3 and 2 SNPs, respectively, which differ between the studies (Table 3).
Table 3. Results of the gene set analysis of the non-homologous end joining pathway with myocardial infarction in GENDER and PROSPER.
| GENDER | PROSPER | |||||||||||||
| Gene | SNPs | Sig. SNPs | P (gene) | Top SNP | MAF | OR | P (SNP) | SNPs | Sig. SNPs | P (gene) | Top SNP | MAF | OR | P (SNP) |
| XRCC5 | 19 | 3 | 0.23 | rs3821107 | 0.24 | 0.79 | 0.04 | 22 | 1 | 0.89 | rs828704 | 0.20 | 0.93 | 0.19 |
| NHEJ1 | 15 | 1 | 0.55 | rs7588654 | 0.03 | 0.65 | 0.14 | 15 | 0 | 1.00 | ||||
| XRCC4 | 29 | 4 | 0.055 | rs13178127 | 0.05 | 2.02 | 0.001 | 30 | 4 | 0.38 | rs35271 | 0.14 | 1.15 | 0.039 |
| RAD50 | 8 | 0 | 1.00 | – | 10 | 1 | 0.52 | rs2237060 | 0.44 | 1.07 | 0.13 | |||
| POLM | 4 | 2 | 0.12 | rs11769882 | 0.25 | 0.79 | 0.040 | 4 | 1 | 0.26 | rs11769882 | 0.22 | 0.93 | 0.14 |
| PRKDC | 14 | 1 | 0.69 | rs7003908 | 0.34 | 1.16 | 0.14 | 14 | 4 | 0.026 | rs10109984 | 0.38 | 1.15 | 0.005 |
| POLL | 3 | 0 | 1.00 | – | 3 | 1 | 0.28 | rs3730477 | 0.21 | 1.09 | 0.14 | |||
| DCLRE1C | 8 | 1 | 0.22 | rs12572872 | 0.23 | 1.24 | 0.06 | 8 | 0 | 1.00 | ||||
| DNTT | 9 | 1 | 0.35 | rs1923703 | 0.12 | 0.80 | 0.14 | 9 | 0 | 1.00 | ||||
| MRE11A | 14 | 3 | 0.0001 | rs535801 | 0.31 | 1.52 | 0.00006 | 13 | 2 | 0.0017 | rs2155209 | 0.35 | 0.86 | 0.002 |
| FEN1 | 5 | 0 | 1.00 | – | 5 | 1 | 0.27 | rs695867 | 0.35 | 0.80 | 0.10 | |||
| LIG4 | 7 | 3 | 0.31 | rs9520823 | 0.30 | 1.21 | 0.07 | 7 | 3 | 0.030 | rs1151403 | 0.42 | 0.87 | 0.003 |
| XRCC6 | 2 | 0 | 1.00 | – | 4 | 1 | 0.45 | rs17002523 | 0.01 | 1.34 | 0.16 | |||
SNPs, total number of SNPs per gene; Sig.SNPs indicate the number of SNPs that passed the test constrains (P<0.2 and R2<0.5) and were thus jointly analyzed in 10,000 permutations; OR odds ratio; P, p-value; MAF, minor allele frequency.
By using imputed data we performed in silico fine mapping of the individual SNPs in the MRE11A genetic region on chromosome 11 (Figure 1). Within the range of 10 Kb around the MRE11A gene genotypic data of 104 SNPs were available. We identified 8 sets of SNPs that were in high LD (R2>0.8) but only one set (10 SNPs) associated with MI in both GENDER and PROSPER (P = 0.0033 and P = 0.0023 respectively). This set contained the top SNP in MRE11A in PROSPER (rs2155209), that was identified in the individual gene analysis (Table 4). This SNP is located in a DNase I hypersensitivity site (UCSC genome browsers database [36]). The promotor 2.0 Prediction Server [37] reported that the region surrounding rs2155209 is not a promotor region. In addition, the is-rSNP algorithm [38] reported that the DNA binding affinity of three transcription factors is significantly affected by rs2155209 (LM221 P = 0.014, estrogen receptor 2 (ESR2) P = 0.028 and LM168 P = 0.037). Unfortunately, rs2155209 is not reported in three publically available eQTL databases (mRNA by SNP browser [39], [40], VarySysDB [41] and the eQTL database of the Pritchard lab [42]).
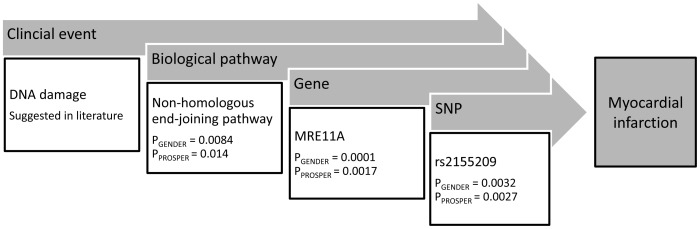
Figure 1. Fine mapping from DNA damage through the identification of an associated DNA repair pathway, the responsible gene in this pathway, to the single nucleotide polymorphism (SNP).
Table 4. Genomic region of MRE11A divided in LD blocks for the association with myocardial infarction.
| GENDER | PROSPER | |||||||
| Set | SNPs | R2 | Tagging SNP | MAF* | OR (95% CI) | P | OR (95% CI) | P |
| 1 | 10 | 0.91 | rs2155209 | 0,36 | 0.74 (0.61–0.91) | 0,0033 | 0.86 (0.78–0.96 | 0,0023 |
| 2 | 10 | 0,90 | rs535801 | 0,31 | 1.52 (1.24–1.87) | 0,000059 | 1.00 (0.90–1.11) | 0.94 |
| 3 | 17 | 0.88 | rs1270146 | 0.43 | 1.32 (1.09–1.60) | 0.0049 | 1.02 (0.93–1.13) | 0.63 |
| 4 | 13 | 0.91 | rs529126 | 0.26 | 1.44 (1.16–1.80) | 0.00090 | 1.02 (0.91–1.12) | 0.71 |
| 5 | 5 | 0.85 | rs499952 | 0.35 | 1.32 (1.08–1.61) | 0.0059 | 1.03 (0.92–1.12) | 0.54 |
| 6 | 4 | 0.91 | rs13447720 | 0.23 | 0.94 (0.75–1.18) | 0.57 | 1.18 (1.05–1.30) | 0.0027 |
| 7 | 18 | 0.91 | rs10765682 | 0.09 | 0.91 (0.65–1.27) | 0.58 | 1.01 (0.87–1.20) | 0.95 |
| 8 | 18 | 1.00 | rs12788248 | 0.01 | 1.22 (0.43–3.49) | 0.71 | 1.02 (0.58–1.72) | 0.95 |
| other** | 9 | – | ||||||
R2 indicates lowest LD between SNPs within the set.
Tagging SNP is a genotyped SNP with the lowest P-value per set.
MAF, minor allele frequency in GENDER.
SNPs not in LD with other SNPs within the gene.
To explore whether the found associations were caused by specific subgroups we analyzed the NHEJ pathway in male and female patients, smokers and non-smokers and in patients with and without diabetes separately. Moreover, in the PROSPER study we also performed the analyses in the pravastatin and placebo group. No clear associations were detected in these subgroups (Table S2).
Discussion
The current study is the first to use a DNA repair pathway approach for the identification of new candidate genes related to cardiovascular outcomes. We show that genetic variation in several of these genes are indeed associated with cardiovascular related endpoints and that the joint analyses of these genetic markers demonstrates a significant association of the NHEJ pathway with prevalent and incident MI in two study populations. Variation in the gene encoding meiotic recombination 11 homolog A (MRE11A) drives the association.
This is the first study that demonstrates a relation between the NHEJ pathway and MI or even with CVD. This particular DNA repair pathway is mainly involved in the repair of double-strand breaks (DSBs), which are considered to have the highest risk of evoking deleterious events, such as chromosomal translocations, cancer and cell death. The main driver of this association, MRE11A, is a highly conserved protein, existing in vivo as a dimer, forms together with RAD50 and NBS1 the MRN complex. [43] The MRN complex has a critical role in the recognition of DNA damage lesions or the chromatin alterations that follow DNA damage [3] and a key role in the cellular response to DSBs. [44] Moreover, the MRN complex has been implicated in telomere maintenance, meiosis, DNA replication and checkpoint activation. [45]–[47] Genetic variation in MRE11A has previously been associated with several types of cancer [48]–[50] and antaxia-telangiectasis-like disease. [51] To our knowledge, no association of this gene with CVD events has yet been described, also not in the previous GWAS on CVD. [23], [52], [53] The SNP rs2155209, significantly associated with MI in both of our study populations, has been associated with an 1.5-fold increased risk of bladder cancer, although the authors of that study suggest that it might has been a false-positive finding. [49] This MRE11A SNP is located in the 3′UTR of the gene, and the possible functional effect has not yet been studied.
When examining the LD structure of MRE11A, we found that the structure is not conform the expected LD block formation, meaning that nearby SNPs are organized into regions of high LD separated by short segments of very low LD. This discontinuous LD structure is not uncommon, and has been described before for this gene. [54] Allen-Brady et al. [54] describe 4 tagging SNPs together accounting for 99% of the genetic variance within the gene region. In the current study the genetic coverage of the gene was more thorough (104 SNPs compared to 11 in the former study), resulting in 8 tagging SNPs. Interestingly, one of the 4 described tagging SNPs, rs556477, is in very high LD (R2 = 0.92) with our top SNP rs2155209. It is unknown whether rs2155209 has any direct functional effects. That this SNP is located in a DNase I hypersensitivity site, which often associated with cis-regulatory sequences, including promoters, insulators, enhancers and locus control regions, increases the likelihood that rs2155209 influences one of these features and thereby exerting its clinical effects, although this remains to be proven. Another method of predicting the possible regulatory abilities of non-coding SNPs is the in silico rSNP algorithm [38]. This approach indicated that rs2155209 affects binding of ERS2. The estrogen receptor 2 belongs to a family of nuclear receptor transcription factors, activating transcription upon binding to specific DNA sequences. Moreover, the SNP was associated with LM221 and LM168, two conserved motifs in the human genome described to be involved in gene regulation, likely serving as insulators. [55] Wet lab confirmation of these bioinformatic predictions will be necessary before definite conclusions can be drawn from these findings.
Cigarette smoking is considered to be an important risk factor in atherosclerotic vascular disease and it is a well-known external factor associated with DNA damage, like single- and double strand breaks and the formation of oxidative DNA-adducts. [56], [57] The association of the NHEJ pathway with MI, demonstrated in the current study, could indicate that this pathway is the underlying mechanism of the strong relation between smoking and CVDs. However, this hypothesis could not be confirmed in subgroup analyses. Whether this absence of association is caused by lack of power of the subgroup analysis, or because the underlying mechanism causing MI is not through smoking, is uncertain. Moreover, the hypothesis that patients with diabetes mellitus have increased oxidative stress which could lead to DNA damage [58], [59], could also not be confirmed in our population. However, considering that the incidence of diabetes was 20% in GENDER and only 10% in PROSPER, there was not enough power to detect a small effect. Furthermore, we cannot exclude that another DNA repair pathway than NHEJ might be responsible for the DNA repair in diabetic patients, but considering the small subgroup size and the fact that the other DNA repair pathways were not significantly associated in the complete populations, we did not perform further analyses for these pathways. The subgroup analysis did demonstrate that the association of the NHEJ pathway with MI was possibly driven by the male subjects, since in PROSPER no association was found in female subjects. Although in GENDER a similar trend was observed, these results were not significant.
The strength of gene set analysis, opposed to GWAS analysis, is that it tests the joint effect of multiple individual SNPs within a larger set. Considering the a priori small effect size of the individual SNPs on complex disease endpoints, like MI, analysis of the joint effect of multiple markers, in this study comprising complete DNA repair pathways, will increase the likelihood of finding biological plausible associations.
Several possible limitations to our study have to be mentioned. For the current study we performed the pathway analysis using the PLINK software. [60] Other software packages have been described, although to date none has been proven to be clearly superior to the others. Gui and colleagues compared 7 tests analyzing the WTCCC Crohn’s Disease dataset. [61] One of their overall conclusions was that the set-based test in PLINK was the most powerful algorithm. Another study, applying PLINK set-based test, Global test, GRASS and SNP ratio test, for the analysis of three pathways regarding human longevity observed similar results with the different tests. [62] Although other software packages could lead to different results, the fact that our fine mapping strategy led to the identification of a single LD block associated in two independent populations, increased the likelihood of a true positive association.
The 5 DNA repair pathways analyzed were derived from the publically available KEGG database [34]. The KEGG database is however not the only database providing biological pathways and there is no consensus on the best database. In our opinion these particular pathways were more elaborately described by KEGG than in other databases (for instance Reactome or BioCarta). Moreover, only the KEGG database provided a description of all 5 DNA repair pathways. Since the overlap of certain pathways of different databases is substantial, we decided only to test the DNA repair pathways described by KEGG. [33] It is important to realize that probably none of these databases provide a perfect representation of the actual biological mechanism, simple because our current knowledge is not that far evolved yet. Likely not all genes incorporated within the current pathways directly influence the actual DNA repair process of interest. These unrelated gene product could therefore interfere with the actual associations, however to what extent this is the case in the current study remains unknown.
As stated above, DNA damage and DNA damage repair are associated with cancer. Since we are interested in the effects of DNA damage repair on clinical events other than cancer, and because the two included study populations are of rather old age, especially PROSPER, we cannot exclude that the competing risk of cancer related mortality and CVD events have led to a selection bias of the patients. Therefore, it could be possible that the role of DNA repair pathways is being underestimated. However, since we cannot correct for this potential selection bias, this remains speculative. Another potential confounder is age. The GENDER study is considerably younger than the PROSPER cohort, possibly explaining part of the different results of both studies. However, since in the set-based analysis of PLINK correction for confounders is not possible, the actual magnitude of the influence of age on the current results remains therefore uncertain.
In conclusion, with this study we demonstrate that genetic variation in the NHEJ pathway of the human DNA repair machinery, and specifically genetic variation in the MRE11A gene, is associated with the occurrence of MI. Results of this study need to be validated by functional studies to further elucidate the precise mechanistic role of NHEJ in atherosclerotic lesion formation.
Supporting Information
Characteristics of the 5 DNA repair pathways.
(XLS)
Subgroup analysis of the non-homologous end-joining pathway in GENDER and PROSPER.
(XLS)
Funding Statement
This work was funded by grants from the Interuniversity Cardiology Institute of the Netherlands (ICIN) http://www.icin.nl/, the European Community Framework FP7 Programme under grant agreement [no HEALTH-F2-2009-223004], the Center for Medical Systems Biology (CMSB) [http://www.cmsb.nl], a center of excellence approved by the Netherlands Genomics Initiative/Netherlands Organisation for Scientific Research (NWO), and the Netherlands Consortium for Healthy Ageing (NCHA) [http://www.healthy-ageing.nl]. JWJ is an established clinical investigator of the Netherlands Heart Foundation (2001D032) [http://www.hartstichting.nl/]. The funders had no role in study design, data collection and analysis, decision to publish or the preparation of the manuscript.
References
- 1. Corral-Debrinski M, Stepien G, Shoffner JM, Lott MT, Kanter K, et al. (1991) Hypoxemia is associated with mitochondrial DNA damage and gene induction. Implications for cardiac disease. JAMA 266: 1812–1816. [PubMed] [Google Scholar]
- 2. Andreassi MG (2003) Coronary atherosclerosis and somatic mutations: an overview of the contributive factors for oxidative DNA damage. Mutat Res 543: 67–86. [DOI] [PubMed] [Google Scholar]
- 3. Mahmoudi M, Mercer J, Bennett M (2006) DNA damage and repair in atherosclerosis. Cardiovasc Res 71: 259–268. [DOI] [PubMed] [Google Scholar]
- 4. Durand E, Scoazec A, Lafont A, Boddaert J, Al HA, et al. (2004) In vivo induction of endothelial apoptosis leads to vessel thrombosis and endothelial denudation: a clue to the understanding of the mechanisms of thrombotic plaque erosion. Circulation 109: 2503–2506. [DOI] [PubMed] [Google Scholar]
- 5. Bauriedel G, Hutter R, Welsch U, Bach R, Sievert H, et al. (1999) Role of smooth muscle cell death in advanced coronary primary lesions: implications for plaque instability. Cardiovasc Res 41: 480–488. [DOI] [PubMed] [Google Scholar]
- 6. Rossi ML, Marziliano N, Merlini PA, Bramucci E, Canosi U, et al. (2004) Different quantitative apoptotic traits in coronary atherosclerotic plaques from patients with stable angina pectoris and acute coronary syndromes. Circulation 110: 1767–1773. [DOI] [PubMed] [Google Scholar]
- 7. Minamino T, Komuro I (2007) Vascular cell senescence: contribution to atherosclerosis. Circ Res 100: 15–26. [DOI] [PubMed] [Google Scholar]
- 8. Durik M, Kavousi M, van dP, I, Isaacs A, Cheng C, et al. (2012) Nucleotide excision DNA repair is associated with age-related vascular dysfunction. Circulation 126: 468–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ozturk S, Demir N (2011) DNA repair mechanisms in mammalian germ cells. Histol Histopathol 26: 505–517. [DOI] [PubMed] [Google Scholar]
- 10. Waters LS, Minesinger BK, Wiltrout ME, D’Souza S, Woodruff RV, et al. (2009) Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev 73: 134–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Voghel G, Thorin-Trescases N, Mamarbachi AM, Villeneuve L, Mallette FA, et al. (2010) Endogenous oxidative stress prevents telomerase-dependent immortalization of human endothelial cells. Mech Ageing Dev 131: 354–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Satoh M, Ishikawa Y, Takahashi Y, Itoh T, Minami Y, et al. (2008) Association between oxidative DNA damage and telomere shortening in circulating endothelial progenitor cells obtained from metabolic syndrome patients with coronary artery disease. Atherosclerosis 198: 347–353. [DOI] [PubMed] [Google Scholar]
- 13. Ragnauth CD, Warren DT, Liu Y, McNair R, Tajsic T, et al. (2010) Prelamin A acts to accelerate smooth muscle cell senescence and is a novel biomarker of human vascular aging. Circulation 121: 2200–2210. [DOI] [PubMed] [Google Scholar]
- 14. Matthews C, Gorenne I, Scott S, Figg N, Kirkpatrick P, et al. (2006) Vascular smooth muscle cells undergo telomere-based senescence in human atherosclerosis: effects of telomerase and oxidative stress. Circ Res 99: 156–164. [DOI] [PubMed] [Google Scholar]
- 15. Gizard F, Heywood EB, Findeisen HM, Zhao Y, Jones KL, et al. (2011) Telomerase activation in atherosclerosis and induction of telomerase reverse transcriptase expression by inflammatory stimuli in macrophages. Arterioscler Thromb Vasc Biol 31: 245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu SC, Wang SS, Wu MZ, Wu DC, Yu FJ, et al. (2005) Activation of telomerase and expression of human telomerase reverse transcriptase in coronary atherosclerosis. Cardiovasc Pathol 14: 232–240. [DOI] [PubMed] [Google Scholar]
- 17. Perez-Rivero G, Ruiz-Torres MP, Rivas-Elena JV, Jerkic M, Diez-Marques ML, et al. (2006) Mice deficient in telomerase activity develop hypertension because of an excess of endothelin production. Circulation 114: 309–317. [DOI] [PubMed] [Google Scholar]
- 18. Poch E, Carbonell P, Franco S, Diez-Juan A, Blasco MA, et al. (2004) Short telomeres protect from diet-induced atherosclerosis in apolipoprotein E-null mice. FASEB J 18: 418–420. [DOI] [PubMed] [Google Scholar]
- 19. Mercer JR, Cheng KK, Figg N, Gorenne I, Mahmoudi M, et al. (2010) DNA damage links mitochondrial dysfunction to atherosclerosis and the metabolic syndrome. Circ Res 107: 1021–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mahabir S, Abnet CC, Qiao YL, Ratnasinghe LD, Dawsey SM, et al. (2007) A prospective study of polymorphisms of DNA repair genes XRCC1, XPD23 and APE/ref-1 and risk of stroke in Linxian, China. J Epidemiol Community Health 61: 737–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bazo AP, Salvadori D Jr, Salvadori RA, Sodre LP, da Silva GN, et al. (2011) DNA repair gene polymorphism is associated with the genetic basis of atherosclerotic coronary artery disease. Cardiovasc Pathol 20: e9–15. [DOI] [PubMed] [Google Scholar]
- 22. Shyu HY, Shieh JC, Lin JH, Wang HW, Cheng CW (2012) Polymorphisms of DNA Repair Pathway Genes and Cigarette Smoking in Relation to Susceptibility to Large Artery Atherosclerotic Stroke among Ethnic Chinese in Taiwan. J Atheroscler Thromb 19: 316–325. [DOI] [PubMed] [Google Scholar]
- 23. Schunkert H, Konig IR, Kathiresan S, Reilly MP, Assimes TL, et al. (2011) Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet 43: 333–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zeller T, Blankenberg S, Diemert P (2012) Genomewide association studies in cardiovascular disease–an update 2011. Clin Chem 58: 92–103. [DOI] [PubMed] [Google Scholar]
- 25. Wang K, Li M, Hakonarson H (2010) Analysing biological pathways in genome-wide association studies. Nat Rev Genet 11: 843–854. [DOI] [PubMed] [Google Scholar]
- 26. Torkamani A, Topol EJ, Schork NJ (2008) Pathway analysis of seven common diseases assessed by genome-wide association. Genomics 92: 265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Agema WR, Monraats PS, Zwinderman AH, de Winter RJ, Tio RA, et al. (2004) Current PTCA practice and clinical outcomes in The Netherlands: the real world in the pre-drug-eluting stent era. Eur Heart J 25: 1163–1170. [DOI] [PubMed] [Google Scholar]
- 28. Shepherd J, Blauw GJ, Murphy MB, Cobbe SM, Bollen EL, et al. (1999) The design of a prospective study of Pravastatin in the Elderly at Risk (PROSPER). PROSPER Study Group. PROspective Study of Pravastatin in the Elderly at Risk. Am J Cardiol 84: 1192–1197. [DOI] [PubMed] [Google Scholar]
- 29. Shepherd J, Blauw GJ, Murphy MB, Bollen EL, Buckley BM, et al. (2002) Pravastatin in elderly individuals at risk of vascular disease (PROSPER): a randomised controlled trial. Lancet 360: 1623–1630. [DOI] [PubMed] [Google Scholar]
- 30. Sampietro ML, Trompet S, Verschuren JJ, Talens RP, Deelen J, et al. (2011) A genome-wide association study identifies a region at chromosome 12 as a potential susceptibility locus for restenosis after percutaneous coronary intervention. Hum Mol Genet 20: 4748–4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Trompet S, de Craen AJ, Postmus I, Ford I, Sattar N, et al. (2011) Replication of LDL GWAs hits in PROSPER/PHASE as validation for future (pharmaco)genetic analyses. BMC Med Genet 12: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR (2010) MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol 34: 816–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M (2012) KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 40: D109–D114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.KEGG website. Available: http://www.genome.jp/kegg/pathway.html. Accessed 1 November 2011.
- 35. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.UCSC genome browsers database. Available: http://genome.ucsc.edu. Accessed 1 August 2012.
- 37. Knudsen S (1999) Promoter2.0: for the recognition of PolII promoter sequences. Bioinformatics 15: 356–361. [DOI] [PubMed] [Google Scholar]
- 38. Macintyre G, Bailey J, Haviv I, Kowalczyk A (2010) is-rSNP: a novel technique for in silico regulatory SNP detection. Bioinformatics 26: i524–i530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, et al. (2007) A genome-wide association study of global gene expression. Nat Genet 39: 1202–1207. [DOI] [PubMed] [Google Scholar]
- 40. Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, et al. (2007) Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 448: 470–473. [DOI] [PubMed] [Google Scholar]
- 41. Shimada MK, Matsumoto R, Hayakawa Y, Sanbonmatsu R, Gough C, et al. (2009) VarySysDB: a human genetic polymorphism database based on all H-InvDB transcripts. Nucleic Acids Res 37: D810–D815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.eQTL database of the Pritchard lab. Available: http://eqtl.uchicago.edu. Accessed 1 August 2012.
- 43. Williams GJ, Lees-Miller SP, Tainer JA (2010) Mre11-Rad50-Nbs1 conformations and the control of sensing, signaling, and effector responses at DNA double-strand breaks. DNA Repair (Amst) 9: 1299–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mirzoeva OK, Petrini JH (2003) DNA replication-dependent nuclear dynamics of the Mre11 complex. Mol Cancer Res 1: 207–218. [PubMed] [Google Scholar]
- 45. Borde V (2007) The multiple roles of the Mre11 complex for meiotic recombination. Chromosome Res 15: 551–563. [DOI] [PubMed] [Google Scholar]
- 46. Borde V, Cobb J (2009) Double functions for the Mre11 complex during DNA double-strand break repair and replication. Int J Biochem Cell Biol 41: 1249–1253. [DOI] [PubMed] [Google Scholar]
- 47. Lamarche BJ, Orazio NI, Weitzman MD (2010) The MRN complex in double-strand break repair and telomere maintenance. FEBS Lett 584: 3682–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Park YB, Chae J, Kim YC, Cho Y (2011) Crystal structure of human Mre11: understanding tumorigenic mutations. Structure 19: 1591–1602. [DOI] [PubMed] [Google Scholar]
- 49. Choudhury A, Elliott F, Iles MM, Churchman M, Bristow RG, et al. (2008) Analysis of variants in DNA damage signalling genes in bladder cancer. BMC Med Genet 9: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Loizidou MA, Michael T, Neuhausen SL, Newbold RF, Marcou Y, et al. (2009) DNA-repair genetic polymorphisms and risk of breast cancer in Cyprus. Breast Cancer Res Treat 115: 623–627. [DOI] [PubMed] [Google Scholar]
- 51. Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, et al. (1999) The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell 99: 577–587. [DOI] [PubMed] [Google Scholar]
- 52. Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, et al. (2007) Genomewide Association Analysis of Coronary Artery Disease. N Engl J Med 357: 443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wellcome Trust Case Control Consortium (2007) Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447: 661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Allen-Brady K, Camp NJ (2005) Characterization of the linkage disequilibrium structure and identification of tagging-SNPs in five DNA repair genes. BMC Cancer 5: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Xie X, Mikkelsen TS, Gnirke A, Lindblad-Toh K, Kellis M, et al. (2007) Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc Natl Acad Sci U S A 104: 7145–7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Thorne D, Wilson J, Kumaravel TS, Massey ED, McEwan M (2009) Measurement of oxidative DNA damage induced by mainstream cigarette smoke in cultured NCI-H292 human pulmonary carcinoma cells. Mutat Res 673: 3–8. [DOI] [PubMed] [Google Scholar]
- 57. Ockene IS, Miller NH (1997) Cigarette Smoking, Cardiovascular Disease, and Stroke: A Statement for Healthcare Professionals From the American Heart Association. Circulation 96: 3243–3247. [DOI] [PubMed] [Google Scholar]
- 58. Arcidiacono B, Iiritano S, Nocera A, Possidente K, Nevolo MT, et al. (2012) Insulin resistance and cancer risk: an overview of the pathogenetic mechanisms. Exp Diabetes Res 2012: 789174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Furukawa S, Fujita T, Shimabukuro M, Iwaki M, Yamada Y, et al. (2004) Increased oxidative stress in obesity and its impact on metabolic syndrome. J Clin Invest 114: 1752–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gui H, Li M, Sham PC, Cherny SS (2011) Comparisons of seven algorithms for pathway analysis using the WTCCC Crohn’s Disease dataset. BMC Res Notes 4: 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Deelen J, Uh HW, Monajemi R, van HD, Thijssen PE, et al. (2011) Gene set analysis of GWAS data for human longevity highlights the relevance of the insulin/IGF-1 signaling and telomere maintenance pathways. Age (Dordr ) In press. 10.1007/s11357-011-9340-3 [doi]. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characteristics of the 5 DNA repair pathways.
(XLS)
Subgroup analysis of the non-homologous end-joining pathway in GENDER and PROSPER.
(XLS)