Abstract
Ascarosides are small-molecule signals that play a central role in C. elegans biology, including dauer formation, aging, and social behaviors, but many aspects of their biosynthesis remain unknown. Using automated 2D NMR-based comparative metabolomics, we identified ascaroside ethanolamides as shunt metabolites in C. elegans mutants of daf-22, a gene with homology to mammalian 3-ketoacyl-CoA thiolases predicted to function in conserved peroxisomal lipid β-oxidation. Two groups of ethanolamides feature β-keto functionalization confirming the predicted role of daf-22 in ascaroside biosynthesis, whereas α-methyl substitution points to unexpected inclusion of methylmalonte at a late stage in the biosynthesis of long-chain fatty acids in C. elegans. We show that ascaroside ethanolamide formation in response to defects in daf-22 and other peroxisomal genes is associated with severe depletion of endocannabinoid pools. These results indicate unexpected interaction between peroxisomal lipid β-oxidation and the biosynthesis of endocannabinoids, which are major regulators of lifespan in C. elegans. Our study demonstrates the utility of unbiased comparative metabolomics for investigating biochemical networks in metazoans.
The free-living nematode Caenorhabditis elegans is used extensively as a model system for the study of aging and metabolism (1–3). Recent analyses of the C. elegans metabolome revealed a family of structurally diverse small molecules, the ascarosides, as products of conserved peroxisomal β-oxidation (Figure 1).(4–6) The ascarosides form a modular compound library, integrating building blocks from lipid, carbohydrate and amino acid metabolism into small molecule signals that regulate key aspects of C. elegans biology.(7–10) Previous work used mass spectrometry-based comparative metabolomics to characterize the functions of three enzymes in ascaroside biosynthesis, the acyl-CoA oxidase ACOX-1, the enoyl-CoA hydratase MAOC-1, and the 3-hydroxyacyl-CoA dehydrogenase DHS-28 (Figure 1b).(4) However, the predicted role of the fourth enzyme in the peroxisomal β-oxidation cascade, which is encoded by the gene daf-22, a close homolog of mammalian 3-keto-acyl-CoA thiolases, could not be confirmed because no characteristic shunt metabolites were found. Given the central role of daf-22 in the proposed peroxisomal β-oxidation pathway, we hypothesized that other, yet unidentified metabolites accumulate in daf-22 mutants, which may help confirm the biochemical function of this enzyme and provide insight into interactions with other biochemical pathways.
Figure 1.
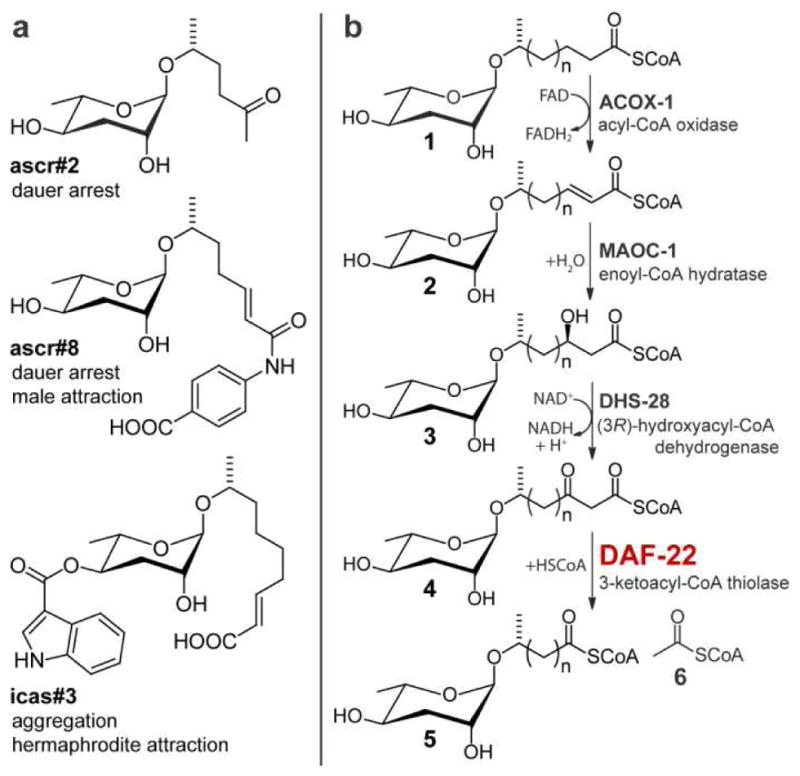
Ascarosides are signaling molecules in C. elegans. (a) Example structures and biological functions of ascaroside-based signaling molecules in C. elegans. (b) Model for peroxisomal biosynthesis of ascaroside side chains and roles of peroxisomal β-oxidation enzymes ACOX-1, MAOC-1, DHS-28, and the putative 3-ketoacyl-CoA thiolase DAF-22.
Therefore, we reinvestigated the daf-22 metabolome using unbiased NMR spectroscopy-based metabolomics. Previously, we had employed manual comparisons of 2D NMR spectra of wild-type and daf-22 metabolomes to identify additional members of the ascaroside family of signaling molecules.(6, 9, 11) These studies highlighted the utility of 2D NMR spectra for comparative metabolomics, but also demonstrated the need for computational tools that would enable statistical analysis of series of spectra from different mutants. For this purpose, we developed multivariate Differential Analysis by 2D NMR Spectroscopy (mvaDANS), which enables automated processing and comparative computational analysis for sets of 2D NMR spectra.
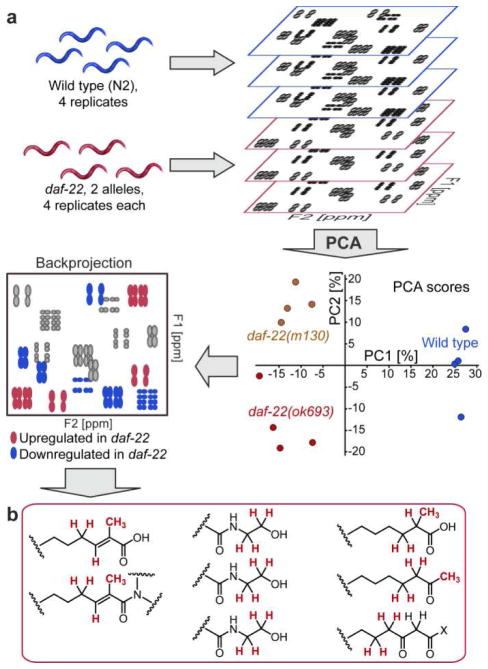
mvaDANS builds on previous efforts to use 2D NMR for metabolomic analysis by incorporating dynamic signal detection and pattern recognition (see Methods).(12–18) Figure 2 summarizes the use of this approach for identifying compounds that are upregulated in daf-22 mutants and thus may represent shunt metabolites or biosynthetic intermediates. We acquired dqfCOSY spectra for metabolome extracts obtained from two different daf-22 alleles (m130, ok693) and wild-type C. elegans, including four independent biological replicates (Figures 2a, S1-S2). Automatic crosspeak identification and binning (Figure S3) prepared the dqfCOSY spectra for statistical analysis via Principal Component Analysis (PCA, see Methods),(19) which clustered the data by genotype (Figures 2a). Coefficients from the PCA loadings were back-projected onto the dqfCOSY spectra, which revealed a large number of crosspeaks that are up- or downregulated in daf-22 mutants relative to wild-type (Figures 2a, S4, and S5).
Figure 2.
mvaDANS reveals structural motifs upregulated in daf-22 mutants. (a) Schematic overview of mvaDANS for identification of up- and downregulated metabolites in daf-22 mutants. In the Principal Component Analysis (PCA), PC 1 separates the four wild-type data sets (blue dots) from both daf-22 mutant data sets (red and brown dots), whereas PC2 separates the two daf-22 alleles. In the back projection (lower left), the coloring of cross speaks corresponds to the PC 1 loading coefficients; blue peaks represent signals downregulated and red peaks signals upregulated in the two daf-22 mutant strains. See Figure S4 for actual spectra. (b) Partial structures inferred from manual analysis of daf-22-specific crosspeaks suggesting α-methyl branched fatty acids, methyl ketones, and β-keto fatty acid derivatives.
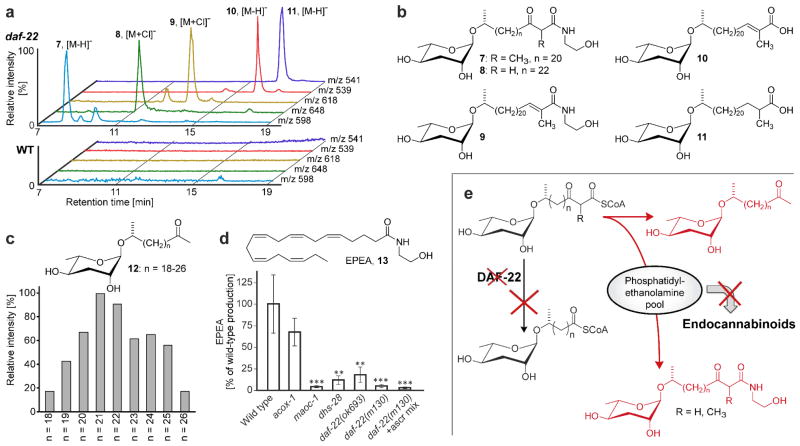
For this study, we restricted analysis to crosspeaks that were strongly featured in spectra from both daf-22 alleles but were very weak or undetectable in wild-type. Furthermore, we excluded crosspeaks also present in the E. coli food source. Manual analysis of the remaining strongly daf-22-upregulated crosspeaks and inputs from additional HMBC spectra suggested a series of partial structures, including methyl ketones, α-methyl branched fatty acids and corresponding ethanolamides as well as β-keto fatty acid derivatives (Figure 2b). Considering the proposed role of daf-22 in ascaroside biosynthesis (Figure 1b), we hypothesized that these structural features belong to accumulated shunt metabolites. Subsequent fractionation of the daf-22 metabolome guided by the daf-22-specific NMR signals led to isolation of six different very long-chain ascarosides (VLCA) that account for all of the predicted structural features (Figure 3, see Supporting Information for full spectroscopic data). None of these daf-22 mutant-specific compounds had previously been reported. Most prominent are a series of (ω-1)-O-ascarosyl 2-ketones (12) as well as (ω-1)-O-ascarosyl 3-keto fatty acid ethanolamides, which include the 2-methyl pentacosanoic acid derivative 7 and the straight-chain heptacosanoic acid derivative 8. Compounds 7 and 8 are accompanied by smaller amounts of the corresponding bis-norhomologues. Similarly, the 2-methyl pentacos-2-enoic acid ethanolamide 9 and the two fatty acids 10 and 11 are accompanied by bis-norhomologues. In contrast, chain lengths of the (ω-1)-O-ascarosyl 2-ketones (12) vary more broadly and are not preferentially odd-numbered (Figure 3c). HPLC-MS analysis confirmed the absence of these components in wild-type samples (Figure 3a).
Figure 3.
Long-chain ascarosides identified in the daf-22-mutant metabolome and interaction with NEA biosynthesis. (a) HPLC-MS analysis confirms long-chain ascarosides and ethanolamides in the daf-22 metabolome that are absent in wild-type. (b) daf-22 mutant-specific VLCAs identified via mvaDANS and subsequent HPLC-MS analysis. (c) Quantitative distribution of daf-22-upregulated ascarosyl methylketones. (d) Mutation of maoc-1, dhs-28, and daf-22 greatly reduces EPEA production and addition of ascarosides (2.5 μM ascr#3 and ascr#9, see reference (4) for structures) to daf-22(m130) cultures does not rescue EPEA production.* P<0.05, ** P<0.01, *** P<0.001. (e) Interactions of peroxisomal β-oxidation and endocannabinoid biosynthesis. Mutation of daf-22 abolishes processing of long-chain ascaroside CoA esters, whose conversion into ethanolamides is associated with reduced endocannabinoid production (13), likely due to depletion of phosphatidylethanolamine pools.
The incorporation of ethanolamine in the daf-22 specific VLCAs was unexpected and suggested the possibility of interactions between peroxisomal β-oxidation and ethanolamide-based signaling pathways. Recently, N-acyl-ethanolamines (NAEs) were identified as endocannabinoid-like signaling molecules in C. elegans and linked to dietary restriction-dependent lifespan regulation.(20) We asked whether the upregulation of VLCAs in daf-22 mutants affects NAE levels. We found that NAE production, including the most active endocannabinoid in C. elegans,(20) eicosapentaenoyl ethanolamide (13, EPEA), and the ligand of mammalian cannabinoid receptors, arachidonoyl ethanolamide (AEA, or anandamide), was dramatically downregulated in both daf-22 alleles (Figures 3d and S6), suggesting a shift of ethanolamine utilization from the NAEs to VLCAs. Similarly, we found greatly reduced EPEA and AEA production in mutants of maoc-1 and dhs-28 (Figures 3d and S6) which act directly upstream of daf-22 (Figure 1b). EPEA-levels in acox-1 mutant worms were not significantly affected and AEA levels only slightly decreased, corresponding to the much weaker effect of the acox-1 mutation on peroxisomal β-oxidation.(4) EPEA and AEA production was not rescued by growing daf-22 worms with addition of synthetic ascarosides (Figures 3d and S6), suggesting that the observed reduction of NAE levels in peroxisomal β-oxidation mutants is not an indirect effect of the lack of ascaroside pheromone. These findings indicate that in C. elegans specific defects in peroxisomal β-oxidation result in depletion of endocannabinoid pools (Figure 3e). It is unknown whether conditions that reduce expression of peroxisomal β-oxidation genes in healthy animals trigger down-regulation of endocannabinoids, which could serve as a regulatory mechanism tying fat metabolism and ascaroside signaling to dietary restriction pathways. Ethanolamides of short-chain ascarosides serve as pheromones in a parasitic nematode species,(21) suggesting that interactions of peroxisomal β-oxidation and ethanolamide metabolism also exist in other nematode genera.
Mammalian homologs of daf-22 are thought to be specifically required for chain shortening of branched-chain fatty acids such as phytanic acid.(22, 23) However, the co-occurrence of straight-chain and methyl-branched derivatives among the daf-22 mutant-specific VLCAs suggests that daf-22 is involved in processing of both branched and straight-chain fatty acids. The chain lengths of the daf-22 mutant-specific VLCAs indicate that 23- to 27-carbon fatty acids may represent endpoints of C. elegans fatty acid biogenesis, in which the last chain-extension step incorporates either malonate or methylmalonate. Notably, our metabolomic analyses provided no evidence for incorporation of methylmalonate at earlier stages in the biosynthesis of these fatty acid derivatives. The chain lengths of the daf-22-mutant upregulated VLCAs in C. elegans are similar to those of very long-chain fatty acids that accumulate in human disorders involving defects in peroxisomal β-oxidation such as Zellweger syndrome.(22, 23) However, human fatty acid biosynthesis is not known to selectively incorporate propionate or methylmalonate in the last chain-extension step to form α-methyl-branched variants.
In a biosynthetic model (Figure 1b, 3e), the identified daf-22 mutant-specific compounds plausibly represent shunt metabolites derived from fatty-acid and β-keto-acid CoA esters, in which the α,β-unsaturation and β-keto functionalities result from upstream action of ACOX-1, MAOC-1, and DHS-28. The daf-22-upregulated long-chain methyl ketones may derive from corresponding β-keto fatty acids, which are highly prone to decarboxylation. Although described in an earlier study,(5) we could not find any evidence for β-keto fatty acids in our metabolite extracts by either NMR spectroscopy or HPLC-MS. The odd-numbered side-chain lengths of the daf-22-upregulated ascarosyl-fatty acids are consistent with the structures of the known end products of peroxisomal β-oxidation, the short-chain ascaroside pheromones, most of which include side chains with 3, 5, 7, or 9 carbons (Figure 1a).(4) These odd-numbered side chains further suggest that ascaroside side-chain biosynthesis starts with a three-carbon (methylmalonate-derived) template or that C. elegans elongates odd-numbered fatty acids obtained from food.
In conclusion, mvaDANS-based comparison of daf-22 and wild-type metabolomes revealed long-chain ascarosyl ethanolamides as an unexpected class of shunt metabolites in daf-22 mutant worms. The abundance of β-keto derivatives and methyl ketones among the identified long-chain ascarosides supports that daf-22 acts as a thiolase in ascaroside pheromone biosynthesis, as had been suspected based on homology to mammalian peroxisomal thiolases. Our results will motivate more detailed inspection of similarities of nematode and mammalian fatty-acid metabolism, especially with regard to possible interactions between endocannabinoid signaling, phosphatidylethanolamine utilization, and peroxisomal β-oxidation. In C. elegans, reduced NAE levels promote dietary-restriction-induced lifespan extension,(20) suggesting that defects in peroxisomal β-oxidation may affect lifespan via changes in NAE levels.
Untargeted comparative metabolomics will form an important tool for reinvestigating metazoan biochemistry. mvaDANS extends the scope of 2D NMR-based metabolomic analysis (24–26) by introducing automated cross-peak identification and integration. Whereas pattern recognition techniques have been applied previously to 2D NMR spectra, these approaches have either relied on manual peak selection,(27) parametric approaches,(28) or used spectral unfolding/refolding techniques.(10, 11) We expect that advances in statistical spectroscopy will continue to accelerate the structural and functional characterization of biogenic small molecules.
Methods
C. elegans Strains and General Culture Methods
Wild-type C. elegans (N2, Bristol), daf-22(m130), daf-22(ok693), acox-1(ok2257), maoc-1(hj13), and dhs-28(hj8) mutant strains were obtained from the Caenorhabditis Genetics Center (CGC). Strains were maintained at 20 °C on NGM plates with bacteria (E. coli OP50) as food.
Preparation of C. elegans Metabolite Extracts
Liquid cultures were grown as previously reported.(3) Briefly, wild-type or daf-22 mutant worms were grown for two generations on NGM plates seeded with OP50. Three crowded 6 cm NGM plates were washed into 100 mL solution of S-medium in a 500 mL Erlenmeyer flask and grown at 22°C and 220 pm. Concentrated OP50 from 1 L bacterial cultures, grown over-night, were given on day 1 and day 3. On day 5 the liquid culture was split into two 500 mL Erlenmeyer flasks and the growth media volume was maintained at 100 mL. An additional 1 L OP50 pellet was given as food upon splitting. The cultures were harvested on day 7 by centrifugation at 4750 rpm. The supernatant was lyophilized and the residue extracted with 95% ethanol (300 mL) at room temperature for 12 h. The resulting suspension was filtered and evaporated in vacuo at room temperature to yield the crude extracts. In this manner four extracts each of wild-type (N2), daf-22(m130), and daf-22(ok693) cultures were prepared. For NAE quantification, mutant strains were grown as above in duplicate. Liquid cultures for ascaroside mixture experiments were prepared as described above with addition of 2.5 μM of ascr#3 and 2.5 μM of ascr#9 at day 0.
In preparation for NMR-spectroscopic analysis, the crude extracts were suspended in 1:8 methanol (MeOH):dichloromethane (DCM), using 7.5 mL of this solvent mixture per 1 gram of crude extract. Following centrifugation, 2 mL aliquots of supernatant of each sample were filtered over 1 g of silica (EMD Chemicals Inc.) using 10 mL of 1:8 MeOH:DCM for elution. The filtrate was evaporated to dryness in vacuo prior to analysis. For NMR spectroscopic analysis, 10–20 mg of the resulting metabolite extracts was used.
Preparation of Bacterial Metabolite Extracts
E. coli OP50 was grown overnight at 37 °C and 220 rpm to an OD of 1.2 in 2 L of Lysogeny broth (LB). The bacterial broth was spun for 20 min at 4750 rpm. The supernatant was then lyophilized, crushed with mortar and pestle and extracted with 150 mL 95% ethanol (2 times). The extract was filtered through a sintered crucible and concentrated in vacuo prior to analysis.
Mass Spectrometric Analysis
Metabolite extracts prepared as described above were resuspended in 1.5 mL methanol, centrifuged, and 30 μL of this sample was injected into HPLC-MS for analysis. HPLC-MS analysis was performed using the Agilent 1100 Series HPLC system with an Agilent Eclipse XDB-C18 column (9.4 × 250 mm, 5 μm particle diameter) connected to the Quattro II mass spectrometer using a 10:1 split. For short chain ascarosides, a 0.1% aqueous acetic acid–acetonitrile gradient was used. The gradient started with 5% acetonitrile for 5 minutes, then increased to 100% acetonitrile over 32 minutes. For long chain ascarosides, an acetonitrile-isopropanol gradient was used, starting with 5% isopropanol in acetonitrile for 5 minutes; then isopropanol was increased to 75% over 32 minutes. Samples were analyzed by HPLC-ESI-MS in negative and positive ion modes using a capillary voltage of 3.5 kV and a cone voltage of −40 V and +20 V, respectively. The elemental composition of the long chain ascarosides was performed by high resolution MS and HPLC-MS using the Xevo G2 QTof.
EPEA and AEA (14) were quantified by HPLC-MS (using the Quattro II) using a water (0.1% acetic acid) – acetonitrile gradient as described above for short chain ascarosides. The retention time of EPEA and AEA were established by injection of an authentic standard (Enzo Life Sciences and Cayman Chemicals, respectively). Relative endocannabinoid content in wild type and mutant strains was quantified by integration of LC-MS signals from corresponding ion traces for (M-H)− or (M+Cl)−. All integration values were normalized by the mass of the liquid culture pellet. All quantitative data reported in Figure 3 were from at least two independent biological repeats.
NMR Spectrometric Analysis
NMR samples sample were prepared by dissolving 10–20 mg portions of metabolite extract (see above) in 0.6 ml of CD3OD. Samples were analyzed using a Varian INOVA 600 MHz NMR spectrometer. Non-gradient phase-cycled dqfCOSY spectra were acquired using the following parameters: 0.8 s acquisition time, 600 complex increments, 16 scans per increment. For processing with mvaDANS, spectra were zero-filled to 2048 × 1024 data points, a sine-square-shaped window function was applied in both dimensions prior to Fourier transformation, and spectra converted to magnitude mode prior to transfer to MATLAB.
For detailed analysis in phase-sensitive mode, dqfCOSY spectra were zero-filled to 8192 × 4096 data points and a cosine bell-shaped window function was applied in both dimensions before Fourier transformation. Gradient and non-gradient HSQCAD and HMBC spectra of purified fractions of daf-22-upregulated metabolites were acquired using 8–32 scans, 0.25 s acquisition time, and 256–500 complex increments. NMR spectra were processed using Bruker TopSpin, Varian VNMR, MestreLabs’ MestReC, and MNova software packages.
Isolation of Major daf-22-Upregulated Metabolites
To 8 g of Celite (prewashed with ethyl acetate) was added a solution of daf-22 metabolite extract from 16 100-ml liquid cultures. After evaporation of the solvent, the material was dry-loaded into an empty 25 g RediSep® Rf loading cartridge. Fractionation was performed using a Teledyne ISCO CombiFlash® system over a RediSep® Rf GOLD 40 g HP Silica Column using a normal phase dichloromethane-methanol solvent system, starting with 6 min of 100% dichloromethane, followed by a linear increase of methanol content up to 10% at 24 min, followed by another linear increase of methanol content up to 40% at 40 min, followed by another linear increase of methanol content up to 95% at 45 min which was then continued to 48 min. This yielded 70 fractions (~20 mL each) which were individually evaporated in vacuo and analyzed by NMR spectroscopy (1H NMR, dqfCOSY) and HPLC-MS. 1H NMR-spectroscopic signals identified via mvaDANS as representing daf-22-upregulated metabolites were detected in fractions 35–45. Isolation of major daf-22-upregulated metabolites was achieved by preparative HPLC using the same HPLC method described above for HPLC-MS analysis of long chain ascarosides. Isolated fractions were characterized further by dqfCOSY, HSQC, and HMBC as well as high-resolution HPLC-MS using the Xevo G2 QTof.
Statistical Analysis
Figure 4d and Figure S6: we used unpaired student’s t-tests with Welch’s correction for comparing the relative abundance of EPEA and AEA in wild type and mutant strains. * P<0.05, ** P<0.01, *** P<0.001.
mvaDANS Analysis
For identification of daf-22-upregulated metabolites via mvaDANS, we acquired dqfCOSY spectra for metabolome extracts obtained from two different daf-22 alleles (m130, ok693) and wild-type C. elegans (Figures 2, S1–S2). dqfCOSY spectra from four independent biological replicates were processed in magnitude mode using the mvaDANS algorithm in Matlab, which incorporated dynamic binning and multivariate pattern recognition analysis (Figure S3). Automatic cross peak identification and binning converted each of the dqfCOSY spectra into a set of crosspeak integrals which were corrected for dilution using Probabilistic Quotient Normalization,(29) mean-centered, and scaled to unit variance. This matrix of bin integrals was then subjected to statistical analysis including two-way hierarchical clustering, Principal Component Analysis (PCA) (Figure 2a),(19) and Partial Least Squares – Discriminant Analysis (PLS-DA) (Figure S7).(30) Coefficients from PCA loadings and PLS-DA predictors were back-projected onto the dqfCOSY spectra to enable the detection of NMR spectroscopic signals representing compounds differentially expressed in the three C. elegans strains (Figures S4, S8).
mvaDANS automation and spectral interpretation
Please see supporting information.
Supplementary Material
Acknowledgments
This work was supported in part by the National Institutes of Health (GM088290 to FCS, and T32GM008500 to YI), and the Vassar College Mary Landon Sague Fellowship (to YI). We thank M. Kukula (BTI) for help with high-resolution MS.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Fielenbach N, Antebi A. C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 2008;22:2149–2165. doi: 10.1101/gad.1701508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 3.Jones KT, Ashrafi K. Caenorhabditis elegans as an emerging model for studying the basic biology of obesity. Dis Model Mech. 2009;2:224–229. doi: 10.1242/dmm.001933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.von Reuss SH, Bose N, Srinivasan J, Yim JJ, Judkins JC, Sternberg PW, Schroeder FC. Comparative metabolomics reveals biogenesis of ascarosides, a modular library of small molecule signals in C. elegans. J Am Chem Soc. 2012;134:1817–1824. doi: 10.1021/ja210202y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butcher RA, Ragains JR, Li W, Ruvkun G, Clardy J, Mak HY. Biosynthesis of the Caenorhabditis elegans dauer pheromone. Proc Natl Acad Sci USA. 2009;106:1875–1879. doi: 10.1073/pnas.0810338106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pungaliya C, Srinivasan J, Fox BW, Malik RU, Ludewig AH, Sternberg PW, Schroeder FC. A shortcut to identifying small molecule signals that regulate behavior and development in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2009;106:7708–7713. doi: 10.1073/pnas.0811918106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butcher RA, Fujita M, Schroeder FC, Clardy J. Small-molecule pheromones that control dauer development in Caenorhabditis elegans. Nat Chem Biol. 2007;3:420–422. doi: 10.1038/nchembio.2007.3. [DOI] [PubMed] [Google Scholar]
- 8.Srinivasan J, Kaplan F, Ajredini R, Zachariah C, Alborn HT, Teal PE, Malik RU, Edison AS, Sternberg PW, Schroeder FC. A blend of small molecules regulates both mating and development in Caenorhabditis elegans. Nature. 2008;454:1115–1118. doi: 10.1038/nature07168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Srinivasan J, von Reuss SH, Bose N, Zaslaver A, Mahanti P, Ho MC, O’Doherty OG, Edison AS, Sternberg PW, Schroeder FC. A modular library of small molecule signals regulates social behaviors in Caenorhabditis elegans. PLoS Biol. 2012;10:e1001237. doi: 10.1371/journal.pbio.1001237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Macosko EZ, Pokala N, Feinberg EH, Chalasani SH, Butcher RA, Clardy J, Bargmann CI. A hub-and-spoke circuit drives pheromone attraction and social behaviour in C. elegans. Nature. 2009;458:1171–1175. doi: 10.1038/nature07886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinette SL, Bruschweiler R, Schroeder FC, Edison AS. NMR in metabolomics and natural products research: Two sides of the same coin. Acc Chem Res. 2011;45:288–297. doi: 10.1021/ar2001606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hedenstrom M, Wiklund-Lindstrom S, Oman T, Lu F, Gerber L, Schatz P, Sundberg B, Ralph J. Identification of lignin and polysaccharide modifications in populus wood by chemometric analysis of 2D NMR spectra from dissolved cell walls. J Mol Plant. 2009;2:933–942. doi: 10.1093/mp/ssp047. [DOI] [PubMed] [Google Scholar]
- 13.Hedenstrom M, Wiklund S, Sundberg B, Edlund U. Visualization and interpretation of OPLS models based on 2D NMR data. Chemometr Intell Lab. 2008;92:110–117. [Google Scholar]
- 14.Robinette SL, Ajredini R, Rasheed H, Zeinomar A, Schroeder FC, Dossey AT, Edison AS. Hierarchical alignment and full resolution pattern recognition of 2D NMR Spectra: Application to nematode chemical ecology. Anal Chem. 2011;83:1649–1657. doi: 10.1021/ac102724x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinette SL, Zhang F, Bruschweiler-Li L, Bruschweiler R. Web server based complex mixture analysis by NMR. Anal Chem. 2008;80:3606–3611. doi: 10.1021/ac702530t. [DOI] [PubMed] [Google Scholar]
- 16.Zhang F, Dossey AT, Zachariah C, Edison AS, Bruschweiler R. Strategy for automated analysis of dynamic metabolic mixtures by NMR. application to an insect venom. Anal Chem. 2007;79:7748–7752. doi: 10.1021/ac0711586. [DOI] [PubMed] [Google Scholar]
- 17.Zhang F, Bruschweiler R. Robust deconvolution of complex mixtures by covariance TOCSY spectroscopy. Angew Chem Int Ed. 2007;46:2639–2642. doi: 10.1002/anie.200604599. [DOI] [PubMed] [Google Scholar]
- 18.Zhang F, Bruschweiler-Li L, Robinette SL, Bruschweiler R. Self-consistent metabolic mixture analysis by heteronuclear NMR. Application to a human cancer cell line. Anal Chem. 2008;80:7549–7553. doi: 10.1021/ac801116u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wold S, Esbensen K, Geladi P. Principal component analysis. Chemometr Intell Lab. 1987;2:37–52. [Google Scholar]
- 20.Lucanic M, Held JM, Vantipalli MC, Klang IM, Graham JB, Gibson BW, Lithgow GJ, Gill MS. N-acylethanolamine signalling mediates the effect of diet on lifespan in Caenorhabditis elegans. Nature. 2010;473:226–229. doi: 10.1038/nature10007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noguez JH, Conner ES, Zhou Y, Ciche TA, Ragains JR, Butcher RA. A novel ascaroside controls the parasitic life cycle of the entomopathogenic nematode Heterorhabditis bacteriophora. ACS Chem Biol. 2012;7:961–966. doi: 10.1021/cb300056q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wanders RJA, Waterham HR. Biochemistry of mammalian peroxisomes revisited. Annu Rev Biochem. 2006;75:295–332. doi: 10.1146/annurev.biochem.74.082803.133329. [DOI] [PubMed] [Google Scholar]
- 23.Wanders RJA. Peroxisomes, lipid metabolism, and peroxisomal disorders. Mol Genet Metab. 2004;83:16–27. doi: 10.1016/j.ymgme.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 24.Wishart DS. Current progress in computational metabolomics. Brief Bioinform. 2007;8:279–293. doi: 10.1093/bib/bbm030. [DOI] [PubMed] [Google Scholar]
- 25.Nicholson JK, Connelly J, Lindon JC, Holmes E. Metabonomics: a platform for studying drug toxicity and gene function. Nature Rev Drug Discov. 2002;1:153–161. doi: 10.1038/nrd728. [DOI] [PubMed] [Google Scholar]
- 26.Gartland KP, Beddell CR, Lindon JC, Nicholson JK. Application of pattern recognition methods to the analysis and classification of toxicological data derived from proton nuclear magnetic resonance spectroscopy of urine. Mol Pharmacol. 1991;39:629–642. [PubMed] [Google Scholar]
- 27.Van QN, Issaq HJ, Jiang Q, Li Q, Muschik GM, Waybright TJ, Lou H, Dean M, Uitto J, Veenstra TD. Comparison of 1D and 2D NMR spectroscopy for metabolic profiling. J Proteome Res. 2008;7:630–639. doi: 10.1021/pr700594s. [DOI] [PubMed] [Google Scholar]
- 28.Chylla RA, Hu K, Ellinger JJ, Markley JL. Deconvolution of two-dimensional NMR spectra by fast maximum likelihood reconstruction: application to quantitative metabolomics. Anal Chem. 2011;83:4871–4880. doi: 10.1021/ac200536b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dieterle F, Ross A, Schlotterbeck G, Senn H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal Chem. 2006;78:4281–4290. doi: 10.1021/ac051632c. [DOI] [PubMed] [Google Scholar]
- 30.de Jong S. SIMPLS: An alternative approach to partial least squares regression. Chemometr Intell Lab. 1993;18:251–263. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.