Abstract
We demonstrate label-free detection of lipid vesicles and polystyrene beads freely diffusing in aqueous solution using surface enhanced Raman scattering (SERS). The signals observed enable real-time identification and monitoring of individual particles interacting with the SERS substrate. SERS is demonstrated as a label-free method capable of monitoring transient species in solution on the millisecond time scale.
Label-free detection and characterization of molecular aggregates is important for understanding the role of macromolecular assemblies in biomedical, environmental, and materials applications. Raman scattering is appealing due to its universality and the chemical specificity of the detected signal. Surface enhanced Raman scattering (SERS) is an ultrasensitive detection strategy with wide ranging applications.1–3 Increased understanding of the electromagnetic origins of SERS have resulted in a number of nanomaterials, SERS substrates4 and Raman tags,1, 5 that enable ultra-sensitive analysis. Raman tags, formed by attaching molecules with bright Raman signals to nanoparticles, have been shown to be competitive with fluorescence labels for trace detection.6 SERS substrates, arrays of nanostructures on a surface, have been shown capable of detecting subtle variation in the chemical composition of complicated samples, including virus genotypes.7
Enhanced Raman signals enable time resolved measurements of diffusion and reorientation of nanoparticles;8, 9 however, reports of dynamics from materials other than noble metal nanoparticles are lacking. The combination of high electric fields associated with an excited localized surface plasmon resonance (LSPR) and re-radiation of molecular scattering by nanoparticles combine to generate enhancements of up to ~1011.3, 10, 11 Enhancements approaching this limit are sufficient to generate detectable Raman scattering from single molecules.10, 12
While these enhancements are typically associated with nanometer gaps, here we present results demonstrating the enhancements present above a SERS substrate are capable of providing chemically specific signals for monitoring lipid vesicles and polymer nanoparticles freely diffusing on the millisecond time scale. Recent reports suggest that SERS enhancements can be quite substantial at the surface of nanostructures. Van Duyne and coworkers reported an average enhancement factor from nanoparticle clusters that was insensitive to the number of particles comprising the cluster,13 which appears contrary to the model where hotspots dominate.14 Moskovits and colleagues examined nanoparticles in solution and reported average enhancement factors ranging from 106 –108.9 These enhancement factors are comparable to those in single molecule imaging experiments obtained from nanoparticle aggregates.15 Significant enhancements outside of gap regions provide new routes to label-free, high-sensitivity detection.
In the present study, we used SERS substrates prepared by thermal evaporation of silver (Ag, Sigma-Aldrich, 99.999%) onto an anodized aluminium (AAO) filter (Whatman Anodisc 13) with 0.1µm pores as described previously.16 The substrates were incorporated into a commercial flow cell (Bioptechs FCS3) by affixing the metal film to a glass microaqueduct slide. The flow channel was defined by a 0.1mm thick gasket with a 1mm channel, cut to accommodate the SERS substrate in the chamber. The AAO template used in substrate fabrication was removed by flowing 0.05M NaOH through the chamber.
Raman measurements were performed using a previously described home-built spectrometer,16 consisting of a 632.8 nm He-Ne laser, an Olympus BX51 microscope, and an Andor i303 spectrograph and CCD camera. An Olympus water immersion objective (60×, 1.2 NA) was used for excitation and collection of the Raman signal. The flow cell was positioned on the microscope stage and the pump was stopped during spectral acquisitions.
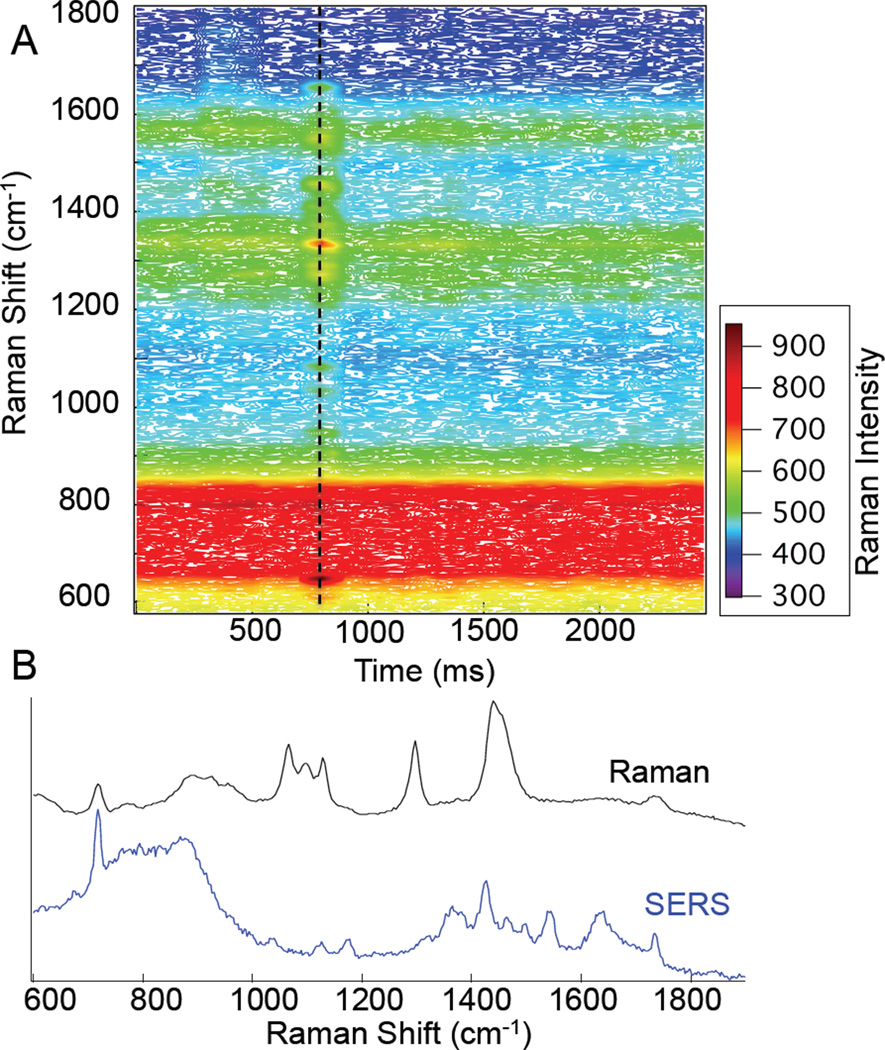
Shown in Figure 1 are the time resolved Raman spectra observed from a 1% (wt) solution of dipalmitoylphosphatidylcholine (DPPC) single shell vesicles. DPPC vesicles were prepared by vesicle extrusion using a 1µm polycarbonate filter (Whatman, Nuclepore). The spectra were acquired sequentially in 50ms intervals over several seconds. At the 750 ms time point, a transient (50 ms) spectrum is observed with Raman peaks attributable to the DPPC lipid headgroup. In particular, peaks are observed at 718, 1129, and 1733 cm−1. These peaks are assigned to the choline headgroup, C-C stretches, and carbonyl stretching, respectively.17
Figure 1.
Map of Raman intensity vs. time for 1µm DPPC vesicles. Spectra (b) show a comparison of a bulk DPPC Raman spectrum (black) and the 50 ms spectrum acquired at t= 750ms (blue). The reference spectrum has been scaled for clarity.
The SERS spectrum observed (Fig 1B) is different from the conventional Raman spectrum of DPPC. Clearly shown in the reference spectrum are the peaks associated with the choline symmetric stretch (718 cm−1), backbone vibrations (1066, 1100, and 1128 cm−1), C-H twist (1298 cm−1), C-H bend (1440 cm−1), and carbonyl stretch (1733 cm−1). The headgroup and interfacial residues dominate the SERS spectrum, while the conventional Raman spectrum primarily consists of vibrations associated with the lipid hydrocarbon acyl chains. These differences are expected, as the portion of the lipids that will interact with the SERS substrate are the hydrophilic residues on the periphery.18 Since lipid headgroups are commonly associated with biological signalling, the ability to distinguish liposomes on this basis is highly beneficial.
The duration of the Raman signal observed in Figure 1 is consistent with Stokes-Einstein diffusion:
| (Eq. 1) |
where D is the diffusion constant, kB is the Boltzmann constant, T is temperature, η is the solution viscosity, and r is the radius of the particle. For the DPPC vesicles used in this study, we calculate the diffusion constant and predict the time for the vesicle to traverse the diffraction limited focus. A particle with r = 0.5µm corresponds to 110ms, which is easily resolved using our 50ms acquisitions. The DPPC signal extends across 3 pixels (~150 ms) in good agreement with the calculation. We previously reported acquisition times of 10ms from this SERS substrate in air. In aqueous solution, the enhancement is sufficient to detect these transient events.
There is an expected difference in sensitivity of the SERS substrate in solution associated with plasmon coupling. We previously demonstrated that the LSPR appears to red shift in water.16 Our current results indicate that the LSPR is in fact very broad, as would be expected for highly coupled nanostructures.19 Indeed we have also observed significant SERS enhancement using 532 nm excitation with these substrates in air. The breadth of the plasmon is responsible for a low, constant, background that enhances modes uniformly across the Raman spectrum.
Shown in Figure 1, the SERS signal has sufficient sensitivity for single particle detection; however, the probability of a single particle interacting with the surface in the detection area is low. To overcome this, one can increase analyte concentration, which defeats high sensitivity applications, or one can increase the probability of analytes interacting in the detection area.
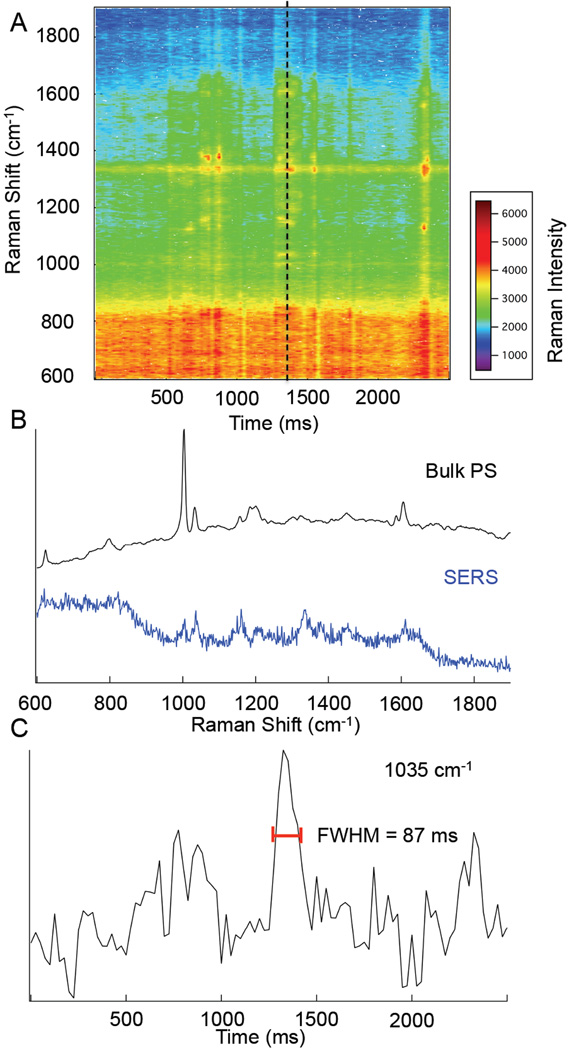
By minimizing the dimensions of the flow cell, we are able to increase the number of particles interacting with the surface as shown in Fig. 2. Solutions of 10% (wt) polystyrene (PS) beads (Sigma-Aldrich) were prepared in nanopure water (18.2 MΩ cm). In this experiment, a drop of PS microspheres was deposited onto the SERS substrate and sealed with a coverglass. This sandwich configuration is expected to have an approximate thickness of a couple micrometers, and promote interaction with the surface.
Figure 2.
Map of Raman intensity vs. time for 0.3 µm polystyrene beads diffusing in a few micrometer gap over the SERS substrate. Spectra (b) show a comparison of a bulk polystyrene Raman spectrum (black) and the 25 ms spectrum collected at t= 1325ms (blue). A profile (c) of the Raman signal at 1035 cm−1 is shown to show the duration of signal.
In Fig. 2C, the signal fluctuations suggest multiple interactions with the surface. The Stokes Einstein diffusion time for the 300 nm particles used is 33 ms. There is a prominent signal observed at t = 1300 ms with a Δt = 87 ms. This longer signal duration may be indicative of adsorption or hindered diffusion, but nonetheless demonstrates that the particles can be detected despite their size inhibiting access to gaps or tight crevices.
The Raman peaks observed agree with the Raman spectrum of bulk PS that shows the ring stretching modes (624, 1032, 1202, and 1605 cm−1), the ring breathing modes (799, and 1004 cm−1), CH in-plane bending modes (1157 and 1184 cm−1), methylene deformation (1450 cm−1), and the tangential ring stretch (1587 cm−1).20
Particles as small as 100nm polystyrene (PS) beads have been detected in solution.† A key advantage of using Raman to characterize particles is the spectra, such as those preceding the PS particle (Fig. S2) can distinguish between contaminants and analytes, or different types of particles in the case of heterogeneous mixtures.
From Eq. 1, the expected diffusion times are comparable to the spectral acquisitions. The detection of transient events provides an interesting comment on signal averaging for the detection of trace analytes. As shown in figures 1 and 2, much of the time we are not detecting the molecule of interest. By acquiring for longer periods we collect noise from periods without analyte that overwhelm the signal that persists for a short duration. We calculate a S/N ratio of 4.4 for 50 ms acquisitions of DPPC vesicles and and S/N of 3.0 for 25 ms acquisitions of PS beads, respectively. If we co-add the spectra over a period of 1s, we obtain S/N of 0.26 and 0.22 for the DPPC and PS samples. The detected PS signal thus suggests that faster acquisitions are advantageous for the detection of smaller particles.
From the measurements, it is clear that we can detect single particles, but it is not clear what is the minimum number of molecules necessary. From geometric consideration of the number of lipids in the outer shell interacting with the surface, a few thousand molecules can be readily detected. This agrees with a Langmuir adsorption analysis.† It is important to note the observed signal arises only from the SERS enhancement and does not include resonance enhancement, such as that used in single molecule detection of molecules like rhodamine dyes.
For assemblies, such as vesicles and polystyrene beads, the number density is actually quite high, supplying thousands of molecules to the surface simultaneously and facilitating data acquisitions on the millisecond time scale. The observed S/N ratio in the SERS signal suggests that this substrate can detect smaller aggregates relevant to heterogeneous chemical phenomena. In fact, the principal challenge does not appear to be sensitivity, but rather the detection frequency. Further experiments will elucidate the utility of this SERS flow detector to enable label-free chemical detection of macromolecular assemblies.
We have demonstrated the label-free detection of a variety of samples, single shell lipid vesicles and various size PS beads, diffusing over a SERS substrate in aqueous solution. The signal enhancements provide a chemically specific method of identifying and classifying sub-micron particles in solution with signal acquisitions on the millisecond time scale.
Supplementary Material
Acknowledgments
This research was supported by the National Institutes of Health Award R00 RR024367 and the University of Notre Dame.
Footnotes
This article is part of the ChemComm ‘Emerging Investigators 2013’ themed issue.
Electronic Supplementary Information (ESI) available: Experimental, peak fitting, and the Langmuir linear regression details are available. See DOI: 10.1039/b000000x/
Notes and references
- 1.Graham D, Stevenson R, Thompson DG, Barrett L, Dalton C, Faulds K. Faraday Discuss. 2011;149:291–299. doi: 10.1039/c005397j. [DOI] [PubMed] [Google Scholar]
- 2.Larmour IA, Faulds K, Graham D. Chemical Science. 2010:151–160. [Google Scholar]
- 3.Stiles PL, Dieringer JA, Shah NC, Van Duyne RR. Annu Rev Anal Chem. 2008;1:601–626. doi: 10.1146/annurev.anchem.1.031207.112814. [DOI] [PubMed] [Google Scholar]
- 4.Camden JP, Dieringer JA, Zhao J, Van Duyne RP. Accounts Chem. Res. 2008;41:1653–1661. doi: 10.1021/ar800041s. [DOI] [PubMed] [Google Scholar]
- 5.Cao YWC, Jin RC, Mirkin CA. Science. 2002;297:1536–1540. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]
- 6.McGuinness CD, Macmillan AM, Karolin J, Smith WE, Graham D, Pickup JC, Birch DJS. Analyst. 2007;132:633–634. doi: 10.1039/b706409h. [DOI] [PubMed] [Google Scholar]
- 7.Hoang V, Tripp RA, Rota P, Dluhy RA. Analyst. 2010;135:3103–3109. doi: 10.1039/c0an00453g. [DOI] [PubMed] [Google Scholar]
- 8.Laurence TA, Braun G, Talley C, Schwartzberg A, Moskovits M, Reich N, Huser T. J. Am. Chem. Soc. 2009;131:162–169. doi: 10.1021/ja806236k. [DOI] [PubMed] [Google Scholar]
- 9.Laurence TA, Braun GB, Reich NO, Moskovits M. Nano Lett. 2012;12:2912–2917. doi: 10.1021/nl3005447. [DOI] [PubMed] [Google Scholar]
- 10.Kneipp J, Kneipp H, Kneipp K. Chemical Society Reviews. 2008;37:1052–1060. doi: 10.1039/b708459p. [DOI] [PubMed] [Google Scholar]
- 11.Moskovits M. Journal of Raman Spectroscopy. 2005;36:485–496. [Google Scholar]
- 12.Dieringer JA, Wustholz KL, Masiello DJ, Camden JP, Kleinman SL, Schatz GC, Van Duyne RP. J. Am. Chem. Soc. 2008;131:849–854. doi: 10.1021/ja8080154. [DOI] [PubMed] [Google Scholar]
- 13.Wustholz KL, Henry A-I, McMahon JM, Freeman RG, Valley N, Piotti ME, Natan MJ, Schatz GC, Duyne RPV. J. Am. Chem. Soc. 2010;132:10903–10910. doi: 10.1021/ja104174m. [DOI] [PubMed] [Google Scholar]
- 14.Fang Y, Seong N-H, Dlott DD. Science. 2008;321:388–392. doi: 10.1126/science.1159499. [DOI] [PubMed] [Google Scholar]
- 15.Willets KA, Stranahan SM, Weber ML. J. Phys. Chem. Lett. 2012;3:1286–1294. doi: 10.1021/jz300110x. [DOI] [PubMed] [Google Scholar]
- 16.Asiala SM, Schultz ZD. Analyst. 2011;136:4472–4479. doi: 10.1039/c1an15432j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schultz ZD, Levin IW. Annu Rev Anal Chem. 2011;4:343–366. doi: 10.1146/annurev-anchem-061010-114048. [DOI] [PubMed] [Google Scholar]
- 18.Pavel I, McCarney E, Elkhaled A, Morrill A, Plaxco K, Moskovits M. The Journal of Physical Chemistry C. 2008;112:4880–4883. doi: 10.1021/jp710261y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halas NJ, Lal S, Chang WS, Link S, Nordlander P. Chem. Rev. 2011;111:3913–3961. doi: 10.1021/cr200061k. [DOI] [PubMed] [Google Scholar]
- 20.Kellar EJC, Galiotis C, Andrews EH. Macromolecules. 1996;29:3515–3520. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.