Abstract
GRL-0519 (1) is a potent antiviral inhibitor of HIV-1 protease (PR) possessing tris-tetrahydrofuran (tris-THF) at P2. The high resolution X-ray crystal structures of inhibitor 1 in complexes with single substitution mutants PRR8Q, PRD30N, PRI50V, PRI54M, and PRV82A were analyzed in relation to kinetic data. The smaller valine side chain in PRI50V eliminated hydrophobic interactions with inhibitor and the other subunit consistent with 60-fold worse inhibition. Asn30 in PRD30N showed altered interactions with neighboring residues and 18-fold worse inhibition. Mutations V82A and I54M showed compensating structural changes consistent with 6-7-fold lower inhibition. Gln8 in PRR8Q replaced the ionic interactions of wild type Arg8 with hydrogen bond interactions without changing the inhibition significantly. The carbonyl oxygen of Gly48 showed two alternative conformations in all structures likely due to the snug fit of the large tris-THF group in the S2 subsite in agreement with high antiviral efficacy of 1 on resistant virus.
Keywords: HIV / AIDS, aspartic protease, X-ray crystallography, drug resistance
INTRODUCTION
HIV/AIDS is a life-threatening disease that interferes with the immune system, and approximately 1.8 million people died of AIDS-related illnesses in 20101. Although there is no effective vaccine available2, highly active antiretroviral therapy (HAART), which employs a combination of different antiretroviral drugs, improves the lives of AIDS patients3. HIV-1 protease (PR) is critical for viral particle maturation since it cleaves the viral precursor polypeptides Gag and Gag-Pol into the mature structural and enzymatic proteins4, 5. Thus PR is an effective target for antiviral drugs, however, the most severe challenge to the long-term efficacy of protease inhibitors (PIs) in HAART is the emergence of drug resistant mutants of PR6.
HIV-1 PR is catalytically active as a homodimer. Structural regions critical for PR activity and stability are the dimer interface including the catalytic Asp25 from each subunit and the flexible flaps comprising residues 45 to 557,8. To date, there are nine approved clinical PIs. The first clinical inhibitors, such as saquinavir (SQV), were designed to bind tightly in the active site cavity of the wild type enzyme; however, their binding affinity can be readily lowered by mutations. Analysis of the structural and biochemical properties of PR mutants suggests that resistant mutations act by multiple mechanisms, including mutations in the binding site that directly lower inhibitor affinity, mutations at the dimer interface that destabilize the catalytically active dimer, and flap mutations that alter the conformational flexibility7. Drug resistant PR mutants exhibit decreased binding affinity for inhibitors while maintaining the critical PR function in viral replication9. Two clinical PIs, darunavir (DRV) and amprenavir (APV), contain tetrahydrofuran (THF) in the P2 group; APV has a single THF while DRV incorporates bis-THF. The bis-THF of DRV introduces more hydrogen bonds with PR main chain atoms, and DRV has demonstrated high potency and clinical efficacy on resistant viral infections10, 11. Recently, a third THF ring was added to enlarge P2 and fit better in the S2 binding pocket of PR, leading to the novel PI called GRL-0519 (1) (Figure 1A). The incorporation of the third ring endows inhibitor 1 with excellent antiviral activity on drug resistant virus12.
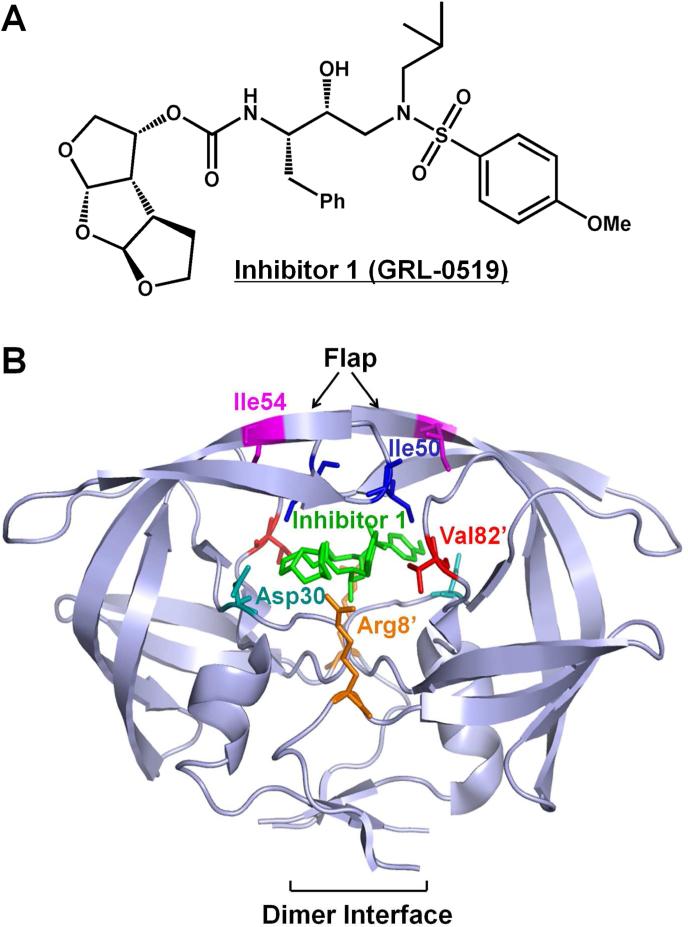
Figure 1.
(A) The chemical structures of protease inhibitor compound 1. (B) The structure of HIV-1 PRWT/inhibitor 1. The HIV-1 protease dimer is shown in light blue cartoon representation. The inhibitor 1 and wild-type residues at the mutation sites are indicated by differently colored sticks. The same residues on the two subunits are shown in the same color with only one of them labeled.
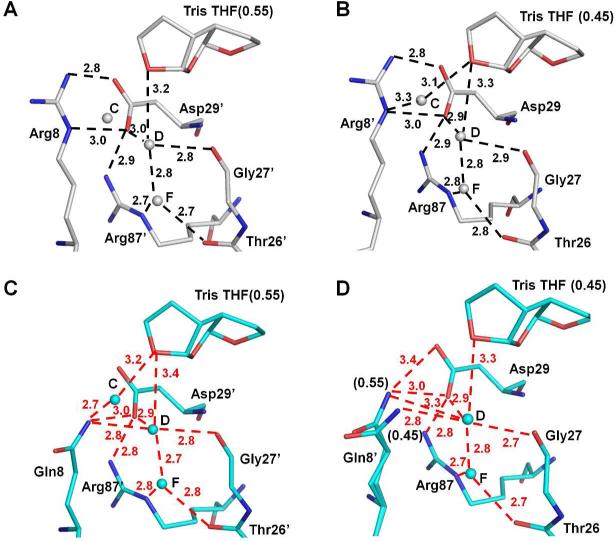
The crystal structure of inhibitor 1 complexed with wild type PR (PRWT) was reported previously12. The third THF ring showed new water molecule-mediated hydrogen bonds with conserved PR residues Gly27, Asp29 and Arg8’. In order to study the molecular basis for the potency of inhibitor 1 against drug resistant viral strains, crystal structures of inhibitor 1 complexes with PR mutants bearing single substitutions of R8Q, D30N, I50V, I54M and V82A (PRR8Q, PRD30N, PRI50V, PRI54M and PRV82A) were analyzed. The location of these mutations in the PR dimer is indicated in Figure 1B. These mutations, with the exception of R8Q, are common in drug resistant clinical isolates13. R8Q was one of the first resistant mutants identified in the laboratory for an investigational inhibitor14. In the wild type enzyme, Arg8 forms an ionic interaction with Asp29’ in the other subunit as an important component of the dimer interface15,16. This intersubunit ionic interaction was eliminated in the mutant with the single substitution of R8Q17. Moreover, in the PRWT-inhibitor 1 complex, the side chain of Arg8 forms a water molecule-mediated hydrogen bond with the third THF of inhibitor 112. Therefore, it is of particular interest to test how the R8Q mutation affects the binding of inhibitor 1. D30N is a major mutation that is associated with resistance to nelfinavir (NFV)18. Asp30 forms hydrogen bond interactions with the bis-THF of DRV, thus mutation of this residue may alter the inhibitor binding. Mutations of the flap residues such as Ile50 and Ile54 can alter the conformational dynamics of this region, thereby affecting the binding affinity for inhibitors19-22. Ile50 is located at the tip of the flap where its side chain forms hydrophobic interactions with inhibitors. Mutation of I50V to a shorter side chain is expected to reduce the binding affinity for inhibitors. Indeed, PR with I50V mutation exhibits significantly reduced inhibition by indinavir, SQV, and DRV21-23. I50V also has a significant effect in destabilizing the PR dimer21. Mutations of Val82 are found frequently in resistant virus13. The mutation V82A in the active site cavity can eliminate interactions with inhibitor, and also exhibits a shift of its main chain atoms to adapt to inhibitor24-26. Here, the inhibitor 1 complexes with PR mutants PRR8Q, PRD30N, PRI50V, PRI54M and PRV82A are analyzed in relation to the PRWT-inhibitor 1 complex and the inhibition values.
RESULTS
Compound 1 inhibition of PRWT and selected mutants
The kinetic parameters and inhibition constants (Ki) of compound 1 for PRWT and selected mutants PRR8Q, PRD30N, PRI50V, PRI54M and PRV82A are shown in Table 1. The catalytic efficiency (kcat/Km) of PRV82A showed 2.7-fold increase, PRI54M was essentially identical and PRR8Q had a slight decrease relative to PRWT. The lowest catalytic efficiency of 10% of PRWT was observed for PRD30N and PRI50V. The relative catalytic efficiency of PRD30N measured here is consistent with previous reports using a different substrate23. The relative activities of PRI50V and PRV82A are slightly different from those reported earlier; however, the trends are identical, with decreased kcat/Km for PRI50V and increased kcat/Km for PRV82A. Compound 1 showed a range of inhibition of up to 60-fold for the various mutants relative to the wild type enzyme. The inhibition constant of PRR8Q for compound 1 was not significantly different from the value of 0.5 nM for PRWT. The PRI54M and PRV82A showed 7- and 6-fold increased Ki for compound 1 relative to PRWT. Again, the mutants PRD30N and PRI50V showed the most significant changes with 18- and 60-fold worse inhibition, respectively.
Table 1.
Kinetic parameters for substrate hydrolysis and inhibition of compound 1
| Protease | Km (mM) | kcat (min-1) | kcat/Km (min-1·μM-1) | Relative kcat/Km | Ki (nM) | Relative Ki |
|---|---|---|---|---|---|---|
| PRWTa | 30±5 | 190±20 | 6.5±1.30 | 1.0 | 0.50±0.06 | 1 |
| PRR8Q | 38±2 | 161±4 | 4.2±0.25 | 0.6 | 1.20±0.08 | 2 |
| PRD30N | 76±8 | 48±2 | 0.6±0.72 | 0.1 | 8.9±0.4 | 18 |
| PRI50Va | 109±8 | 68±5 | 0.6±0.03 | 0.1 | 30.9±0.6 | 60 |
| PRI54Ma | 41±5 | 300±40 | 7.3±0.80 | 1.1 | 3.5±0.2 | 7 |
| PRV82Ab | 29±3 | 512±26 | 17.7±2.00 | 2.7 | 3.2±0.2 | 6 |
Crystallographic analysis of inhibitor 1 complexes
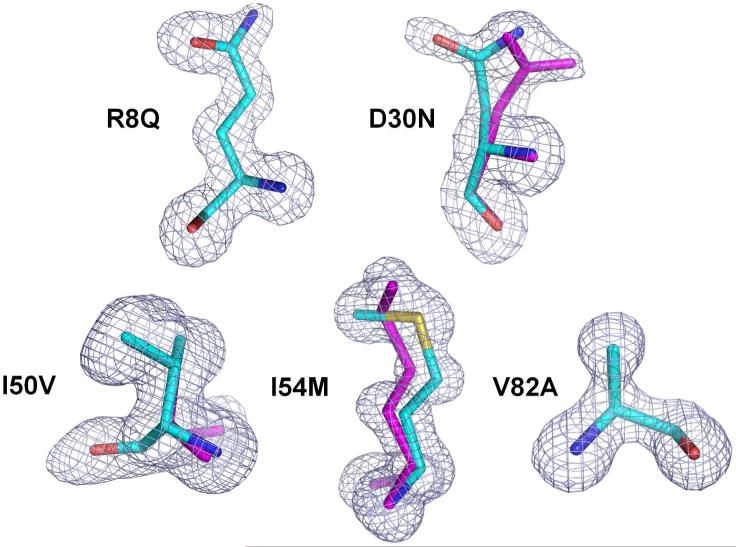
Crystal structures of PR mutants PRR8Q, PRD30N, PRI50V, PRI54M and PRV82A were obtained in complex with inhibitor 1 and the structures were refined at resolutions of 1.06-1.49 Å. The crystallographic data collection and refinement statistics are summarized in Table 2. All five complexes were determined in space group P21212 and refined with anisotropic B-factors, solvent molecules, and hydrogen atoms to the R-factors of 13.8-16.3. The asymmetric unit contains one PR dimer of residues labeled 1–99 and 1’–99’ for each monomer. Well defined electron density was observed for the PR residues, as illustrated for the mutated residues in Figure 2. The mutated residues, except for Ala82, show alternative conformations. The side chain of mutated residue Asn30/Asn30’ in both subunits of PRD30N-inhibitor 1 structure has two alternative conformations with relative occupancies of 0.55/0.45. Alternative conformations of the side chains of mutated Met54 and Gln8 with relative occupancies of 0.58/0.42 and 0.55/0.45, respectively, were observed in one subunit in the PRI54M and PRR8Q-complexes. Mutated residue Val50 showed alternative conformations for both main chain and side chain atoms in both subunits of PRI50V-inhibitor 1 with relative occupancies of 0.7/0.3. Inhibitor 1 has two alternative conformations related by 180° in all five complexes with relative occupancies listed in Table 2.
Table 2.
Crystallographic Statistics for PR Mutants in Complex with Inhibitor 1.
| Inhibitor 1 complex | PRR8Q | PRD30N | PRI50V | PRI54M | PRV82A |
|---|---|---|---|---|---|
| Space group | P21212 | P21212 | P21212 | P21212 | P21212 |
| Unit cell dimensions: (Å) | |||||
| a | 58.37 | 58.72 | 58.59 | 58.78 | 58.61 |
| b | 85.96 | 86.24 | 85.95 | 85.84 | 86.15 |
| c | 46.18 | 45.90 | 46.07 | 46.02 | 46.33 |
| Resolution range (Å) | 50.0-1.46 | 50.0-1.49 | 50.0-1.22 | 50.0-1.06 | 50.0-1.29 |
| Unique reflections | 40,949 | 37,721 | 64,041 | 103,379 | 59, 015 |
| Rmerge (%) overall (final shell) | 5.3 (49.7) | 6.2 (42.1) | 5.3 (47.4) | 6.3 (56.8) | 5.6 (40.7) |
| I/σ(I) overall (final shell) | 30.8 (2.6) | 25.5 (2.1) | 31 (2.1) | 26.7 (2.1) | 18.9 (2.2) |
| Completeness (%) overall (final shell) | 99.2 (94.8) | 97.0 (76.3) | 91.5(65.4) | 97.4 (85.6) | 99.0 (92.7) |
| Data range for refinement (Å) | 10-1.46 | 10-1.49 | 10-1.22 | 10-1.06 | 10-1.29 |
| R (%) | 15.8 | 15.6 | 16.5 | 14.9 | 16.3 |
| Rfree (%) | 21.0 | 21.4 | 20.3 | 17.8 | 20.9 |
| No. of solvent atoms (total occupancies) | 175(170.5) | 189 (144.2) | 251(237.6) | 248(234.5) | 182(179.0) |
| RMS deviation from ideality | |||||
| Bonds(Å) | 0.010 | 0.010 | 0.014 | 0.016 | 0.012 |
| Angle distance (Å) | 0.029 | 0.030 | 0.034 | 0.038 | 0.031 |
| Average B-factors (Å2) | |||||
| Main-chain atoms | 15.0 | 17.5 | 12.9 | 12.6 | 13.0 |
| Side-chain atoms | 21.1 | 24.1 | 19.6 | 19.1 | 19.8 |
| Solvent | 31.6 | 33.7 | 30.8 | 28.8 | 30.5 |
| Inhibitor | 22.6 | 25.3 | 19.5 | 17.1 | 19.3 |
| Relative Occupancy of 1 | 0.55/0.45 | 0.55/0.45 | 0.7/0.3 | 0.53/0.47 | 0.6/0.4 |
Figure 2.
Fo-Fc omit maps (contoured at 3.0 σ) for the mutated residues. The magenta sticks indicate the alternative conformations of Asn30, Met54 and Val50.
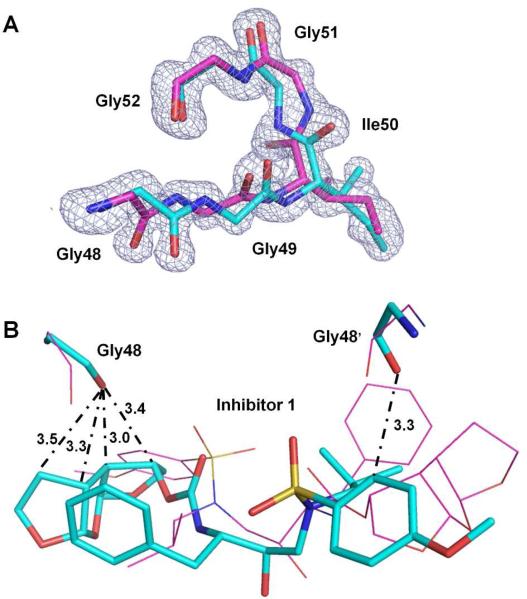
The residues 48-51/52 at the tip of the flaps show alternative conformations in all the complexes, which was associated with approximately 180° rotation of the peptide bond between Ile50 and Gly51 and alternative conformations of the carbonyl group of Gly48 in both subunits. The electron density map of residues Gly48-Gly52 from the atomic resolution (1.06 Å) structure of PRI54M-inhibitor 1 illustrates clearly the alternative conformations (Figure 3A). Corresponding regions of the other mutant structures are shown in Figure S1. Similar disorder in the flaps was observed in the wild type complex with inhibitor 1. However, alternative conformations are unusual for Gly48 in complexes with other inhibitors. Here, in each conformation of the flap residues, the carbonyl oxygen of Gly48 forms several C-H...O interactions with the tris-THF P2 group and Gly48’ forms a C-H...O interaction with the P2’ aniline in the corresponding conformation of inhibitor 1 (Figure 3B). The two conformations of Gly48 may arise from steric hindrance due to the tight fit of the tris-THF group in the S2 subsite.
Figure 3.
(A) Fo-Fc omit map (contoured at 3.0 σ) for the flap residues Gly48 - Gly52 in complex PRI54M–inhibitor 1 (1.06Å). Carbon atoms are colored cyan and magenta for the major and minor conformations, respectively. (B) Interactions of Gly48/48’ with inhibitor 1 in complex PRI50V–inhibitor 1 (0.7/0.3 occupancy). The major and minor conformations of Gly48/48’ and the inhibitor 1 are represented by sticks with carbon atoms colored cyan and lines with carbon atoms colored magenta, respectively. C-H...O interactions between the major conformations are indicated by dash-dot lines with distances in Å. C-H...O interactions between the minor conformations are the same but not shown here for clarity.
The mutant dimers superimpose on the PRWT-inhibitor 1 complex (PDB ID: 3OK9) with pairwise root mean square deviations (rmsd) of 0.13-0.24 Å on 198 Cα atoms. Both alternative inhibitor conformations are considered, except for the minor conformation of inhibitor in PRI50V-inhibitor 1, which has the low relative occupancy of 0.3. Hydrogen bond interactions of inhibitor 1 with all five mutants are similar except for variation in interactions of residue 30 (Table S1). The side chain of Asn30/30’ in PRD30N forms a hydrogen bond interaction with the first THF oxygen of inhibitor 1 as observed for the major conformation of inhibitor 1 in the PRWT complex; however, this interaction was not observed for Asp30/Asp30’ in the other four mutants. Interatomic distances for hydrogen bond interactions (O-H...O, N-H...O, etc.) are considered in the range of 2.4-3.4 Å, C-H...O interactions at 3.0-3.7 Å separation, C-H...π interactions for distances of <4.0 Å, and van der Waals contacts when distances are 3.8-4.2 Å, as described previously27. Structural changes are described for each mutant separately or in related pairs in the following sections.
Influence of D30N on PR structure and its interaction with inhibitor 1
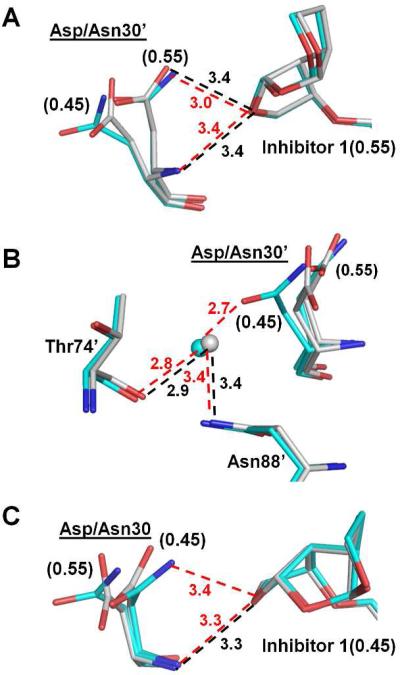
Residue 30 is located at one end of the active site cavity and interacts with the inhibitor, although the side chain frequently has alternate conformations. In PRWT–inhibitor 1, the side chain and main chain amide of the major conformation of Asp30’ form hydrogen bond interactions with the first THF ring of inhibitor 1 (Figure 4A). The main chain of residue 30’ lies in essentially the same position in PRD30N–inhibitor 1 and PRWT–inhibitor 1, thus the hydrogen bond interaction of the main chain amide of Asp30’ with O26 of inhibitor 1 is preserved in the mutant (Figure 4A). In addition, the major conformation of the side chain (occupancy of 0.55) of residue 30’ was similar in both structures and retained the hydrogen bond interaction with inhibitor. However, the minor conformation (occupancy of 0.45) of the side chain differs in the mutant and the wild type enzyme, especially the position of Oδ1 atom, due to the Cγ movement of approximately 0.9 Å as well as rotation of the side chain (approximately 90°) of Asn30’ compared to Asp30’ of PRWT. As a result, the side chain of Asn30’ in the mutant forms a hydrogen bond with a water molecule, which further interacts with the side chain of Asn88’ as well as the main chain of Thr74’ (Figure 4B). This water molecule also forms hydrogen bond interactions with the main chain and side chain of Thr31’ in both wild type and mutant (not shown in Figure 4B). In the other subunit (1-99), the Asn30 side chain in PRD30N forms a hydrogen bond with the minor conformation of inhibitor 1 , which was not observed in PRWT (Figure 4C). The interactions of Asn30 with neighboring residues are similar to those shown in Figure 4C for Asn30’. It seems that the observed 18-fold increase in the inhibition constant of the inhibitor 1 for PRD30N does not arise from changes in the direct interactions between residue 30 and the inhibitor. Instead, the elimination of the negative charge of Asp30 and alterations within an internal network of interactions may provide indirect effects on inhibition. Similar changes in interactions of residues 30 and 88 have been reported in mutants containing D30N and N88D and also proposed to effect inhibition28, 29.
Figure 4.
Structural changes of PRD30N. Carbon atoms are colored gray for PRWT–inhibitor 1 and cyan for PRD30N–inhibitor 1. Hydrogen bond interactions in PRWT–inhibitor 1 and PRD30N–inhibitor 1 are represented by black and red dashed lines, respectively, with distances in Å. Water molecules are shown as gray and cyan spheres for PRWT–inhibitor 1 and PRD30N–inhibitor 1, respectively. Occupancies for alternative conformations are labeled in parentheses for Asp/Asn30’. (A) Interactions of Asp/Asn30’ with the major conformation of inhibitor 1. (B) Interactions of Asp/Asn30’ with the neighboring residues. (C) Interactions of Asp/Asn30 with the minor conformation of inhibitor 1.
Mutations I54M and V82A produce compensating shifts in the 80s loop residues
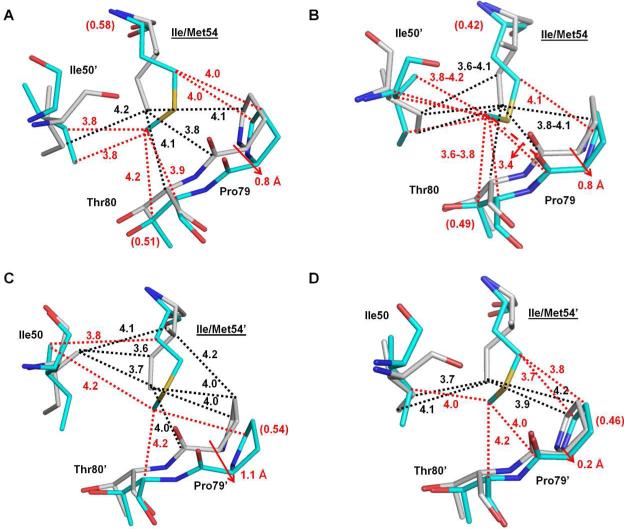
Ile54, which is located in the flap region, does not contact inhibitors directly, but it forms many hydrophobic interactions with surrounding residues, including residues in the 80s loop (residues 78-82) and Ile50’. In the subunit comprising residues 1-99, Met54 had two alternative conformations as shown in Figure 2. Compared with Ile54 in PRWT, both the major (Figure 5A) and minor (Figure 5B) conformations of Met54 in PRI54M formed one more inter-subunit van der Waals contact with the major and minor conformations of Ile50’. The 80s loop in PRI54M has shifted to accommodate the larger side chain of Met54 compared with Ile54 in PRWT. Pro79 has shifted away from residue 54 by about 0.8Å at the Cα atom to maintain their van der Waals interactions. The major conformation of Met54 (relative occupancy of 0.58) formed the same number of van der Waals contacts with Pro79 (Figure 5A). In addition, it formed one new van der Waals interaction with the major conformation of Thr80. The minor conformation of Met54 (relative occupancy of 0.42) formed the same number of van der Waals contacts with the proline ring (Figure 5B); however, it gained a new C-H...O interaction with the main-chain carbonyl oxygen atom of Pro79 and one more van der Waals interaction with the minor conformation of Thr80 compared with Ile54 in PRWT. In the other subunit 1’-99’, Met54’ had a single conformation and formed one less inter-subunit van der Waals contact with both the major (Figure 5C) and minor (Figure 5D) conformations of Ile50 compared with Ile54’ in PRWT. Unlike subunit 1-99, the 80's loop residue Pro79’ in subunit 1’-99’ exhibited two alternative conformations. A larger shift of approximately 1.1Å was observed for the major conformation of Pro79’ whereas the minor conformation showed an insignificant change of 0.2 Å. Although Met54’ had less van der Waals interactions with the major conformation of Pro79’, it gained a new one with Thr80’ (Figure 5C). The interactions of residue 54’ with the minor conformations of proline were identical for PRWT and the mutant (Figure 5D). However, it gained new interactions with the C atoms of Pro79’ and Thr80’. Taken together, there are either two conformations of Met54 or the 80s loop residues to accommodate the large side chain of the mutated Met54. Ile50 exhibits two alternative conformations in all structures as mentioned previously. Met54 in PRI54M has a similar number of intersubunit van der Waals contacts with Ile50’ as does Ile54 in PRWT (Figures 5A-D). The separation of Pro79 and residue 54 is increased in the mutant so that their van der Waals interaction is retained. The new interactions between Met54 and Thr80 may help maintain the conformation of the rest of the 80s loop residues despite the shift of Pro79/79’. As shown in Figure 5E, the hydrophobic interactions of Val82 and 82’ with the P1 and P1’ groups of inhibitor 1 are retained in PRWT and PRI54M. Therefore, the similar catalytic efficiency of PRI54M and PRWT may reflect the similar internal contacts of residue 54. Moreover, the hydrogen bond between the major conformation of inhibitor 1 and Asp30 in PRWT was not observed in PRI54M, which may contribute to the 7-fold lower inhibition for PRI54M.
Figure 5.
Structural changes of PRI54M and PRV82A. Carbon atoms are colored gray for PRWT–inhibitor 1 and cyan for PRI54M–inhibitor 1 or PRV82A–inhibitor 1. Interactions are indicated by black lines for PRWT–inhibitor 1 and red lines for the mutants. Van der Waals and C-H...π interactions are indicated by dotted lines. (A-B) Interactions of the major (A) and minor (B) conformations of Ile/Met54 with Ile50’ and 80s loop residues in subunit 1-99. (C-D) Interactions of Ile/Met54’ with Ile50 and the major (C) and minor (D) conformation of 80's loop residues in subunit 1’-99’. Relative occupancies for alternative conformations of residues are shown in parentheses. Red arrows indicate the shifts of the mutants relative to PRWT. (E) Interactions of Val82/82’ with inhibitor 1 in PRWT and PRI54M. (F) Interactions of Val/Ala82/82’ with inhibitor 1 in PRWT and PRV82A.
Val82/82’ has extensive C-H...π interactions with the P1 phenyl group and a van der Waals contact with the P1’ group of inhibitor 1 in PRWT (Figure 5F). Mutation V82A introduces the smaller Ala side chain, leading to the loss of C-H...π interactions with the P1 phenyl group of inhibitor 1 compared with those in the wild type complex. However, the main chain of Ala82’ has shifted toward the inhibitor by around 0.5 Å compared to the wild type complex, which preserves the hydrophobic interaction with the P1’ of inhibitor 1 (Figure 5F). The interactions of Val82/82’ with the major and minor conformations of the inhibitor were identical, thus only those with the major conformation of the inhibitor 1 are shown. Similar to PRI54M, the hydrogen bond between the major conformation of inhibitor 1 and Asp30 in PRWT was also not observed in PRV82A. The absence of a hydrogen bond plus fewer interactions of Ala82 with the P1 phenyl of inhibitor 1 may be responsible for the decreased inhibitory activity of inhibitor 1 for PRV82A. However, the compensation mechanisms observed in PRI54M and PRV82A may explain why there was only 6-7 fold decrease of the inhibition constants for these two mutants relative to PRWT.
Mutations I50V and R8Q alter the dimer interface and interactions with inhibitor 1
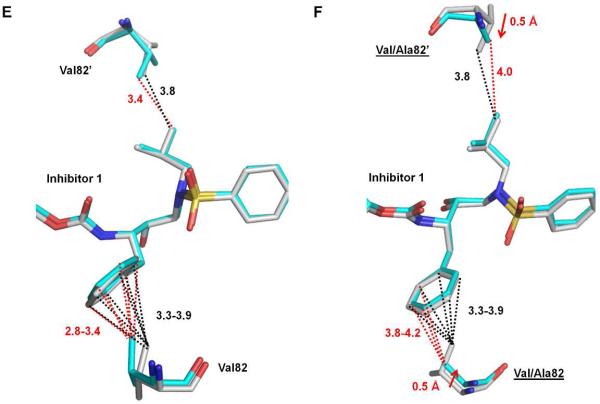
The side chain of Ile50’ in PRWT had extensive interactions with inhibitor 1, including C-H...O and C-H...π interactions (Figure 6A). However, the interactions with the P2’ group of inhibitor were eliminated for the small Val50’ side chain in the mutant PRI50V and only two C-H...O interactions with the sulfonamide oxygen were maintained. Similarly, the two van der Waals interactions between Ile50 and the P2 tris-THF group of inhibitor 1 were lost in the other subunit of PRI50V (Figure 6B). Similar to PRI54M and PRV82A, the hydrogen bond between the major conformation of inhibitor 1 and Asp30 in PRWT was also absent in PRI50V. Taken together, the considerably reduced interactions of mutant PRI50V with inhibitor 1 are consistent with the significantly decreased inhibition (60-fold) for PRI50V compared to the PRWT.
Figure 6.
Structural changes of PRI50V. (A) Interactions of Ile/Val50’ with inhibitor 1. (B) Interactions of Ile/Val50 with inhibitor 1. (C) Intersubunit interactions of Ile/Val50’ with surrounding residues. Carbon atoms are colored gray for PRWT–inhibitor 1 and cyan for PRI50V–inhibitor 1. Interactions in PRWT–inhibitor 1 and PRI50V–inhibitor 1 are represented by black lines and red lines, respectively. C-H...O and van der Waals/C-H...π interactions are indicated by dash-dot lines and dotted lines, respectively, with distances in Å.
The mutation of Ile50 to Val50 also affects the inter-subunit interactions. In PRWT, the Ile50’ side chain had hydrophobic interactions with the side chains of Ile84 and Ile47, and the same inter-subunit interactions were also observed for Ile50. However, the interactions of Val50’ with both Ile84 and Ile47 are eliminated in the PRI50V mutant as illustrated in Figure 6C. The loss of the interaction between Val50 and Ile47’ was also observed in the other subunit. These findings are consistent with our previous analysis of PRI50V structures21, and the overall decreased interaction between subunits may account for the 10 fold decrease of its catalytic efficiency as well as likely contributing to the significantly worse inhibition relative to PRWT.
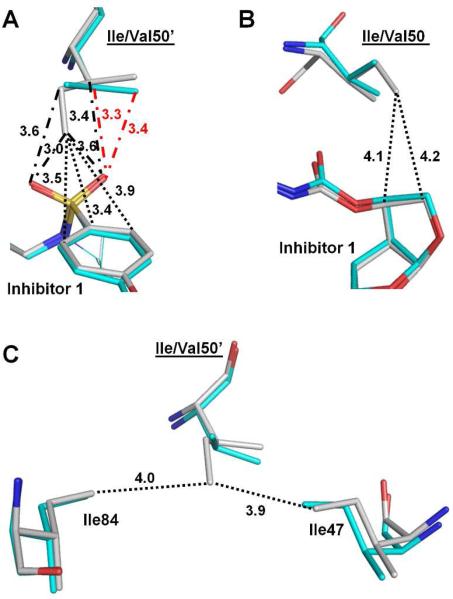
In contrast to mutant PRI50V, mutation of R8Q has altered the type of interaction rather than eliminated interactions. In the wild type PR, Arg8’ forms an ionic interaction with Asp29 in both subunits (Figure 7A & B). However, this inter-subunit ionic interaction is eliminated in PRR8Q (Figure 7C & D). In one subunit of PRR8Q, a direct hydrogen bond is formed between the side chains of Gln8 and Asp29’ (Figure 7C). The side chain of Gln8 in PRR8Q has shifted away from Arg8 in PRWT by approximately 0.9 Å, thus gaining new hydrogen bond interactions with conserved water molecules C and D and a water molecule-mediated interaction with the tris-THF of inhibitor. In the other subunit, Gln8’ has two alternative conformations with relative occupancies of 0.55/0.45 (Figure 7D). The side chain of Gln8’ in the major conformation forms two direct and one water molecule-mediated hydrogen bond with Asp29, whereas that of the minor conformation forms one water molecule-mediated hydrogen bond with Asp29. Overall, direct as well as water molecule-mediated inter-subunit hydrogen bonds between Gln8 and Asp29’ were observed in both subunits, suggesting that these new interactions might compensate for the loss of the inter-subunit ionic interactions in the wild type enzyme (and many other mutants).
Figure 7.
Structural changes of PRR8Q. (A-B) Interactions of Arg8/8’ in PRWT–inhibitor 1. (C-D) Interactions of Gln8/8’ in PRR8Q–inhibitor 1. Carbon atoms are colored gray for PRWT–inhibitor 1 and cyan for PRR8Q–inhibitor 1. Water molecules are shown as gray and cyan spheres for PRWT–inhibitor 1 and PRR8Q–inhibitor 1, respectively. The hydrogen bond interactions are indicated by the dashed lines with distances in Å. Occupancies for alternative conformations of inhibitor 1 and Gln8’ are indicated in parentheses. Hydrogen bond interactions in RR8Q–inhibitor 1 are colored red. Interactions between water molecules C and D in all figures (7A-7D) are omitted for clarity.
The conserved water molecule D also has a hydrogen bond interaction with the oxygen of the third THF ring of inhibitor 1 in both PRWT and PRR8Q (Figure 7). This water molecule mediates a hydrogen bond interaction between the inhibitor 1 and Gln8/8’, which is not seen for Arg8/8’ in PRWT (Figure 7C & D). Moreover, conserved water molecule C mediates an additional hydrogen bond between the major conformation of inhibitor 1 and the side chain of Gln8, which resembles the interaction between the minor conformation of inhibitor 1 and Arg8’ in PRWT (Figure 7B).
In summary, although PRR8Q has lost the strong inter-subunit ionic interaction between Arg8 and Asp29’, compensation is provided by new direct and water molecule-mediated hydrogen bonds. Meanwhile, even though the hydrogen bond between the major conformation of inhibitor 1 and Asp30 in PRWT was not seen in PRR8Q, the water molecule-mediated hydrogen bond interaction between inhibitor 1 and Gln8/8’ was enhanced to some extent. Therefore, there is little absolute change in the interaction of PRR8Q with inhibitor 1. In contrast, PRI50V showed significant loss of interactions with inhibitor 1. These structural changes explain why inhibitor 1 has 60-fold worse inhibition for PRI50V while its inhibition for PRR8Q was not affected significantly.
DISCUSSION AND CONCLUSIONS
Inhibitor 1 is a novel inhibitor incorporating a unique tris-THF group at P2 to target drug resistant HIV-1 PR mutants. Previously inhibitor 1 was reported to possess potent antiviral activity against wild type HIV-1 and multidrug-resistant strains 12. Here, structural and kinetic analyses are described for inhibitor 1 with selected PR mutants. The single mutants showed various effects. Substitution of the smaller side chains in PRI50V and PRV82A resulted in the loss of their interactions with inhibitor; however, mutation V82A caused a shift of its main chain atoms to compensate for the loss. Mutation to larger side chain in PRI54M pushed the Pro79 away from residue 54 maintaining their interactions; moreover, new interactions were formed between Met54 and Thr80. The elimination of the interactions caused by the change of the residue size in PRI50V and PRV82A may be related to their worse inhibition (Ki) by inhibitor 1. In addition, the mechanisms PRV82A and PRI54M adopted to compensate for the loss of the interactions may account for the moderate decrease (6~7-fold) in Ki for PRV82A and PRI54M compared to significant reduction (60-fold) in Ki for PRI50V. Moreover, inter-subunit interactions were lost in PRI50V, which is expected to contribute to the worse inhibition. Mutations in PRD30N and PRR8Q eliminated the negative and positive charges of Asp30 and Arg8, respectively. In PRR8Q, new hydrogen bond interactions of Gln8 replaced the inter-subunit ionic interaction of Arg8 with Asp29’. Meanwhile, Gln8 formed new water molecule-mediated hydrogen bonds with inhibitor 1, which may explain why inhibitor 1 has similar inhibition for PRR8Q and PRWT. In terms of hydrogen bonds with inhibitor 1, PRWT and all five mutants are similar except for variation in interactions with residue 30 due to the mobility of its side chain. The variation in Asp30/30’ and its interaction with inhibitor did not appear to be a major factor in the Ki for these mutants.
PRD30N differs from the other mutants since the major changes are loss of negative charge for Asp30 and altered internal interactions of residue 30/30’ to account for 18-fold decrease of its Ki value for inhibitor 1. D30N mutation is the major mutation associated with resistance to NFV13, which has been suggested to arise from the change of conformational entropy upon inhibitor binding29. Our structural analysis also showed that mutation D30N introduced changes in its interactions with neighboring residues Thr74 and Asn88. In PRD30N, the side chain of Asn30 formed water molecule-mediated hydrogen bond interactions with both Thr74 and Asn88, whereas, these interactions were absent in PRWT-inhibitor 1. Moreover, the proteolytic activity is sensitive to mutations of residues Gly86-Arg87-Asn8816, 30. Considering the positive association of mutation D30N and N88D31, it would be interesting to study how N88D influences the structure of D30N in complex with this new inhibitor 1.
Compound 1 is an inhibitor designed to fill the S2 binding pocket of PR by incorporating a third THF ring in the P2 group relative to DRV (Figure 1A). As analyzed in its complex with PRWT12 (Figure 7A-B), the oxygen of the third THF ring of inhibitor 1 gains a hydrogen bond interaction with a conserved water molecule (D in Figure 7A) and helps stabilize the inter-subunit interaction of Arg8’ with Asp29 and other surrounding residues. In the mutant PRR8Q, the interaction of the newly incorporated ring of inhibitor 1 with this conserved network was maintained, which is consistent with almost identical inhibition for PRWT and PRR8Q. Mutation R8Q, however, has rarely been found in resistant clinical isolates. DRV has a Ki value of 6.6 nM for mutant PRD30N and up to 18 nM for PRI50V consistent with resistance to virus with these mutations22, 23. Similarly, our study suggests that viral strains containing D30N or I50V are likely to show resistance to inhibitor 1 given their Ki values of 8.9 and 30.9 nM for PRD30N and PRI50V, respectively. Inhibitor 1 shows moderate effects on the other two mutants, PRV82A and PRI54M, similar to their effects with DRV22, 26. Therefore, mutations D30N, I50V, I54M, and V82A exhibit similar effects on both inhibitors, although inhibitor 1 is less effective than DRV on PRI50V. Mutation R8Q exhibits higher sensitivity to compound 1 than other mutations studied here, suggesting that compound 1 may be a good inhibitor against viral strains bearing R8Q mutation. Drug resistant clinical isolates generally have more than one mutation, however, and previous studies of single and double mutations indicate that the changes in structure and other properties of the respective single mutants may not be preserved in the double mutants29, 32. More importantly, the influence of drug resistant mutants on the viral infectivity depends not only on the inhibitor sensitivity, but also on the protease activity, especially for those mutations lacking direct interactions with the inhibitors33. Their study showed that mutant PRI50V had the most negative effect on viral infectivity. Our previous studies suggested that the loss of dimer stability due to I50V mutation was a major contributor to the diminished catalytic activity and inhibition of PRI50V21. Mutant PRD30N also showed ~50% loss of infectivity33 in agreement with our previous observation of altered activity on different precursor cleavage sites17. Therefore, a single mutation may have a variety of effects on inhibition, dimer stability, catalytic activity and viral replication. Finally, since inhibitor 1 possesses the large tris-THF P2 group, our structural analysis suggests it may fit better with the mutants that have an expanded S2/S2’ pocket, such as the extreme multiple mutant PR2028.
EXPERIMENTAL SECTION
General
Inhibitor 1 has shown analytical purity of >98% by HPLC12. The structure was confirmed by 1H and 13C NMR spectral analysis, and high resolution mass spectrometry. HRMS (ESI) [M+Na]+ calcd for C30H40N2O9SNa: 627.2352, found: 627.2359.
Protein expression and purification
In order to prevent PR autoproteolysis, a PR clone (Genbank HIVHXB2CG) engineered with five mutations (Q7K, L33I, L63I, C67A and C95A) was used as a template. Mutants (R8Q, D30N, I50V, I54M and V82A) were generated by using the Quick-Change mutagenesis kit (Stratagene, La Jolla, CA). The expression, purification and refolding were performed as previously described34, 35.
Enzyme kinetics
Kinetic parameters were determined by a fluorescence assay. The fluorogenic substrate was Abz-Thr-Ile-Nle-p-nitro-Phe-Gln-Arg-NH2, where Abz is anthranilic acid and Nle is norleucine (Bachem, King of Prussia, PA, USA), with the sequence derived from the p2/NC cleavage site of the Gag polyprotein. 10 μl protease (final concentration of 70-120 nM) diluted in 98 μl reaction buffer (100 mM Mes, pH 5.6, 400 mM sodium chloride, 1 mM EDTA and 5% glycerol) and 2 μl dimethylsulfoxide or inhibitor (dissolved in dimethylsulfoxide) were incubated at 37 °C for 5 min. The reaction was initialized by adding 90 μl substrate. The reaction was monitored over 5 min in the POLARstar OPTIMA microplate reader at wavelengths of 340 and 420 nm for excitation and emission. Data analysis was performed using the program sigmaplot 9.0 (SPSS Inc., Chicago, IL, USA). Km and kcat values were obtained by standard data fitting with the Michaelis–Menten equation. The Ki value was obtained from the IC50 values estimated from an inhibitor dose–response curve using the equation Ki = (IC50) [E]/2)/(1 + [S]/Km), where [E] and [S] are the PR and substrate concentrations.
Crystallographic Analysis
Crystals were grown by the hanging drop vapor diffusion method using protein solutions preincubated with inhibitor, which was dissolved in dimethylsulfoxide, in a molar ratio of 1:5-10. The final crystallization drop was 1.6-2 μl with reservoir solution and protein in a 1:1 ratio by volume. Crystallization conditions for different complexes were as follows: 22%-24% saturated Ammonium Sulfate, 130-135 mM Sodium Phosphate, 0.05 M Sodium Citrate, pH 6.1 for PRR8Q; 1.46 M NaCl, 0.1 M Sodium Citrate, pH 5.0 for PRD30N; 1.26-1.46 M NaCl, 0.06 M Sodium Acetate, pH 5.0-5.4 for PRI50V; 0.6-0.93 M NaCl, 0.06 M Sodium Acetate, pH 4.6-5.0 for PRI54M; and 10% Ammonium Sulfate, 0.05M Citrate-Phosphate, pH 5.6 for PRV82A. The crystals for X-ray data collection were soaked in the reservoir solution with 20-30% glycerol as cryoprotectant for ~1 min and frozen immediately in liquid nitrogen. X-ray data for all the complexes were collected on the SER CAT beamline at the Advanced Photon Source, Argonne National Laboratory. Data were processed using HKL200036. The structures were solved by molecular replacement using PHASER37 in the CCP4i suite of programs38. Crystal structures were refined with SHELX9739. Manual adjustments and rebuilding were performed using the molecular graphics program COOT40. Figures of the structures were prepared with PYMOL (http://www.pymol.org).
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported in part by the Georgia State University Molecular Basis of Disease Fellowships (H.Z. and C.-H.S.), the Georgia State University Research Program Enhancement Award in Bioinformatics (C.-H.S.), and the National Institutes of Health grants GM062920 (IW) and GM053386 (AKG). X-ray data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) beamlines 22ID (R8Q, D30N, I50V, I54M) and 22BM (V82A) at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions may be found at www.ser cat.org/members.html. Use of the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science, under Contract No. W-31-109-Eng-38.
ABBREVIATIONS USED
- HAART
highly active antiretroviral therapy
- PRWT
mature HIV-1 protease
- PI
clinical inhibitor of PR
- DRV
darunavir
- NFV
nelfinavir
- tris-THF
tris-tetrahydrofuran
- PRR8Q
PR with R8Q mutation
- PRD30N
PR with D30N mutation
- PRI50V
PR with I50V mutation
- PRI54M
PR with I54M mutation
- PRV82A
PR with V82A mutation
Footnotes
ANCILLARY INFORMATION
Supporting Information Available
Hydrogen bonds between protease mutants and inhibitor 1 are listed, and figures of electron density for alternate conformations of residues 48-52 for all mutants. This material is available free of charge via the Internet at http://pubs.acs.org.
Accession codes
Crystallographic data are available for inhibitor 1 complexes with: PRR8Q (PDB ID: 4HEG), PRD30N (PDB ID: 4HDB), PRI50V (PDB ID: 4HDP), PRI54M (PDB ID: 4HE9), and PRV82A (PDB ID: 4HDF).
The authors declare no competing financial interests.
REFERENCES
- 1.UNAIDS report on the global AIDS epidemic. Geneva: 2010. [Google Scholar]
- 2.Girard MP, Bansal GP. HIV/AIDS vaccines: a need for new concepts? Int Rev Immunol. 2008;27:447–471. doi: 10.1080/08830180802432160. [DOI] [PubMed] [Google Scholar]
- 3.Brenner BG, Turner D, Wainberg MA. HIV-1 drug resistance: can we overcome? Expert Opin Biol Ther. 2002;2:751–761. doi: 10.1517/14712598.2.7.751. [DOI] [PubMed] [Google Scholar]
- 4.Darke PL, Nutt RF, Brady SF, Garsky VM, Ciccarone TM, Leu CT, Lumma PK, Freidinger RM, Veber DF, Sigal IS. HIV-1 protease specificity of peptide cleavage is sufficient for processing of gag and pol polyproteins. Biochem Biophys Res Commun. 1988;156:297–303. doi: 10.1016/s0006-291x(88)80839-8. [DOI] [PubMed] [Google Scholar]
- 5.Oroszlan S, Luftig RB. Retroviral proteinases. Curr Top Microbiol Immunol. 1990;157:153–185. doi: 10.1007/978-3-642-75218-6_6. [DOI] [PubMed] [Google Scholar]
- 6.Richman DD, Morton SC, Wrin T, Hellmann N, Berry S, Shapiro MF, Bozzette SA. The prevalence of antiretroviral drug resistance in the United States. AIDS. 2004;18:1393–1401. doi: 10.1097/01.aids.0000131310.52526.c7. [DOI] [PubMed] [Google Scholar]
- 7.Weber IT, Agniswamy J. HIV-1 Protease: Structural Perspectives on Drug Resistance. Viruses-Basel. 2009;1:1110–1136. doi: 10.3390/v1031110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weber IT, Wang Y-F. Aspartic Acid Proteases as Therapeutic Targets. Wiley-VCH Verlag GmbH & Co. KGaA; 2010. HIV-1 Protease: Role in Viral Replication, Protein–Ligand X-Ray Crystal Structures and Inhibitor Design. pp. 107–137. [Google Scholar]
- 9.Miller V. International perspectives on antiretroviral resistance. Resistance to protease inhibitors. J Acquir Immune Defic Syndr. 2001;26(Suppl 1):S34–50. doi: 10.1097/00042560-200103011-00005. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh AK, Kincaid JF, Cho W, Walters DE, Krishnan K, Hussain KA, Koo Y, Cho H, Rudall C, Holland L, Buthod J. Potent HIV protease inhibitors incorporating high affinity P2-ligands and (R)-(hydroxyethylamino)sulfonamide isostere. Bioorg Med Chem Lett. 1998;8:687–690. doi: 10.1016/s0960-894x(98)00098-5. [DOI] [PubMed] [Google Scholar]
- 11.Surleraux DL, Tahri A, Verschueren WG, Pille GM, de Kock HA, Jonckers TH, Peeters A, De Meyer S, Azijn H, Pauwels R, de Bethune MP, King NM, Prabu-Jeyabalan M, Schiffer CA, Wigerinck PB. Discovery and selection of TMC114, a next generation HIV-1 protease inhibitor. J Med Chem. 2005;48:1813–1822. doi: 10.1021/jm049560p. [DOI] [PubMed] [Google Scholar]
- 12.Ghosh AK, Xu CX, Rao KV, Baldridge A, Agniswamy J, Wang YF, Weber IT, Aoki M, Miguel SG, Amano M, Mitsuya H. Probing multidrug-resistance and protein-ligand interactions with oxatricyclic designed ligands in HIV-1 protease inhibitors. ChemMedChem. 5:1850–1854. doi: 10.1002/cmdc.201000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson VA, Calvez V, Gunthard HF, Paredes R, Pillay D, Shafer R, Wensing AM, Richman DD. 2011 update of the drug resistance mutations in HIV-1. Top Antivir Med. 2011;19:156–164. [PMC free article] [PubMed] [Google Scholar]
- 14.Ho DD, Toyoshima T, Mo H, Kempf DJ, Norbeck D, Chen CM, Wideburg NE, Burt SK, Erickson JW, Singh MK. Characterization of human immunodeficiency virus type 1 variants with increased resistance to a C2-symmetric protease inhibitor. J Virol. 1994;68:2016–2020. doi: 10.1128/jvi.68.3.2016-2020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weber IT. Comparison of the crystal structures and intersubunit interactions of human immunodeficiency and Rous sarcoma virus proteases. J Biol Chem. 1990;265:10492–10496. [PubMed] [Google Scholar]
- 16.Ishima R, Gong Q, Tie Y, Weber IT, Louis JM. Highly conserved glycine 86 and arginine 87 residues contribute differently to the structure and activity of the mature HIV-1 protease. Proteins. 78:1015–1025. doi: 10.1002/prot.22625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahalingam B, Louis JM, Reed CC, Adomat JM, Krouse J, Wang YF, Harrison RW, Weber IT. Structural and kinetic analysis of drug resistant mutants of HIV-1 protease. Eur J Biochem. 1999;263:238–245. doi: 10.1046/j.1432-1327.1999.00514.x. [DOI] [PubMed] [Google Scholar]
- 18.Jarvis B, Faulds D. Nelfinavir. A review of its therapeutic efficacy in HIV infection. Drugs. 1998;56:147–167. doi: 10.2165/00003495-199856010-00013. [DOI] [PubMed] [Google Scholar]
- 19.Liu F, Kovalevsky AY, Louis JM, Boross PI, Wang YF, Harrison RW, Weber IT. Mechanism of drug resistance revealed by the crystal structure of the unliganded HIV-1 protease with F53L mutation. J Mol Biol. 2006;358:1191–1199. doi: 10.1016/j.jmb.2006.02.076. [DOI] [PubMed] [Google Scholar]
- 20.Pazhanisamy S, Stuver CM, Cullinan AB, Margolin N, Rao BG, Livingston DJ. Kinetic characterization of human immunodeficiency virus type-1 protease resistant variants. J Biol Chem. 1996;271:17979–17985. doi: 10.1074/jbc.271.30.17979. [DOI] [PubMed] [Google Scholar]
- 21.Liu F, Boross PI, Wang YF, Tozser J, Louis JM, Harrison RW, Weber IT. Kinetic, stability, and structural changes in high-resolution crystal structures of HIV-1 protease with drug-resistant mutations L24I, I50V, and G73S. J Mol Biol. 2005;354:789–800. doi: 10.1016/j.jmb.2005.09.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu F, Kovalevsky AY, Tie Y, Ghosh AK, Harrison RW, Weber IT. Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir. J Mol Biol. 2008;381:102–115. doi: 10.1016/j.jmb.2008.05.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kovalevsky AY, Tie Y, Liu F, Boross PI, Wang YF, Leshchenko S, Ghosh AK, Harrison RW, Weber IT. Effectiveness of nonpeptide clinical inhibitor TMC-114 on HIV-1 protease with highly drug resistant mutations D30N, I50V, and L90M. J Med Chem. 2006;49:1379–1387. doi: 10.1021/jm050943c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahalingam B, Wang YF, Boross PI, Tozser J, Louis JM, Harrison RW, Weber IT. Crystal structures of HIV protease V82A and L90M mutants reveal changes in the indinavir-binding site. Eur J Biochem. 2004;271:1516–1524. doi: 10.1111/j.1432-1033.2004.04060.x. [DOI] [PubMed] [Google Scholar]
- 25.Tie Y, Kovalevsky AY, Boross P, Wang YF, Ghosh AK, Tozser J, Harrison RW, Weber IT. Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir. Proteins. 2007;67:232–242. doi: 10.1002/prot.21304. [DOI] [PubMed] [Google Scholar]
- 26.Tie Y, Boross PI, Wang YF, Gaddis L, Hussain AK, Leshchenko S, Ghosh AK, Louis JM, Harrison RW, Weber IT. High resolution crystal structures of HIV-1 protease with a potent non-peptide inhibitor (UIC-94017) active against multi-drug-resistant clinical strains. J Mol Biol. 2004;338:341–352. doi: 10.1016/j.jmb.2004.02.052. [DOI] [PubMed] [Google Scholar]
- 27.Kovalevsky AY, Liu F, Leshchenko S, Ghosh AK, Louis JM, Harrison RW, Weber IT. Ultra-high resolution crystal structure of HIV-1 protease mutant reveals two binding sites for clinical inhibitor TMC114. J Mol Biol. 2006;363:161–173. doi: 10.1016/j.jmb.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agniswamy J, Shen CH, Aniana A, Sayer JM, Louis JM, Weber IT. HIV-1 protease with 20 mutations exhibits extreme resistance to clinical inhibitors through coordinated structural rearrangements. Biochemistry. 2012;51:2819–2828. doi: 10.1021/bi2018317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kozisek M, Bray J, Rezacova P, Saskova K, Brynda J, Pokorna J, Mammano F, Rulisek L, Konvalinka J. Molecular analysis of the HIV-1 resistance development: enzymatic activities, crystal structures, and thermodynamics of nelfinavir-resistant HIV protease mutants. J Mol Biol. 2007;374:1005–1016. doi: 10.1016/j.jmb.2007.09.083. [DOI] [PubMed] [Google Scholar]
- 30.Ishima R, Ghirlando R, Tozser J, Gronenborn AM, Torchia DA, Louis JM. Folded monomer of HIV-1 protease. J Biol Chem. 2001;276:49110–4916. doi: 10.1074/jbc.M108136200. [DOI] [PubMed] [Google Scholar]
- 31.Mitsuya Y, Winters MA, Fessel WJ, Rhee SY, Hurley L, Horberg M, Schiffer CA, Zolopa AR, Shafer RW. N88D facilitates the co-occurrence of D30N and L90M and the development of multidrug resistance in HIV type 1 protease following nelfinavir treatment failure. AIDS Res Hum Retroviruses. 2006;22:1300–1305. doi: 10.1089/aid.2006.22.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahalingam B, Boross P, Wang YF, Louis JM, Fischer CC, Tozser J, Harrison RW, Weber IT. Combining mutations in HIV-1 protease to understand mechanisms of resistance. Proteins. 2002;48:107–116. doi: 10.1002/prot.10140. [DOI] [PubMed] [Google Scholar]
- 33.Henderson GJ, Lee SK, Irlbeck DM, Harris J, Kline M, Pollom E, Parkin N, Swanstrom R. Interplay between single resistance-associated mutations in the HIV-1 protease and viral infectivity, protease activity, and inhibitor sensitivity. Antimicrob Agents Chemother. 2012;56:623–633. doi: 10.1128/AAC.05549-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wondrak EM, Louis JM. Influence of flanking sequences on the dimer stability of human immunodeficiency virus type 1 protease. Biochemistry. 1996;35:12957–12962. doi: 10.1021/bi960984y. [DOI] [PubMed] [Google Scholar]
- 35.Mahalingam B, Louis JM, Hung J, Harrison RW, Weber IT. Structural implications of drug-resistant mutants of HIV-1 protease: high-resolution crystal structures of the mutant protease/substrate analogue complexes. Proteins. 2001;43:455–464. doi: 10.1002/prot.1057. [DOI] [PubMed] [Google Scholar]
- 36.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Vol. 276. Elsevier; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 37.McCoy AJ, Grosse Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 38.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr D Biol Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 39.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 40.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.Chang YC, Yu X, Zhang Y, Tie Y, Wang YF, Yashchuk S, Ghosh AK, Harrison RW, Weber IT. Potent antiviral HIV-1 protease inhibitor GRL-02031 adapts to the structures of drug resistant mutants with its P1'-pyrrolidinone ring. J Med Chem. 2012;55:3387–3397. doi: 10.1021/jm300072d. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.