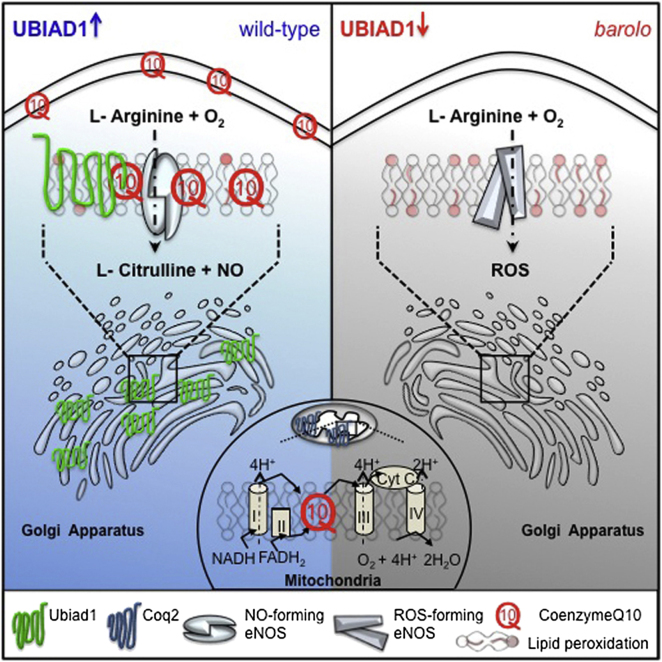
Summary
Protection against oxidative damage caused by excessive reactive oxygen species (ROS) by an antioxidant network is essential for the health of tissues, especially in the cardiovascular system. Here, we identified a gene with important antioxidant features by analyzing a null allele of zebrafish ubiad1, called barolo (bar). bar mutants show specific cardiovascular failure due to oxidative stress and ROS-mediated cellular damage. Human UBIAD1 is a nonmitochondrial prenyltransferase that synthesizes CoQ10 in the Golgi membrane compartment. Loss of UBIAD1 reduces the cytosolic pool of the antioxidant CoQ10 and leads to ROS-mediated lipid peroxidation in vascular cells. Surprisingly, inhibition of eNOS prevents Ubiad1-dependent cardiovascular oxidative damage, suggesting a crucial role for this enzyme and nonmitochondrial CoQ10 in NO signaling. These findings identify UBIAD1 as a nonmitochondrial CoQ10-forming enzyme with specific cardiovascular protective function via the modulation of eNOS activity.
Graphical Abstract
Highlights
► UBIAD1 is a Golgi prenyltransferase ► UBIAD1 contributes to the nonmitochondrial pool of CoQ10 ► UBIAD1 protects cardiovascular tissues from NOS-dependent oxidative damage ► UBIAD1 is a target for therapeutic strategies by limiting the side effects of statins
UBIAD1 is identified as a nonmitochondrial CoQ10 biosynthetic enzyme required for oxidative damage protection. Through CoQ10 synthesis, UBIAD1 modulates endothelial nitric oxide synthase activity and nitric oxide signaling necessary for cardiovascular development and homeostasis.
Introduction
Reactive oxygen species (ROS) play an important role in signal transduction and physiological regulation of the cardiovascular system by facilitating various biological responses, including senescence in heart and endothelial cells (ECs) (Lander et al., 1996; Finkel, 2003; Finkel and Holbrook, 2000; Hare and Stamler, 2005; D’Autréaux and Toledano, 2007). In cardiovascular tissues, ROS are normally produced in response to RTK and GPC receptor signaling, as well as during mechanical (shear) stress. ROS help regulate cardiovascular homeostasis (Colavitti et al., 2002; Werner, 2004; Ushio-Fukai, 2006). An antioxidant network is required to balance these ROS and prevent oxidative damage, which may contribute to cardiovascular pathologies (Finkel, 2003; Giordano, 2005; Kuster et al., 2010). Although several enzymes responsible for ROS production in ECs have been identified, many of the molecules involved in the endogenous antioxidant network are unknown.
A molecule known to play an important antioxidant role in the cardiovascular system is CoQ10 (also known as Coenzyme Q10 or ubiquinone) (Pepe et al., 2007; Kumar et al., 2009). CoQ10 is the only endogenously synthesized lipid-soluble antioxidant (Crane, 2007; Bentinger et al., 2007). It is available in the membranes of the Golgi, where it is present at an even higher concentration than in the mitochondria, as well as in plasma membranes (Kalen et al., 1987; Crane, 2001; Bentinger et al., 2008). Although the biosynthesis of CoQ10 in mitochondria has been studied in detail, only a limited amount of data is available in regards to the synthesis of cellular membrane CoQ10. Several in vivo experiments suggest that CoQ10 synthesis may also occur in the Golgi and endoplasmic reticulum membranes, providing the cellular membrane CoQ10 pool (Kalén et al., 1990). However, although hypothesized, a “nonmitochondrial” CoQ10 biosynthetic enzyme has not been identified (Bentinger et al., 2010). Identification of such an enzyme could be extremely useful in order to devise strategies to counteract the excess of ROS responsible for cellular oxidative damage (such as lipid peroxidation, protein nitrosylation, and DNA oxidation) and therefore balance redox signaling especially in cardiovascular tissues.
CoQ10 is a mobile lipophilic electron carrier that is critical for electron transfer both in the mitochondrial membrane for respiratory chain activity as well as in Golgi and plasma membranes for NAD(P)H-oxidoreductase-dependent reactions such as in NO synthesis (Navas et al., 2007). Endothelial nitric oxide synthase (eNOS) is a critical regulator of cardiovascular functions by generating nitric oxide (NO), which is an important mediator of cardiovascular homeostasis (Alp and Channon, 2004; Förstermann and Sessa, 2012). As in the case with nonmitochondrial CoQ10, eNOS is specifically localized in the Golgi and plasma membrane of heart and endothelial cells, and it can be differentially regulated in these two cellular compartments (Fulton et al., 2002). Different reports have suggested that CoQ10 may improve endothelial dysfunction by “recoupling” eNOS and modulating NO-related signaling (Chew and Watts, 2004; Tsai et al., 2012).
Here, we identify UBIAD1 as an enzyme for CoQ10 synthesis at the level of Golgi membranes and show that it is critical for oxidative stress protection. In particular, UBIAD1 protects cardiovascular tissues from eNOS-dependent oxidative stress. We propose a functional link between UBIAD1, CoQ10, and NO signaling during cardiovascular development and homeostasis.
Results
Oxidative Stress and Cardiovascular Failure Characterize barolo Mutants
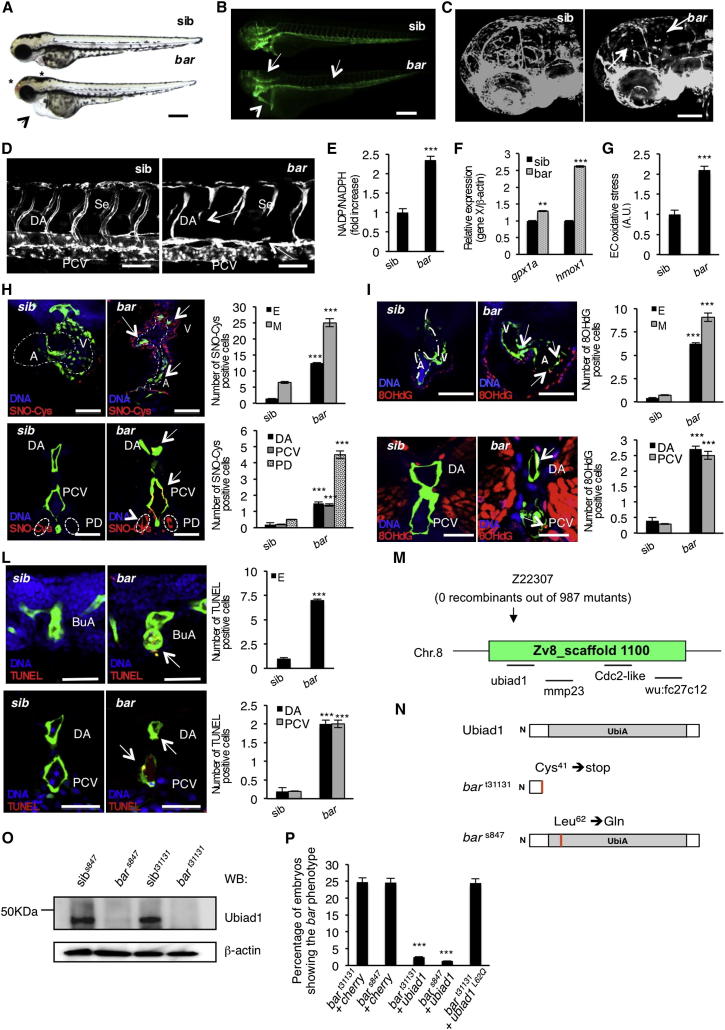
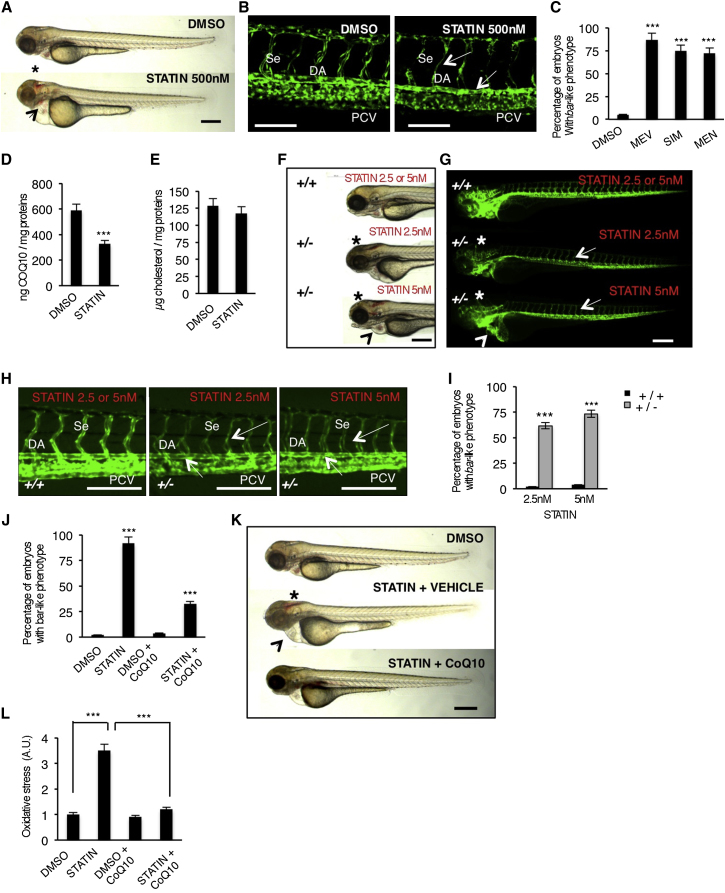
By forward genetic screens for cardiovascular mutants (2005 Tuebingen screen; Jin et al., 2007), we identified mutations in the barolo (bar) gene (bart31131 and bars847). These mutations are recessive and show complete penetrance and expressivity. bar mutants show distinct cranial vascular hemorrhages and pericardial edema in the context of wild-type body morphology (Figure 1A). To examine whether this phenotype is associated with cardiovascular defects, we crossed the mutations into Tg(kdrl:GFP)s843 (Figure 1B). The hemorrhagic phenotype in bar mutants is accompanied by endothelial regression and fragmentation in the cranial and trunk vascular compartments (Figures 1C and 1D). Time-lapse analyses of trunk vasculature indicate that the bar phenotype starts around 36 hr postfertilization (hpf) (Movies S1, S2, S3, and S4) and that it is not due to general loss of blood flow. The cardiac edema of bar mutants is caused by a gradual breakdown of endocardial and myocardial cells in the heart, leading to the collapse of the heart between 60 and 72 hpf (data not shown). All these features lead to the complete cardiac and vascular organ failure by 72 hpf in bar mutant embryos.
Figure 1.
Loss of Ubiad1 Induces Cardiovascular Failure in Zebrafish Embryos by Increasing Oxidative Stress
(A and B) Siblings (sib) and barolot31131 (bar) mutant embryos at 65 hpf. (A) Vascular hemorrhages (asterisks) and heart failure (arrowhead) in bar. (B) Tg(kdrl:GFP)s843bar show vascular integrity defects (arrows) and collapsed endocardium (arrowhead). Scale bar, 300 μm.
(C) Tg(Fli1a:GFP)y1bar show severe loss of cranial network vasculature and fragmented endothelium (arrow) at 65 hpf.
(D) Confocal images of trunk vasculature of Tg(kdrl:GFP)s843bar report loss of ECs at the level of Se, DA (arrows), and PCV. Scale bar, 50 μm.
(E) NADP+/NADPH ratio at 60 hpf. Increased ratio is indicative of oxidative stress state.
(F) Real-time PCR analysis show upregulation of the antioxidant genes glutathione peroxidase1a (gpx1a) and heme-oxigenase1 (hmox1) in bar at 56 hpf. Data are means ± SD.
(G) Quantification of oxidative stress level in ECs derived from Tg(kdrl:GFP)s843 embryos at 56 hpf.
(H) Immunofluorescence analyses for S-nitroso-cysteine (SNO-Cys, red) and DNA (blue) in Tg(kdrl:GFP)s843bar and sib embryos at 60 hpf. Upper left: heart transverse sections; upper right: quantification of SNO-Cys positive cells. Lower left: trunk vasculature transverse sections; lower right: quantification of SNO-Cys positive cells. Scale bar, 75 μm. Specific SNO-Cys signals were also detectable in pronephros and myocardium of bar (arrowhead).
(I) Immunofluorescence analyses for 8-hydroxy-2′deoxyguanosine (8OHdG, red) and DNA (blue) in Tg(kdrl:GFP)s843bar and sib at 56 hpf. Upper left: heart transverse sections; upper right: quantification of 8OHdG positive cells. Scale bars, 50 μm. Lower left: trunk vasculature transverse sections; lower right; quantification of 8OHdG positive. Scale bar, 20 μm. bar show specific 8OHdG staining in endocardial and endothelial cells (arrows) and myocardium (arrowhead).
(L) TUNEL-positive (red) cells show apoptosis in endocardium and endothelium (arrows). Upper left: heart transverse sections of Tg(kdrl:GFP)s843bars847 and sib; upper right: quantification of TUNEL-positive cells. Lower left: transverse sections of trunk vasculature; lower right: quantification of positive cells in DA and PCV. Scale bars, 20 μm.
(M) Genetic map of the zebrafish barolo locus. The SSLP markers (z22307) was used to identify the “zero recombinant” region by analysis of 987 diploid mutants.
(N) Schematic representation of zebrafish Ubiad1. UbiA:UbiA domain.
(O) Western blot analysis of protein extracts from bars847 and bart31131 and respective siblings at 72 hpf.
(P) Embryos from bars847 and bart31131 heterozygote intercrosses were injected with mRNA encoding for wild-type ubiad1 mRNA, ubiad1L62Q or fluorescent protein cherry (ctrl). bars847 as well as bart31131 were not rescued by microinjection of ubiad1L62Q mRNA.
A, atrium; V, ventricle; DA, dorsal aorta; PCV, posterior cardinal vein; BuA bulbus arteriosus; PD, pronephric ducts; E, endocardium; M myocardium. All data are means ± SEM. ∗∗p < 0.01, ∗∗∗p < 0.001.
See also Figure S1, Table S1, and Movies S1, S2, S3, and S4.
Cardiovascular failure is related to altered redox signaling and an increased production of ROS (Hare and Stamler, 2005). bar mutants compared to wild-type siblings show altered redox balance and oxidative stress features evaluated as an increase of the NADP/NADPH ratio and as an overexpression of antioxidant genes, respectively (Figures 1E and 1F). Increase of ROS production occurs specifically in cardiovascular tissues as demonstrated by measuring oxidizing species in ECs of bar mutants using the selective and stable ROS detector CellROX (Figure 1G). We also measured S-nitrosylation levels on heart and blood vessel sections of bar mutants. bar mutants show higher levels of S-nitrosylated proteins in heart and blood vessel cells as well as in pronephros as measured by an antibody against S-nitroso-cysteine (SNO-Cys) (Figure 1H). A high level of S-nitrosylated proteins is normally associated with altered redox signaling and cellular protection against oxidative stress (Stamler et al., 2001; Hess et al., 2005; Sun and Murphy, 2010).
We found that cardiovascular tissues from bar mutants are positive for 8-hydroxy-2′-deoxyguanosine (8-OHdG), a free radical-induced oxidative lesion in DNA (Figure 1I). Analyses of heart and blood vessel sections show that both myocardial and endothelial tissues of bar mutants exhibit ROS-mediated DNA damage compared to controls. Indeed, endothelial and endocardial cells in bar mutants were positive for TUNEL staining and show typical morphological features of apoptotic cells at this stage (Figure 1L). In summary, both mutant embryos carrying the barolo mutation show increased oxidative stress, DNA damage, and cell death specifically in blood vessels and the heart.
We determined that bar encodes Ubiad1 (UbiA-domain containing protein 1) (Figure 1M). Sequence analyses show that the bart31131 mutant allele contains a nonsense mutation (123 T > A) in ubiad1 introducing a premature stop codon at amino acid position 41, likely generating a null allele. The second allele of bar (bars847) bears a base change (185 T > A) leading to the substitution of a conserved leucine residue to glutamine at amino acid position 62 (Ubiad1L62Q) (Figure 1N). bars847 shows the same phenotypical features as bart31131 except the phenotype is delayed by 12 hr (Figures S1A–S1E and S1T available online). Both bar allele mutants do not express Ubiad1 (Figure 1O) and are fully rescued by Ubiad1 mRNA expression. Ubiad1L62Q mRNA does not rescue the bart31131 mutant indicating that this amino acid mutation is sufficient to knock out Ubiad1 activity (Figure 1P). To confirm that Ubiad1 regulates cardiovascular protection and survival, we knocked down Ubiad1 expression using an antisense morpholino oligonucleotide. Ubiad1 morphants show the same hemorrhagic phenotype and cardiovascular defects as bar mutants (Figures S1F–S1I).
Figure S1.
Barolo Alleles Phenotypes Are Characterized by Oxidative Stress Leading to Progressive Cardiovascular Failure, Related to Figure 1
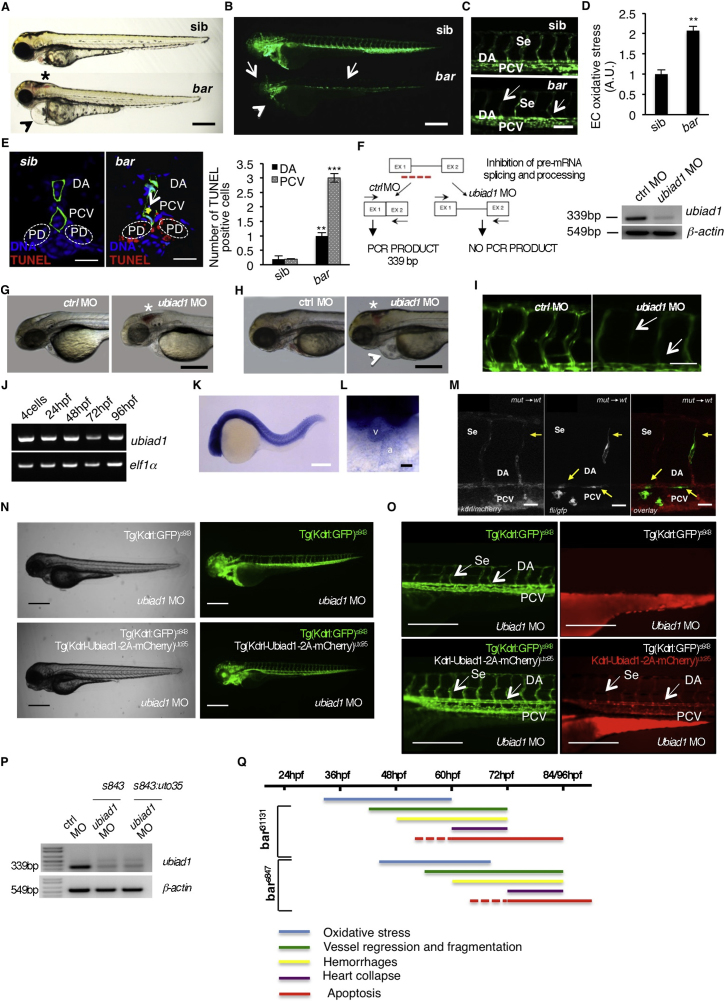
(A) Wild-type siblings s847 (sib) and barolos847 (bar) mutant embryos at 72 hpf. Vascular hemorrhages (asterisk) and heart failure (arrowhead) are visible in bars847 mutants. Scale bar, 300 μm.
(B) Fluorescent micrographs of Tg(kdrl:GFP)s843 siblings (sib) and barolos847 (bar) mutants at 72 hpf. bars847 mutants show vascular integrity defects and regression both in brain and trunk vasculature (arrows). Endothelial cell regression is observed throughout the vascular network of bars847 mutants at 72 hpf. The collapsed endocardium is also evident (arrowhead). Scale bar, 300 μm.
(C) Fluorescent micrographs representing magnification of trunk vasculature of Tg(kdrl:GFP)s843 sib s847 (sib) and barolos847 (bar) mutants at 72 hpf. In bars847 mutant’s endothelial cells are lost at the level of intersegmental vessels (Se), dorsal aorta (DA) and posterior cardinal vein (PCV) (arrows). Scale bar, 50 μm.
(D) Histogram shows relative quantifications of oxidative stress in primary endothelial cells derived from Tg(kdrl:GFP)s843 sib s847 (sib) and bars847 (bar) mutant at 65 hpf. ROS levels in endothelial cells of zebrafish embryos were measured by flow cytometric analyses using the specific CellROX probe on Kdrl:GFP+ cells. ROS level is significantly higher in bars847 mutants than respective siblings. Results are shown as a mean of n = 3 independent experiments. Data are means ± SD.
(E) Staining of Tg(kdrl:GFP)s843 sibs847 (sib) and barolos847 (bar) mutant embryos at 80 hpf for TUNEL (red) and DNA (blue). Left: confocal transverse sections of trunk vasculature at the level of the 10th somite; right: histograms report numbers of dorsal aorta and posterior cardinal vein cells positive for TUNEL staining. Data are means ± SD; ∗∗p < 0.01, ∗∗∗p < 0.001. TUNEL-positive cells were also present in pronephric ducts of bar mutants. Scale bars, 100 μm.
(F) Validation of in vivo ubiad1 knockdown by morpholino injection. Left: schematic diagram shows the molecular mechanism of ubiad1splice-morpholino (ubiad1 MO; dashed line). ubiad1 MO targeting the splicing site between exon1 (EX1) and intron1 blocks ubiad1 pre-mRNA maturation and leads to ubiad1 non-coding mRNA. Molecular mechanism of control non-targeting morpholino (ctrl MO) is also indicated. Primers used for RT-PCR validation are localized on exons 1 and 2 (EX1-EX2; black arrows). Right: RT-PCR analysis was performed on cDNA from zebrafish embryos 72 hr post microinjection. Amplification of 339 bp is possible only for cDNA from ctrl-injected embryos, whereas no amplification is expected from ubiad1MO-injected embryos. Compared to ctrl MO, ubiad1 MO show reduced 339 bp PCR products.
Representative bright-field images of ubiad1 morphants (ubiad1 MO) and relative control (ctrl MO) at 48 hpf (G) and 72 hpf (H). ubiad1 morphants such as bar mutants are characterized by brain hemorrhages (asterisk) and stringy heart morphology (arrowhead). Scale bar, 100 μm.
(I) Fluorescent micrographs of trunk vasculature (between 14th and 18th somites) of Tg (kdrl:GFP)s843ubiad1 morphants (ubiad1 MO) and control (ctrl MO) injected embryos at 72 hpf. Knockdown of ubiad1 led to loss of intersegmental vessels integrity (arrows) as observed in bar mutants. Scale bar, 75 μm.
(J) RT-PCR analyses for ubiad1 mRNA expression and ef1α (as control) on cDNA from wild-type embryos at different developmental stages. Samples are from 4-cell stage to 96 hpf stage.
(K) Whole-mount ISH analyses of wild-type embryo at 24 hpf show ubiad1 mRNA expression throughout the embryo. Scale bar, 250 μm.
(L) Magnification on heart of whole-mount ISH analyses of wild-type embryo at 48 hpf. Specific expression of ubiad1 mRNA can be detected in the developing heart. A, atrium; V, ventricle. Scale bar, 50 μm.
(M) Cell transplantation experiments in barolo mutant embryos. Confocal fluorescent image shows chimeric vessels made of endothelial cells from Tg(fli:GFP)y1bart31131 mutant embryos (donor) that have been transplanted at pre-dome stage into Tg(kdrl: mCherry)uto2 wild-type (wt) embryos (acceptor) and visualized at 48 hpf. Transplanted mutant endothelial cells (green fluorescence) show retracting and collapsed morphology (arrows), also at the level of intersegmental vessel (Se) and dorsal aorta (DA) suggesting ubiad1 acts cell autonomously. Left: mCherry channel corresponding to Tg(kdrl: mCherry)uto2 wild-type cells. Middle: GFP channel corresponding to Tg(fli:GFP)y1bart31131 mutant endothelial cells. Right: Merge image of mCherry and GFP channels. Se, intersomitic vessels; DA, dorsal aorta; PCV, posterior cardinal vein. Scale bar, 20 μm.
(N) Bright field (left) and corresponding fluorescent micrographs (right) of Tg(kdrl:GFP)s843 and Tg [(kdrl-Ubiad1-2A-mCherry)uto35; (kdrl:GFP)s843 ] wild-type embryos injected with ubiad1 splice-morpholino (ubiad1 MO). After morpholino injection Tg(kdrl:GFP)s843 embryos show characteristic barolo phenotype (upper). On the contrary, after ubiad1 morpholino injection Tg [(kdrl-Ubiad1-2A-mCherry)uto35; (kdrl:GFP)s843 ] show wild-type phenotype (lower). Scale bar, 400 μm.
(O) Magnification of trunk vasculature of embryos showed in (N). Green fluorescence shows endothelial cells. Red fluorescence, which is exclusive of uto35 line, indicates expression of exogenous Ubiad1 in endothelial cells (lower right). Injection of ubiad1 morpholino in Tg(kdrl:GFP)s843 induce bar phenotype characterized by intersegmental vessels (Se) disintegration and collapsed dorsal aorta (DA) (upper left; arrows). Injection of ubiad1 morpholino in Tg [(kdrl-Ubiad1-2A-mCherry)uto35;(kdrl:GFP)s843 ] do not exert any vascular phenotype (lower left; arrowheads) indicating that endothelial-specific expression of Ubiad1 is sufficient to prevent barolo cardiovascular phenotype. All together these data support that Ubiad1 acts cell autonomously in vascular cells. Scale bar, 150 μm.
(P) Validation of ubiad1 knockdown by morpholino injection, related to embryos in (N and O). RT-PCR analysis was performed on cDNA from zebrafish embryos at 72 hpf. Amplification of 339bp is possible only for cDNA from ctrl MO-injected embryos (ctrl MO), whereas no amplification is expected from ubiad1MO-injected embryos (ubiad1 MO). Compared to ctrl MO, ubiad1 MO show reduced 339bp PCR products in Tg(kdrl:GFP)s843 (s843) as well as in Tg [(kdrl-Ubiad1-2A-mCherry)uto35; (kdrl:GFP)s843 ] (s843:uto35) morphants. RT-PCR strategy is showed in (F).
(Q) Chronological documentation of barolo phenotypes onset. bart31131 mutant embryos phenotype starts with an increase of oxidative stress (detectable from 32 hpf until 60 hpf) that causes a progressive cardiovascular breakdown characterized by vessels regression and fragmentation (starting from 40 hpf), hemorrhages (from 48 hpf) and heart failure (from 60 hpf). barolo cardiovascular degeneration ends up with heart and vascular cells apoptosis that become evident at molecular levels starting from 60 hpf. Definitive signs of apoptosis can be scored at 72 hpf.
bars847 shows the same progressive cardiovascular failure features of as bart3113, but delayed of 12 hr in the same experimental conditions. Both barolo alleles are not able to survive over 96 hpf.
Analyses of ubiad1 mRNA expression in developing zebrafish embryos reveal a ubiquitous expression at 24 hpf in addition to a distinct expression in the heart at 48 hpf (Figures S1J–S1L), in agreement with mammalian expression data (Nakagawa et al., 2010). In addition, cell transplantation and tissue-specific rescue experiments showed that Ubiad1 functions autonomously within endothelial and endocardial cells (Figures S1M–S1P and Table S1).
These data indicate that loss of ubiad1 triggers a progressive cardiovascular failure by increasing oxidative stress and apoptosis in heart and blood vessels suggesting a role for Ubiad1 as a controller of redox state and ROS levels in vertebrate cardiovascular tissues (Figure S1Q). Thereby, bar represents a unique model to study oxidative stress conditions and redox signaling during zebrafish development.
UBIAD1 Is a CoQ10 Biosynthetic Enzyme
Ubiad1 is conserved among species and contains an UbiA prenyltransferase domain, present also in the vertebrate protein Coq2. So far, the mitochondrial enzyme Coq2 is the only vertebrate enzyme known to synthesize CoQ10 through the prenylation of 4-hydroxybenzoic acid by oligoprenyl diphosphates (Forsgren et al., 2004). The CoQ10 molecule is composed of a redox active benzoquinone ring, which is connected to a polyisoprenoid side chain of variable length (typically 9/10 units in vertebrates) (Crane, 2001). As with Coq2, we speculate that Ubiad1 might catalyze the biosynthesis of CoQ10. We measured CoQ10 levels in bar mutants and ubiad1 morphants by HPLC-MS (Figures 2A and 2B). The absence of Ubiad1 in zebrafish larvae significantly reduces CoQ10 levels (Figure 2C), whereas cholesterol levels are not significantly affected (Figure S2A). By the time course of synthesis analyses, we found that CoQ10 increases during zebrafish embryonic development and is almost entirely produced by the zygote without any contribution from the yolk sac (Figure S2B). On the other hand, a significant amount of embryonic cholesterol is provided by the yolk sac throughout early development (Figure S2C). These data suggest that de novo zygotic synthesis of CoQ10 by Ubiad1 (and/or Coq2) is required for zebrafish development.
Figure 2.
Ubiad1 Is a Prenyltranferase Required for CoQ10 Biosynthesis in Vertebrates
(A) Schematic representation of the 13C-based metabolomic approach used to demonstrate that Ubiad1 is a 4-OH-benzoic acid-prenyltranferase enzyme. Coenzyme Q10 (CoQ10) biosynthesis starts with formation of a hydroxy-benzoic acid head group (4-OH-benzoic acid) and a lipophilic polyisoprenoid tail (polyprenyl-PP). The aromatic precursor of the benzoquinone ring (4-OH-benzoic acid) derives from tyrosine. Synthesis of the polyisoprenoid tail takes place in the cytoplasm starting from acetyl-CoA through the mevalonate pathway (which is shared with the cholesterol and dolichol biosynthetic pathways). The polyisoprenoid tail is assembled by polyprenyl diphosphate synthase, which is responsible for determining the number of isoprene units (designated as n). Next is the condensation of the polyisoprenoid chain with the benzoquinone ring to form the 4-hydroxy-3-polyprenyl benzoic acid intermediate (PPHB) by a 4-OH-benzoic-polyprenyl-tranferase (UbiA-containing enzyme). The subsequent reactions (O-methylations and C-methylations, hydroxylations and decarboxylations) modify the structure of the aromatic ring, but some of the enzymes involved are still unknown. Exogenous 4-OH-benzoic acid-13C6 containing heavy carbon element (13C) is administered to embryos and cells so that de novo synthesis of PPHB and therefore CoQ10 can be monitored by HPLC-MS analyses. Loss of UbiA-containing enzymes (red) blocks the condensation of the polyisoprenoid chain with the benzoquinone ring and then formation of CoQ-13C6. 13C6-labeled compounds used or detected in this work are shown in green.
(B) High-resolution MS spectrum of lipid extracts from wild-type zebrafish embryos. The MS spectrum of the HPLC fraction containing CoQ10 reveals the analyte as [CoQ10-Na+] at m/z = 885.6751.
(C) HPLC-MS analyses show that bar mutants are characterized by a reduced level of endogenous CoQ10 in comparison to their siblings.
(D) Exogenous supplement of CoQ10, but not of VitaminK2, can efficiently rescue bar mutants. CoQ10F1; liposomal CoQ10 preparation 0.6 mM; CoQ10F2, LiQsorb Liposomal CoQ10 Gel 0.6 mM; COQ10 F3, SANOMIT nanoparticles CoQ10 liquid, 6.9 mM; VitK; Liposomal Vitamin K2 0.3 mM.
(E and F) Levels of 13C6-labeled CoQ10 (CoQ10-13C6) and 13C6-labeled CoQ9 (CoQ9-13C6) are detected by HPLC-MS in bar mutants and siblings at 72 hpf. Loss of Ubiad1 expression protein significantly reduces de novo CoQ10-13C6 synthesis by 78% (E) and de novo CoQ9-13C6 synthesis by 66.5% (F). All data are means ± SEM. ∗∗p < 0.01, ∗∗∗p < 0.001.
See also Figure S2.
Figure S2.
UBIAD1 Regulates CoQ10 but Not Cholesterol or Vitamin K2 Levels in Zebrafish Embryos and Human Endothelial Cells, Related to Figure 2
(A) Histograms show levels of cholesterol detected by HPLC-UV analysis in bart31131 and respective siblings (sib) or ubiad1 morphants and relative controls (ctrl MO). The level of total cholesterol is not affected among all conditions. The results are shown as a mean of n = 6 independent experiments for each condition. Error bars indicate SD of measurements.
(B) Histograms show levels of CoQ10 determined by HPLC-UV analysis during zebrafish development starting from 24 hpf to 72 hpf embryos. Whole embryos (black) and deyolk embryos (gray) lipid extracts were subjected to chemical quantitative analyses. Equal amount of CoQ10 is detected in both compartments suggesting CoQ10 is produced entirely by zygotic tissues. Each sample is a pool of 25 zebrafish embryos at the same developmental stage. Error bars indicate SD of measurements.
(C) Histograms show levels of cholesterol determined by HPLC-UV analysis during zebrafish development starting from 24 hpf to 72 hpf embryos. Whole embryos (black) and deyolk embryos (gray) lipid extracts were subjected to chemical quantitative analyses. Cholesterol is higher in extracts containing the whole embryo suggesting a large amount of cholesterol is present in the yolk sac. Each sample is a pool of 25 zebrafish embryos at the same developmental stage. Error bars indicate SD of measurements.
Analysis of lipid extracts from zebrafish embryos for Vitamin K levels.
(D) Mass spectrum corresponding to the peak in chromatogram (E) at retention time 4.0 min and corresponding to Vitamin K2 standard. Vitamin K2 is detected as 445 m/z [molecule-H+].
(E) SIM chromatogram of a 1 mM standard mix of Vitamin K2. Select Ion Monitoring (SIM) (m/z = 445) allows us to have a maximum sensibility toward Vitamin K2. Peak corresponds to Vitamin K2 standard. Vitamin K2 retention time is 4.0 min.
(F) TIC chromatogram of 1 mM standard mix shown in (E). The arrow shows the peak of Vitamin K2.
(G) TIC chromatogram of lipid extract from bart31131sibling. No Vitamin K2 peak is evident. This sample was also recorded for SIM-HPLC-MS chromatograms too, but there is no variation to control (data not shown).
(H) TIC chromatogram of lipid extract from bart31131 mutant embryos. No Vitamin K2 peak is evident. This sample was also recorded for SIM-HPLC-MS chromatograms, but there is no variation to control (data not shown).
(I) TIC chromatogram of lipid extract from UBIAD1overespression human primary endothelial cells. No Vitamin K2 peak is evident. This sample was also recorded for SIM-HPLC-MS chromatograms too, but there is no variation to control (data not shown).
(J) Full mass chromatogram of human primary endothelial cells (HUAEC) lipid extracts incubated with 4-hydroxy-benzoic acid-13C6 for 48 hr. Channel I: CoQ10 (m/z = 885.7, retention time: 10.46); Channel II: CoQ9 (m/z = 8117.6, retention time: 7.78); Channel III: CoQ10-13C6 (m/z = 891.7, retention time: 10.46 min).
(K) Mass spectrum chromatogram of human primary endothelial cells (HUAEC) lipid extracts incubated with hydroxyl-4-benzoic acid-13C6 for 48h. Chromatogram corresponds to the MS picks of CoQ10 and 13C6-labeled CoQ10 (CoQ10-13C6). CoQ10 is detected as m/z 885.6804 [molecule-Na+] and CoQ10-13C6 is detected as m/z 891.7011 [molecule-Na+], respectively. Endothelial cells metabolize 4-hydroxy-benzoic acid-13C6 into CoQ10-13C6.
To test whether it is the absence of CoQ10 synthesis in zebrafish embryos that causes the bar phenotype, we injected CoQ10 in single-cell stage bar mutants (Figure 2D). Three different CoQ10 formulations rescue bar mutants, suggesting that the intake of CoQ10 in Ubiad1 deficient embryos may restore normal cardiovascular development. A recent paper reports that UBIAD1 is required for Vitamin K2 production (Nakagawa et al., 2010); however, our data indicate that a lack of Vitamin K2 is not responsible for the cardiovascular phenotype in bar mutants (Figures 2D and S2D–S2K).
To provide biochemical evidence that Ubiad1 is able to synthesize CoQ10, we performed metabolic studies in zebrafish embryos by analyzing the ability of bar mutants to convert hydroxy-4-benzoic acid to CoQ10 (Figure 2E). We confirmed that administration of 13C6-labeled hydroxy-4-benzoic acid (hydroxy-4-benzoic acid-13C6) is metabolized to CoQ10-13C6 in zebrafish embryos by injecting this C13-labeled precursor in zebrafish embryos at one-cell stage. After 72 hpf of incubation, lipids were extracted and analyzed by HPLC-MS to quantify the amount of CoQ10-13C6 formed (Figure 2E). bar mutants show a reduced CoQ10-13C6 production compared to wild-type siblings. CoQ9-13C6 production was also impaired (Figure 2F), indicating that zebrafish larvae can synthesize both CoQ9 and CoQ10 as can human cells (Turunen et al., 2004). Therefore, Ubiad1 is a 4-OH-benzoic acid-prenyl-tranferase enzyme required for CoQ10 and CoQ9 biosynthesis in vivo.
Ubiad1 and HMG-CoA Reductase inhibitors act in the Mevalonate Pathway for CoQ10-Dependent Cardiovascular Oxidative Stress Protection.
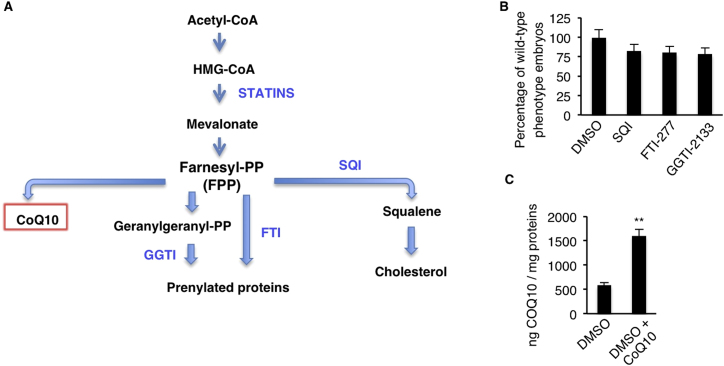
The rate-limiting reaction in the biosynthesis of CoQ10 is the transfer of the polyprenyl-pyrophosphate (polyprenyl-PP) chain, derived from mevalonate, on to 4-OH-benzoic acid, derived from tyrosine metabolism (Bentinger et al., 2008) (Figure 2A). We reduced the pool of polyprenyl-PP available for CoQ10 synthesis by treating zebrafish embryos with statins, inhibitors of its precursor (e.g., mevalonate) (Figure S3A). By blocking HMG-CoA reductase, statins impair the mevalonate and FPP synthesis, which is mandatory to synthetize the polyprenyl-PP chain (Figure 2A). Wild-type embryos (ubiad1+/+) treated with statin developed a bar-like phenotype (Figures 3A–3C). Statin treatments efficiently reduced CoQ10 synthesis without interfering with cholesterol levels, probably because statin treatment blocks de novo cholesterol biosynthesis but does not interfere with the maternally provided cholesterol (Figures 3D and 3E). Zebrafish embryos treated with the squalene synthase inhibitor (SQI) or with farnesyl or geranyl-geranyl tranferase inhibitors (FTI or GGTI) did not exhibit a bar-like phenotype (Figure S3B), indicating that the absence of CoQ10 is the cause of the statin-induced cardiovascular oxidative phenotype. Furthermore, while treatment with a low-dose of statin did not produce any phenotype in wild-type (ubiad1+/+) embryos, it caused a bar-like phenotype in embryos heterozygous for ubiad1 (ubiad1+/−) (Figures 3F–3I). The statin-dependent phenotype in embryos was efficiently rescued by exogenous administration of liposomal CoQ10 (Figures 3J and 3K and S3C). To address the role of the mevalonate pathway in protection from oxidative stress in vivo, we measured ROS levels in zebrafish embryos treated with HMG-CoA reductase inhibitors. A ROS level increase is detected in statin-treated zebrafish embryos, and this oxidative stress was fully rescued by CoQ10 treatment (Figure 3L). These data suggest that HMG-CoA reductase and Ubiad1 are on the same metabolic pathway to produce CoQ10 as a potent antioxidant molecule which protects cardiovascular tissues during development.
Figure S3.
Statin-Dependent barolo Phenotype Is Caused by CoQ10, but Not Cholesterol or Protein Prenylation, Deficiency, Related to Figure 3
(A) Schematic representation of mevalonate biosynthetic pathway and inhibitors used in this work to block final products of this metabolic pathway: CoQ10, prenylated protein and cholesterol. Mevalonate-derived farnesyl-pyrophasphates (Farnesyl PP: FPP) is the key intermediate for protein isoprenylation, cholesterol and CoQ10 biosynthesis. Specific inhibitors used in this work are indicated in blue and are: statins (HMG-CoA reductase inhibitor), FTI (Farnesyl Tranferase Inhibitor), GGTI (Geranyl-Geranyl Transferase Inhibitor) and SQI (Squalene synthase Inhibitor). Statins were used to block mevalonate pathway and FPP synthesis. FTI and GGTI were used to specifically block protein isoprenylation, while SQI was used to block cholesterol synthesis.
(B) Histograms show the percentage of embryos treated with SQI, FTI-277 and GGTI-2133 inhibitors and displaying wild-type morphology. Inhibition of protein isoprenilation by treatment with FTI and GTI or inhibition of de novo biosynthesis of cholesterol by SQI treatment does not phenocopy bar mutants or perturb embryos development. FTI, farnesyltransferase inhibitor, GGTI, geranylgeranyl transferase inhibitor, SQI, squalene synthase inhibitor.
(C) Histograms show HPLC-MS quantifications of CoQ10 from lipid extracts of wild-type zebrafish embryos treated with the SANOMIT nanoparticles CoQ10 liquid (COQ10 F3; 6.95 mM) and relative control. Compared to control sample, SANOMIT treated embryos show higher amount of CoQ10 indicating that CoQ10 is delivered and correctly adsorbed by developing embryos.
All data are means ± SEM. ∗∗p < 0.01.
Figure 3.
Block of Mevalonate Pathway Causes Cardiovascular Failure in Zebrafish Embryos by Reducing CoQ10 Synthesis
(A) Wild-type (WT) embryos at 72 hpf treated from 54 hpf with statin (mevastatin) or DMSO. Statin treatments induce a bar-like phenotype which is characterized by hemorrhages (asterisk) and heart failure (arrowhead). Scale bar, 300 μm.
(B) Three dimensional projections of trunk vessels at 72 hpf of DMSO and statin-treated zebrafish embryos. Statin treatments induce specific endothelial vessels regression and fragmentation in DA and Se (arrows). Scale bar, 100 μm.
(C) Quantification of bar-like phenotype after statin treatments. MEV, mevastatin 500 nM; SIM, simvastatin 500 nM; MEN, mevinolin 500 nM.
(D and E) Levels of CoQ10 (D) and cholesterol (E) detected by HPLC-UV analyses in statin-treated embryos.
(F and G) Bright-field (F) and fluorescent (G) images of Tg(kdrl:GFP)s843ubiad1+/+ and ubiad1+/− embryos at 65 hpf treated with mevastatin (2.5 nM and 5 nM) from 32 to 50 hpf. Treatment with a low dose of statin caused hemorrhages (asterisks) and altered heart morphology (arrowheads) in ubiad1+/−, but not in ubiad1+/+ embryos. Although treatment with a low dose of statin did not induce specific alterations of the main vasculature, ubiad1+/− embryos showed head vasculature, Se, and DA regression; scale bar, 300 μm.
(H) Images of Tg(kdrl:GFP)s843ubiad1+/+ and ubiad1+/− embryos trunk vasculature show thinner and collapsed Se and DA morphology in statin-treated ubiad1+/− but not in ubiad1+/+ embryos. Scale bar, 75 μm.
(I) Penetrance of bar-like phenotype at 65 hpf after treatment with mevastatin. ubiad1+/− embryos are significantly more sensitive to develop a bar-like phenotype than normal embryos (ubiad1+/+).
(J) CoQ10 supplemented embryos show less susceptibility to statin treatments in terms of developing a bar-like phenotype as indicated by histograms showing the percentage of statin-treated embryos having bar-like phenotype after exogenous CoQ10 delivery.
(K) CoQ10 supplementation before statin treatment prevents cardiac edema (arrowhead) and brain hemorrhages (asterisk) in barolo. Images of vehicle- and CoQ10-injected embryos after statin treatment. Scale bar, 300 μm.
(L) Histograms show oxidative stress in embryos after statin treatment with or without CoQ10.
All data are means ± SEM. ∗∗∗p < 0.001.
See also Figure S3.
UBIAD1 Synthesizes CoQ10 in the Golgi Compartment
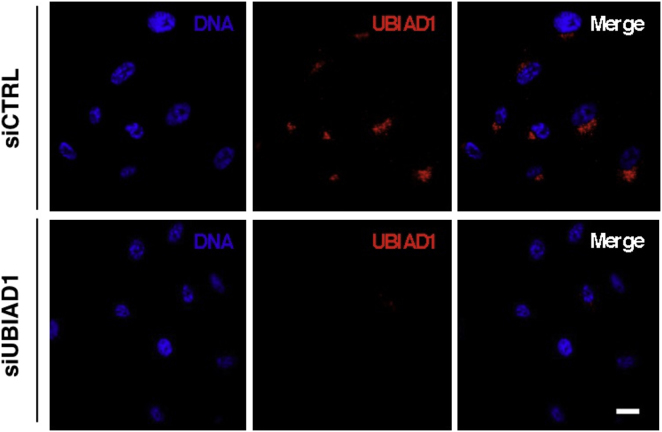
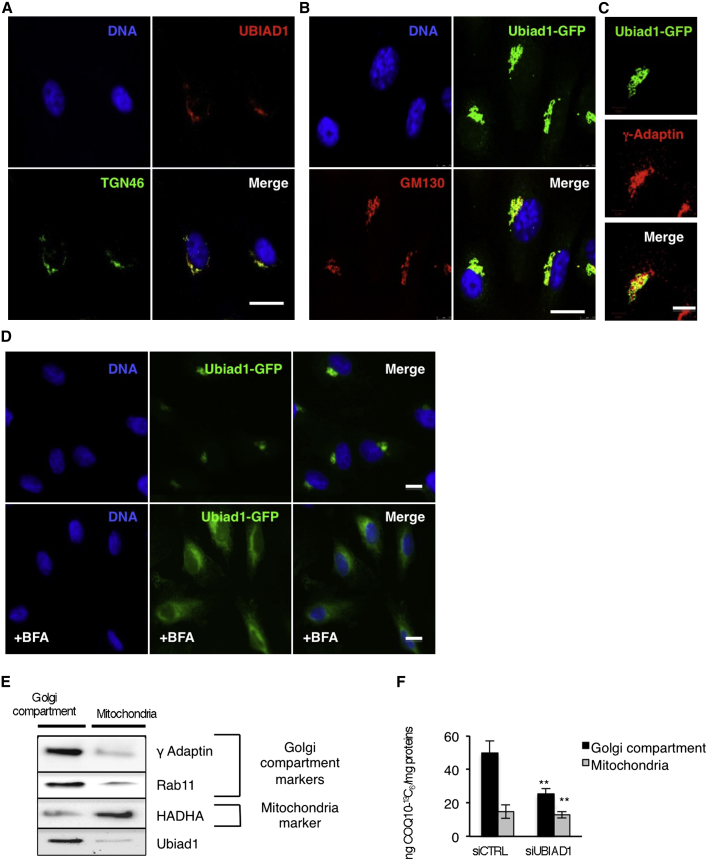
To determine whether Ubiad1 could be an enzyme responsible for cellular CoQ10 production, we examined its subcellular localization. Ubiad1 is conserved among species and is predicted to be a transmembrane protein (Nickerson et al., 2010). Using human primary ECs, we found that UBIAD1 localizes in an asymmetric perinuclear region that resembles the Golgi compartment (Figure S4A). Colocalization studies using the Golgi-specific marker TGN46 demonstrate that endogenous UBIAD1 is in the proximity of the Golgi (Figure 4A). Also, green fluorescent protein (GFP)-tagged zebrafish Ubiad1 expressed in human ECs localizes in the same perinuclear region compartment and colocalizes with the Golgi markers, GM-130 and γ-adaptin (Figures 4B and4C). Further, we observe a rapid redistribution of Ubiad1-GFP proteins in cells treated with brefeldin A, supporting its Golgi localization (Figure 4D).
Figure S4.
UBIAD1 Is Localized in the Golgi Compartment, Related to Figure 4
Confocal images of human primary endothelial cells (HUAEC) transfected with control siRNA (siCTRL) and UBIAD1 siRNA (siUBIAD1) and stained for DNA (blue) and UBIAD1 (red). Compared to control, siUBIAD1-silenced cells lose the Golgi compartment fluorescence signal conforming the specificity of the 9D4 antibody and the Golgi-localization of UBIAD1. Scale bar, 20 μm.
Figure 4.
UBIAD1 Is a Prenyltransferase Responsible for CoQ10 Synthesis in the Golgi Compartment
(A and B) Confocal images of human ECs showing Ubiad1 colocalization with Golgi markers. (A) DNA (blue), human Ubiad1(red), and TGN46 (green). (B) DNA (blue), zebrafish Ubiad1-GFP (green), GM130 (red). Scale bar, 20 μm.
(C) High-magnification confocal images of Golgi compartment showing colocalization between Ubiad1-GFP (green) and the Golgi marker γ-adaptin (red). Scale bar, 10 μm.
(D) Fluorescence images of human ECs transfected with Ubiad1-GFP and treated or not with brefeldin A (+BFA). After BFA treatment, Ubiad1 localization becomes diffuse throughout the cell. Scale bar, 10 μm.
(E) Golgi compartment (Golgi, ER, and endosome fractions) and mitochondrial fractions from human ECs were analyzed by western blot with the following antibodies: γ-Adaptin and Rab11, as marker for Golgi/ER/endosome fractions, HADHA (Hydroxyacyl-CoA Dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA Hydratase [trifunctional protein], Alpha subunit) as marker for mitochondrial fractions.
(F) Levels of 13C6-isotope labeled CoQ10 (CoQ10-13C6) detected by HPLC-MS. Compared to control (siCTRL) the level of CoQ10-13C6 present in cells silenced for UBIAD1 (siUBIAD1) is significantly reduced in Golgi compartment but not in mitochondria. Data are means ± SD. ∗∗p < 0.01.
See also Figure S4.
We hypothesized that since UBIAD1 is localized in the Golgi compartment, this enzyme might be a nonmitochondrial CoQ10 prenyltransferase. To test this, we performed subcellular fractionation experiments on human ECs and detected UBIAD1 only in Golgi compartments and not in mitochondria (Figure 4E). We analyzed de novo CoQ10 synthesis in these subcellular fractions by incubating ECs with the hydroxy-4-benzoic acid-13C6 precursor and observed a decrease in CoQ10-13C6 production in Golgi compartments compared to mitochondrial fractions when UBIAD1 expression is reduced (Figure 4F). These data indicate that UBIAD1 is a CoQ10 biosynthetic enzyme located in the Golgi membrane compartment where large amounts of cellular CoQ10 are normally synthesized (Kalén et al., 1990; Swiezewska et al., 1993).
SCCD Mutations Protect Cardiovascular Tissues from Oxidative Stress by CoQ10 Production
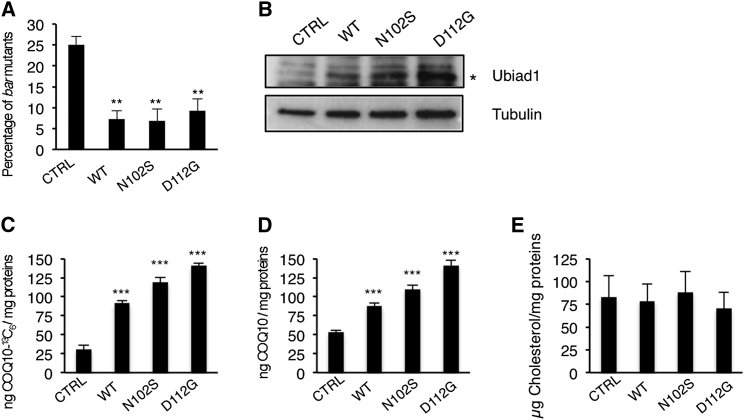
Mutations in the UBIAD1 gene are linked to the Schnyder Crystalline Corneal Syndrome (SCCD; OMIM 121800) (Weiss et al., 2007; Orr et al., 2007; Yellore et al., 2007). Two common UBIAD1 mutations found in SCCD patients lead to N102S and D112G substitutions. We generated human UBIAD1 constructs carrying these mutations (UBIAD1-N102S and UBIAD1-D112G) and found that they were able to rescue the loss of Ubiad1 during zebrafish development. We tested these variants for levels of expression and CoQ10 synthesis in human ECs. Compared to wild-type UBIAD1 protein, SCCD variants were expressed at higher levels (Figure 5B). Furthermore, human ECs expressing the UBIAD-SCCD mutant proteins produced higher CoQ10 levels compared to wild-type protein (Figures 6C and 6D). Intriguingly, SCCD mutations did not affect cholesterol synthesis in human ECs (Figure 5E). Although it remains unclear why SCCD mutation would results in the SCCD phenotype, these findings suggest that the SCCD variants are effective in CoQ10 production.
Figure 5.
SCCD UBIAD1 Variants Rescue barolo Mutants and Positively Regulate CoQ10 Synthesis
(A) wild-type UBIAD1 (WT) or SCCD UBIAD1 isoforms (N102S; D112G) rescue bar mutants at a significant extent.
(B) Western blot analysis of protein extracts from HUAEC transfected with plasmids encoding for wild-type UBIAD1 (WT) or SCCD UBIAD1 isoforms (N102S; D112G).
(C–E) HUAEC were transfected with plasmids encoding for wild-type UBIAD1 (WT) or SCCD UBIAD1 isoforms (N102S; D112G) and total lipid extracts were quantitated for CoQ10-13C6 (C) and CoQ10 by HPLC-MS (D) and for cholesterol by HPLC-UV (E). Expression of all UBIAD1 proteins rises levels of CoQ10 as well as de novo synthetized CoQ10-13C but does not change cholesterol levels in human ECs.
All data are means ± SEM. ∗∗p < 0.01, ∗∗∗p < 0.001.
Figure 6.
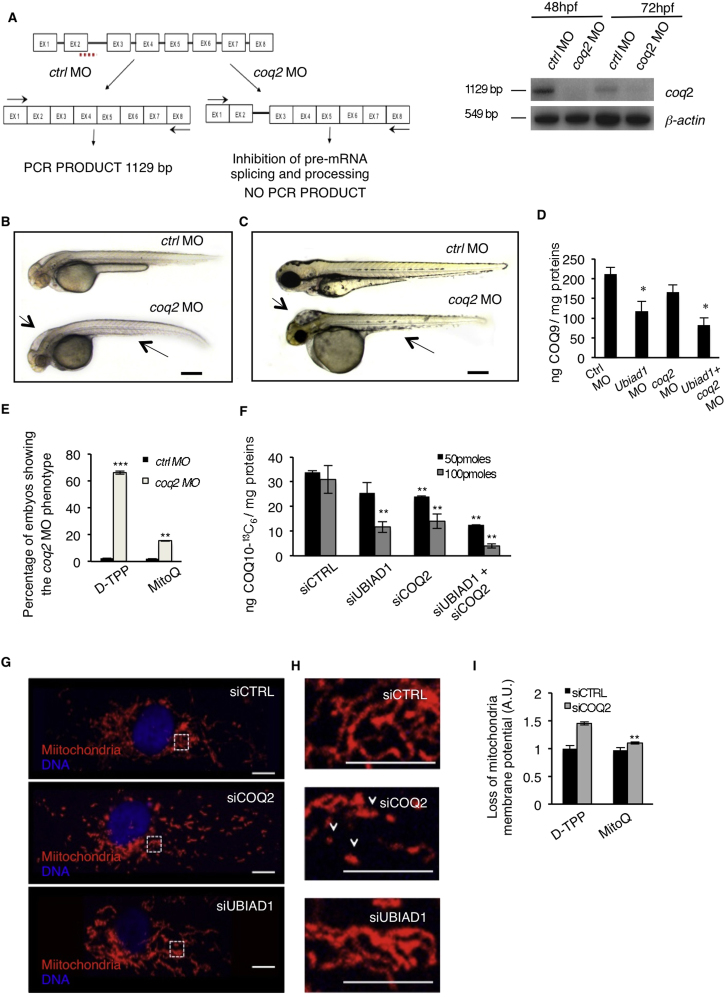
Ubiad1 and Coq2 Play Different Functions during Development
(A) Se integrity is affected in Tg(kdrl:GFP)s843 embryos injected with ubiad1 morpholino (ubiad1 MO) but not in coq2 morphants (coq2 MO). Scale bar, 100 μm.
(B) Oxidative stress levels in entire ubiad1 morphants (ubiad1 MO), coq2 morphants (coq2 MO), and relative control (ctrl MO).
(C) CoQ10 levels detected by HPLC-MS are significantly lower in ubiad1 morphants (ubiad1 MO), coq2 morphants (coq2 MO), ubiad1 and coq2 double morphants (ubiad1+ coq2 MO) than control morphants (ctrl MO).
(D) Embryos from bar heterozygote intercrosses were injected with coq2 mRNA and mitochondria-targeted CoQ10 analog called MitoQ (10 μM) or cherry mRNA and Decyl-TPP (10 μM) as respective controls. Phenotypes were scored at 72 hpf. bar phenotype is not caused by lack of mitochondria CoQ10.
(E) Silencing of both CoQ10 biosynthetic enzymes (siUBIAD1 and siCOQ2) cause increase of total cellular ROS in ECs.
(F) Levels of CoQ10 detected by HPLC-MS in total lipid extracts from HUAEC silenced for UBIAD1 (siUBIAD1), COQ2 (siCOQ2), or both (siUBIAD1+siCOQ2). UBIAD1 and COQ2 were silenced with 50 pmoles (black bars) or with 100 pmoles (gray bars). Levels of CoQ10 are significantly reduced in cells silenced for UBIAD1 or COQ2 expression.
(G) Quantitative PCR analyses of UBIAD1 and COQ2 mRNA levels in HUAEC cells.
(H) Quantification of loss of mitochondrial membrane potential (Δψm) in HUAEC silenced for UBIAD1 (siUBIAD1) or COQ2 (siCOQ2).
(I) Quantification of lipid peroxidation by MDA adducts levels in HUAEC silenced for UBIAD1 (siUBIAD1) or COQ2(siCOQ2).
All data are means ± SD. ∗∗p < 0.01, ∗∗∗p < 0.001.
See also Figure S5.
Ubiad1 and Coq2 Play Different Roles in CoQ10-Mediated Cardiovascular Oxidative Protection
CoQ10 is a mobile lipophilic electron carrier critical for electron transfer by the mitochondrial inner membrane respiratory chain (Mitchell, 1961; Turunen et al., 2004; Duncan et al., 2009). Primary CoQ10 deficiencies have been associated with mutations in mitochondrial CoQ10 biosynthetic genes, such as COQ2 (Salviati et al., 2005; Quinzii et al., 2006). In order to investigate whether the Golgi-localized Ubiad1 and the mitochondrial-localized Coq2 can have different functions in vivo, we knocked-down Coq2 expression during zebrafish development by morpholino (MO) injections. Similar to Drosophila sbo/coq2 mutants (Liu et al., 2011), coq2 morphants are characterized by a general developmental delay, small body, and severe hindbrain edema (Figures S5A–S5C). These phenotypes do not occur in bar mutants or ubiad1 morphants, and, although a strong increase in oxidative stress occurs in coq2 morphants (Figure 6B), they did not show blood vessel regression (Figure 6A). Therefore, loss of Coq2 leads to oxidative stress, but it does not significantly affect vascular integrity and survival. We measured CoQ9 and CoQ10 levels, and as expected, they dropped in coq2 and ubiad1 morphants, compared to controls, and in double morphants (Figures 6C and S5D). Loss of Ubiad1 and Coq2 in vivo unequivocally causes a decrease in CoQ10 levels that possibly increases oxidative damage in different subcellular compartments leading to different embryonic phenotypes.
Figure S5.
UBIAD1 Functions Differently from Mitochondrial COQ2, Related to Figure 6
(A) Validation of in vivo coq2 knockdown by morpholino injection. Left: schematic diagram shows the molecular mechanism of coq2 splice- morpholino (coq2 MO; dashed line). coq2 MO targeting the splicing site between exon2 (EX2) and intron2 (IN2) blocks coq2 pre-mRNA maturation and leads to coq2 non-coding mRNA. Molecular mechanism of control non-targeting morpholino (ctrl MO) is also indicated. Primers used for RT-PCR validation are localized on exons 1 and exon 8 (EX1, EX8; black arrows). RT-PCR analysis was performed on cDNA from zebrafish embryos at 48h and 72 hr post microinjection. Amplification of 1129bp is possible only for cDNA from ctrl MO-injected embryos, whereas no amplification is expected from coq2 MO-injected embryos. Compared to control morphants, coq2 MO show reduced PCR products.
Representative bright field pictures of coq2 morphants (coq2 MO) and controls (ctrl MO) at 48 hpf (B) and 72 hpf (C). At 48 hpf coq2 MO show hindbrain edema (arrow), and abnormal yolk sac morphology. Scale bar, 500 μm. At 72 hpf coq2 morphants show severe brain edema (arrow) and severe development delayed. These are typical features of reduced energy metabolism possibly due to primary CoQ10 deficiency. Scale bar, 300 μm.
(D) Histograms show levels of endogenous CoQ9 detected by HPLC-MS analyses in ubiad1 morphants (ubiad1 MO), coq2 morphants (coq2 MO), double morphants (ubiad1+ coq2 MO). Compared to control (ctrl MO), the CoQ9 levels are significantly lower in ubiad1 morphants. coq2 morphants show a reduced decreased in CoQ9 levels compared to ubiad morphants. Such difference is under investigation. Results are shown as a mean of n = 3 independent experiments for each condition.
(E) Mitochondria-targeted CoQ10 (MitoQ) efficiently rescue coq2 morphants. Histogram shows the percentage of embryos showing the coq2 morpholino phenotype when injected with mitochondria-targeted CoQ10 (MitoQ; 10 μM) or its respective control (D-TPP). Supplementation of MitoQ but not of D-TPP clearly rescues the absence of coq2 in zebrafish embryos. Results are shown as a mean of n = 3 independent experiments for each condition.
(F) Histograms show levels of 13C6-labeled CoQ10 (CoQ10-13C6) detected by HPLC-MS in total lipid extracts from human primary endothelial cells (HUAEC) silenced for UBIAD1 (siUBIAD1), COQ2 (siCOQ2) or both (siUBIAD1+siCOQ2). UBIAD1 and COQ2 were silenced with 50 pmoles (black bars) or with 100 pmoles (gray bars) of respective siRNA. Compared to relative control (siCTRL), the levels of CoQ10-13C6 are significantly reduced in cells silenced for UBIAD1 or COQ2 expression. The combination of the two siRNA further decrease CoQ10 levels. Results are shown as a mean of n = 3 independent experiments for each condition.
(G) Confocal images of human primary endothelial cells (HUAEC) transfected with siRNA for UBIAD1 (siUBIAD1), COQ2 (siCOQ2) or control (siCTRL) and stained for mitochondria (red) and DNA (blue). Regular reticular mitochondrial morphology is detected in cells treated with siUBIAD1 and siCTRL. On the contrary, cells treated with siCOQ2 show less and fragmented mitochondria. Scale bar, 10 μm.
(H) Magnification of the area outlined in (G). siCOQ2-treated cells show few and collapsed mitochondria (arrowhead). In contrast normal reticular mitochondria morphology is evident in siUBIAD1- and siCTRL-treated cells. Scale bar, 0.5 μm.
(I) Histograms show the loss of mitochondria membrane potential (Δψm) of human EC silenced for COQ2 after treatment with the mitochondria-targeted CoQ10 (MitoQ) efficiently rescue coq2 morphants. MitoQ (10nM) but not D-TPP (10nM) supplementation efficiently rescues loss of mitochondrial membrane potential of siCOQ2 ECs. These results indicate that mitochondrial damage in siCOQ2 cells is due to loss of mitochondrial CoQ10. Results are shown as a mean of n = 3 independent experiments for each condition.
All data are means ± SD, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
To test whether Coq2 might compensate for loss of Ubiad1, we performed rescue experiments by injecting mRNA encoding for Coq2 in bar mutants (Figure 6D). Although coq2 mRNA injection could rescue coq2 morphants (data not shown) it did not rescue bar mutants, suggesting that Coq2 does not protect cardiovascular tissues from oxidative stress during zebrafish development. MitoQ is a mitochondria-targeted antioxidant (Murphy and Smith, 2007). Due to its selective accumulation within mitochondria, MitoQ only protects against the mitochondrial oxidative damage that is caused in embryos and cells by blocking mitochondrial CoQ10 synthesis through knocking down Coq2 (Figure S5E). However, MitoQ (which will not interact with the Golgi apparatus) did not rescue bar mutants (Figure 6D), indicating that the two different subcellular pools of CoQ10 contribute in quite distinct ways to the phenotype.
To demonstrate the existence of a functional difference between the subcellular pools of CoQ10, we used human primary ECs where we can silence UBIAD1 and COQ2 genes at the same extent. Their silencing causes an increase in oxidative stress and accumulation of oxidative damage (Figure 6E). To analyze the efficiency of UBIAD1 and COQ2 in total CoQ10 biosynthesis, we performed labeled precursor experiments where 4-hydroxybenzoic acid-13C6 was added to human ECs previously transfected with siRNA for UBIAD1 (siUBIAD1) or for COQ2 (siCOQ2) or both (Figure S5F). Silencing of UBIAD1 or COQ2 individually or together significantly reduced CoQ10-13C6 production. We also confirmed the efficiency of UBIAD1 and COQ2 siRNAs in decreasing endogenous total CoQ10 levels in ECs (Figure 6F). Interestingly, the silencing of the UBIAD1 gene did not significantly interfere with the transcriptional regulation of COQ2 and vice versa, indicating that the effect on CoQ10 levels from the silencing of one gene is not be altered by upregulation of the other (Figure 6G). We then analyzed mitochondrial morphology and viability in ECs that have been silenced for UBIAD1 or COQ2. The absence of COQ2 significantly altered mitochondrial morphology (Figures S5G and S5H) and mitochondrial membrane potential (Figure 6H), which can indeed be rescued by MitoQ (Figure S5I). Mitochondrial damage could not be detected in UBIAD1-silenced cells, indicating that the lack of the mitochondrial pool of CoQ10 in COQ2-silenced cells exclusively impaired mitochondrial function.
High levels of CoQ10 have been found in cellular membranes, including the Golgi and plasma membranes (Kalen et al., 1987; Turunen et al., 2004). The presence of CoQ10 in nonmitochondrial membranes has been associated with its role in protecting membrane lipids from peroxidation as well as from an extracellular-induced ceramide-dependent apoptotic pathway (Navas et al., 2007). We evaluated oxidative degradation of lipids by measuring lipid peroxidation in siUBIAD1-treated cells (Figure 6I). ECs without a cellular membrane pool of CoQ10 showed twice the level of oxidized fatty acid lipids, indicating a functional role for UBIAD1 in protection from oxidative species specifically localized in cellular but not mitochondrial membrane. We conclude that UBIAD1-derived CoQ10 prevents lipid peroxidation and propose a specific role for UBIAD1 in membrane protection from oxidative damage.
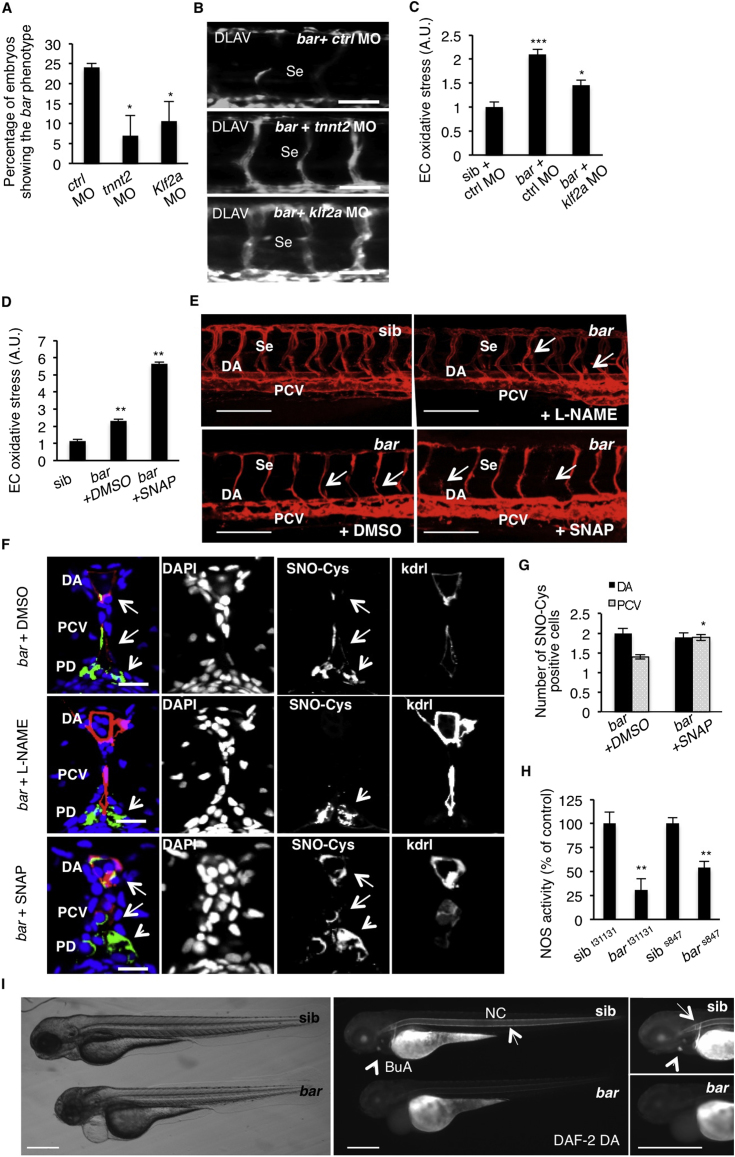
UBIAD1 Prevents a Shear Stress/Klf2a/eNOS-Mediated Oxidative Stress Pathway
A hemodynamic environment, such as shear stress, is known to lead to ROS generation, which is responsible for vascular remodeling and angiogenesis (Matlung et al., 2009). Kruppel-like factor 2 (Klf2) is an immediate mediator of hemodynamic forces created by blood flow and regulates flow-dependent blood vessel integrity both in zebrafish and mice (Lee et al., 2006; Nicoli et al., 2010). By injecting cardiac troponin T2 (tnnt2) morpholino to block blood circulation (Sehnert et al., 2002) and klf2a morpholino (Nicoli et al., 2010) in bar mutants, we detected a significant delay of the bar phenotype as assessed by intersegmental vessels (Se) disintegration (Figures S6A and S6B). klf2a morpholino injection in barolo mutants lead to a partial recovery of oxidative stress in ECs (Figure S6C). These data indicate that a Klf2a-dependent shear stress pathway is required for oxidative stress conditions in bar mutants and that loss of Ubiad1 in vivo can be protected by impairment of this pathway.
Figure S6.
UBIAD1 Regulates a Blood Flow-Dependent NO Signaling and NO-dependent Oxidative Stress, Related to Figure 7
(A) Embryos from bart31131 heterozygote intercrosses were injected at one-cell stage with tnnt2 morpholino (tnnt2 MO) or Kruppel-like factor 2a morpholino (klf2a MO) or control morpholino (ctrl MO). Histogram shows the percentage of larvae showing the bar mutant phenotype at 72 hpf after injection. Compared to control, impairment of the blood flow-Klf2a pathway by morpholino injection significantly delayed the penetrance of the bar phenotype, evaluated as trunk vessels disintegration. Results are shown as a mean of n = 2 independent experiments for each condition.
(B) Fluorescent micrographs of DLAV and Se of Tg(kdrl:GFP)s843bar mutants at 72 hpf after injection of control morpholino (ctrl MO), tnnt2 morpholino (tnnt2 MO), klf2a morpholino (klf2a MO). Knockdown of tnnt2 and klf2a delayed Se and DLAV disintegration in bar mutants. Se, intersegmental vessels; DLAV: dorsal longitudinal anastomotic vessel. Scale bar, 75 μm.
(C) Histograms show oxidative stress levels in endothelial cells (EC) derived from Tg(kdrl:GFP)s843bart31131 after klf2a morpholino injection. barolo mutants were injected with control morpholino (ctrl MO) or klf2a morpholino (klf2a MO). Compared to control-injected embryos, klf2a MO-injected barolo (bar) mutants show reduced EC oxidative stress levels. Results are shown as a mean of n = 2 independent experiments for each condition.
(D) Histograms show oxidative stress levels in endothelial cells derived from Tg(kdrl:GFP)s843 and bart31131 (bar) mutant at 65 hpf after treatment with the NO donor, S-nitroso-N-acetyl-DL-penicillamine (SNAP) or DMSO as control. ROS levels in endothelial cells of zebrafish embryos were measured by flow cytometric analyses using the specific CellROX probe on Kdrl:GFP+ cells. NO overload in barolo cardiovascular tissues significantly enhances oxidative stress level. Results are shown as a mean of n = 2 independent experiments for each condition.
(E) Representative confocal 3D projections of trunk vasculature between 10th and 18th somites of Tg(Kdrl:mCherry)uto2barolos847(bar) mutant at 65 hpf. Embryos were treated from 48 hpf with the NO inhibitor, N- Nitro-L-arginine methyl ester hydrochloride (L-NAME; 500 μM) or NO donor, S-nitroso-N-acetyl-DL-penicillamine (SNAP; 100 μM) or equivalent volume of DMSO as control.
Compared to DMSO-treated embryos, L-NAME treatment efficiently prevents cardiovascular failure as indicated by intact dorsal aorta (DA), posterior cardinal vein (PCV) and intersegmental vessels integrity (Se) (arrows). On the other hand, SNAP treatment accelerates oxidative stress and cardiovascular failure in bar mutant embryos. Scale bar, 150 μM.
(F) Confocal transverse sections of Tg(kdrl:GFP)s843bart31131 trunk vasculature at the level of 10th somite and stained for S-nitroso-cysteine, a biomarker of oxidative damage at 65 hpf. Confocal acquisitions are showed as single channel images and relative merged image: DNA (DAPI, blue), S-nitroso-cysteine (SNO-Cys, red), endothelium (Kdrl; green). bart31131 were previously treated with the NO inhibitor, L-NAME (500 μM) or NO donor, SNAP (100 μM) or DMSO as relative control. Oxidative stress-positive cells are detectable in dorsal aorta (DA) and posterior cardinal vein (PCV) in bart31131 (arrows) and SNAP-treated bar embryos. L-NAME treatment blocks oxidative stress in cardiovascular cells but not in pronephros suggesting a eNOS-dependent mechanism in oxidative stress caused by the loss of Ubiad1. DA, dorsal aorta; PCV posterior cardinal vein. Scale bar, 20 μm.
(G) Histograms show numbers of endothelial cells in dorsal aorta (DA) and posterior caudal vein (PCV) positive for S-nitroso-cysteine (SNO-Cys) in bar mutant embryos treated with DMSO or the NO donor, SNAP (100 μM). Comparative confocal 3D projections were analyzed for SNO-Cys-positive endothelial cells in dorsal aorta and posterior cardinal vein. Results are shown as a mean of n = 3 independent experiments for each condition.
(H) Histograms show NOS activity in protein extracts from barolo mutants (bar) and siblings (sib) at 72 hpf. Evaluation of NOS activity was based on conversion of [3H]-L-arginine to [3H]-L-citrulline. bart31131 and bars843 mutants show significant reduction in NOS activity compared to controls. These results indicate that loss of Ubiad1 impairs NOS enzymatic activity and therefore NO production. Results are shown as a mean of n = 2 independent experiments for each condition.
(I) Representative micrographs of bar mutant embryos (bar) and siblings (sib) stained for nitric oxide (NO) with the green fluorescent probe 4,5 Diaminofluorescein Diacetate (DAF-2DA) at 72 hpf. NO production in the notochord (NC; arrows) and bulbus arteriousus (BuA; arrowheads) are evident in sibling but not in bar mutants. Scale bar, 300 μm. BuA, bulbus arteriosus; NC, notochord.All data are means ± SEM; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
In human endothelial cells KLF2 has been found to be directly responsible for eNOS expression and signaling (Searles, 2006). Recently, a blood-flow-dependent klf2a-NO signaling cascade has been identified in developing zebrafish embryos (Wang et al., 2011). Endothelial nos isoforms have been well characterized in zebrafish development (Fritsche et al., 2000; Pelster et al., 2005; North et al., 2009; Wang et al., 2011). Inhibition of nos1 (the zebrafish ortholog of mammalian endothelial Nos) expression significantly rescued the bar mutant phenotype as well as endothelial regression (Figures 7A and 7B). Since abrogation of nos1 expression may interfere with HSC development and blood flow we treated bar mutants at the onset of oxidative stress (32 hpf) with the selective inhibitor of eNOS activity, N-nitro-L-arginine methyl ester (L-NAME) (North et al., 2009). L-NAME treatment promoted a full morphological and functional cardiovascular recovery of a statistically significant number of bar mutants (Figures 7C and 7D and S6E). This recovery was carefully evaluated for ROS level and altered redox signaling (SNO-Cys staining) in ECs. Blocking of eNOS activity by L-NAME treatment significantly reduced oxidative stress in endothelial cells of bar embryos (Figure 7E) and restored normal redox signaling (Figures 7F and S6E). Pronephric duct cells affected by oxidative stress in bar mutants did not improve after L-NAME treatments (Figure S6F). We also evaluated the effect of the NO donor S-nitroso-N-acetyl-penicillamine (SNAP) on bar mutants. SNAP treatments did aggravate the bar phenotype by increasing oxidative species in these mutant embryos (Figures S6D–S6F). To test whether the source of ROS in the absence of UBIAD1 was caused by eNOS, we silenced eNOS in primary human ECs. ROS levels could be fully normalized by reducing eNOS expression in siUBIAD1-treated cells (Figure 7G).
Figure 7.
NOS Activity in Cardiovascular Tissues Is Regulated by UBIAD1 and CoQ10
(A) Knockdown of endothelial nitric oxide synthase 1 by morpholino (nos1 MO) reduces the penetrance of bar phenotype at 72 hpf.
(B) Knockdown of nos1 rescue endothelial integrity defects of bar mutants. Images of Se of bar at 72 hpf injected with nos1 morpholino (bar + nos1MO) or control morpholino (bar + ctrl MO) Scale bar, 50 μm.
(C) Bright-field images (left) and fluorescent micrographs showing the trunk vasculature (right) of Tg(kdrl:GFP)s843bar treated from 48 hpf with the specific eNOS inhibitor L-NAME (500 μM). Heart failure (arrowhead) and endothelial regression (arrows) were fully rescued by L-NAME treatment. Scale bar, 100 μm.
(D) Penetrance of the bar phenotype at 65 hpf is significantly decreased by inhibition of eNOS activity with L-NAME treatment from 32 hpf.
(E) Oxidative stress level in ECs derived from Tg(kdrl:GFP)s843bar mutant embryos is decreased by L-NAME treatment.
(F) eNOS inhibition by L-NAME treatment significantly reduces ECs positive for S-nitroso-cysteine (SNO-Cys) in DA and PCV of bar embryos.
(G) Silencing of eNOS (sieNOS) rescues oxidative stress induced by the lack of UBIAD1 (siUBIAD1). Silencing eNOS alone did not decrease ROS level in ECs.
(H) Silencing of UBIAD1 gene (siUBIAD1) causes a significant decrease of eNOS activity in ECs evaluated as [3H]-L-citrulline.
(I) Schematic representation of Ubiad1 molecular function in CoQ10 production and maintenance of nitrix oxide (NO) signaling. In wild-type cells, Ubiad1 localizes in the Golgi compartment and produces CoenzymeQ10 (CoQ10), an antioxidant molecule, important to counteract oxidative damage in particular in cellular membranes (cytosol and plasma membrane). In the Golgi compartment CoQ10 as an electron carrier might also play a fundamental role as a cofactor for eNOS activity by maintaining its “coupled” conformation and allowing normal NO production. On the other hand, lack of UBIAD1 and lowering of CoQ10, as occurs in the cardiovascular tissues of bar mutants, might “uncouple” eNOS causing loss of NO production and consequently reactive oxygen species overload leading to cellular oxidative damage (e.g., lipid peroxidation). Thereby, the “barolo” phenotypes can be rescued by impairment of eNOS activity or expression.
Data are means ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
See also Figure S6.
To verify that the NO-forming activity of eNOS was compromised due to the lack of Ubiad1 and Golgi-CoQ10, we measured NOS activity in bar mutants and human ECs silenced for UBIAD1. The lack of Ubiad1 drastically reduced the formation of NO in zebrafish embryos (Figure S6H) as well as in human ECs (Figure 7H).
We measured tissue-specific NO levels in bar mutants by imaging DAF-2DA staining in live embryos. NO accumulation was reduced in the notochord (NC) and was completely absent in the bulbus arteriosus (BuA) of bar mutants compared with controls (Figure S6I). All together we show a specific role for Ubiad1 on regulation of eNOS-mediated NO formation in endothelial and cardiovascular cells in vivo.
These data indicate that the primary source of ROS causing cardiovascular oxidative stress in bar mutant as well as in ECs lacking UBIAD1 is dependent on eNOS activity. Therefore, we propose an unexpected functional link in the Golgi compartments between UBIAD1 and COQ10 in regulating eNOS activity and eventually NO signaling during cardiovascular development and homeostasis.
Discussion
Ubiad1 Is a Prenyltranferase Enzyme Required for CoQ10 Biosynthesis in Golgi Membranes
CoQ10 is an important cellular and mitochondrial redox component and the only endogenously produced lipid-soluble antioxidant (Bentinger et al., 2010). Although the genes encoding for the CoQ10 biosynthetic enzymes have been identified in bacteria and yeast, there is still only limited information about these synthetic enzymes in vertebrates (Turunen et al., 2004). The rate-limiting enzyme for the biosynthesis of CoQ10 is the enzyme that catalyzes the condensation of the polyisoprenoid chain with the benzoquinone ring. So far, the mitochondrial COQ2 enzyme has been considered the only prenyltransferase able to catalyze this reaction (Trevisson et al., 2011). Here, we identified UBIAD1 as a vertebrate CoQ10 prenyltransferase. UBIAD1 contains an UbiA prenyltransferase domain also present in vertebrate COQ2. Although COQ2 encodes a mitochondrial prenyltransferase, we found that UBIAD1 resides in the Golgi compartment where it produces CoQ10. While the presence of CoQ10 in nonmitochondrial membranes was previously explained by the existence of specific mechanisms for its redistribution within the cell (Crane and Morre, 1977; Jonassen and Clarke, 2000), our data now formally demonstrate that CoQ10 are synthetized in the Golgi compartment.
In favor of our hypothesis of a Golgi-synthetized CoQ10, it has been recently reported that COQ6, COQ7, and COQ9, which are critical enzymes for CoQ10 maturation, are also localized in the Golgi compartment (Heeringa et al., 2011). Similarly, these authors suggested that the Golgi-localized pool of CoQ10 may function in specific cells as an essential antioxidant for plasma membrane lipids that are normally derived from the Golgi compartment.
In this work, we demonstrated that the two distinct ubiquinone prenyltransferases act to resolve different molecular functions inside the cells and in living organisms. We found that COQ2-mediated CoQ10 production is mainly for mitochondrial respiratory chain function and energy production, whereas UBIAD1-mediated CoQ10 production is important for membrane redox signaling and protection from lipid peroxidation.
Ubiad1 and CoQ10: An Antioxidant System Controlling Redox Cellular Membrane in Cardiovascular Tissues
Oxidative damage is caused by an imbalance between the production of ROS and the antioxidant network. Although ROS are predominantly implicated in causing cell damage and premature aging via oxidation of DNA, lipids, and proteins, they also play a major physiological role in several aspects of intracellular signaling and regulation, especially in cardiovascular tissues (Hare and Stamler, 2005; Kuster et al., 2010). Therefore, heart, endothelial, and vascular smooth-muscle cells need an efficient antioxidant network to balance ROS levels. We have shown a set of genetic and cellular data that reveal an unexpected role for UBIAD1 as an essential antioxidant gene with important functions in the protection of heart and endothelial cells from oxidative stress at the level of cellular membranes by producing CoQ10 in the Golgi for distribution to nonmitochondrial membranes throughout the cell. In addition to its crucial role in oxidative phosphorylation, CoQ10 plays another vital role in cellular function as an antioxidant molecule. The antioxidant nature of CoQ10 is derived from its function as an electron carrier: in this role, CoQ10 continually shuttles between oxidized and reduced forms. As it accepts electrons, it becomes reduced. As it gives up electrons, it becomes oxidized. In its reduced ubiquinol form, the CoQ10 molecule will quite easily give up one electron, and thus act as an antioxidant. In such a way, CoQ10 inhibits lipid peroxidation by acting as a chain breaking antioxidant. Moreover, CoQH2 reduces the initial lipid peroxyl radical, with concomitant formation of ubisemiquinone and an alkyl peroxide. This quenching of the initiating peroxyl radicals thereby prevents the propagation of lipid peroxidation and protects not only lipids, but also proteins from oxidation. In addition, the reduced form of CoQ10 might also contribute to the stabilization of the plasma membrane, regenerating antioxidants such as α-tocopherol. A crucial role in all these processes is played by NAD(P)H-dependent reductase(s) acting at the plasma membrane to regenerate the reduced ubiquinol form of CoQ10, contributing to the maintainance of its antioxidant properties (Navas et al., 2007). This finding also opens an interesting link among cellular redox-state and metabolic pathways such as the mevalonate pathway.
An Essential Function for Ubiad1/CoQ10 in Regulation of NO Signaling
We have demonstrated that UBIAD1 protects cardiovascular tissue from ROS-mediated oxidative stress by producing CoQ10 located in Golgi and plasma membranes. Major enzymatic pathways responsible for the generation of ROS in cardiovascular tissues are mainly NADPH oxidases and eNOS (Browning et al., 2012). By using drug inhibition and gene inactivation approaches, we identify eNOS dysfunction as the primary cause of ROS increase in bar mutant and UBIAD1-silenced human ECs. The NO synthetized by eNOS is an essential factor for cardiovascular development and homeostasis in vertebrates (Alp and Channon, 2004; Pelster et al., 2005; North et al., 2009; Wang et al., 2011). It has been suggested that CoQ10 might have a positive role in modulating NO-related pathways by recoupling eNOS in endothelial cells (Stuehr et al., 2001; Chew and Watts, 2004; Tsai et al., 2012). eNOS is a “L-arginine, NADPH:oxygen oxidoreductases, NO-forming enzyme” (EC 1.14.13.39) that couples reduction of molecular oxygen to L-arginine oxidation and generation of L-citrulline and NO. eNOS controls the flow of electrons donated by NADPH to flavins FAD and FMN in the reductase domain of one monomer through BH4 to the ferrous-dioxygen complex (Fe) in the oxygenase domain. When NADPH and BH4 cofactors are limiting, electron transfer becomes uncoupled from L-arginine oxidation, the ferrous–dioxygen complex dissociates, and superoxide (⋅O2−) is generated from the oxygenase domain (Schmidt and Alp, 2007).
Our data indicate that barolo mutants lacking the Golgi pool of CoQ10 die due to the accumulation of oxidative damage in cardiovascular tissues caused by ROS produced by eNOS. We suggest a mechanism whereby UBIAD1 is required in the Golgi compartment to produce CoQ10 as an important cofactor for eNOS-mediated NO production (Figure 7I). This model could fit with the oxido-reductive properties of CoQ10 and eNOS: the electron flux inside eNOS that is mandatory to produce NO from L-arginine might require CoQ10, together with NADPH and BH4. Such function would not be very different from what CoQ10 does in the mitochondrial electron transport chain (ETC) coupling electron transfer between an electron donor (such as NADH) and an electron acceptor (such as O2) with the transfer of H+ ions across the mitochondrial inner membrane. In the Golgi membranes, the flow of electrons within eNOS transfers electrons from NADPH to the flavins FAD and FMN, which have the capacity to reduce molecular oxygen. If the ETC is disturbed, such as in the absence of UBIAD1/CoQ10, the ferrous-dioxygen complex dissociates, and O−2 is generated from the FMN oxygenase domain (Liu et al., 2002; Rigoulet et al., 2011). In cardiovascular tissues, the CoQ10 produced by UBIAD1 in the Golgi compartment may be an important cofactor to maintain eNOS in a coupled conformation (e.g., required to produce physiological NO) and, eventually, quench leaking-uncoupled electrons. We speculate that when UBIAD1/CoQ10 in the Golgi compartment is absent or decreased (bar mutants and siUBIAD1-treated cells) eNOS switches to an “uncoupled” conformation, producing oxidative species instead of NO and causing cardiovascular oxidative damage (Figure 7I). Further experiments need to be done to assess this hypothesis as well as the possibility that other NOS (e.g., nNOS) could be affected by the lack of Ubiad1 in vivo.
In summary, our study places UBIAD1 in a pathway with important therapeutic implications for cardiovascular failure, such as the opportunity to decrease oxidative damage and counteract some of the side effects of statins. In addition, pharmacological or genetic stimulation of UBIAD1, as a CoQ10 biosynthetic enzyme, represents a promising therapeutic approach for antioxidant-related diseases such as aging and cancer.
Experimental Procedures
Zebrafish Strains, Mapping, Genotyping
Embryos and adult fishes were raised and maintained under standard laboratory conditions. bars847 and bart31131 mutants were generated by ENU mutagenesis as previously described (Jin et al., 2007).
Oxidative Stress Analyses in Zebrafish Embryos and Cells
Oxidative damage was detected by immunofluorescence on agarose sections of zebrafish embryos with the following markers: anti-8-hydroxy-2′-deoxyguanosine (1:500, Abcam) and anti-S-nitroso-cysteine (1:250, Sigma).
ROS levels were quantitated in isolated human (HUAEC) and zebrafish (Tg(Kdrl:GFP)s843) endothelial cells with CellROX Deep Red Reagent (Invitrogen; 2.5 μM) and analyzed by FACS.
Generation of Human SCCD Mutation in UBIAD1
Constructs containing the SCCD mutation N102S and D112G were introduced in pCS2+ plasmids and used for expression in zebrafish embryos as well as human ECs.
NO Levels and NOS Activity in Zebrafish Embryos and Human ECs
NOS activity was assayed using the NOS Activity Assay Kit (Cayman Chemicals). NO levels were detected using the DAF-2 DA (Calbiochem).
Cell Culture Experiments
Human primary EC were purchased from Lonza and cultured according to the manufacturer’s protocols (Lonza). Cells were electroporated with siRNA (Dharmacon).
HPLC Analyses
Samples for HPLC-MS analysis were zebrafish embryos (n = 25) or human HUAEC (2 × 106 cells). CoQ10 and CoQ9 levels were detected by HPLC-MS or HPLC-UV (as indicated). Total cholesterol from zebrafish embryos was analyzed by HPLC-UV. 4-Hydroxy-benzoic acid-13C6 was stocked in Dulbecco's modified Eagle's medium (DMSO) and dissolved in cell culture medium (1 mM) or microinjected in zebrafish embryo at one cell stage (2 mM). ECs or embryos were extracted after 48 or 72 hr, respectively, after labeled precursor administration. Extracts were quantified by HPLC-MS as described.
Exogenous CoQ10 and Vitamin K Administration
All chemicals and solvents for liposomal CoQ10 and Vitamin K formulation were purchased from Sigma Aldrich and Avanti Polar Lipids. Liposomal SINAMIT was a gift from Dr. Franz Enzmann (MSE Pharmazeutika GmbHT), liposomal LiQsorb was from Tishcon Corp. MitoQ was synthetized as previously reported (Murphy and Smith, 2007). All CoQ10 and vitamin K formulations were tested for toxicity and microinjected at specific concentrations into zebrafish embryos at one cell stage.
For detailed procedure of refer to Extended Experimental Procedures.
Extended Experimental Procedures.
Zebrafish Strains, Mapping, and Genotyping
Following zebrafish lines were used for these studies: bars847, bart31131, wild-type AB/TAB, Tg (kdrl:GFP)s843, Tg (fli1a:EGFP)y1, Tg(Kdrl:mCherry)uto2
Bulked segregate analysis was performed as previously described (Michelmore et al., 1991). Fine mapping of the t31131 mutation was performed with marker z22307 close to the mutation on contig: Zv8_scaffold 1100 of the zebrafish genome. Sequencing the coding region of the ubiad1 gene (ubiad1, LOC554810) revealed a t > a mutation at nucleotide 123 in bart31131 and a t > a mutation at nucleotide position 185 in bars847.
Genotyping were performed by two sequential PCR, which products were analyzed by sequencing. We used following primers: first PCR: FW: 5′-CCTGTGTGTGTTGTGATCG-3′; RW: 5′-TAGGTGTTGACCAGGTTTCC-3′, second PCR: FW 5′-TGTAAAACGACGGCCAGTCTGGATGCAGGAGATGAAG-3′; RW: 5′- AGGAAACAGCTATGACCATCCAGCTTGTAGGCCAGAG-3′.
Morpholino Microinjections
Morpholinos or mRNAs were injected at one cell-stage in different strains and phenotype was assayed between 48–72 hpf as described. Gene knockdown in zebrafish embryos was performed by microinjection of the following morpholinos: Control (5′-CCTCTTACCTCAGTTACAATTTATA-3′; 0.2 mM), ubiad1 (5′- GAAGCCAATCGGTATATTCACCTCC-3′; 0.2 mM), tnnt2a (5′- CATGTTTGCTCTGATCTGACACGCA-3′; 0.2 mM; Sehnert et al., 2002), klf2a (5′-GGACCTGTCCAGTTCATCCTTCCAC-3′; 0.2 mM; Nicoli et al., 2010), coq2 (5′-GTGTGAAATACAGAAAGCTCACCTA-3′; 0.2 mM), nos1 (5′- TTAATGACATCCCTCACCTCTCCAC-3′; 0.2 mM; North et al., 2009). Morpholinos were synthetized from GeneTools and dissolved in nuclease-free water. Primers for testing ubiad1 morpholino were designed on zebrafish ubiad1 (GeneBank accession number: NM_001199726) and are FW: 5′- CCGCAGGACGTGGTGATGTTTG-3′; REW: 5′- GTCTGAGTCCATGTCCCGCG-3′). Primers for testing coq2 morpholino were designed on zebrafish coq2 (GeneBank accession number: NM_001089486) and are: FW: 5′-TTCAAAATATGTCACGGTGCT-3′; RW: 5′-GTCCTCGGGTCTGTTGATGT-3′. As control β-actin was detected with following primers: FW: 5′- GTATCCACGAGACCACCTTCA-3′; RW: 5′-GAGGAGGGCAAAGTGGTAAAC-3′.
mRNA Microinjection
Embryos from bar heterozygote intercrosses were injected at one-cell stage with 80pg of mRNA (mMessage Machine, Ambion) encoding for the indicated proteins. We also included a control mRNA for fluorescent protein H2B-cherry (50pg) in each injection.
Transplantation Experiments
Transplantation experiments were performed as previously described (Ho and Kane, 1990). Donor embryos of the genotype Tg(fli1a:GFP)y1 were injected with 70 kDa Tetramethyl Rhodamine (TAMRA; Molecular Probes) at the one-cell stage and used for transplantations at the predome stage. Acceptor embryos were from wild-type or Tg(Kdrl:mCherry)uto2 lines as specifically indicated.
Generation of the Zebrafish Transgenic Line Tg(kdrl:Ubiad1-2A-mCherry)uto 35 and Analysis of Ubiad1 Endothelial Cell Autonomous Effect
The Tol2-based kdrl:Ubiad1-2A-mCherry construct has been assembled by using the Tol2 Kit and three-fragment gateway recombination cloning strategy (Kwan et al., 2007). For 5′ entry cloning, the 6.4Kb Kdrl promoter was amplified from genomic DNA of wild-type zebrafish by PCR with following primers containing appropriate attB4 and attB1 sites (5′-GGGGACAACTTTGTATAGAAAAGTTGAAGCTTTTTCTTTTATTTAA-3′ and 5′-GGGGACTGCTTTTTTGTACAA ACTTGTTTGTCTGTTAAAATAACGT-3′, respectively). The PCR product was then cloned into pDONRP4-P1R by BP reaction to obtain p5E-Kdrl. For the middle entry cloning zebrafish ubiad1 was amplified with following primers containing appropriate attB1 and attB2 sites: 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTAGGAGATGAAGCCGGCTGCGCTTTC-3′ and 5′- GGGGACCACTTTGTACAAGAAAGCTGGGTAAGAAATTCACAATAACGGCAGGCT-3′, respectively) and cloned into pDONR221 by BP reaction. The 3′ entry clone was p3E-2A-mCherrypA. Entry vectors were assembled together with the pDestTol2pA2 by LR reaction to create the expression vector pDestTol2-Kdrl-Ubiad1-2A-mCherry-pA. The pDestTol2-Kdrl-Ubiad1-2A-mCherrypA was mixed with mRNA for Tol2 transposase and microinjected into one-cell stage wild-type embryos. Injected embryos were raised to adulthood and founders were screened for red fluorescence in the vasculature. The transgene integration was also confirmed by PCR on genomic DNA with forward primer on ubiad1 sequence (5′- TGCACAGCAACAACACGGGG −3′) and reverse primer on mCherry sequence (5′- CGAACTCGTGCCGTTCACGG −3′). As control β-actin gene was detected with following primers: FW: 5′- GTATCCACGAGACCACCTTCA-3′; RW: 5′-GAGGAGGGCAAAGTGGTAAAC-3′. The transgenic fish line nomenclatures uto35 was approved by the Zebrafish Nomenclature Committee of ZFIN (http://zfin.org). Microinjection of control and ubiad1 was performed as previously described in intercrossed from Tg(Kdrl:GFP)s843 and Tg(kdrl:Ubiad1-2A-cherry)uto 35 lines. After MO injection, double Tg[(Kdrl: GFP)s843:(kdrl:Ubiad1-2A-cherry)uto 35] or single Tg(Kdrl: GFP)s843 were scored for embryos showing the barolo phenotype as measurement of ubiad1 morpholino penetrance.
Whole-Mount In Situ Hybridization
Whole mount in situ hybridization was performed as previously described (Santoro et al., 2009). The following oligonucleotides were used for antisense digoxigenin labeled probe: FW: 5′-CCGCAGGACGTGGTGATGTTTG-3′, RW: 5′- CGCGGGACATGGACTCAGAC-3′.
TUNEL Assay
The TUNEL cell death assay was carried out using the “In Situ Cell Death Detection Kit” TMR Red (Roche) as previously reported (Santoro et al., 2007). Zebrafish sections were analyzed by confocal microscopy. Acquisitions were quantitated with appropriate image processing tools as described in the “Image acquisition section.”
Western Blot Analyses on Zebrafish Embryos
Zebrafish embryos were collected and deyolked in a solution of PBS1X supplemented with complete protease inhibitors cocktail (Roche). For Ubiad1 detection, embryos were lysed in RIPA buffer. 50 μg of total extracts were not boiled and loaded on 12% SDS-PAGE denaturing gel. Western blot analyses were performed with mouse anti-UBIAD1 (9D4; 1:500).
Extraction of Single Cells from Zebrafish Embryos for Quantitation of Cellular ROS Level
Tg (kdrl:GFP)s843 embryos were dissociated into single cells by following procedure (modified from Covassin et al., 2006). Embryos (n = 20) were collected and deyolked with forceps in sterile PBS supplemented with complete protease inhibitor cocktail (Roche). Embryos were then transferred into a 24 wells plate with 500 μl of Protease solution (0.25% trypsin, 1 mM EDTA, pH 8.0, sterile PBS) and 30 μl of Collagenase P (100 mg/ml, Roche). Embryos were incubated at 28°C for 30’ and homogenized every 10’with a 200 μl pipette tip. Lysis was blocked with 500 μl of FBS (Invitrogen) and cells were collected into a prechilled Eppendorf tubes and centrifugated at 450rcf / 4°C for 5′. Supernatant was removed and cells were washed with 1 ml of HBSS (GIBCO). Cells were then incubated with 1 ml of cold HBSS containing CellROX probe for 10’ and analyzed at appropriate fluorescence wavelength on FACSCalibur (Becton Dickinson) according to manufacture’s protocol. All procedures including the CellROX probe were carried out on dark.
Analysis of NADP/NADPH Ratio in Zebrafish Embryos
The NADP/NADPH ratio assay was performed on zebrafish tissue extracts using the NADP/NADPH assay kit (Abcam). Samples were extracted in 400 μl of the recommended extraction buffer and 50 μl were processed following instructions. OD450 measurements were made on a Glomax Multi Detection System (Promega) plate-reader. OD450 measurements were converted to nmol/sample using a standard curve and values were used for ratio as previously reported (O’Neill and Reddy, 2011).
Chemical Treatments
Chemicals for zebrafish treatments were dissolved in sterile water or dimethyl sulfoxide (DMSO). Zebrafish embryos were treated with the following drugs: Statins (Mevastatin, Simvastatin and Mevinolin, 5 or 500nM; Sigma), from 48 to 72 hpf or as otherwise indicated; Squalene Inhibitor (SQI, 10 μM; Calbiochem) from 32 hpf to 72 hpf, GGTI-2133 and FTI-277 (10–50 μM, Sigma) from 54 hpf to 72 hpf; Nω-Nitro-L-arginine methyl ester hydrochloride (L-NAME) (500 μM, Sigma) from 32 to 65 hpf or as otherwise indicated, S-nitroso-N-acetyl-DL-penicillamine (SNAP) (100 μM, Sigma) from 48 hpf to 65 hpf.
Zebrafish and Human UBIAD1 Constructs
Zebrafish ubiad1 (GeneBank accession number: XM_681613.2) and human UBIAD1 (GeneBank accession number NM_013319.2) cDNAs were amplified using the following primers: FW: 5′-ATGAAGCCGGCTGCGCTTTC-3′; RW: 5′-TCACAATAACGGCAGGCT-3′; FW: 5′-ATGGCGGCCTCTCAGGTCCTG-3′; RW: 5′-TTAAATTTTGGGCAGACTGCCTGCTGG-3′ respectively and cloned in PCS2+ vector. All constructs were then amplified and subcloned into pGEM T-easy vector with following primers: human UBIAD1 forms:
FW: 5′-AAAGAATTCTCCATGGCGGCCTCTCAG-3′, RW: 5′-GAAAGATCTAATTTTGGGCAGACTGCC-3′, zebrafish ubiad1: FW: 5′ - AAAGAATTCGAGATGAAGCCGGCTGCG-3′, RW: 5′-TTTGGATCCCAATAACGGCAGGCTGCC-3′. Constructs were then cloned into pEGFP-N3 vector with restriction enzymes. All constructs were tested for comparable levels of expression and toxicity in cells.
Human SCCD Mutant UBIAD1 Constructs
Human SCCD mutant isoforms UBIAD1-N102S and UBIAD1-D112G were generated with QuickChange site-directed mutagenesis kit (Stratagene) with following primers: D112G FW: 5′- CTTTTCCAAGGGCATTGGCCACAAAAAGAGTGATG-3′, RW: 5′- CATCACTCTTTTTGTGGCCAATGCCCTTGGAAAAG-3′) and N102S: FW:5′-CGGGGCCGGTAATTTGGTCAGCACTTACTATGACTTTTCC-3′, RW:5′-GGAAAAGTCATAGTAAGTGCTGACCAAATTACCGGCCCCG-3′.
All constructs were then amplified and subcloned into pGEM T-easy vector with following primers: human UBIAD1 forms: FW: 5′-AAAGAATTCTCCATGGCGGCCTCTCAG-3′, RW: 5′-GAAAGATCTAATTTTGGGCAGACTGCC-3′, zebrafish ubiad1: FW: 5′ - AAAGAATTCGAGATGAAGCCGGCTGCG-3′, RW: 5′-TTTGGATCCCAATAACGGCAGGCTGCC-3′. Constructs were then sub-cloned into pCS2+ vector with specific restriction enzymes. All constructs were tested for comparable levels of expression and toxicity in cells.
Transfection of Human Primary Endothelial Cells
4.0 x105 human primary EC (P3-P4) were transfected with 50 or 100 pmoles of control-, UBIAD1-siRNA and COQ2-siRNA (siGENOME, Dharmacon) or 3 μg of expression constructs using Amaxa protocols (Nucleofector kit: VPB-1002, Nucleofector program: U-001). pCDNA-GFP (3 μg) was used as control of electroporation each time.
Subcellular Fractionation Experiments in Endothelial Cells
HUAEC (5 × 105 cells) were transfected with siRNA (60 pmoles) as previously stated. 2 × 107 cells were trypsinized and pelleted by centrifugation at 800rpm for 5′ and washed twice with cold PBS. Pellet was resuspended with mitochondria isolation reagent A (Mitochondria Isolation Kit Thermo Scientific), vortexed at 15Hz for 5 s and incubated on ice for 5 min. Cell suspension was then transferred to a prechill Dounce Grinder (Sigma) and homogenized on ice (30 strokes/3 times). Lysed cells were transferred to a new tube and mixed with 800 μl of mitochondria isolation reagent C (Mitochondria Isolation Kit; Thermo Scientific). Cell lysate was centrifuged at 700 g for 10 min at 4°C to separate membrane fractions (pellet) from mitochondria and cytosol (supernatant). Membrane fraction was then suspended in reagent A and centrifuged at 500 g for 10 min at 4°C (Lindner, 2001). Pellet containing Golgi/ER/endosome fractions were then subjected to protein or lipid extractions as described. Supernatant containing mitochondria and cytosol fractions was transferred to a new tube and centrifuged at 3,000 g for 15 min at 4°C. The upper phase was discarded (cytosol) and mitochondria pellet was washed twice with 500 μl reagent C and centrifuged at 12,000 g for 5 min at 4°C (Rice and Lindsay, 1997). Pellet containing mitochondria was subjected to protein or lipid extractions as described.
Western Blot Analyses on Human Endothelial Cells
For western blot analysis whole-cell extracts or subcellular fractions were lysed in RIPA Buffer and different amount of total protein or fractions were loaded on 4%–15% SDS-PAGE denaturing gel under reducing conditions. For Ubiad1 analyses samples were not boiled before loading on gel. Subsequent procedures were performed as described in Santoro et al., 2007. Western blot analyses were performed with following primary antibodies: mouse anti-UBIAD1 (9D4; 1:500), goat anti TERE1 (UBIAD1)-N16 (1:250; SantaCruz), mouse anti βactin (1:5,000; Sigma), mouse anti Rab11 (1:1,000; BD Biosciences), rabbit anti HADHA (Hydroxyacyl-CoA Dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA Hydratase (trifunctional protein), Alpha subunit) (1:5,000; Abcam).
UBIAD1 Monoclonal Antibody Production
Monoclonal antibody against UBIAD1 (clone 9D4) was produced in our laboratory by immunizing mice with recombinant GST-Ubiad1 (aa 96-126) fusion protein and its reactivity was characterized in ELISA, completion with peptide, western blot and immunofluorescence as described in the text.
UBIAD1 Localization in Endothelial Cells
For UBIAD1 localization studies human primary endothelial cells were transfected (where specified) and plated on fibronectin-coated glass slides. After 48 hr cells were fixed in 4% paraformaldehyde and incubated with the following primary antibodies: mouse anti UBIAD1 (1:5, 9D4), rabbit anti TGN46 (1:25, Abcam), mouse anti γ−Adaptin (1:100; BD Biosciences), mouse anti GM130 (1:100, BD Biosciences). Secondary antibodies were from Alexa Fluor (Invitrogen). Nuclei were stained with Hoechst. Localization studies were confirmed by treatment of cells with Brefeldin A (Sigma, 36μM) for 3h in cell culture medium.
Mitochondria Staining in Cultured Endothelial Cells
Mitochondria morphology was detected on human primary ECs by immunofluorescence with mouse anti-60 kDa mitochondrial protein antibody (1:100; Abcam). Hoechst was used for nuclei staining. ECs were then analyzed at confocal microscopy.
HPLC Analyses
Samples (cells or zebrafish embryos) were lysed in 500μl water-ethanol 2:3 and a 5μl lysate aliquot was analyzed for protein content according to the Bradford assay. Lysates were extracted twice with 500μl of hexane and the resulting organic phase was evaporated dry under reduced pressure. The residue was dissolved in 200 μl of methanol, sonicated and lastly centrifuged (12,000 rpm, 5 min). The upper layer was then immediately subjected to HPLC analyses. A standard amount of CoQ6 (Spectra2000) was added to the cell or embryo lysates as an internal standard for the assessment of the recovery percentage. Quantitation of CoQ10, CoQ9, cholesterol, Vitamin K and CoQ10-13C6, CoQ9-13C6 analytes was done by external calibration curves and corrected to % recovery evaluated on CoQ6.
HPLC-UV analyses for CoQ10 and cholesterol from zebrafish embryo extracts were performed on a Waters Alliance System (separations module: 2695 model) equipped with a photodiode array detector (model 2998) and a Waters Symmetry C18 reverse phase column (4.6 mm × 75 mm, 3.5 μm); pure methanol was used for the separation of lipid extracts (flow rate: 1 ml/min; run time: 45 min). HPLC-MS analyses for CoQ10 and cholesterol from zebrafish embryo extracts were performed on a liquid-chromatography/electrospray ionization tandem mass spectrometry system (HPLC-ESI-MS, Waters 515 HPLC pump-3100 mass detector); 5 mM of ammonium formate was added to the eluent (100% methanol) to promote the ionization of the analytes (Ruiz-Jiménez et al., 2007).
HPLC MS/MS analyses for Vitamin K2 from zebrafish embryo and cell extracts were done on API3200 HPLC-MS system equipped with APCI ion source (Applied Biosystems). Waters Symmetry C18 reverse phase column (4.6 mm × 75 mm, 3.5 μm particle size) was used with 100% methanol as the mobile phase (isocratic conditions, time course 60 min). Acquisitions were recorded in TIC (Total Ion Current) and SIM (Select Ion Monitoring) mode.
HPLC-MS analyses for CoQ10 and CoQ10-13C6 from cell extracts were done on an Ultimate 3000 HPLC (Dionex) coupled to a high resolving power mass spectrometer LTQ Orbitrap (Thermo Scientific), equipped with an atmospheric pressure interface and an ESI ion source. Samples were analyzed using an RP C4 column (Phenomenex Jupiter, 150 × 2 mm, 5 μm particle size, Phenomenex) at a 200 μL/min flow rate. The isocratic mobile phase composition was 95% of methanol and 5% of 10 mM aqueous formic acid. The injection volume was 20 μL. The tuning parameters adopted for the ESI source were: source voltage 4.5 kV, capillary voltage 48.00 V, and tube lens voltage 180 V. The heated capillary temperature was maintained at 265°C. The mass accuracy of the recorded ions (versus the calculated ones) was ± 5 mmu (milli-mass units). Analyses were run using both full MS (300–1,000 m/z range) and MS/MS acquisition in the positive ion mode. Analytes were quantified in the full MS mode as [M+Na]+. Five-points calibration curves were done by standard addition of the analytes on a cell lysate matrix in the range 0–500 ng/ml.
If not otherwise stated all chemicals used for extractions and HPLC analyses were from Sigma.
Exogenous CoQ10 and Vitamin K2 Administration
The composition of liposomes used for rescue tests was typically: 65% POPC (2-Oleoyl-1-palmitoyl-sn-glycero-3-phosphocholine), 5% DPPG (1,2-Dipalmitoyl-sn-glycero-3-phosphoglycerol), 25% cholesterol, 5% ubiquinone or menaquinone. A mixture of the appropriate amounts of phospholipids and cholesterol was dissolved in chloroform and the solution evaporated to dryness under reduced pressure (20 mbar) on a rotary evaporator (temperature of the bath 30°C). The lipid film was subsequently flowed with nitrogen for 2 hr to remove residual traces of organic solvents. The film was then heated to 65°C and hydrated with the dialysis buffer for 10 min. The resulting lipid suspension was sonicated two times for 20 s at a frequency of 20 kHz and finally dialysed overnight. The mean hydrodynamic diameter of the liposomes was determined by dynamic light scattering by means of a Malvern ZS Nanosizer (Malvern Instrumentation), while the efficiency of incorporation of ubiquinone or menaquinone into the liposomes was assessed by analytical HPLC-UV. Liposome suspensions typically contain 0.3-0.6 mM of CoQ10 or vitamin K2. Mitochondria-targeted CoQ10 (MitoQ) and relative control compound Decyl-TPP (D-TPP) were synthetized as previously reported (Murphy and Smith, 2007) and dissolved in DMSO at appropriate working concentrations. Embryos were injected at one-single cell stage with the indicated amount of MitoQ and D-TPP. Primary endothelial cells (HUAEC) were silenced for appropriate gene and treated for 24 hr with MitoQ or D-TPP. Then, cells were analyzed by FACS analysis for mitochondrial membrane potential as previously described.
Mitochondrial Membrane Potential (ψ) Analyses
For mitochondria membrane potential analysis (ψ) 105 cells were incubated with JC-1 probe (Invitrogen, 4.5 μg/ml) for 15’ at 37°C/ 5% CO2. Then cells were washed twice with PBS1X, trypsinzed, collected on prechilled tubes and immediately analyzed on FACSCalibur (Beckton-Dickinson). All procedures were performed in the dark. JC1 stock solution (5 mg/ml) was diluted at intermediate stock solution (50 μg/ml) and vortex for several minutes to allow complete dissolution.
Lipid Peroxidation Analyses
Detection of lipid peroxidation was achieved in human primary endothelial cells (HUAEC) by measuring malondialdehyde-proteine (MDA) adducts with OxiSelect MDA Adduct ELISA Kit (CellBiolabs). 106 cells were trypsinized and centrifuged at room temperature for 800rpm /10’. Pellet was suspended with sterile PBS, supplemented with Complete Protease Inhibitor Cocktail (Roche) and lysed by micro-homogenization. Cell lysates were analyzed for protein content according to the Bradford assay and diluted as appropriate according to instructions. Enzyme reaction was stopped after 30’ and OD450 nm measurements were immediately read on Glomax Multi Detection System (Promega).
Quantitative Real-Time PCR Analyses
Total RNA was isolated with PureLink Micro-to-Midi Total RNA Purification System (Invitrogen) and cDNA was made with RT High Capacity kit (Applied Biosystems) according to the manufacturer’s protocol. Quantitative real-time RT-PCR on human cell samples was performed with ABI Prism 7300 real-time PCR System (Applied Biosystems) using Platinum Quantitative PCR SuperMix-UDG with ROX (Invitrogen). Following genes were analyzed with respective primers: human UBIAD1 (GeneBank accession number NM_013319.2; FW: 809-832; RW: 874-895) and probe #36 (Roche); human COQ2 (GeneBank accession number NM_015697.6; FW:937-959; RW: 1009-1030) and probe #37 (Roche). 18S rRNA was used as internal control.
Zebrafish genes for qRT-PCR analysis were: gpx1a (GeneBank accession number: NM_001007281.2) and hmox1 (GeneBank accession number: NM_001127516). Genes were selected as reported to be upregulated under oxidative stress conditions (Malek et al., 2004;) and qRT-PCR assay was performed by COGENTEC with following protocol: 5 ng of cDNA was amplified (in triplicate) in a reaction volume of 15 uL containing the following reagents: 7.5ul of TaqMan PCR Mastermix 2x No UNG (Life Technologies, Foster City, CA), 0.75 ul of TaqMan Gene expression assay 20x (Life Technologies, Foster City, CA). Real-time PCR was carried out on the 7500 Sequence Detector System (Life Technologies), using a pre-PCR step of 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 60 s at 60°C. Preparations with RNA template without reverse transcriptase were used as negative controls. Samples were amplified with following Q-PCR assay (Life Technologies, Foster City, CA): gpx1a: Dr03071768_m1; hmox1: Dr03434096_m1; β-actin: Dr03432610_m1. β-actin gene was included as control housekeeping gene.
NOS Activity and NO Levels Analyses in Human Primary Endothelial Cells and in Zebrafish Embryos
The NOS enzymatic activity was quantified using NOS Activity Assay Kit (Cayman Chemicals; No.781001). The assay is based on the biochemical conversion of L-Arginine to L-Citrulline by the eNOS and nNOS enzymes. Samples were human primary endothelial cells (HUAEC) or zebrafish embryos (n = 100) at 72 hpf. Human primary endothelial cells (6∗106) were suspended with short treatment of trypsin and washed twice with PBS 1X. Cells were then centrifuged at 800 rpm for 10 min and suspended with 100 μl of Homogenization Buffer (Tris-HCl [pH 7.4]; 10 mM EDTA, 10 mM EGTA), microhomogenizated and sonicated. Samples lysates were centrifuged at 13,000 rpm for 10 min at 4°C. The supernatant was separated from pellet and quantified for proteins. Samples (50 μg) were incubated with a reaction mix containing 25 mM Tris-HCl [pH 7.4], 1 mM NADPH (Sigma, N1630), 0.6 mM calcium cloride, 0.1 μM calmodulin, 3 μM tetrahydrobiopterin (BH4), 1 μM flavin adenine mononucleotide, 1 μM flavin adenine mononucleotide and [3H]-Arginine monohydrocloride (1 mCi/ml; Perkin Elmer, NET1123001MC). Samples were incubated at 37°C for 15 min and stopped by adding 400 μl of stop-buffer. [3H]-Arginine was separated from [3H]-Citrulline by passing samples through the equilibrated resin provided by the kit. Radioactivity was quantitated by liquid scintillation counting. Activities were calculated on percentage of Citrulline formed in the reaction in relation to total possible counts and expressed as arbitrary parameter (percentage of activity to control sample).
Live Tg(kdrl:mCherry)uto2 siblingss847 (sib) and bar s847 (bar) mutant embryos were incubated at 56 hpf with 20 μM solution of DAF-2 DA (Calbiochem) in fish water, pH 7.0, in dark light at 28°C for 12 hr. After incubation, embryos were washed twice in fish water, pH 7.0, and analyzed at 72 hpf at stereo fluorescence microscopy.
Statistical Analyses
All experiments were performed at least as three independent times for each conditions, and the error bars represent the mean of ± the standard error of the mean, unless otherwise stated. Statistical significance was performed by a Student’s test, as appropriate, and significance is reported in accordance with p value (∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001).
Microscopy and Image Analysis
Images were acquired with TCSII SP5X confocal microscope, MZ16 FA stereomicroscope equipped with DCF300FY camera (Leica) or AZ100 stereomicroscope equipped with AxioCam MRm camera (Zeiss) and ApoTome AxioObserver Z1 (Zeiss). LAS AF and AxioVision software were used for analysis and image processing. Confocal acquisitions were analyzed for quantification of colocalization with ImageJ software applying Colocalization Finder PlugIn (French et al., 2008).
Time-Lapse Experiments
Siblings and barolo mutant embryos in Tg(kdrl:GFP)s843 background were embedded in 0.5% low melting agarose in presence of PTU in 8-well microscopic chambers (Ibidi) with glass bottom. Embryos were imaged with a 20x dry objective lens (Leica N/A 0.5) with a confocal scanning microscope Leica TCSII SP5. z Stacks were acquired at 512 × 512 resolution, scan speed of 400 Hz, and 3 micron z step size. Images were taken every 20 min for total movie duration of 12 hr. Videos were processed with ImageJ software (Abramoff et al., 2004) and shown at 2 frames/s.
Acknowledgments
We thank Federica Logrand, Marco Vincenti, Annalisa Camporeale, and Ellen Jane Corcoran for technical assistances as well as Pier Paolo Pandolfi, Guido Serini, Massimiliano Mazzone, Franz Enzmann and Silvio Aime for discussion, technical support and reagents. This work was supported by a HFSP Career Developmental Award (MMS), Marie Curie Action IRG 247852 (MMS), Telethon GGP10185(MMS), AIRC-MFAG 8911(MMS) and FP7-NMP-2007-214539 (JB). VM and MMS planned and discussed the whole project. RP positionally cloned the t31131 allele in JB’s lab. Independently, MMS identified the s847 allele in DYRS’ lab. RP performed ubiad1 expression analyses and cell transplantation experiments in zebrafish. MPM provided the MitoQ. ET produced the Ubiad1 MAb. VM and MMS performed all other experiments together with VC, GD, CM, EDL and LS. MMS wrote the manuscript and all authors commented on it. The authors declare no conflict of interests.
Published: January 31, 2013
Footnotes
Supplemental Information includes Extended Experimental Procedures, six figures, one table, and four movies and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2013.01.013.
Supplemental Information
Wild-type siblings embryos were analyzed for endothelial cell development at the level of the trunk vasculature from 32 to 48 hpf. Movies are z stack time-lapse of Tg(kdrl:GFP)s843 siblings between 32 and 48 hpf is shown.
barolo (bar) mutant embryos were analyzed for endothelial cell disintegration at the level of the trunk vasculature. z Stack time-lapse movies of Tg(kdrl:GFP)s843barolot31131 (bar) mutants between 32 and 48 show vascular integrity disintegration in the trunk vasculature (arrows) starting at 32 hpf. Endothelial cell regression is observed throughout the vascular network starting in intersegmental vessels (Se) and, later, also in dorsal aorta (DA).
Wild-type siblings embryos were analyzed for endothelial cell development at the level of the trunk vasculature. z Stack time-lapse movie of Tg(kdrl:GFP)s843 siblings between 48 and 60 hpf is shown.
barolo (bar) mutant embryos were analyzed for endothelial cell disintegration at the level of the trunk vasculature. z Stack time-lapse movies of Tg(kdrl:GFP)s843barolot31131 (bar) mutants between 48 and 60 hpf show vascular integrity disintegration in the trunk vasculature (arrows) progressively through 60 hpf. Endothelial cell regression is observed throughout the vascular network starting in intersegmental vessels (Se) and later also in dorsal aorta (DA).
References
- Alp N.J., Channon K.M. Regulation of endothelial nitric oxide synthase by tetrahydrobiopterin in vascular disease. Arterioscler. Thromb. Vasc. Biol. 2004;24:413–420. doi: 10.1161/01.ATV.0000110785.96039.f6. [DOI] [PubMed] [Google Scholar]
- Bentinger M., Brismar K., Dallner G. The antioxidant role of coenzyme Q. Mitochondrion. 2007;7(Suppl):S41–S50. doi: 10.1016/j.mito.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Bentinger M., Tekle M., Brismar K., Chojnacki T., Swiezewska E., Dallner G. Stimulation of coenzyme Q synthesis. Biofactors. 2008;32:99–111. doi: 10.1002/biof.5520320112. [DOI] [PubMed] [Google Scholar]
- Bentinger M., Tekle M., Dallner G. Coenzyme Q—biosynthesis and functions. Biochem. Biophys. Res. Commun. 2010;396:74–79. doi: 10.1016/j.bbrc.2010.02.147. [DOI] [PubMed] [Google Scholar]
- Browning E.A., Chatterjee S., Fisher A.B. Stop the flow: a paradigm for cell signaling mediated by reactive oxygen species in the pulmonary endothelium. Annu. Rev. Physiol. 2012;74:403–424. doi: 10.1146/annurev-physiol-020911-153324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew G.T., Watts G.F. Coenzyme Q10 and diabetic endotheliopathy: oxidative stress and the ‘recoupling hypothesis’. QJM. 2004;97:537–548. doi: 10.1093/qjmed/hch089. [DOI] [PubMed] [Google Scholar]
- Colavitti R., Pani G., Bedogni B., Anzevino R., Borrello S., Waltenberger J., Galeotti T. Reactive oxygen species as downstream mediators of angiogenic signaling by vascular endothelial growth factor receptor-2/KDR. J. Biol. Chem. 2002;277:3101–3108. doi: 10.1074/jbc.M107711200. [DOI] [PubMed] [Google Scholar]
- Crane F.L. Biochemical functions of coenzyme Q10. J. Am. Coll. Nutr. 2001;20:591–598. doi: 10.1080/07315724.2001.10719063. [DOI] [PubMed] [Google Scholar]
- Crane F.L. Discovery of ubiquinone (coenzyme Q) and an overview of function. Mitochondrion. 2007;7(Suppl):S2–S7. doi: 10.1016/j.mito.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Crane F.L., Morre D.J. Evidence for coenzyme Q function in Golgi membranes. In: Folkers K., Yamamura Y., editors. Biomedical and Clinical Aspects of Coenzyme Q. Elsevier; Amsterdam: 1977. pp. 3–14. [Google Scholar]
- D’Autréaux B., Toledano M.B. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- Duncan A.J., Bitner-Glindzicz M., Meunier B., Costello H., Hargreaves I.P., López L.C., Hirano M., Quinzii C.M., Sadowski M.I., Hardy J. A nonsense mutation in COQ9 causes autosomal-recessive neonatal-onset primary coenzyme Q10 deficiency: a potentially treatable form of mitochondrial disease. Am. J. Hum. Genet. 2009;84:558–566. doi: 10.1016/j.ajhg.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T. Oxidant signals and oxidative stress. Curr. Opin. Cell Biol. 2003;15:247–254. doi: 10.1016/s0955-0674(03)00002-4. [DOI] [PubMed] [Google Scholar]
- Finkel T., Holbrook N.J. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- Forsgren M., Attersand A., Lake S., Grünler J., Swiezewska E., Dallner G., Climent I. Isolation and functional expression of human COQ2, a gene encoding a polyprenyl transferase involved in the synthesis of CoQ. Biochem. J. 2004;382:519–526. doi: 10.1042/BJ20040261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förstermann U., Sessa W.C. Nitric oxide synthases: regulation and function. Eur. Heart J. 2012;33:829–837. doi: 10.1093/eurheartj/ehr304. 837a–837d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French A.P., Mills S., Swarup R., Bennett M.J., Pridmore T.P. Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat. Protoc. 2008;3:619–628. doi: 10.1038/nprot.2008.31. [DOI] [PubMed] [Google Scholar]
- Fritsche R., Schwerte T., Pelster B. Nitric oxide and vascular reactivity in developing zebrafish, Danio rerio. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2000;279:R2200–R2207. doi: 10.1152/ajpregu.2000.279.6.R2200. [DOI] [PubMed] [Google Scholar]
- Fulton D., Fontana J., Sowa G., Gratton J.P., Lin M., Li K.X., Michell B., Kemp B.E., Rodman D., Sessa W.C. Localization of endothelial nitric-oxide synthase phosphorylated on serine 1179 and nitric oxide in Golgi and plasma membrane defines the existence of two pools of active enzyme. J. Biol. Chem. 2002;277:4277–4284. doi: 10.1074/jbc.M106302200. [DOI] [PubMed] [Google Scholar]
- Giordano F.J. Oxygen, oxidative stress, hypoxia, and heart failure. J. Clin. Invest. 2005;115:500–508. doi: 10.1172/JCI200524408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare J.M., Stamler J.S. NO/redox disequilibrium in the failing heart and cardiovascular system. J. Clin. Invest. 2005;115:509–517. doi: 10.1172/JCI200524459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeringa S.F., Chernin G., Chaki M., Zhou W., Sloan A.J., Ji Z., Xie L.X., Salviati L., Hurd T.W., Vega-Warner V. COQ6 mutations in human patients produce nephrotic syndrome with sensorineural deafness. J. Clin. Invest. 2011;121:2013–2024. doi: 10.1172/JCI45693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess D.T., Matsumoto A., Kim S.O., Marshall H.E., Stamler J.S. Protein S-nitrosylation: purview and parameters. Nat. Rev. Mol. Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- Jin S.W., Herzog W., Santoro M.M., Mitchell T.S., Frantsve J., Jungblut B., Beis D., Scott I.C., D’Amico L.A., Ober E.A. A transgene-assisted genetic screen identifies essential regulators of vascular development in vertebrate embryos. Dev. Biol. 2007;307:29–42. doi: 10.1016/j.ydbio.2007.03.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonassen T., Clarke C.F. Genetic Analysis of Coenzyme Q Biosynthesis. In: Kagan V.E., Quinn P.J., editors. Coenzyme Q: Molecular Mechanisms in Health and Disease. CRC Press LLC; Boca Raton, FL: 2000. pp. 185–208. [Google Scholar]
- Kalen A., Appelkvist E.L., Dallner G. Biosynthesis of ubiquinone in rat liver. Acta Chem. Scand. A. 1987;41:70–72. doi: 10.3891/acta.chem.scand.41b-0070. [DOI] [PubMed] [Google Scholar]
- Kalén A., Appelkvist E.L., Chojnacki T., Dallner G. Nonaprenyl-4-hydroxybenzoate transferase, an enzyme involved in ubiquinone biosynthesis, in the endoplasmic reticulum-Golgi system of rat liver. J. Biol. Chem. 1990;265:1158–1164. [PubMed] [Google Scholar]
- Kumar A., Kaur H., Devi P., Mohan V. Role of coenzyme Q10 (CoQ10) in cardiac disease, hypertension and Meniere-like syndrome. Pharmacol. Ther. 2009;124:259–268. doi: 10.1016/j.pharmthera.2009.07.003. [DOI] [PubMed] [Google Scholar]
- Kuster G.M., Häuselmann S.P., Rosc-Schlüter B.I., Lorenz V., Pfister O. Reactive oxygen/nitrogen species and the myocardial cell homeostasis: an ambiguous relationship. Antioxid. Redox Signal. 2010;13:1899–1910. doi: 10.1089/ars.2010.3464. [DOI] [PubMed] [Google Scholar]
- Lander H.M., Milbank A.J., Tauras J.M., Hajjar D.P., Hempstead B.L., Schwartz G.D., Kraemer R.T., Mirza U.A., Chait B.T., Burk S.C. Redox regulation of cell signaling. Nature. 1996;30:380–381. doi: 10.1038/381380a0. [DOI] [PubMed] [Google Scholar]
- Lee J.S., Yu Q., Shin J.T., Sebzda E., Bertozzi C., Chen M., Mericko P., Stadtfeld M., Zhou D., Cheng L. Klf2 is an essential regulator of vascular hemodynamic forces in vivo. Dev. Cell. 2006;11:845–857. doi: 10.1016/j.devcel.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Liu Y., Fiskum G., Schubert D. Generation of reactive oxygen species by the mitochondrial electron transport chain. J. Neurochem. 2002;80:780–787. doi: 10.1046/j.0022-3042.2002.00744.x. [DOI] [PubMed] [Google Scholar]
- Liu J., Wu Q., He D., Ma T., Du L., Dui W., Guo X., Jiao R. Drosophila sbo regulates lifespan through its function in the synthesis of coenzyme Q in vivo. J. Genet. Genomics. 2011;38:225–234. doi: 10.1016/j.jgg.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Matlung H.L., Bakker E.N., VanBavel E. Shear stress, reactive oxygen species, and arterial structure and function. Antioxid. Redox Signal. 2009;11:1699–1709. doi: 10.1089/ars.2008.2408. [DOI] [PubMed] [Google Scholar]
- Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature. 1961;191:144–148. doi: 10.1038/191144a0. [DOI] [PubMed] [Google Scholar]
- Murphy M.P., Smith R.A.J. Targeting antioxidants to mitochondria by conjugation to lipophilic cations. Annu. Rev. Pharmacol. Toxicol. 2007;47:629–656. doi: 10.1146/annurev.pharmtox.47.120505.105110. [DOI] [PubMed] [Google Scholar]
- Nakagawa K., Hirota Y., Sawada N., Yuge N., Watanabe M., Uchino Y., Okuda N., Shimomura Y., Suhara Y., Okano T. Identification of UBIAD1 as a novel human menaquinone-4 biosynthetic enzyme. Nature. 2010;468:117–121. doi: 10.1038/nature09464. [DOI] [PubMed] [Google Scholar]
- Navas P., Villalba J.M., de Cabo R. The importance of plasma membrane coenzyme Q in aging and stress responses. Mitochondrion. 2007;7(Suppl):S34–S40. doi: 10.1016/j.mito.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Nickerson M.L., Kostiha B.N., Brandt W., Fredericks W., Xu K.P., Yu F.S., Gold B., Chodosh J., Goldberg M., Lu W. UBIAD1 mutation alters a mitochondrial prenyltransferase to cause Schnyder corneal dystrophy. PLoS ONE. 2010;5:e10760. doi: 10.1371/journal.pone.0010760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoli S., Standley C., Walker P., Hurlstone A., Fogarty K.E., Lawson N.D. MicroRNA-mediated integration of haemodynamics and Vegf signalling during angiogenesis. Nature. 2010;464:1196–1200. doi: 10.1038/nature08889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North T.E., Goessling W., Peeters M., Li P., Ceol C., Lord A.M., Weber G.J., Harris J., Cutting C.C., Huang P. Hematopoietic stem cell development is dependent on blood flow. Cell. 2009;137:736–748. doi: 10.1016/j.cell.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill J.S., Reddy A.B. Circadian clocks in human red blood cells. Nature. 2011;469:498–503. doi: 10.1038/nature09702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr A., Dubé M.P., Marcadier J., Jiang H., Federico A., George S., Seamone C., Andrews D., Dubord P., Holland S. Mutations in the UBIAD1 gene, encoding a potential prenyltransferase, are causal for Schnyder crystalline corneal dystrophy. PLoS ONE. 2007;2:e685. doi: 10.1371/journal.pone.0000685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelster B., Grillitsch S., Schwerte T. NO as a mediator during the early development of the cardiovascular system in the zebrafish. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2005;142:215–220. doi: 10.1016/j.cbpb.2005.05.036. [DOI] [PubMed] [Google Scholar]
- Pepe S., Marasco S.F., Haas S.J., Sheeran F.L., Krum H., Rosenfeldt F.L. Coenzyme Q10 in cardiovascular disease. Mitochondrion. 2007;7(Suppl):S154–S167. doi: 10.1016/j.mito.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Quinzii C., Naini A., Salviati L., Trevisson E., Navas P., Dimauro S., Hirano M. A mutation in para-hydroxybenzoate-polyprenyl transferase (COQ2) causes primary coenzyme Q10 deficiency. Am. J. Hum. Genet. 2006;78:345–349. doi: 10.1086/500092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigoulet M., Yoboue E.D., Devin A. Mitochondrial ROS generation and its regulation: mechanisms involved in H(2)O(2) signaling. Antioxid. Redox Signal. 2011;14:459–468. doi: 10.1089/ars.2010.3363. [DOI] [PubMed] [Google Scholar]
- Salviati L., Sacconi S., Murer L., Zacchello G., Franceschini L., Laverda A.M., Basso G., Quinzii C., Angelini C., Hirano M. Infantile encephalomyopathy and nephropathy with CoQ10 deficiency: a CoQ10-responsive condition. Neurology. 2005;65:606–608. doi: 10.1212/01.wnl.0000172859.55579.a7. [DOI] [PubMed] [Google Scholar]
- Santoro M.M., Samuel T., Mitchell T., Reed J.C., Stainier D.Y. Birc2 (cIap1) regulates endothelial cell integrity and blood vessel homeostasis. Nat. Genet. 2007;39:1397–1402. doi: 10.1038/ng.2007.8. [DOI] [PubMed] [Google Scholar]
- Schmidt T.S., Alp N.J. Mechanisms for the role of tetrahydrobiopterin in endothelial function and vascular disease. Clin. Sci. 2007;113:47–63. doi: 10.1042/CS20070108. [DOI] [PubMed] [Google Scholar]
- Searles C.D. Transcriptional and posttranscriptional regulation of endothelial nitric oxide synthase expression. Am. J. Physiol. Cell Physiol. 2006;291:C803–C816. doi: 10.1152/ajpcell.00457.2005. [DOI] [PubMed] [Google Scholar]
- Sehnert A.J., Huq A., Weinstein B.M., Walker C., Fishman M., Stainier D.Y. Cardiac troponin T is essential in sarcomere assembly and cardiac contractility. Nat. Genet. 2002;31:106–110. doi: 10.1038/ng875. [DOI] [PubMed] [Google Scholar]
- Stamler J.S., Lamas S., Fang F.C. Nitrosylation. the prototypic redox-based signaling mechanism. Cell. 2001;106:675–683. doi: 10.1016/s0092-8674(01)00495-0. [DOI] [PubMed] [Google Scholar]
- Stuehr D., Pou S., Rosen G.M. Oxygen reduction by nitric-oxide synthases. J. Biol. Chem. 2001;276:14533–14536. doi: 10.1074/jbc.R100011200. [DOI] [PubMed] [Google Scholar]
- Sun J., Murphy E. Protein S-nitrosylation and cardioprotection. Circ. Res. 2010;106:285–296. doi: 10.1161/CIRCRESAHA.109.209452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewska E., Dallner G., Andersson B., Ernster L. Biosynthesis of ubiquinone and plastoquinone in the endoplasmic reticulum-Golgi membranes of spinach leaves. J. Biol. Chem. 1993;268:1494–1499. [PubMed] [Google Scholar]
- Trevisson E., DiMauro S., Navas P., Salviati L. Coenzyme Q deficiency in muscle. Curr. Opin. Neurol. 2011;24:449–456. doi: 10.1097/WCO.0b013e32834ab528. [DOI] [PubMed] [Google Scholar]
- Tsai K.L., Huang Y.H., Kao C.L., Yang D.M., Lee H.C., Chou H.Y., Chen Y.C., Chiou G.Y., Chen L.H., Yang Y.P. A novel mechanism of coenzyme Q10 protects against human endothelial cells from oxidative stress-induced injury by modulating NO-related pathways. J. Nutr. Biochem. 2012;23:458–468. doi: 10.1016/j.jnutbio.2011.01.011. [DOI] [PubMed] [Google Scholar]
- Turunen M., Olsson J., Dallner G. Metabolism and function of coenzyme Q. Biochim. Biophys. Acta. 2004;1660:171–199. doi: 10.1016/j.bbamem.2003.11.012. [DOI] [PubMed] [Google Scholar]
- Ushio-Fukai M. Localizing NADPH oxidase-derived ROS. Sci. STKE. 2006;2006:re8. doi: 10.1126/stke.3492006re8. [DOI] [PubMed] [Google Scholar]
- Wang L., Zhang P., Wei Y., Gao Y., Patient R., Liu F. A blood flow-dependent klf2a-NO signaling cascade is required for stabilization of hematopoietic stem cell programming in zebrafish embryos. Blood. 2011;118:4102–4110. doi: 10.1182/blood-2011-05-353235. [DOI] [PubMed] [Google Scholar]
- Weiss J.S., Kruth H.S., Kuivaniemi H., Tromp G., White P.S., Winters R.S., Lisch W., Henn W., Denninger E., Krause M. Mutations in the UBIAD1 gene on chromosome short arm 1, region 36, cause Schnyder crystalline corneal dystrophy. Invest. Ophthalmol. Vis. Sci. 2007;48:5007–5012. doi: 10.1167/iovs.07-0845. [DOI] [PubMed] [Google Scholar]
- Werner E. GTPases and reactive oxygen species: switches for killing and signaling. J. Cell Sci. 2004;117:143–153. doi: 10.1242/jcs.00937. [DOI] [PubMed] [Google Scholar]
- Yellore V.S., Khan M.A., Bourla N., Rayner S.A., Chen M.C., Sonmez B., Momi R.S., Sampat K.M., Gorin M.B., Aldave A.J. Identification of mutations in UBIAD1 following exclusion of coding mutations in the chromosome 1p36 locus for Schnyder crystalline corneal dystrophy. Mol. Vis. 2007;13:1777–1782. [PubMed] [Google Scholar]
Supplemental References
- Abramoff M.D., Magalhaes P.J., Ram S.J. Image Processing with ImageJ. Biophotonics International. 2004;7:36–42. [Google Scholar]
- Covassin L., Amigo J.D., Suzuki K., Teplyuk V., Straubhaar J., Lawson N.D. Global analysis of hematopoietic and vascular endothelial gene expression by tissue specific microarray profiling in zebrafish. Dev. Biol. 2006;299:551–562. doi: 10.1016/j.ydbio.2006.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho R.K., Kane D.A. Cell-autonomous action of zebrafish spt-1 mutation in specific mesodermal precursors. Nature. 1990;348:728–730. doi: 10.1038/348728a0. [DOI] [PubMed] [Google Scholar]
- Kwan K.M., Fujimoto E., Grabher C., Mangum B.D., Hardy M.E., Campbell D.S., Parant J.M., Yost H.J., Kanki J.P., Chien C.-B. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- Lindner R. One-step separation of endocytic organelles, Golgi/trans-Golgi network and plasma membrane by density gradient electrophoresis. Electrophoresis. 2001;22:386–393. doi: 10.1002/1522-2683(200102)22:3<386::AID-ELPS386>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Malek R.L., Sajadi H., Abraham J., Grundy M.A., Gerhard G.S. The effects of temperature reduction on gene expression and oxidative stress in skeletal muscle from adult zebrafish. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2004;138:363–373. doi: 10.1016/j.cca.2004.08.014. [DOI] [PubMed] [Google Scholar]
- Michelmore R.W., Paran I., Kesseli R.V. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA. 1991;88:9828–9832. doi: 10.1073/pnas.88.21.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice J.E., Lindsay J.G. Subcellular fraction of mitochondria. In: Graham J.M., Rickwood D., editors. subcellular Fractionation: A practical approach. IRL Press; Oxford: 1997. pp. 107–142. [Google Scholar]
- Ruiz-Jiménez J., Priego-Capote F., Mata-Granados J.M., Quesada J.M., Luque de Castro M.D. Determination of the ubiquinol-10 and ubiquinone-10 (coenzyme Q10) in human serum by liquid chromatography tandem mass spectrometry to evaluate the oxidative stress. J. Chromatogr. A. 2007;1175:242–248. doi: 10.1016/j.chroma.2007.10.055. [DOI] [PubMed] [Google Scholar]
- Santoro M.M., Pesce G., Stainier D.Y. Characterization of vascular mural cells during zebrafish development. Mech. Dev. 2009;126:638–649. doi: 10.1016/j.mod.2009.06.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Wild-type siblings embryos were analyzed for endothelial cell development at the level of the trunk vasculature from 32 to 48 hpf. Movies are z stack time-lapse of Tg(kdrl:GFP)s843 siblings between 32 and 48 hpf is shown.
barolo (bar) mutant embryos were analyzed for endothelial cell disintegration at the level of the trunk vasculature. z Stack time-lapse movies of Tg(kdrl:GFP)s843barolot31131 (bar) mutants between 32 and 48 show vascular integrity disintegration in the trunk vasculature (arrows) starting at 32 hpf. Endothelial cell regression is observed throughout the vascular network starting in intersegmental vessels (Se) and, later, also in dorsal aorta (DA).
Wild-type siblings embryos were analyzed for endothelial cell development at the level of the trunk vasculature. z Stack time-lapse movie of Tg(kdrl:GFP)s843 siblings between 48 and 60 hpf is shown.
barolo (bar) mutant embryos were analyzed for endothelial cell disintegration at the level of the trunk vasculature. z Stack time-lapse movies of Tg(kdrl:GFP)s843barolot31131 (bar) mutants between 48 and 60 hpf show vascular integrity disintegration in the trunk vasculature (arrows) progressively through 60 hpf. Endothelial cell regression is observed throughout the vascular network starting in intersegmental vessels (Se) and later also in dorsal aorta (DA).