Abstract
Inhibitors of neuronal nitric oxide synthase have been proposed as therapeutics for the treatment of different types of neurological disorders. On the basis of a cis-3,4-pyrrolidine scaffold, a series of trans-cyclopropyl- and methyl-containing nNOS inhibitors have been synthesized. The insertion of a rigid electron-withdrawing cyclopropyl ring decreases the basicity of the adjacent amino group, which resulted in decreased inhibitory activity of these inhibitors compared to the parent compound. Nonetheless, three of them exhibited double-digit nanomolar inhibition with high nNOS selectivity on the basis of in vitro enzyme assays. Crystal structures of nNOS and eNOS with these inhibitors bound provide a basis for detailed structure-activity relationship (SAR) studies. The conclusions from these studies will be used as a guide in the future development of selective NOS inhibitors.
Keywords: Neuronal nitric oxide synthase, inhibition, isozyme selectivity, cyclopropyl analogues, X-ray crystallography
Nitric oxide (NO) has a wide variety of functions in the body, the most well-studied of which are its regulatory action on smooth muscle relaxation, cytotoxic activity in the immune system, neurotransmission, brain development and protection, and maintenance of synaptic plasticity.1 The production of NO is mediated via the activity of nitric oxide synthase (NOS),2, 3 a family of homodimeric heme-containing monooxygenases, which metabolizes L-arginine and O2 to L-citrulline and NO.4 A considerable body of research has shown that overproduction of cerebral NO by the neuronal isoform of NOS (nNOS) is a general pathological phenomenon for various neurological disorders such as Parkinson’s,5 Alzheimer’s,6 Huntington’s,7 headaches,8 and cerebral palsy.5–10 This has led to the search for and development of selective inhibitors of nNOS over endothelial NOS (eNOS), the isozyme that is responsible for the regulation of blood pressure, and inducible NOS (iNOS), the isozyme that is critical for immune responses, as therapeutic agents for the treatment of neurological disorders.11
In our on-going effort to develop novel nNOS inhibitors, we recently reported a series of cis-3,4-pyrrolidine-based inhibitors.12–14 In this family, several inhibitors have shown remarkable neuroprotective properties in a preclinical rabbit model for cerebral palsy.13 Compound 1as an example, not only is neuroprotective but is the most selective nNOS inhibitor over eNOS and iNOS yet reported.14 Despite its promising inhibitory activity, further application of 1 has been impeded by several of its structural characteristics. First, the flexible m-fluorophenyl ethanamino tail produces multiple rotatable bonds in the inhibitor. In addition, the benzylic position of the m-fluorophenyl ring is highly susceptible to metabolic oxidation.15 Also, the two positive charges of 1 at physiological pH, derived from the two secondary amino groups, decreases the chance of 1 to penetrate the blood-brain barrier (BBB).13 These considerations prompted the development of new pyrrolidine nNOS inhibitors with a potentially more desirable pharmacokinetic and pharmacodynamic profile.

Different strategies have been applied to modify the chemical structure of 1.16–20 Herein we describe the design and synthesis of a new series of inhibitors (2), with a structurally constrained cyclopropyl ring inserted in the position adjacent to the amino group of the ethanamino tail. Conformational restriction using cyclopropyl fragments is a strategy that has been widely used in modern drug design to create novel inhibitors for a variety of enzymes.21–23 Introduction of the cyclopropyl group (2) can potentially enhance inhibitory activity by stabilizing and thereby reducing the energetic penalty in binding to the enzyme active site and thus improve selectivity.24, 25 In addition, the insertion of a cyclopropyl fragment can block the potential metabolic oxidation at the benzylic position of the m-fluorophenyl ring.15 Furthermore, the electron-withdrawing character of the cyclopropyl ring decreases the basicity of the adjacent amino group. The calculated pKa value of the amino group in the lipophilic tail of 2 is ~7.4, which is significantly lower than 8.9 in 1.26 As a result, pseudo-monocationic molecule 2 may have improved BBB permeability.15 Here we report structure-activity relationship studies on these cyclopropyl containing inhibitors to determine if enhanced potency and selectivity can be attained prior to consideration of pharmacokinetic property assessment.
Chemistry
The key amine building blocks (3a–e) were synthesized as shown in Schemes 1 and 2. Starting from 1-substituted-3-vinylbenzenes 4a–cRh (II)-catalyzed cyclopropanation produced 5a–c as cis/trans mixtures in good yields.27 Ethyl esters 5a–c were treated with NaOCH3 in refluxing EtOH to induce epimerization, generating the thermodynamically more stable trans isomers, which were hydrolyzed in aqueous LiOH to yield 6a–c in good yields.27 Carboxylic acids 6a–c were converted to Boc-protected amines (7a–c) through Curtius rearrangement reactions in reasonable yields.28 Finally, the Boc-protecting groups of 7a–c were removed in trifluoroacetic acid (TFA) to provide (±)-trans-3a–c as TFA salts in high yields.
Scheme 1.
Synthesis of 3a–c a
a Reagents and conditions: (a) EtO2CCHN2Rh2(OAc)4, toluene, 80–85 °C, 2 h; (b) (i) NaOCH3 in EtOH (1 M), reflux, 40 h, (ii) LiOH, MeOH/H2O, 70 °C, 16 h, 75–80% for two steps; (c) diphenyl phosphorazidate, triethylamine, t-BuOH, 85 °C, 48 h, 75–82%; (d) TFA/CH2Cl2 (1:2), room temp, 45 min.
Scheme 2.
Resolution of enantiomers 3d and 3e a
a Reagents and conditions: (a) (S)-(-)-camphanic chloride, CH2Cl2, TEA, room temp, 30 min, 91% for two diastereomers; (b) 12 N HCl, EtOH (2:1), reflux, 72 h, 67–70%.
The two enantiomers of 3a were resolved in two steps. First, 3a was treated with (S)-(-) camphanic chloride in the presence of triethylamine (TEA) to produce two separable diastereomers 8a and 8b. Then the amide bonds of 8a and 8b were hydrolyzed in concentrated HCl to generate single enantiomers 3d and 3e in good yields.
The syntheses of inhibitors 2a–d began with (R,R)-9a.29 Compound 9a underwent reductive amination with amines 3a–c and 3e using NaBH(OAc)3 as a reducing reagent to generate 10a–d in good yields. Next, the three Boc-protecting groups of 10a–d were removed in HCl to yield inhibitors 2a–d in high yields.
The synthesis of inhibitors 2e,f began with aldehyde 9b (Scheme 4).13 Reductive amination of 9b with amine 3a or 3e provided the corresponding secondary amine, which was further protected with a Boc-protecting group to give 10e,f in reasonable yields. Next, the cyclopropyl group was reduced, along with deprotection of the Bn-protecting group, by catalytic hydrogenation using Pd(OH)2 as a catalyst at 60 °C to generate 11e,f. Finally, the three Bocprotecting groups were removed in HCl to provide inhibitors 2e,f as tri-HCl salts in good yields.
Scheme 4.
Synthesis of inhibitors 2e–fa
a Reagents and conditions: (a) (i) 3a or 3e, TEA, NaHB(OAc)3, room temp, 3 h, (ii) (Boc)2O, TEA, MeOH, room temp, 6 h, 60% for two steps; (b) H2Pd(OH)2/C, 60 °C, 30 h; (c) N HCl/MeOH (2:1), room temp, 16 h, 25% for two steps. The R/S notations shown indicate the chirality of the two chiral centers of the cyclopropyl ring; the pyrrolidine ring has (R,R) stereochemistry in all of the compounds.
Structure activity studies
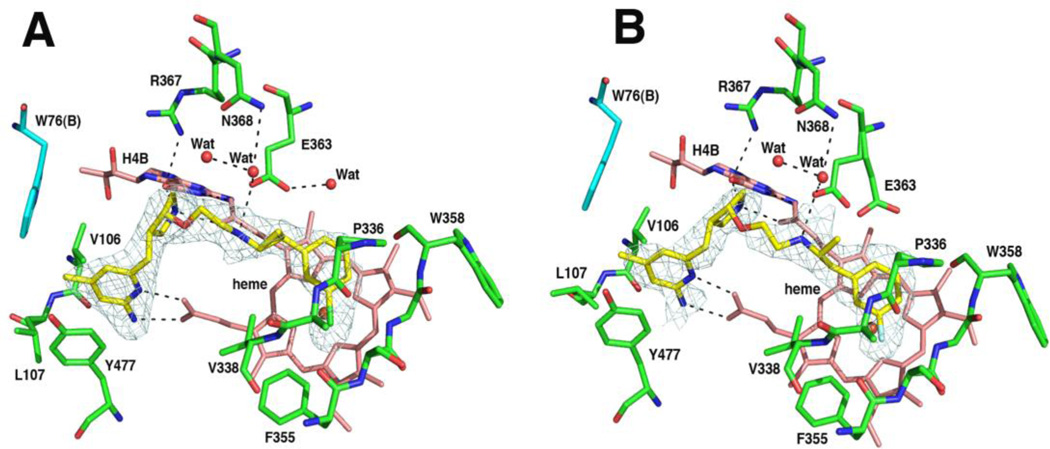
Nitric oxide hemoglobin capture assays were performed with 2a through 2f against three NOS isoforms to obtain the in vitro inhibitory potency and isoform selectivity for this series of compounds (Table 1). Crystal structures of nNOS and eNOS with these inhibitors bound were also determined, which provides the basis for structure activity relationship (SAR) studies. Consistent with the binding mode of (3R,4R)-1,14 all six inhibitors in this study bind to both nNOS and eNOS in a flipped orientation. As shown in Figure 1, the two nitrogen atoms of aminopyridine donate two hydrogen bonds to heme propionate D. The pyrrolidine ring nitrogen is positioned between heme propionate A and the C=O group of the pterin, thereby forming hydrogen bonds to both. This binding mode leaves the cyclopropyl and phenyl rings of the inhibitor tail right above the heme and provides the primary differences between the 1 and 2 interactions with the local surroundings.
Table 1.
Ki a Values of Inhibitors for Rat nNOS, Bovine eNOS, and Murine iNOS
 | |||||
|---|---|---|---|---|---|
| selectivityb | |||||
| Compound | nNOS (μM) | eNOS (μM) | iNOS (μM) | ||
| n/e | n/i | ||||
| (±) 2a | 0.170 | 51 | 16 | 300 | 94 |
| 2b | 0.175 | 31 | 12 | 177 | 69 |
| (±) 2c | 0.140 | 45 | 10 | 320 | 72 |
| (±) 2d | 0.071 | 26 | 0.86 | 370 | 12 |
| (±) 2e | 0.046 | 68 | 10.2 | 1500 | 220 |
| 2f | 0.070 | 40 | 7.6 | 570 | 110 |
The Ki values were calculated based on the directly measured IC50 values, which represent at least duplicate measurements with standard deviations of ±10%.
The ratio of Ki (eNOS or iNOS) to Ki (nNOS).
Figure 1.
The nNOS active site structure with (A) 2b (1.85Å, PDB code 3RQJ), (B) 2c (2.21Å, 3RQK), or (C) 2d (1.93Å, 3RQL) bound. The omit Fo – Fc difference density contoured at 3 σ level is shown around the bound inhibitor. Note that the two alternate Glu592 side chain rotamers are observed for 2b, but only the new rotamer is seen for 2c and 2d. Significant hydrogen bonds are depicted with dashed lines. The heme pyrrole ring positions are labeled in panel A. All structural figures were made with PyMol (www.pymol.org).
The introduction of a cyclopropyl ring adjacent to the amine in the tail end of the inhibitor was designed to lower the pKa (pKa = 7.4, calculated by ACD/Labs 7.0) of this otherwise basic group (1pKa = 8.9, calculated by ACD/Labs 7.0) to improve membrane permeability of the inhibitor. The rigidity along the inhibitor tail results in the cyclopropyl ring forcing the amino group to face more exclusively toward the active site Glu residue (Glu592 in nNOS or Glu363 in eNOS). This amino group of the inhibitor recruits the Glu residue as its hydrogen-bonding partner, forcing the Glu side chain into an alternate rotamer position (Figure 1). The alternate Glu side chain rotamer also was observed in the eNOS-1 complex structure but not in the nNOS-1 structure.14 The more flexible tail in 1 allows the amino nitrogen to point in various directions. And, therefore, there is not enough driving force to bring the Glu side chain into an alternate rotamer in nNOS.
We also have tested the impact of chirality at the cyclopropyl ring on inhibitor binding potency. The nNOS complex structures show that when a racemic compound mixture is used in crystal soaking, the ‘ring up’ (R,S) enantiomer binds exclusively (Figures 1B and 1C), although the ‘ring down’ (S,R) enantiomer (2b) can still bind if it is the only isomer used for crystal soaking (Figure 1A). The different stereochemistry around the cyclopropyl ring results in slightly different orientations (more than 12°) for the tail phenyl ring, which is more parallel to the heme plane in the ‘ring down’ (S,R) enantiomer than in the ‘ring up’ (R,S) isomer (Figure 2A). Moreover, in the ‘ring down’ (S,R) enantiomer both the cyclopropyl (3.1 Å) and phenyl (2.7 Å) rings are farther away from the native Glu592 side chain position and, thus, can tolerate both the native and alternate Glu592 rotamer positions. In contrast, the cyclopropyl or phenyl ring in the ‘ring up’ (R,S) enantiomer, as in 2c or 2dis so close to the native Glu592 side chain, being 2.5 Å to cyclopropyl and 2.3 Å to phenyl ring in 2c assuming the Glu592 is in its native position, that the alternate Glu592 rotamer is favored (Figures 1B and 1C). Therefore, two factors can potentially influence the Glu592 side chain conformation: 1) a H-bond (or salt bridge) with the inhibitor amino group and 2) a steric repulsion between the phenyl ring and Glu592. The cyclopropyl-containing inhibitors reported here either force the amino group to face Glu592 or bring the tail phenyl ring closer to Glu592, resulting in alternate Glu592 side chain conformations.
Figure 2.
(A) The active site structure of nNOS-2b (green, 3RQJ) with the molecule of 2d (gray, 3RQL) overlaid in order to illustrate the chirality difference at the cyclopropyl ring. Two alternate Glu592 side chain rotamers are shown. (B) The active site structure of nNOS-2e (gray, 3RQM) with the molecule of 2f (green, 3RQN) overlaid. The two compounds in the structures show different stereochemistry (S or R) at the position of the methyl group.
Overall the 'ring up' (R,S) enantiomer fits better into the nNOS active site. The alternate rotamer of Glu592 plays an important role here since in the better binding 'ring up' (R,S) mode Glu592 is in a better position to H-bond with the tail amine group of inhibitors 2c and 2d (Figure 1). Another factor to consider is how the halogen/methyl group of the phenyl ring fits into the hydrophobic pocket defined by Val567 and Phe584 (Figure 1). The larger methyl group and chloro atom relative to the smaller fluoro atom pushes the phenyl ring about 0.5 Å closer to Glu592, which forces Glu592 to adopt primarily the alternate rotamer conformation. As noted earlier, this alternate rotamer makes a better hydrogen bond with the inhibitor. Therefore, it is both the size of the substituent attached to the phenyl ring and the chirality of the inhibitor that contribute to differences in binding affinity. It is probably the resulting difference in hydrogen bond strength from the tail amino group to the alternate Glu592 side chain position that is the main reason why 2d shows a two-fold better binding affinity to nNOS than that of either 2a or 2c (Table 1).
Introduction of a cyclopropyl ring next to the amino group in the tail decreases the charge, which results in decreased potency; the analogous compounds with a methyl group at this position, such as 2e and 2fshow slightly better binding affinity to nNOS (Table 1). The methyl group should increase the basicity of the amino group in these two compounds compared to the cyclopropyl-containing counterparts. Although the tail amino groups in both the methyl and cyclopropyl inhibitors superimpose well, and both are in position to H-bond with Glu592 (Figure 2), the higher basicity of the tail amino group (pKa = 8.0, calculated by ACD/Labs 12.0), resulting from the methyl group compared to the cyclopropyl group, very likely contributes to the tighter binding of the methyl inhibitors. The higher affinity of 2e over 2f also is readily understood on the basis of amine-Glu592 interactions (Figure 3). In 2f the amino nitrogen is pointing away from the Glu592 side chain because of the R-chirality at the methyl position in this inhibitor. The S-stereochemistry seen in the nNOS-2e structure must be the dominant one preferred by the enzyme because it is the only enantiomer observed in the structure, even though 2e used for the crystal soak is a racemic (R/S) mixture. This is consistent with the lower Ki value towards nNOS measured for 2e versus 2f (Table 1). The R-configuration can only be observed when the enantiomer pure compound (2f) is used.
Figure 3.
The active site of nNOS with (A) 2e (1.95Å, 3RQM) or (B) 2f (1.95Å, 3RQN) bound. The omit Fo – Fc difference density contoured at 3 σ level is shown around the bound inhibitor. Significant hydrogen bonds are depicted with dashed lines. In panel A, two partially occupied water molecules are also shown which are complimentary to the two alternate Glu592 side chain positions.
To explore the structural basis for isoform selectivity of these cyclopropyl-containing inhibitors, crystal structures of eNOS with 2d or 2e bound were also determined as shown in Figure 4. Both inhibitors show the same binding mode in eNOS as that observed in nNOS structures (see Figures 1C and 3A). For eNOS-2d the bulky chlorophenyl ring also forces the Glu363 side chain into its alternate rotamer conformation to avoid the potential clashes with Glu363 in its native rotamer conformation. For eNOS-2ethe smaller fluorophenyl ring and the absence of the cyclopropyl ring allow both Glu363 rotamer conformations to exist. In both cases, the enzyme-inhibitor interactions right above the heme in eNOS replicate what we have seen in nNOS. However, the binding affinity of these inhibitors to eNOS is significantly lower than that to nNOS. Isoform selection has been attributed primarily to two amino acid differences in the active site of NOS, Asp597 in nNOS vs. Asn368 in eNOS and Met336 in nNOS vs. Val106 in eNOS. Binding free energy calculations19 indicated that although Asp597 (or Asn368) is not directly involved in binding of these pyrrolidine inhibitors, it does impose a significant electrostatic influence on how tight these inhibitors fit into the NOS active site. On the other side of the binding site, the Met336 (Val106) residue does contact the aminopyridine ring of the inhibitors. One of the valine methyl groups actually makes close contact with the inhibitor, thereby pushing the aminopyridine ring of inhibitor farther away from the Val106 position in eNOS compared to the aminopyridine position seen in nNOS. This different position of the aminopyridine ring in eNOS also pushes the Tyr477 side chain away relative to the Tyr706 position in nNOS, and as a result, Tyr706 in nNOS makes better stacking interactions with the aminopyridine ring of the inhibitor than does Tyr477 in eNOS. Taken together these differences observed between eNOS and nNOS provide a structural basis for the relatively poor binding of these inhibitors to eNOS.
Figure 4.
The eNOS active site with (A) 2d (2.08Å, 3RQO) or (B) 2e (2.35Å, 3RQP) bound. The omit Fo – Fc difference density contoured at 3 σ level is shown around the bound inhibitor. Significant hydrogen bonds are depicted with dashed lines.
In summary, the cyclopropyl containing pyrrolidine inhibitors reported in this study show a decrease in potency with both nNOS and eNOS compared with parental inhibitor 1. This may result in part from the lowered basicity of the tail amino group. The structures here demonstrate the importance of the hydrogen bond strength of this amino group to the alternate side chain rotamer of the active site Glu residue. Weakening of this hydrogen bond results in a loss in potency toward nNOS, leading to poorer isoform selectivity of these more rigid cyclopropylcontaining inhibitors. These observations indicate that other chemical modifications are needed to retain the inhibitory potency in future inhibitor design.
Experimental Section
General Method A: Rh (II)-catalyzed cyclopropanation
To a solution of styrene derivative 4a–c (20 mmol) in dry toluene (40 mL) was added Rh2(OAc)4 (0.4 mmol) catalyst. The resulting mixture was heated at 80 °C for 30 min, then ethyl diazoacetate (10 mmol) was added dropwise at the same temperature over a period of 1 h. The reaction mixture was allowed to stir at 85 °C for an additional 2 h and then cooled to room temperature. The solvent was removed by rotary evaporation, and the resulting oil was purified by flash chromatography (1–10% ethyl acetate in hexanes) to generate 5a–c as mixtures of cis/trans isomers.
General Method B: Epimerization and hydrolysis
To a solution of 5a–c (10 mmol) in EtOH (10 mL) was added NaOCH3 (40 mL) portionwise. The reaction solution was heated under reflux for 40 h and then concentrated by rotary evaporation. The resulting residue was partitioned between CH2Cl2 (200 mL) and H2O (100 mL). The aqueous layer was extracted with CH2Cl2 (2 × 100 mL). The combined organic layers were dried over Na2SO4 and concentrated. The crude ethyl ester was taken up in MeOH (70 mL), to which was added LiOH (345 mg, 15 mmol) and H2O (70 mL). The reaction was heated at 70 °C for 16 h. After cooling to room temperature, MeOH was removed by rotary evaporation. The resulting aqueous solution was acidified with concentrated HCl to pH 1 and then extracted with ethyl acetate (3 × 150 mL). The combined organic layers were dried over Na2SO4, and concentrated. The crude product was purified by flash chromatography to yield 6a–c (75–80%) as white solids.
General Method C: Curtius rearrangement
To a solution of 6a–c (2.0 mmol) in dry t-BuOH (0.3 M) was added diphenyl phosphorazidate (DPPA, 2.2 mmol) and TEA (3.0 mmol). The reaction solution was heated at 85 °C for two days then cooled to room temperature and concentrated. The resulting solution was partitioned between ether (50 mL) and NaHCO3 (50 mL). The aqueous layer was extracted with ether (2 × 50 mL). The combined organic layers were dried over Na2SO4 and concentrated. The crude product was purified by flash chromatography to yield 7a–c (75–82%) as white solids.
General Method D: Boc-deprotection of 7a–c
To a solution of 7a–c (1.0 mmol) in CH2Cl2 (5 mL) was added TFA (5 mL). The reaction mixture was stirred at room temperature for 30 min. The solvent was removed by rotary evaporation. The yellow oil was dried under vacuum for 24 h to give crude amines 3a–c as yellow oils, which were used in the next step without further purification.
General Method E: Reductive amination
To a solution of aldehyde (0.1 mmol) in CH2Cl2 (3 mL) was added amine (0.11 mmol), followed by TEA (0.2 mmol), and NaBH(OAc)3 (0.12 mmol). The mixture was stirred at room temperature for an additional 3 h and then concentrated. The crude product was purified by flash column chromatography (ethyl acetate/hexanes, 2:1–4:1) to give the product as a colorless oil.
General Method F: Boc-deprotection of 10a–d and 11e–f
To a solution of tri-Boc-protected inhibitor (0.2 mmol) in MeOH (0.5 mL) was added 6 N HCl (1.0 mL). The reaction mixture was allowed to stand at room temperature for 16 h and then concentrated. The resulting pale yellow oil was evacuated for 30 h to give the inhibitors (95–99%).
2-(3-Fluorophenyl)cyclopropanecarboxylic acid (6a)
Compound 6a was synthesized using general method A and B (75%): 1H NMR (500 MHz, CDCl3) δ 0.87–0.91 (dd, J = 6.0, 13.5 Hz, 1H), 1.21–1.29 (m, 1H), 1.39–1.43 (ddd, J = 5.0, 6.5, 8.0 Hz, 1H), 1.67–1.72 (m, 1H), 1.90–1.94 (m, 1H), 2.58–2.63 (m, 1H), 6.79–6.82 (dd, J = 2.0, 5.5 Hz, 1H), 6.90–6.95 (m, 2H), 7.24–7.28 (m, 1H), 8.90–11.00 (br s, 1H); 13C NMR (125 MHz, CDCl3) δ 17.8, 24.4, 26.9, 31.8, 113.3, 113.5, 113.8, 114.0, 122.30, 122.32, 130.2, 130.3, 142.4, 142.5, 162.2, 164.2, 179.8; LCQ-MS (M - H+) calcd for C10H8FO2 179, found 179.
2-m-Tolylcyclopropanecarboxylic acid (6b)
Compound 6b was synthesized using general method A and B (77%): 1H NMR (500 MHz, CDCl3) δ 1.30–1.40 (ddd, J = 4.5, 7.0, 7.5 Hz, 1H), 1.60–1.65 (dd, J = 5.0, 9.0 Hz, 1H), 1.85–1.90 (ddd, J = 4.5, 5.0, 7.5 Hz, 1H), 2.30 (s, 3H), 2.50–2.60 (ddd, J = 4.5, 7.0, 9.0 Hz, 1H), 6.85–6.95 (m, 1H), 7.00–7.05 (m, 1H), 7.15–7.22 (m, 2H), 9.00–11.00 (br s, 1H); 13C NMR (125 MHz, CDCl3) δ 17.5, 21.4, 24.0, 27.1, 123.2, 127.0, 127.4, 128.4, 138.2, 139.4, 180.1; LCQ-MS (M - H+) calcd for C11H13O2 177, found 177.
2-(3-Clorophenyl)cyclopropanecarboxylic acid (6c)
Compound 6c was synthesized using general method A and B (80%): 1H NMR (500 MHz, CDCl3) δ 1.30–1.40 (ddd, J = 2.0, 3.5, 7.0 Hz, 1H), 1.60–1.65 (dd, J = 5.0, 9.0 Hz, 1H), 1.85–1.91 (m, 1H), 2.50–2.60 (m, 1H), 6.85–7.02 (m, 1H), 7.05–7.10 (m, 1H), 7.15–7.22 (m, 2H), 9.00–11.00 (br s, 1H); 13C NMR (125 MHz, CDCl3) δ 17.8, 24.4, 26.9, 31.8, 113.3, 113.5, 113.8, 114.0, 122.30, 122.32, 130.2, 130.3, 142.4, 142.5, 162.2, 164.2, 179.8; LC-MS (M - H+) calcd for C10H10ClO2 197, found 197.
tert-Butyl 2-(3-fluorophenyl)cyclopropylcarbamate (7a)
Compound 7a was synthesized using general method C (75%): 1H NMR (500 MHz, CDCl3) δ 1.10–1.25 (m, 1H), 1.40–1.55 (m, 10H), 2.00–2.10 (br s, 1H), 2.73 (br s, 1H), 5.04 (br s, 1H), 6.82–6.88 (m, 2H), 6.92–6.94 (d, J = 7.5 Hz, 1H), 7.20–7.25 (dd, J = 7.5, 14.0 Hz, 1H); 13C NMR (125 MHz, CDCl3) δ 16.6, 25.1, 28.6, 32.9, 113.0, 113.2, 113.4, 113.6, 122.4, 129.9, 130.0, 130.1, 143.77, 143.83, 162.2, 164.1; LCQ-MS (M + H+) calcd for C14H19FNO2 252, found 252.
tert-Butyl 2-m-tolylcyclopropylcarbamate (7b)
Compound 7b was synthesized using general method C (82%): 1H NMR (500 MHz, CDCl3) δ 1.00–1.20 (m, 1H), 1.46 (s, 9H), 1.95–2.05 (ddd, J = 3.0, 6.5, 9.5 Hz, 1H), 2.31 (s, 3H), 2.74 (br s, 1H), 4.85 (br s, 1H), 6.91–6.93 (m, 2H), 6.97–6.99 (d, J = 7.5 Hz, 1H), 7.13–7.16 (dd, J = 7.5, 8.0 Hz, 1H); 13C NMR (125 MHz, CDCl3) δ 16.3, 21.4, 24.5, 28.4, 32.6, 120.2, 120.3, 123.4, 126.1, 126.8, 127.2, 128.2, 130.1, 137.9, 140.6; LCQMS (M + H+) calcd for C15H21NO2 248, found 248.
tert-Butyl 2-(3-chloroluorophenyl)cyclopropylcarbamate (7c)
Compound 7c was synthesized using general method C (77%): 1H NMR (500 MHz, CDCl3) δ 1.14–1.17 (dd, J = 6.5, 7.0 Hz, 2H), 1.45 (s, 9H), 1.99–2.03 (ddd, J = 2.5, 7.5, 10.5 Hz, 1H), 2.72 (br s, 1H), 4.88 (br s, 1H), 6.95–7.02 (d, J = 7.0 Hz, 1H), 7.10–7.25 (m, 3H); 13C NMR (125 MHz, CDCl3) δ 16.3, 24.5, 28.4, 32.6, 120.1, 120.3, 124.8, 126.2, 126.6, 129.5, 129.9, 134.1, 142.9; LCQ-MS (M + H+) calcd for C14H19ClNO2 268, found 268.
Compounds 8a and 8b
To a solution of amine 3a (450 mg, 3.0 mmol) in CH2Cl2 was added camphanic chloride (650 mg, 3.0 mmol) followed by TEA (510 µL, 3.75 mmol). The reaction was allowed to stir at room temperature for 30 min and then concentrated. The resulting oil was purified by flash chromatography (ethyl acetate/hexanes, 1:4) to generate 8a (445 mg, 45%): 1H NMR (500 MHz, CDCl3) δ 0.92 (s, 3H), 1.12 (s, 6H), 1.19–1.22 (m, 1H), 1.28–1.30 (m, 2H), 1.65–1.75 (m, 1H), 1.85–2.00 (m, 2H), 2.05–2.15 (m, 1H), 2.50–2.60 (m, 1H), 2.90–2.98 (m, 1H), 6.68 (br s, 1H), 6.84–6.90 (m, 2H), 6.95–6.97 (d, J = 7.5 Hz, 1H), 7.21–7.25 (m, 1H); 13C NMR (125 MHz, CDCl3) δ 9.7, 15.8, 16.5, 16.7, 24.67, 24.78, 29.0, 30.3, 31.6, 54.0, 55.3, 92.4, 113.1, 113.27, 113.32, 113.5, 122.30, 122.32, 129.8, 129.9, 142.6, 142.7, 162.0, 163.9, 168.3, 178.2; LC-TOF (M + H+) calcd for C19H23FNO3 332.1662, found 332.1673.
8b (360 mg, 36%): 1H NMR (500 MHz, CDCl3) δ 0.91 (s, 3H), 1.11 (s, 6H), 1.20–1.35 (m, 3H), 1.65–1.75 (m, 1H), 1.85–2.00 (m, 2H), 2.00–2.10 (m, 1H), 2.50–2.58 (m, 1H), 2.89–2.95 (m, 1H), 6.67 (br s, 1H), 6.84–6.90 (m, 2H), 6.95–6.97 (d, J = 8.0 Hz, 1H), 7.21–7.24 (m, 1H); 13C NMR (125 MHz, CDCl3) δ 9.7, 16.0, 16.5, 16.7, 24.61, 24.63, 29.0, 30.3, 31.5, 54.0, 55.3, 92.38, 92.40, 113.1, 113.29, 113.32, 113.4 113.5, 113.6, 122.30, 122.32, 129.8, 129.9, 142.66, 142.72, 162.0, 163.9, 168.2, 168.3, 178.2; LC-TOF (M + H+) calcd for C19H23FNO3 332.1662, found 332.1677.
(1S,2R)-2-(3-Fluorophenyl)cyclopropanamine (3d)
To a solution of 8a (330 mg, 1.0 mmol) in EtOH (5 mL) was slowly added 12 N HCl (10 mL). The resulting mixture was heated under reflux for 72 h and then cooled to room temperature. The solvent was removed by rotary evaporation, and the resulting crude material was purified with flash chromatography (2–5% MeOH in CH2Cl2) to give 3d as a yellow oil (105 mg, 70%): 1H NMR (500 MHz, CDCl3) δ 1.10–1.30 (br s, 1H), 1.40–1.60 (br s, 1H), 2.30–2.50 (br s, 1H), 2.70–2.90 (br s, 1H), 6.70–6.72 (m, 1H), 6.72–6.77 (m, 1H), 6.89–7.00 (m, 1H), 7.10–7.30 (m, 1H), 7.80–8.20 (br s, 2H); 13C NMR (125 MHz, CDCl3) δ 13.2, 21.5, 31.6, 113.5, 113.7, 114.2, 114.4, 122.29, 122.31, 130.4, 130.5, 140.1, 140.2; LC-TOF (M + H+) calcd for C9H11FN 152.0876, found 152.0870.
(1R,2S)-2-(3-Fluorophenyl)cyclopropanamine (3e)
Compound 3e was synthesized using a similar procedure to that of 3d using 8b as a starting material (67%): 1H NMR (500 MHz, CDCl3) δ 1.10–1.30 (br s, 1H), 1.40–1.60 (br s, 1H), 2.30–2.50 (br s, 1H), 2.70–2.90 (br s, 1H), 6.70–6.72 (m, 1H), 6.72–6.77 (m, 1H), 6.89–7.00 (m, 1H), 7.10–7.30 (m, 1H), 7.80–8.20 (br s, 2H); 13C NMR (125 MHz, CDCl3) δ 13.2, 21.5, 31.6, 113.5, 113.7, 114.2, 114.4, 122.29, 122.31, 130.4, 130.5, 140.1, 140.2; LC-TOF (M + H+) calcd for C9H11FN 152.0876, found 152.0870.
(3R,4R)-tert-Butyl 3-((6-(bis(tert-butoxycarbonyl)amino)-4-methylpyridin-2-yl)methyl)-4-(2-((1S,2R/1R,2S)-2-(3-fluorophenyl)cyclopropylamino)ethoxy)pyrrolidine-1-carboxylate (10a)
Compound 10a was synthesized using general method D (81%): 1H NMR (500 MHz, CDCl3) δ 0.95–1.00 (m, 1H), 1.06–1.10 (m, 1H), 1.40–1.46 (m, 27H), 1.86–1.90 (m, 1H), 2.26–2.33 (m, 3H), 2.35–2.40 (m, 1H), 2.60–2.75 (m, 1H), 2.76–2.85 (m, 1H), 2.86–2.92 (m, 2H), 2.95–2.98 (m, 1H), 3.05–3.13 (m, 1H), 3.24–3.30 (m, 1H), 3.30–3.35 (m, 1H), 3.36–3.51 (m, 1H), 3.57–3.75 (m, 1H), 3.77–3.85 (m, 1H), 6.85–6.92 (m, 2H), 6.93–7.20 (m, 4H); 13C NMR (100 MHz, CDCl3) δ 17.4, 17.5, 17.6, 18.7, 20.9, 21.0, 22.2, 24.9, 25.0, 25.3, 26.2, 27.9, 28.5, 34.6, 34.7, 35.6, 41.3, 41.4, 41.5, 41.6, 42.5, 43.1, 48.7, 48.8, 49.1, 50.3, 52.0, 68.4, 68.5, 76.7, 78.6, 79.2, 79.3, 79.4, 82.8, 82.9, 119.5, 119.6, 122.7, 124.0, 124.07, 124.10, 124.12, 124.3, 125.5, 125.6, 125.7, 125.81, 125.83, 125.86, 125.94, 128.5, 128.6, 129.4, 129.5, 129.6, 132.0, 132.1, 134.1, 134.2, 143.6, 144.6, 144.7, 149.6, 151.47, 151.53, 151.8, 154.8, 159.7; LC-TOF (M+H+) calcd for C43H60F3N4O8 685.3977, found 685.3991.
(3R,4R)-tert-Butyl 3-((6-(bis(tert-butoxycarbonyl)amino)-4-methylpyridin-2-yl)methyl)-4-(2-((1S,2R)-2-(3-fluorophenyl)cyclopropylamino)ethoxy)pyrrolidine-1-carboxylate (10b)
Compound 10b was synthesized using general method D (82%): 1H NMR (500 MHz, CDCl3) δ 0.95–1.01 (m, 1H), 1.10–1.15 (m, 1H), 1.40–1.45 (m, 27H), 1.89–1.92 (m, 1H), 2.28–2.32 (m, 3H), 2.33–2.37 (m, 1H), 2.60–2.75 (m, 1H), 2.76–2.83 (m, 1H), 2.85–2.93 (m, 2H), 2.95–3.00 (m, 1H), 3.05–3.15 (m, 1H), 3.27–3.31 (m, 1H), 3.32–3.34 (m, 1H), 3.35–3.55 (m, 1H), 3.57–3.74 (m, 1H), 3.75–3.85 (m, 1H), 6.65–6.72 (m, 1H), 6.80–6.89 (m, 2H), 6.90–6.95 (m, 2H), 7.15–7.20 (m, 1H); 13C NMR (100 MHz, CDCl3) δ 14.2, 17.4, 17.5, 21.0, 22.7, 24.7, 24.8, 27.9, 28.5, 29.7, 31.6, 34.6, 34.7, 36.6, 41.4, 41.5, 42.5, 43.1, 44.7, 48.7, 48.8, 49.1, 50.3, 50.9, 60.4, 68.3, 78.6, 79.2, 79.3, 79.4, 82.8, 82.9, 112.2, 112.4, 112.6, 119.5, 119.6, 121.6, 122.7, 128.5, 128.6, 129.6, 129.7, 131.9, 132.0, 132.1, 132.2, 149.6, 151.5, 151.8, 154.6, 154.8, 159.0, 159.1, 162.0, 163.9; LCTOF (M+H+) calcd for C37H54FN4O7 685.3977, found 685.3979.
(3R,4R)-tert-Butyl 3-((6-(bis(tert-butoxycarbonyl)amino)-4-methylpyridin-2-yl)methyl)-4-(2-((1S,2R/1R,2S)-2-m-tolylcyclopropylamino)ethoxy)pyrrolidine-1-carboxylate (10c)
Compound 10c was synthesized using general method D (87%): 1H NMR (500 MHz, CDCl3) δ 0.90–0.97 (m, 1H), 1.00–1.05 (m, 1H), 1.40–1.45 (m, 27H), 1.84–1.91 (m, 1H), 2.27–2.32 (m, 6H), 2.33–2.40 (m, 1H), 2.60–2.75 (m, 1H), 2.75–2.83 (m, 1H), 2.85–2.93 (m, 2H), 2.95–3.00 (m, 1H), 3.05–3.15 (m, 1H), 3.25–3.30 (m, 1H), 3.31–3.34 (m, 1H), 3.35–3.55 (m, 1H), 3.57–3.74 (m, 1H), 3.75–3.85 (m, 1H), 6.85–6.95 (m, 4H), 6.92–7.01 (m, 2H), 7.10–7.20 (m, 2H); 13C NMR (100 MHz, CDCl3) δ 13.7, 14.1, 14.2, 17.2, 17.3, 17.4, 19.1, 20.9, 21.0, 21.1, 21.4, 22.2, 24.96, 24.99, 25.0, 25.5, 27.9, 28.5, 29.7, 30.6, 34.6, 34.7, 41.0, 41.1, 42.6, 43.2, 48.8, 48.9, 49.0, 49.1, 50.3, 60.4, 64.4, 68.4, 78.6, 79.2, 79.3, 79.4, 82.8, 82.9, 119.5, 119.6, 122.7, 122.8, 122.9, 126.3, 126.5, 126.6, 126.7, 126.8, 128.17, 128.24, 128.3, 128.5, 128.6, 132.0, 132.1, 137.8, 137.9, 141.3, 142.3, 149.6, 151.4, 151.5, 151.8, 154.8, 159.1, 159.2, 171.2; LC-TOF (M+H+) calcd for C38H57N4O7 681.4227, found 681.4224.
(3R,4R)-tert-Butyl 3-((6-(bis(tert-butoxycarbonyl)amino)-4-methylpyridin-2-yl)methyl)-4-(2-((1S,2R/1R,2S)-2-(3-clorophenyl)cyclopropylamino)ethoxy)pyrrolidine-1-carboxylate (10d)
Compound 10d was synthesized using general method D (87%): 1H NMR (500 MHz, CDCl3) δ 0.92–0.99 (m, 1H), 1.07–1.11 (m, 1H), 1.40–1.45 (m, 27H), 1.85–1.90 (m, 1H), 2.29–2.33 (m, 3H), 2.34–2.38 (m, 1H), 2.60–2.75 (m, 1H), 2.76–2.83 (m, 1H), 2.85–2.93 (m, 2H), 2.95–3.00 (m, 1H), 3.05–3.15 (m, 1H), 3.25–3.30 (m, 1H), 3.31–3.34 (m, 1H), 3.35–3.52 (m, 1H), 3.57–3.74 (m, 1H), 3.75–3.85 (m, 1H), 6.85–6.92 (m, 2H), 6.93–7.05 (m, 2H), 7.08–7.20 (m, 4H); 13C NMR (100 MHz, CDCl3) δ 17.4, 17.5, 17.6, 18.7, 20.9, 21.0, 22.2, 24.9, 25.0, 25.3, 26.2, 27.9, 28.5, 34.6, 34.7, 35.6, 41.3, 41.4, 41.5, 41.6, 42.5, 43.1, 48.7, 48.8, 49.1, 50.3, 52.0, 68.4, 68.5, 76.7, 78.6, 79.2, 79.3, 79.4, 82.8, 82.9, 119.5, 119.6, 122.7, 124.0, 124.07, 124.10, 124.12, 124.3, 125.5, 125.6, 125.7, 125.81, 125.83, 125.86, 125.94, 128.5, 128.6, 129.4, 129.5, 129.6, 132.0, 132.1, 134.1, 134.2, 143.6, 144.6, 144.7, 149.6, 151.47, 151.53, 151.8, 154.8, 159.7; LC-TOF (M+H+) calcd for C37H53ClN4O7 701.3681, found 701.3884.
6-(((3R,4R)-4-(2-((1S,2R/1R,2S)-2-(3-Fluorophenyl)cyclopropylamino)ethoxy)pyrrolidin-3-yl)methyl)-4-methylpyridin-2-amine (2a)
Compound 2a was synthesized using general method E as a mixture of two diastereomers (99%): 1H NMR (500 MHz, D2O) δ 1.30–1.40 (m, 1H), 1.41–1.48 (m, 1H), 2.10–2.20 (m, 3H), 2.30–2.50 (m, 1H), 2.51–2.77 (m, 2H), 2.80–2.90 (m, 1H), 2.91–3.03 (m, 1H), 3.04–3.40 (m, 3H), 3.41–3.72 (m, 2H), 3.73–3.88 (m, 1H), 3.90–4.11 (m, 1H), 4.40–4.50 (m, 1H), 6.30–6.60 (m, 2H), 6.70–6.90 (m, 2H), 7.00–7.20 (m, 2H); 13C NMR (125 MHz, D2O) δ 12.6, 12.9, 20.7, 20.9, 21.2, 21.4, 29.1, 37.7, 37.8, 38.6, 41.5, 45.3, 47.3, 47.6, 47.7, 49.6, 63.8, 64.5, 78.3, 78.5, 109.0, 110.5, 113.2, 113.4, 113.8, 114.0, 114.3, 122.4, 127.2, 128.1, 129.2, 130.1, 136.4, 140.7, 140.9, 145.9, 146.6, 153.0, 154.0, 158.3, 161.8, 163.8; LCTOF (M+H+) calcd for C21H30FN4O2 385.2404, found 385.2393.
6-(((3R,4R)-4-(2-((1S,2R)-2-(3-Fluorophenyl)cyclopropylamino)ethoxy)pyrrolidin-3-yl)methyl)-4-methylpyridin-2-amine (2b)
Compound 2b was synthesized using general method E (95%): 1H NMR (500 MHz, D2O) δ 1.30–1.40 (m, 1H), 1.41–1.46 (m, 1H), 2.18 (s, 3H), 2.19–2.20 (m, 1H), 2.40–2.50 (m, 1H), 2.55–2.70 (m, 2H), 2.81–3.00 (m, 3H), 3.19–3.23 (dd, J = 3.5, 13.5 Hz, 1H), 3.30–3.40 (m, 2H), 3.49–3.52 (m, 1H), 3.55–3.61 (m, 1H), 3.75–3.85 (m, 1H), 4.06 (br s, 1H), 6.38 (s, 1H), 6.54 (s, 1H), 6.80–6.95 (m, 3H), 7.15–7.25 (dd, J = 5.5, 10.5 Hz, 1H); 13C NMR (125 MHz, D2O) δ 12.6, 20.4, 21.0, 28.8, 30.1, 38.4, 41.3, 47.1, 47.5, 49.3, 64.3, 78.3, 110.3, 112.9, 113.1, 113.6, 113.8, 114.0, 122.1, 128.9, 129.0, 130.4, 130.5, 131.8, 131.9, 133.1, 140.6, 140.7, 145.6, 153.9, 158.1; LC-TOF (M+H+) calcd for C21H30FN4O2 385.2404, found 385.2384.
4-Methyl-6-(((3R,4R)-4-(2-((1S,2R/1R,2S)-2-m-tolylcyclopropylamino)ethoxy)pyrrolidin-3-yl)methyl)pyridin-2-amine (2c)
Inhibitor 2c was synthesized using general method E as a mixture of two diastereomers (90%): 1H NMR (500 MHz, D2O) δ 1.10–1.30 (m, 1H), 1.35–1.45 (m, 1H), 2.10–2.15 (m, 3H), 2.15–2.20 (m, 3H), 2.20–2.50 (m, 1H), 2.50–2.80 (m, 2H), 2.81–3.00 (m, 2H), 3.19–3.25 (m, 1H), 3.30–3.40 (m, 2H), 3.47–3.52 (m, 1H), 3.55–3.70 (m, 1H), 3.71–3.85 (m, 1H), 4.00–4.15 (m, 1H), 6.35–6.60 (m, 2H), 6.85–6.90 (m, 2H), 6.91–7.15 (m, 2H); 13C NMR (125 MHz, D2O) δ 12.2, 12.3, 15.1, 20.2, 20.4, 20.5, 20.6, 20.7, 21.0, 21.6, 23.2, 28.8, 28.9, 30.1, 30.5, 31.8, 37.4, 38.5, 41.2, 41.3, 47.0, 47.2, 47.5, 49.2, 49.3, 63.6, 64.5, 78.1, 78.3, 110.2, 110.3, 113.9, 114.0, 122.9, 123.1, 123.3, 126.3, 126.5, 126.6, 126.7, 126.8, 127.5, 127.6, 127.7, 128.6, 128.7, 128.78, 128.82, 128.9, 129.0, 131.8, 131.9, 137.9, 138.1, 138.7, 138.9, 139.0, 145.59, 145.62, 153.8, 158.1; LC-TOF (M+H+) calcd for C21H30FN4O2 381.2654, found 381.2653.
6-(((3R,4R)-4-(2-((1S,2R/1R,2S)-2-(3-Clorophenyl)cyclopropylamino)ethoxy)pyrrolidin-3-yl)methyl)-4-methylpyridin-2-amine (2d)
Compound 2d was synthesized using general method E as a mixture of two diastereomers (91%): 1H NMR (500 MHz, D2O) δ 1.10–1.30 (m, 1H), 1.35–1.50 (m, 1H), 2.15–2.20 (m, 3H), 2.25–2.30 (m, 1H), 2.35–2.50 (m, 1H), 2.55–2.70 (m, 2H), 2.70–2.77 (m, 1H), 2.81–3.00 (m, 2H), 3.19–3.25 (m, 1H), 3.30–3.40 (m, 1H), 3.49–3.52 (m, 1H), 3.55–3.70 (m, 1H), 3.75–3.85 (m, 1H), 4.05 (br s, 0.5H), 4.12 (br s, 0.5H), 6.34 (s, 0.5H), 6.39 (s, 0.5H), 6.51 (s, 0.5H), 6.53 (s, 0.5H), 6.95–7.01 (m, 1H), 7.05–7.20 (m, 3H); 13C NMR (125 MHz, D2O) δ 12.3, 12.4, 12.6, 20.3, 20.4, 20.5, 21.1, 28.8, 28.9, 30.6, 37.4, 38.6, 41.2, 41.3, 47.0, 47.1, 47.2, 47.5, 49.2, 49.3, 63.5, 64.5, 78.2, 78.4, 110.31, 110.34, 113.9, 114.0, 124.7, 124.75, 124.82, 126.1, 126.16, 126.23, 126.7, 126.9, 127.0, 130.0, 130.1, 130.2, 133.8, 133.9, 134.0, 140.2, 140.7, 145.52, 145.54, 153.8, 158.1; LC-TOF (M+H+) calcd for C22H30ClN4O 401.2108, found 401.2093.
(3R,4R)-tert-butyl 3-((6-(benzyl(tert-butoxycarbonyl)amino)-4-methylpyridin-2-yl)methyl)-4-(2-((tert-butoxycarbonyl)((1S,2R/1R,2S)-2-(3-fluorophenyl)cyclopropyl)amino)ethoxy)pyrrolidine-1-carboxylate (10e)
To a solution of aldehyde 9b (100 mg, 0.18 mmol) in CH2Cl2 (2 mL) was added 3a (60 mg, 0.37 mmol) followed by TEA (125 µL, 0.9 mmol). The mixture was allowed to stir at room temperature for 5 min before NaBH(OAc)3 (50 mg, 0.23 mmol) was added. The reaction mixture was stirred for an additional 3 h then was partitioned between ethyl acetate (50 mL) and brine (25 mL). The organic layer was dried over Na2SO4 and concentrated to yield crude secondary amine. To a solution of the resulting crude amine in MeOH (1.5 mL) was added (Boc)2O (120 mg, 0.56 mmol) and Et3N (75 µL, 0.56 mmol). The reaction mixture was stirred at room temp for 6 h and was partitioned between ethyl acetate (50 mL) and brine (20 mL). The organic layer was dried over Na2SO4, and the solvents were removed by rotary evaporation. The resulting material was purified by flash column chromatography (ethyl acetate/hexanes, 1:4–1:2) to yield 10e (70 mg, 60%) as a colorless oil (71%): 1H NMR (500 MHz, CDCl3) δ 1.15–1.21 (m, 2H), 1.35–1.50 (m, 28H), 1.65–1.80 (br s, 1H), 2.10–2.20 (br s, 1H), 2.25–2.33 (m, 3H), 2.50–2.80 (m, 3H), 2.80–2.97 (m, 1H), 3.00–3.10 (m, 1H), 3.15–3.20 (m, 1H), 3.25–3.50 (m, 3H), 3.55–3.70 (m, 3H), 4.05–4.10 (m, 1H), 5.18 (s, 2H), 6.55–6.65 (m, 1H), 6.70–6.90 (m, 3H), 7.10–7.30 (m, 6H), 7.40–7.45 (m, 1H); 13C NMR (125 MHz, CDCl3) δ 14.1, 14.3, 14.5, 19.5, 21.2, 21.4, 23.0, 28.5, 28.6, 28.9, 30.0, 30.7, 31.7, 34.7, 34.7, 40.2, 40.3, 42.9, 49.1, 49.6, 50.3, 50.4, 51.1, 60.6, 64.6, 78.1, 79.4, 79.7, 80.2, 81.3, 81.5, 112.9, 113.0, 117.2, 117.3, 120.2, 122.2, 126.7, 127.0, 127.2, 127.3, 128.3, 130.1, 140.2, 148.8, 148.8, 154.2, 154.7, 154.7, 155.1, 157.9, 162.1, 164.2, 171.4; LC-TOF (M+H+) calcd for C44H60FN4O7 775.4441, found 775.4432.
(3R,4R)-tert-butyl 3-((6-(benzyl(tert-butoxycarbonyl)amino)-4-methylpyridin-2-yl)methyl)- 4-(2-((tert-butoxycarbonyl)((1S,2R)-2-(3-fluorophenyl)cyclopropyl)amino)ethoxy)pyrrolidine-1-carboxylate (10f)
Compound 10f was synthesized, using a procedure similar to that for 10e (60%), as a colorless oil (71%): 1H NMR (500 MHz, CDCl3) δ 1.10–1.22 (m, 2H), 1.35–1.50 (m, 28H), 1.65–1.80 (br s, 1H), 2.10–2.20 (br s, 1H), 2.20–2.35 (m, 3H), 2.40–2.80 (m, 3H), 2.80–2.95 (m, 1H), 2.97–3.10 (m, 1H), 3.16–3.21 (m, 1H), 3.22–3.44 (m, 3H), 3.45–3.70 (m, 3H), 4.04–4.10 (m, 1H), 5.10–5.25 (br s, 2H), 6.55–6.65 (m, 1H), 6.65–6.95 (m, 3H), 7.10–7.27 (m, 6H), 7.31–7.45 (m, 1H); 13C NMR (125 MHz, CDCl3) δ 14.0, 14.4, 14.5, 19.4, 21.3, 21.4, 22.9, 28.4, 28.7, 28.8, 30.0, 30.9, 31.8, 34.8, 34.9, 40.0, 40.1, 42.9, 49.1, 49.5, 50.2, 50.5, 51.1, 60.7, 64.6, 78.2, 79.4, 79.7, 80.2, 81.4, 81.5, 112.9, 113.0, 117.2, 117.3, 120.2, 122.1, 126.8, 127.1, 127.2, 127.3, 128.3, 130.0, 140.1, 148.7, 148.8, 154.1, 154.6, 154.7, 155.0, 157.9, 162.2, 164.1, 171.4, 171.5; LC-TOF (M+H+) calcd for C44H60FN4O7 775.4441, found 775.4441.
6-(((3R,4R)-4-(2-(((R/S)-1-(3-fluorophenyl)propan-2-yl)amino)ethoxy)pyrrolidin-3-yl)methyl)-4-methylpyridin-2-amine (2e)
To a solution of 10e (0.2 mmol) in EtOH (20 mL) was added Pd(OH)2/C (100 mg). The reaction vessel was charged with H2, heated at 60 °C for 30 h, then cooled to room temperature. The catalyst was removed by filtration, and the resulting solution was concentrated by rotary evaporation. The crude material was purified by flash column chromatography (ethyl acetate/hexanes, 1:4–1:2) to yield 11e as a white foamy solid. To a solution of the resulting 11e in MeOH (0.5 mL) was added 6 N HCl (1.0 mL). The reaction mixture was allowed to stand at room temperature for 16 h. The solvent was removed by rotary evaporation. The crude product was recrystallized using cold diethyl ether to provide 2e as a pale yellow solid (20 mg, 25%): 1H NMR (500 MHz, D2O) δ 1.18–1.20 (m, 3H), 2.20 (s, 3H), 2.60–2.72 (m, 2H), 2.75–2.90 (m, 2H), 3.00–3.10 (m, 2H), 3.15–3.33 (m, 3H), 3.34–3.42 (m, 1H), 3.44–3.60 (m, 3H), 3.70–3.80 (m, 1H), 4.09 (s, 1H), 6.46–4.67 (m, 1H), 6.56 (s, 1H), 6.90–7.10 (m, 3H), 7.20–7.25 (m, 1H); 13C NMR (125 MHz, D2O) δ 15.2, 15.5, 21.3, 29.2, 38.4, 38.5, 41.6, 41.7, 44.2, 44.4, 47.3, 49.5, 55.6, 64.0, 64.4, 78.4, 110.6, 114.3, 114.4, 114.5, 116.2, 116.4, 125.4, 125.6, 130.8, 130.9, 138.3, 145.9, 146.0, 154.1, 158.4, 161.9, 163.9; LC-TOF (M+H+) calcd for C22H32FN4O 387.2560, found 385.2556.
6-(((3R,4R)-4-(2-(((S)-1-(3-fluorophenyl)propan-2-yl)amino)ethoxy)pyrrolidin-3-yl)methyl)-4-methylpyridin-2-amine (2f)
Compound 2f was synthesized using a procedure similar to that for 2e (25%): 1H NMR (500 MHz, D2O) δ 1.28–1.30 (d, J = 6.5 Hz, 3H), 2.31 (s, 3H), 2.75–2.85 (m, 2H), 2.85–3.00 (m, 2H), 3.10–3.20 (m, 2H), 3.30–3.37 (m, 2H), 3.37–3.46 (m, 1H), 3.46–3.55 (m, 1H), 3.55–3.70 (m, 3H), 3.80–3.90 (m, 1H), 4.20 (s, 1H), 6.57 (s, 1H), 6.67 (s, 1H), 7.00–7.05 (m, 1H), 7.06–7.09 (m, 1H), 7.09–7.13 (d, J = 7.5 Hz, 1H), 7.33–7.40 (dd, J = 7.5, 14.5 Hz, 1H); 13C NMR (125 MHz, D2O) δ 15.0, 21.0, 29.0, 38.2, 41.4, 43.9, 47.0, 49.3, 55.4, 64.2, 78.1, 110.4, 114.0, 114.1, 114.3, 115.9, 116.1, 125.18, 125.20, 130.6, 130.7, 130.07, 130.13, 145.7, 153.9, 158.1, 161.7, 163.6; LC-TOF (M+H+) calcd for C22H32FN4O 387.2560, found 385.2547.
Inhibitory Activity Assay
The three isozymes, murine macrophage iNOS, rat nNOS, and bovine eNOS, were recombinant enzymes, overexpressed in E. coli and isolated as reported.30 IC50 values for inhibitors 2a–f were measured for the three different isoforms of NOS using L-arginine as a substrate. The formation of nitric oxide was measured using a hemoglobin capture assay described previously.31 All NOS isozymes were assayed at room temperature in a 100 mM Hepes buffer (pH 7.4) containing 10 µM L-arginine, 1.6 mM CaCl211.6 µg /mL calmodulin, 100 µM DTT, 100 µM NADPH, 6.5 µM H4B, 3.0 µM oxyhemoglobin (for iNOS assays, no Ca2+ and calmodulin were added). The assay was initiated by the addition of enzyme, and the initial rates of the enzymatic reactions were determined by monitoring the formation of NOhemoglobin complex at 401 nm for 60 sec. The corresponding Ki values of inhibitors were calculated from the IC50 values using equation 1 with known Km values (rat nNOS, 1.3 µM; iNOS, 8.3 µM; eNOS, 1.7 µM).
| (1) |
Crystal Structure Determination
The preparation of protein samples of nNOS and eNOS heme domain and the procedures for crystal growth and inhibitor soaks were the same as reported.19 The cryogenic (100K) X-ray diffraction data were collected remotely at various beamlines at Stanford Synchrotron Radiation Light Source (SSRL) through the data collection control software Blu-Ice32 and crystal mounting robot.
Raw data frames were indexed, integrated, and scaled using HKL2000.33 The reflection files were then concerted to the mtz format using routines Scalepack2mtz and Truncate in the CCP4 suite.34 The binding of each inhibitor was revealed by the initial difference Fourier maps calculated with REFMAC.35 The inhibitor molecules were then modeled in COOT36 and refined using REFMAC. Water molecules were added in REFMAC and inspected visually through COOT. The TLS37 protocol was implemented in the final stage of refinements with each subunit as one TLS group. The omit Fo – Fc electron density map was calculated at the end of the refinement with the coefficient DELFWT in a mtz file generated by a round of TLS refinement fed with an inhibitor-free coordinate file. The refined structures were validated in COOT before deposition in the RCSB protein data bank. The crystallographic data collection and structure refinement statistics are summarized in Table 2 with PDB accession codes included.
Table 2.
Crystallographic Data Collection and Refinement Statistics
| Data set1 | nNOS-2b | nNOS-2c | nNOS-2d | nNOS-2e | nNOS-2f | eNOS-2d | eNOS-2e |
|---|---|---|---|---|---|---|---|
| Data collection | |||||||
| PDB code | 3RQJ | 3RQK | 3RQL | 3RQM | 3RQN | 3RQO | 3RQP |
| Space group | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 |
| Cell dimensions | |||||||
| a, b, c (Å) | 51.8, 110.7, 164.3 | 52.0, 111.5, 164.4 | 51.7, 110.8, 163.9 | 51.9, 110.4, 164.0 | 52.1, 110.9, 164.2 | 58.1, 106.6, 156.5 | 57.9, 106.6, 156.9 |
| Resolution (Å) | 1.85 (1.88−1.85) | 2.20 (2.24−2.20) | 1.93 (1.96−1.93) | 1.95 (1.98−1.95) | 1.95 (1.98−1.95) | 2.08 (2.12−2.08) | 2.35 (2.39−2.35) |
| Rsym or Rmerg | 0.044 (0.63) | 0.076 (0.54) | 0.052 (0.59) | 0.061 (0.59) | 0.056 (0.55) | 0.072 (0.66) | 0.086 (0.67) |
| I / σI | 36.2 (2.5) | 25.6 (2.1) | 29.8 (2.3) | 22.9 (2.0) | 27.0 (2.6) | 22.8 (2.1) | 13.8 (1.5) |
| No. unique reflections | 82,378 | 47,658 | 71,869 | 69,867 | 70,044 | 59,039 | 41,301 |
| Completeness (%) | 99.6 (99.7) | 97.9 (97.5) | 99.7 (100.0) | 99.7 (99.5) | 99.9 (100.0) | 99.6 (99.9) | 99.5 (99.2) |
| Redundancy | 4.1 (4.1) | 3.8 (3.7) | 4.1 (4.0) | 3.8 (3.6) | 4.1 (4.1) | 3.7 (3.8) | 3.6 (3.5) |
| Refinement | |||||||
| Resolution (Å) | 1.85 | 2.21 | 1.93 | 1.95 | 1.95 | 2.08 | 2.35 |
| No. reflections used | 78,043 | 45,045 | 68,065 | 66,358 | 66,498 | 55,805 | 39,184 |
| Rwork / Rfree2 | 0.189/0.221 | 0.200/0.258 | 0.190/0.235 | 0.173/0.208 | 0.173/0.209 | 0.174/0.221 | 0.178/0.228 |
| No. atoms | |||||||
| Protein | 6,677 | 6,662 | 6,697 | 6,672 | 6,698 | 6,416 | 6,462 |
| Ligand/ion | 185 | 185 | 188 | 179 | 185 | 193 | 203 |
| Water | 328 | 170 | 312 | 538 | 396 | 403 | 255 |
| R.m.s. deviations | |||||||
| Bond lengths (Å) | 0.015 | 0.015 | 0.017 | 0.013 | 0.013 | 0.015 | 0.014 |
| Bond angles (°) | 1.508 | 1.535 | 1.637 | 1.372 | 1.352 | 1.557 | 1.522 |
See Table 1 for nomenclature and chemical formula of inhibitors.
Rfree was calculated with the 5% of reflections set aside throughout the refinement. For each NOS isoform the set of reflections for the Rfree calculation were kept the same for all data sets according to those used in the data of the starting model.
Scheme 3.
Synthesis of 2a–d.a
a Reagents and conditions: (a) amine hydrochloride, TEA, NaHB(OAc)3, room temp, 3 h, 81–87%; (d) 6 N HCl in MeOH (2:1), room temp, 16 h, 90–99%. The R/S notations shown indicate the chirality of the two chiral centers of the cyclopropyl ring; the pyrrolidine ring has (R,R) stereochemistry in all of the compounds.
Acknowledgments
The authors are grateful for financial support from the National Institutes of Health (GM49725 to RBS and GM57353 to TLP). We thank Dr. Bettie Sue Siler Masters (NIH grant GM52419, with whose laboratory P.M. and L.J.R. are affiliated). B.S.S.M. also is grateful to the Welch Foundation for a Robert A. Welch Distinguished Professorship in Chemistry (AQ0012). P.M. is supported by grant 0021620849 from MSMT of the Czech Republic. We also thank the staff at SSRL for their assistance during the remote X-ray diffraction data collections.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alderton WK, Cooper CE, Knowles RG. Biochem. J. 2001;357:593. doi: 10.1042/0264-6021:3570593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marletta MA. J. Biol. Chem. 1993;268:12231. [PubMed] [Google Scholar]
- 3.Marletta MA. Cell. 1994;78:927. doi: 10.1016/0092-8674(94)90268-2. [DOI] [PubMed] [Google Scholar]
- 4.Hall AV, Antoniou H, Wang Y, Cheung AH, Arbus AM, Olson SL, Lu WC, Kau CL, Marsden PA. J. Biol. Chem. 1994;269:33082. [PubMed] [Google Scholar]
- 5.Rosen GM, Tsai P, Weaver J, Porasuphatana S, Roman LJ, Starkov AA, Fiskum G, Pou S. J. Biol. Chem. 2002;277:40275. doi: 10.1074/jbc.M200853200. [DOI] [PubMed] [Google Scholar]
- 6.Zhang L, Dawson VL, Dawson TM. Pharmacol. Ther. 2006;109:33. doi: 10.1016/j.pharmthera.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 7.Dorheim MA, Tracey WR, Pollock JS, Grammas P. Biochem. Biophys. Res. Commun. 1994;205:659. doi: 10.1006/bbrc.1994.2716. [DOI] [PubMed] [Google Scholar]
- 8.Norris PJ, Waldvogel HJ, Faull RLM, Love DR, Emson PC. Neuronsicence. 1996;72:1037. doi: 10.1016/0306-4522(95)00596-x. [DOI] [PubMed] [Google Scholar]
- 9.Sims NR, Anderson MF. Neurochem. Int. 2002;40:511. doi: 10.1016/s0197-0186(01)00122-x. [DOI] [PubMed] [Google Scholar]
- 10.Calabrese V, Mancuso C, Calvani M, Rizzarelli E, Butterfield DA, Stella AM. Nat. Rev. 2007;8:766. doi: 10.1038/nrn2214. [DOI] [PubMed] [Google Scholar]
- 11.Hobbs AJ, Higgs A, Moncada S. Annu. Rev. Pharmacol. Toxicol. 1999;39:191. doi: 10.1146/annurev.pharmtox.39.1.191. [DOI] [PubMed] [Google Scholar]
- 12.Delker SL, Ji H, Li H, Jamal J, Fang J, Xue F, Silverman RB, Poulos TL. J. Am. Chem. Soc. 2010;132:5437. doi: 10.1021/ja910228a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ji H, Tan S, Igarashi J, Li H, Derrick M, Martásek P, Roman LJ, Vásquez-Vivar J, Poulos TL, Silverman RB. Ann. Neurol. 2009;65:209. doi: 10.1002/ana.21555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.(a) Lawton GR, Ranaivo HR, Wing LK, Ji H, Xue F, Martesek P, Roman LJ, Watterson DM, Silverman RB. Bioorg. Med. Chem. 2009;17:2371. doi: 10.1016/j.bmc.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ji H, Delker SL, Li H, Martasek P, Roman LJ, Poulos TL, Silverman RB. J. Med. Chem. 2010;53:7804. doi: 10.1021/jm100947x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silverman RB. San Diego: Academic Press; 2004. [Google Scholar]
- 16.Xue F, Fang J, Lewis WW, Martasek P, Roman LJ, Silverman RB. Bioorg. Med. Chem. Lett. 2010;20:554. doi: 10.1016/j.bmcl.2009.11.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xue F, Huang, Li HJ, Ji H, Fang J, Poulos TL, Silverman RB. Bioorg. Med. Chem. 2010;18:6526. doi: 10.1016/j.bmc.2010.06.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delker SL, Xue F, Li H, Jamal J, Silverman RB, Poulos TL. Biochemistry. 2010;49:10803. doi: 10.1021/bi1013479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xue F, Li H, Fang J, Poulos TL, Silverman RB. J. Am. Chem. Soc. 2010;132:14229. doi: 10.1021/ja106175q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xue F, Delker SL, Li H, Fang J, Jamal J, Silverman RB, Poulos TL. J. Med. Chem. 2011;54:2039. doi: 10.1021/jm101071n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fishlock D, Perdicakis B, Montgomery HJ, Guillemette JG, Jervisb E, Lajoiea GA. Bioorg. Med. Chem. 2003;11:869. doi: 10.1016/s0968-0896(02)00554-0. [DOI] [PubMed] [Google Scholar]
- 22.Yamanoi K, Ohfune Y. Tetrahedron Lett. 1988;29:1181. [Google Scholar]
- 23.Shimamoto K, Ishida M, Shinozaki H, Ohfune Y. J. Org. Chem. 1991;56:4167. [Google Scholar]
- 24.Kozikowski A, editor. New York: Raven Press; 1993. [Google Scholar]
- 25.Wermuth CG, editor. San Diego: Academic Press; 1996. [Google Scholar]
- 26.Perrin CL, Fabian MA, Rivero IA. Tetrahedron. 1999;55:5773. [Google Scholar]
- 27.Pryde DC, Cook AS, Burring DJ, Jones LH, Foll S, Platts MY, Sanderson V, Corless M, Stobie A, Middleton DS, Foster L, Barker L, Graaf PVD, Stacey P, Kohl C, Coggon S, Beaumont K. Bioor. Med. Chem. 2007;15:142. doi: 10.1016/j.bmc.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 28.Tchilibon S, Kim S-K, Gao Z-G, Harris BA, Blaustein JB, Gross AS, Duong HT, Melman N, Jacobson KA. Bioorg. Med. Chem. 2004;12:2021. doi: 10.1016/j.bmc.2004.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xue F, Kraus JM, Jansen Labby K, Ji H, Mataka J, Xia G, Li H, Delker SL, Roman LJ, Martásek P, Poulos TL, Silverman RB. J. Med. Chem. 2011;54:6399. doi: 10.1021/jm200411j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.(a) Hevel JM, White KA, Marletta MA. J. Biol. Chem. 1991;266:22789. [PubMed] [Google Scholar]; (b) Roman LJ, Sheta EA, Martásek P, Gross SS, Liu Q, Masters BSS. Proc.Natl. Acad. Sci. U.S.A. 1995;92:8428. doi: 10.1073/pnas.92.18.8428. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Martasek P, Liu Q, Roman LJ, Gross SS, Sessa WC, Masters BSS. Biochem. Biophys. Res. Commun. 1996;219:359. doi: 10.1006/bbrc.1996.0238. [DOI] [PubMed] [Google Scholar]
- 31.Hevel JM, Marletta MA. Method Enzymol. 1994;233:250. doi: 10.1016/s0076-6879(94)33028-x. [DOI] [PubMed] [Google Scholar]
- 32.McPhillips TM, McPhillips SE, Chiu HJ, Cohen AE, Deacon AM, Ellis PJ, Garman E, Gonzalez A, Sauter NK, Phizackerley RP, Soltis SM, Kuhn P. J Synchrotron Radiat. 2002;9:401. doi: 10.1107/s0909049502015170. [DOI] [PubMed] [Google Scholar]
- 33.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 34.Collaborative Computational Project Number 4, the CCP4 Suite: Programs for Protein Crystallography. Acta cryst. 1994;D50:760. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 35.Murshudov GN, Vagin AA, Dodson EJ. Acta Cryst. 1997;D53:240. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K. Acta Cryst. 2004;D60:2126. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.Winn MD, Isupov MN, Murshudov GN. Acta Cryst. 2001;D57:122. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]