Abstract
When coexpressed with its cognate amber suppressing , a pyrrolysyl-tRNA synthetase mutant N346A/C348A is able to genetically incorporate twelve meta-substituted phenylalanine derivatives into proteins site-specifically at amber mutation sites in Escherichia coli. These genetically encoded noncanonical amino acids resemble phenylalanine in size and contain diverse bioorthogonal functional groups such as halide, trifluoromethyl, nitrile, nitro, ketone, alkyne, and azide moieties. The genetic installation of these functional groups in proteins provides multiple ways to site-selectively label proteins with biophysical and biochemical probes for their functional investigations. We demonstrate that a genetically incorporated trifluoromethyl group can be used as a sensitive 19F NMR probe to study protein folding/unfolding, and that genetically incorporated reactive functional groups such as ketone, alkyne, and azide moieties can be applied to site-specifically label proteins with florescent probes. This critical discovery allows the synthesis of proteins with diverse bioorthogonal functional groups for a variety of basic studies and biotechnology development using a single recombinant expression system.
Introduction
Site-selective modification is an important biotechnological strategy to introduce new functionalities to proteins. Examples include the PEGylation of therapeutic proteins to prolong their in vivo half lives,(1) fluorescent labeling of proteins for protein folding/unfolding analysis,(2) linking proteins with photocrosslinkers for protein-protein/DNA interaction studies,(3, 4) and introducing NMR, EPR, and IR probes for protein functional investigations.(5-7) Traditionally, protein modifications use the high reactivity of cysteine and lysine side chains.(8, 9) However, this approach lacks selectivity. Modern techniques that attempt to achieve site-selective protein modification include the genetic noncanonical amino acid (NAA) incorporation,(10) native chemical ligation, expression protein ligation,(11, 12) enzymatic and chemical modifications of peptide tags,(13-15) and specific modifications of protein N- and C-termini.(16, 17) These techniques, in general, seek to introduce functional groups that do not exist in the 20 canonical amino acids (CAAs). Some of these functional groups can undergo bioorthogonal reactions for further modifications. Using orthogonal aminoacyl-tRNA synthetases (aaRSs) from different cell origins and their corresponding suppressing tRNAs, more than 70 NAAs have been genetically incorporated into proteins in E. coli, Saccharomyces cerevisiae, and mammalian cells.(18-21) These NAAs contain most functional groups that are chemically stable in an aqueous environment. Genetic incorporation of these NAAs has been used to synthesize proteins containing biophysical probes for structural and functional investigations, as well as proteins with different biochemical functionalities.
Although powerful, the genetic NAA incorporation approach usually requires using a uniquely evolved aaRS for a specific NAA. AaRS-tRNACUA pairs with different origins might have to be used for different cell strains due to the non-orthogonal nature of some aaRS-tRNACUA pairs toward other endogenous aaRS-tRNA pairs in these hosts. A single aaRS that allows genetically encoding NAAs with diverse bioorthogonal functional groups in both prokaryotic and eukaryotic cells will relieve the burden of evolving aaRSs and searching for aaRS-tRNACUA pairs with different origins for use in different cell strains. It will also provide a single recombinant expression system for genetic incorporation of diverse NAAs so that parallel studies can be carried out rapidly. One system that meets this standard is the wild-type pair. It has been demonstrated that wild-type PylRS has a relatively broad substrate spectrum. However, all NAAs that are recognized by wild-type PylRS are much larger than the 20 CAAs.(22-24) They are not optimal choices for protein modifications when it is necessary to minimize the extent of structural perturbations. In addition, NAAs with functional groups such as ketone, halide, nitro, and nitrile moieties have not been successfully incorporated into proteins using wild-type PylRS. Schultz and coworkers recently demonstrated that a mutant Methanocaldococcus jannaschii tyrosyl-tRNA synthetase pair that was originally evolved for para-cyano-phenylalanine is able to mediate genetic incorporation of multiple para-substituted phenylalanine derivatives into proteins in E. coli.(25) However, this pair can only be used in bacteria due to its non-orthogonal nature of the pair in eukaryotic cells.(26)
In one of our previous studies, we described a rationally designed PylRS mutant N346A/C348A.(27) When coexpressed with , this enzyme mediates genetic incorporation of six tyrosine derivatives with large O-alkyl substituents into proteins at amber mutation sites in E. coli. Here, we reveal that the same enzyme-tRNA pair allows genetic incorporation of twelve phenylalanine derivatives with small meta-substituents and various bioorthogonal functional groups in E. coli. This new discovery allows the synthesis of diversely functionalized proteins using a single recombinant protein expression system in E. coli. Given the orthogonal nature of the pair in S. cerevisiae, Caenorhabditis elegans and mammalian cells, the pair can potentially be transferred into these cellular systems as well.(28-32)
Results
Genetic Incorporation of nine meta-substituted phenylalanine derivatives
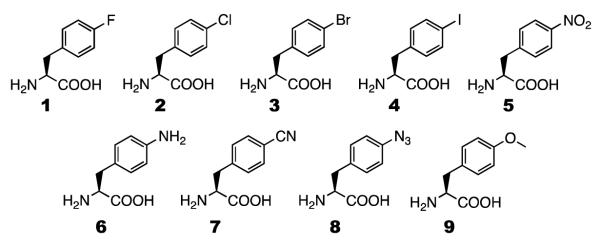
In a previous study, we show that, in comparison to wild-type PylRS, N346A/C348A has an enhanced recognition of phenylalanine and mediates its incorporation at an amber mutation site in E. coli.(27) E. coli BL21 cells transformed with two plasmids pEVOL-pylT-N346A/C348A and pEVOL-pylT-sfGFP2TAG that carry genes coding N346A/C348A, and superfolder green fluorescent protein (sfGFP) with an amber mutation at its S2 position were not able to express sfGFP in GMML (a minimal medium supplemented with 1% glycerol and 0.3 mM leucine). However, supplementing GMML with 2 mM phenylalanine induced sfGFP expression with a yield of 1.5 mg/L. When six tyrosine derivatives with large O-alkyl groups were used to supplement GMML to a final concentration of 2 mM, they all led to much higher sfGFP expression levels than that for phenylalanine, indicating that N346A/C348A might have higher binding affinities toward these NAAs. In contrast, when 2 mM phenylalanine derivatives with small para-substituents (2-9 shown in Scheme 1) were used to supplement GMML, sfGFP expression levels were much lower than for phenylalanine. Except for 9, the addition of all other NAAs led to negligible sfGFP expression levels. Following this previous study, recognition of 1 by N346A/C348A was recently tested. However, the same transformed E. coli BL21 cells could not grow at all in GMML supplemented with 2 mM 1. Apparently 1 is highly toxic to E. coli BL21 cells.
Scheme 1.
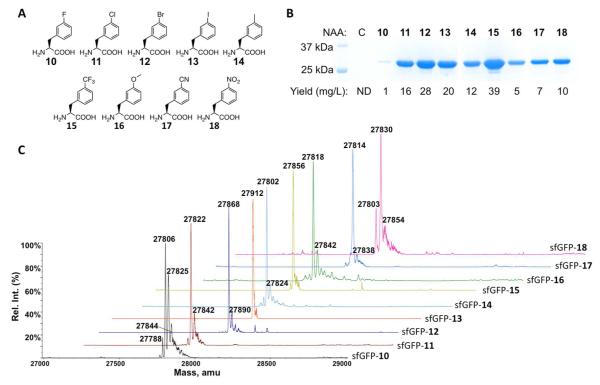
Given that N346A/C348A has relatively high substrate promiscuity, four meta-halo-phenylalanine derivatives were also tested as substrates of N346A/C348A. Similar to the results shown in the previous study, E. coli BL21 cells transformed with pEVOL-pylT-N346A/C348A and pEVOL-pylT-sfGFP2TAG displayed undetectable sfGFP expression in GMML; however, providing 2 mM meta-chloro-phenylalanine, meta-bromo-phenylalanine, or meta-iodo-phenylalanine (11, 12, and 13 in Figure 1A) in GMML induced sfGFP overexpression (Figure 1B). Expression levels under these conditions are all one order of magnitude higher than using 2 mM phenylalanine as a substrate of N346A/C348A. All three purified sfGFP variants had molecular weights determined by electrospray ionization mass spectrometry (ESI-MS) analysis that agreed well with their theoretical molecular weights (Figure 1C and Table 1). ESI-MS spectra of all three proteins also showed a small side peak corresponding to 20-24 Da more than the expected molecular weight. These small side peaks possibly originated from the sodium adduct ions that are commonly found in the ESI-MS analysis. Similar peaks were also found in other sfGFP proteins described later in this study. The uptake of meta-fluoro-phenylalanine (10 in Figure 1A) by N346A/C348A was also tested. Although providing 2 mM 10 in GMML induced sfGFP expression, the yield was significantly lower than for 11-13. Given that 10 could be misincorporated at phenylalanine sites, this low sfGFP expression yield might be due to a toxic effect of 10.(33) Indeed, the harvested cell mass obtained from GMML supplemented with 10 was considerably less than from GMML supplemented with either of 11-13. In addition, the ESI-MS spectrum of the purified sfGFP incorporated with 10 displayed several side peaks that indicate misincorporation of 10 at phenylalanine sites (Figure 1C and Table 1). Two mass peaks at 27,825 Da and 27,844 Da clearly indicate the presence of one and two additional residues of 10 at phenylalanine sites. A small peak at 27,788 Da matches the exact molecular weight of the sfGFP protein with a phenylalanine residue at the S2 amber mutation site. This indicates that 10 is not fully able to inhibit the binding of phenylalanine to N346A/C348A even though the recognition of phenylalanine by N346A/C348A is considered weak and the concentration of phenylalanine in E. coli grown in minimal media is only about 20 μM.(34) Mass peaks that indicate the incorporation of phenylalanine at S2 of sfGFP could not be identified in ESI-MS spectra of sfGFP proteins incorporated with other NAAs shown in this study.
Figure 1.
(A) Structures of NAAs 10-18. (B) Site-specific incorporation of 10-18 into sfGFP at its S2 position. C indicates a control experiment without the addition of any NAA. ND stands for non-detected. (C) Deconvoluted ESI-MS spectra of sfGFP variants incorporated with 10-18. In sfGFP-X, X is one of 10-18 incorporated at the S2 position.
Table 1. Calculated and detected molecular weights of sfGFP variants with different NAAs incorporated at S2.
| NAA | Calculated mass (Da) | Detected mass (Da)a |
|---|---|---|
| 10 | 27806 | 27806, 27825, 27844 |
| 11 | 27822 | 27822 |
| 12 | 27867 | 27868 |
| 13 | 27914 | 27912 |
| 14 | 27802 | 27802 |
| 15 | 27856 | 27856 |
| 16 | 27818 | 27818 |
| 17 | 27812 | 27813 |
| 18 | 27829 | 27829 |
| 19 | 27830 | 27829 |
| 20 | 27813 | 27814 |
| 21 | 27833 | 27830 |
with an error of ±1 Da.
Following the success with 11-13, recognition of five other commercially available phenylalanine derivatives with small meta-substituents (14-18 in Figure 1A) by N346A/C348A were tested. Growing E. coli BL21 cells transformed with pEVOL-pylT-N346A/C348A and pET-pylT-sfGFP2TAG in GMML supplemented with 2 mM of either of 14-18 led to overexpression of sfGFP (Figure 1B). The major detected molecular weights of purified sfGFP variants determined by the ESI-MS analysis agreed well with their theoretic molecular weights (Figure 1C and Table 1). In most spectra, a minor mass peak that is at 20-24 Da higher than expected was detected. As discussed previously, these peaks are possibly from sodium adduct ions. The spectrum of sfGFP incorporated with 18 also displayed a minor peak at 27,803 Da. Although this peak matches the molecular weight of sfGFP incorporated with tyrosine at its S2 position (theoretical molecular weight: 27,804 Da), a tyrosine residue at S2 is unlikely. The previous study of N346A/C348A showed that this enzyme was not able to mediate genetic incorporation of tyrosine at an amber mutation site even when 2 mM tyrosine was provided in GMML.(27) Since E. coli contains a nitroreductase enzyme,(35) it is possible that 18 in sfGFP was reduced by this enzyme to meta-amino-phenylalanine (the theoretical molecular weight of sfGFP with meta-amino-phenylalanine incorporated at its S2 position is 27,802 Da). 18 could also be reduced to meta-amino-phenylalanine that is then incorporated into sfGFP. To rule out this possibility, E. coli BL21 cells were grown in GMML supplemented with 2 mM meta-amino-phenylalanine. However, sfGFP was expressed at a very low and barely detectable level under this condition. Among all nine meta-substituted phenylalanine derivatives, 15 had the highest corresponding sfGFP expression yield. Among the NAAs that has been tested so far, 15 contains the most hydrophobic meta-substituent, a property that likely contributes to its strong interaction with the hydrophobic active site of N346A/C348A and therefore leads to its high incorporation rate. In comparison to all O-alkyl tyrosine derivatives and all para-substituted phenylalanine derivatives that were tested previously, most meta-substituted phenylalanine derivatives gave higher incorporation levels.
Using 15 as a sensitive 19F NMR probe to study protein folding/unfolding
Among 10-18, 12 and 13 could potentially undergo Suzuki-Miyaura cross-coupling reactions for site-selective modifications,(36) while 15 has three equivalent fluorine atoms that can serve as a sensitive 19F NMR probe for protein folding/unfolding and structural dynamics analysis,(37) the nitrile group in 17 is a strong IR probe,(7) and 18 can be used as a distance probe due to its ability to quench the intrinsic fluorescence of a tryptophan residue in a protein.(38) In this study, we chose to demonstrate the application of 15 in protein folding/unfolding analysis. Mehl and coworkers previously showed that para-trifluoromethyl-phenylalanine could be genetically incorporated into enzymes in E. coli using an evolved M. jannaschii tyrosyl-tRNA synthetase pair, and that genetically incorporated para-trifluoromethyl-phenylalanine residues in enzymes could be used as 19F NMR probes to sense the binding of substrates, inhibitors, and cofactors.(37) The same technique was also extended to side chain relaxation analysis of a SH3 domain.(39) In these studies, para-trifluoromethyl-phenylalanine was incorporated at the protein surface, which potentially avoided disruption of protein folding due to the steric hindrance that might be introduced by the incorporated para-trifluoromethyl-phenylalanine residue. Folding disruption might be a concern when para-trifluoromethyl-phenylalanine is incorporated in a protein interior. Given that 15 is an isomer of para-trifluoromethyl-phenylalanine, it is a useful alternative of para-trifluoromethyl-phenylalanine for protein folding/unfolding and structural dynamics analysis and might be a solution when the incorporation of para-trifluoromethyl-phenylalanine into a protein interior disrupts folding. In the following experiments, 15 was genetically incorporated into sfGFP at both its surface and its interior. The resulting proteins were used to undergo unfolding analysis using 19F NMR.
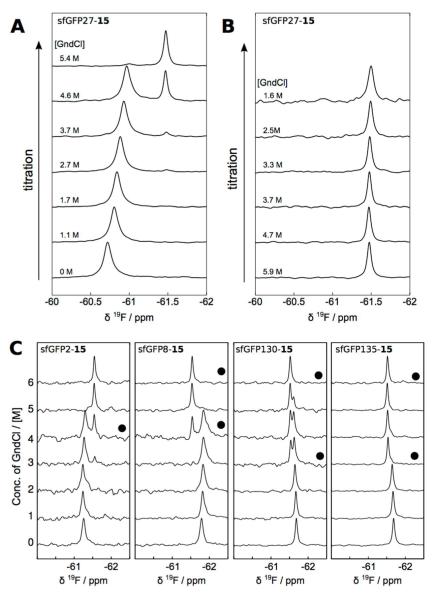
To express sfGFP incorporated with 15 at F27 (sfGFP27-15), a plasmid pBAD-sfGFP27TAG that carried a sfGFP gene with an amber mutation at F27 was constructed. F27 is located at the second β-strand of sfGFP. Its side chain faces towards the protein interior and is surrounded by several hydrophobic residues (SI Figure 1). After unfolding, sfGFP is expected to completely expose F27 to a hydrophilic environment. We anticipated that 15 at F27 of sfGFP would show a large chemical shift change during the unfolding process. Together with pEVOL-pylT-N346A/C348A, pBAD-sfGFP27TAG was used to transform E. coli Top10 cells. The transformed cells were then grown in lysogenic broth (LB) media supplemented with 1 mM 15 and induced with the addition of 0.2% arabinose. This procedure resulted in sfGFP27-15 expression at an expression level of 79 mg/L (SI Figure 2). Only a minimal amount of sfGFP was expressed in LB without a NAA supplement. The purified sfGFP27-15 had a fluorescent spectrum and intensity similar to wild-type sfGFP, and a sample of 15N-sfGFP-15 also showed a similar, but not identical, fingerprint as wild-type sfGFP in a [15N,1H] correlation spectrum (SI Figures 3 and 4).(40) Differences included small shifts in various peaks such as e.g. those near 10 ppm/112 ppm and the appearance of a weak peak near 10.2 ppm/128 ppm. The relatively close agreement of the spectra suggests that replacing F27 with 15 does not significantly affect the protein folding and chromophore formation processes.
Purified sfGFP27-15 was then used to undergo unfolding analysis in the presence of guanidinium chloride (GndCl). In the titration of sfGFP27-15 against GndCl, 19F NMR signals of sfGFP27-15 were detected (Figure 2A). At low denaturant concentration, the chemical shift of folded sfGFP27-15 was dependent on the GndCl concentration. This concentration dependence may likely be explained by a local structural change prior to denaturation. The appearance of the peak at 61.5 ppm indicates that the unfolding transition in this titration occurred at around 4.6 M GndCl, which based on the time of 1 h between titrated points appears consistent with an unfolding hysteresis observed in ref. (40). Likewise, a stepwise titration of decreasing GndCl indicates the presence of unfolded sfGFP27-15 to a much lower GndCl concentration of 1.6 M (Figure 2B). Similarly, sfGFP proteins with 15 incorporated at S2, F8, F130, and N135 positions were recombinantly expressed (SI Figure 2) and used to undergo unfolding studies in the presence of GndCl and determined by 19F NMR (Figure 2C). The locations of the mutations have been chosen such that S2 and F8 are at the beginning and in the first α-helix of the protein (F27 is in a β-sheet) and F130 and N135 are located in a loop, with the latter more exposed to the solvent than the former. All proteins were expressed as soluble and folded forms. In all mutants, the titration resulted in the appearance of a second peak corresponding to the unfolded form at intermediate concentrations of GdnCl. Mutations located in well-defined secondary structures of sfGFP, including those at F27, S2 and F8, show larger chemical shift differences than those located in loop regions. Further, a gradual chemical shift change, in some cases concomitant with line broadening, can be observed prior to denaturation. This change is also most prominent in the F8 and F27 mutants, which are located in well-structured regions. We also carried out fluorescence equilibrium unfolding analysis for all synthesized 15-containing sfGFP proteins. Except sfGFP8-15, all other 15-containing sfGFP proteins displayed unfolding patterns very similar to sfGFP proteins with phenylalanine at 15-corresponding sites (SI Figure 5-9).
Figure 2.
(A) Unfolding of sfGFP27-15, by stepwise addition of GndCl, characterized by 19F NMR. Titrations were carried out by sequential addition of GndCl, starting from 0 M. (B) Spectra of unfolded sfGFP27-15 upon stepwise reduction of GndCl. Precipitation occurred below 1.6 M GndCl. In A and B, time between each point was 1 h, and measurement temperature was 25 °C. (C) Unfolding of sfGFP2-15, sfGFP8-15, sfGFP130-15, and sfGFP135-15 by stepwise addition of GndCl, characterized by 19F NMR. Time between spectra was 2-3 h, except for the spectra indicated with (●), where additional time of 1-2 days was allowed for transitions to occur.
The significant change in 19F chemical shifts observed in this study illustrates that 15 can be used as a sensitive site-specific probe for protein folding, or potentially for protein-ligand interaction. Since 19F allows for highly receptive, background free NMR detection, and the chemical shift of this nucleus is sensitive to structural changes,(41) 15 inserted at a known position will be particularly useful for measurements with large proteins, or with dilute samples, where chemical shift assignments typically cannot be obtained. The present study also indicates that 15 resembles phenylalanine in size and its incorporation into a protein may not disrupt its folding, although a careful analysis of this property for any particular protein incorporated with 15 is strongly recommended.
Genetic incorporation of three chemically reactive meta-substituted phenylalanine derivatives for site-selective protein modifications
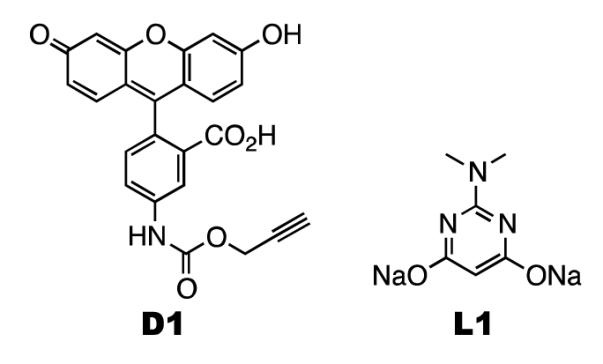
Since we previously showed that a para-iodo-phenylalanine residue that was incorporated into a protein using an evolved pair could be fluorescently labeled using a Suzuki-Miyaura cross-coupling reaction,(42) a similar reaction to label 13 is not repeated here. Instead, we carried out a recently developed copper-free Sonogashira cross-coupling reaction(43) to label 13. Both dye D1 and a palladium ligand L1 shown in scheme 2 were synthesized according to literature procedures.(43) Incubating sfGFP with 13 incorporated at S2 (sfGFP-13) together with dye D1 in the presence of the L1-palladium(II) complex and sodium ascorbate at pH 7 for 3 h led to no fluorescein-labeled sfGFP-13 that could be detected on a denaturing SDS-PAGE gel. To avoid the background fluorescent emission of sfGFP itself, the sample was treated with 10% SDS and boiled for 20 min to totally denature sfGFP before it was analyzed. The same treatment was carried out with all other sfGFP variants that were labeled with small molecule fluorescein dyes, as described later. Prolonging the time of the reaction between sfGFP-13 and D1 to overnight induced a large amount of protein aggregation but no apparently improved fluorescein labeling (data not shown). Since the L1-palladium(II) complex needs to react with 13 to form an intermediate for a further reaction with D1,(43) we think the low concentration of sfGFP-13 makes its reaction with D1 kinetically unfavorable. The protein aggregation might be due to the interaction between exposed sulfur-containing cysteine and methionine residues of sfGFP-13 and the L1-palladium(II) complex.
Scheme 2.
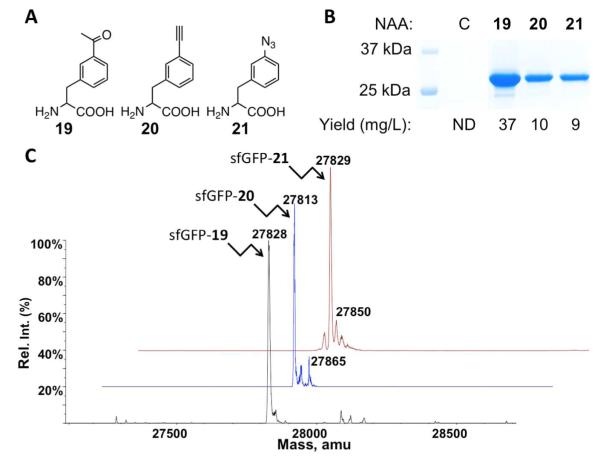
In order to synthesize proteins incorporated with chemically active meta-substituted phenylalanine derivatives for their site-selective modifications, we synthesized three NAAs shown as 19-21 in Figure 3A and tested their recognition by N346A/C348A. These NAAs were synthesized as racemic mixtures and used directly in the following analysis without further purification. Specific recognition of L-amino acids by aminoacyl-tRNA synthetases and the ribosome has been demonstrated elsewhere.(44),(45) E. coli BL21 cells transformed with pEVOL-pylT-N346A/C348A and pET-pylT-sfGFP2TAG were used to test 19-21. The transformed cells displayed undetectable sfGFP expression in GMML; however, providing 19, 20, or 21 in GMML induced sfGFP overexpression (Figure 3B). Since 19-21 are racemic mixtures, 4 mM was added to yield a final concentration of 2 mM for the L-isoform. Expression levels under these conditions are much higher than using 2 mM phenylalanine as a substrate of N346A/C348A. All three purified sfGFP variants had molecular weights determined by ESI-MS that agreed well with their theoretical molecular weights (Figure 3C and Table 1). The sfGFP variant with 20 incorporated at S2 (sfGFP-20) also showed a minor mass peak at 27,865 Da. This peak is possibly from sfGFP-20 with N-terminal acetylation.(46) A minor peak at 27,850 Da was also found in the spectrum of sfGFP with 21 incorporated at S2 (sfGFP-21). As discussed previously, this peak is possibly from the sodium adduct ion of sfGFP-21.
Figure 3.
(A) Structures of 19-21. (B) Site-specific incorporation of 19-21 into sfGFP at its S2 position. C indicates a control experiment without the addition of any NAA. ND stands for non-detected. (C) Deconvoluted ESI-MS spectra of sfGFP variants incorporated with 19-21. In sfGFP-X, X is one of 19-21 represents incorporated at the S2 position.
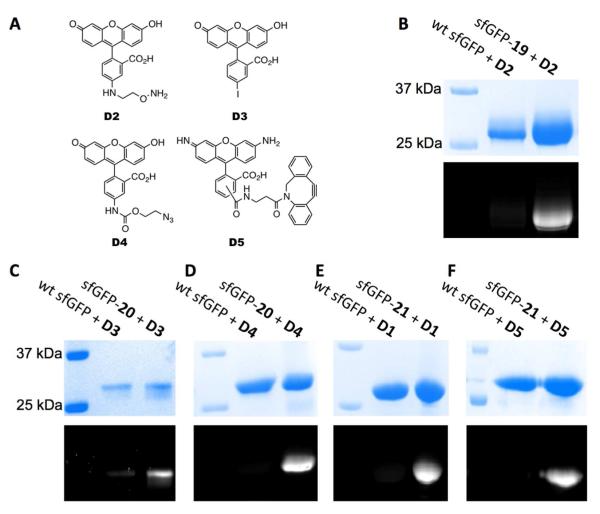
With the sfGFP protein with 19 incorporated at S2 (sfGFP-19), sfGFP-20, and sfGFP-21 in hand, we then proceeded to their labeling with corresponding fluorescein dyes. Besides using the copper-free Sonogashira cross-coupling reaction to label sfGFP-21, the oxime formation reaction to label sfGFP-19 and the alkyneazide Husigen cycloaddition reaction(47, 48) to label sfGFP-20 and sfGFP-21 were also tested. Four additional fluorescein dyes D2-D5 (Figure 4A) were either synthesized or purchased.(43, 49) As expected, sfGFP-19 reacted specifically with dye D2 at pH 4 to give a fluorescein-labeled sfGFP-19 that showed intense fluorescence in a denaturing SDS-PAGE gel under UV irradiation (Figure 4B). As a control, wild-type sfGFP could not be labeled with D2 under the same condition. We also tested the reaction of sfGFP-19 with D2 at pH 7. An overnight incubation gave a negligible fluorescein-labeled sfGFP-19 (data not shown). This is consistent with what was described previously by Brustad et al.(50) Two reactions, the copper-free Sonogashira cross-coupling reaction and the azide-alkyne Husigen cycloaddition reaction were employed to label sfGFP-20. The 3h reaction between sfGFP-20 and dye D3 in the presence of the L1-palladium(II) complex and sodium ascorbate at pH 7 resulted in a detectable fluorescein-labeled protein (Figure 4C). To achieve this labeling, 10 eq. of the catalyst was preincubated with 10 eq. of D3 for 1 h to obtain an activated reagent cocktail. The final labeling reaction was performed with the addition of another 90 eq. of D3 and 1 eq. of sfGFP-20 to this cocktail. Although we have obtained labeling of sfGFP-20 with D3, we did notice that a large portion of sfGFP-20 underwent aggregation during the labeling process. There was a relatively low level of background labeling of wild-type sfGFP with D3 (Figure 4C). Labeling of sfGFP-20 with dye D4 was carried out under an optimized condition that contained 0.1 mM Cu(I):tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA) complex, 0.4 mM additional TBTA, 5 mM sodium ascorbate, and 1 mM NiCl2 for 3h.(51) The finally labeled protein on a denaturing SDS-PAGE gel was intensely fluorescent under UV irradiation (Figure 4D). This same click labeling reaction was also used to label sfGFP-21 specifically with D1 (Figure 4E). We also carried out the copper-free cyclooctyne-azide cycloaddition reaction to label sfGFP-21 with D5. An overnight direct incubation of sfGFP-21 with D5 led to a fluorescein-labeled protein that showed strong fluorescence under UV irradiation. For all three click conditions, similar reactions with wild-type sfGFP were used as controls that did not give any detectable fluorescein-labeled final products. These assembled data demonstrate that diverse bioorthogonal groups installed in proteins using the pair enable a variety of ways for site-specific protein modifications.
Figure 4.
(A) Structures of dyes D2-D5. (B) Specific labeling of sfGFP-19 with D2. (C) Specific labeling of sfGFP-20 with D3. (D) Specific labeling of sfGFP-20 with D4. (E) Specific labeling of sfGFP-21 with D1. (F) Specific labeling of sfGFP-21 with D5. In B-F, the top panels show denaturing SDS-PAGE analysis of sfGFP proteins with Coomassie blue staining and the bottom panels are from fluorescent imaging of the same gels before Coomassie blue staining. Wt sfGFP stands for wild-type sfGFP.
Discussion
Genetic incorporation of meta-substituted phenylalanine derivatives
Using evolved pairs, Schultz and coworkers showed that more than twenty para-substituted phenylalanine derivatives could be genetically incorporated into proteins at amber mutation sites in E. coli.(7, 18, 38, 50, 52-68) Compared to para-substituted phenylalanine derivatives, genetic incorporation of meta-substituted phenylalanine derivatives is far less explored. Zhang et al. demonstrated that a meta-substituted phenylalanine derivative, 19, could be genetically encoded in E. coli using an evolved pair.(69) However, there has been no follow-up study since this work was originally published in 2003. Developing methods for genetic incorporation of other meta-substituted phenylalanine derivatives will not only expand the genetically encoded NAA inventory, leaving more choices for protein engineering when subtle variations are necessary, but also help understand the scope of NAA tolerance of the cellular protein translation machinery. In this study, all twelve meta-substituted phenylalanine derivatives (10-21) have small substituents. Their incorporation into protein interiors is expected not to significantly disturb protein folding. This may allow using these NAAs, e.g. 15 and 17, to probe local environments in the interior of proteins. Although para-substituted phenylalanine derivatives with similar functional groups have been incorporated into proteins in E. coli using multiple evolved pairs, 10-21 may be excellent alternatives when the incorporation of para-substituted phenylalanine derivatives disturbs protein folding. There is also an advantage of using the pair. This pair was derived from the wild-type pair that is orthogonal in E. coli, S. cerevisiae, and mammalian cells. The pair can be potentially transferred into eukaryotic cell systems for the expression of proteins with diverse functional groups that cannot be achieved in E. coli due to protein folding problems or the need of post-translational modifications. So far, evolved pairs can only be used in E. coli due to recognition of by endogenous yeast and mammalian aaRSs (Liu & Schultz, unpublished data). Since genetic incorporation of NAAs has been used in phage display,(70) the current development also provides more chemical moieties that can be loaded into phage-displayed libraries to expand the structural diversity for drug discovery.
Recognition of meta-substituted phenylalanine derivatives by N346A/C348A
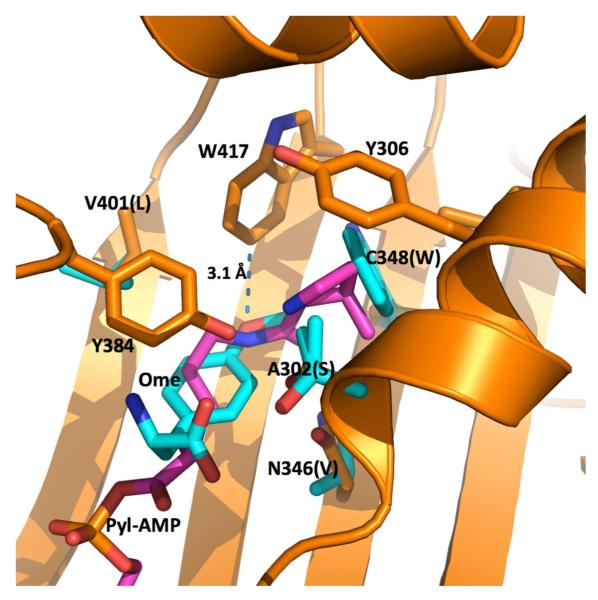
In the previous study, we show that N346A/C348A is able to charge with phenylalanine for genetic incorporation of phenylalanine at an amber stop codon in E. coli.(27) The phenylalanine recognition of N346A/C348A is probably due to the removal of the N346 amide that can potentially prevent the binding of phenylalanine to the active site.(27, 42) Since there is a large hydrophobic pocket at the active site of N346A/C348A, we predicted and demonstrated that tyrosine derivatives with large O-alkyl substituents could be recognized by N346A/C348A and genetically encoded in E. coli using the pair. However, phenylalanine derivatives with small para-substituents could not be incorporated into proteins using the same system. Figure 5 presents the superimposed active site structures of two PylRS variants. One is wild-type PylRS with pyrrolysyl-AMP binding at the active site and the second one is a mutant PylRS (OmeRS) complex with 9.(71, 72) OmeRS was specifically evolved for 9 by Wang and coworkers. Although Y384 is not observed in the crystal structure of the OmeRS complex with 9, it is expected to locate at the top of 9 in a similar way as in the wild-type PylRS complex with pyrrolysyl-AMP. Yokoyama and coworkers have demonstrated that Y384 is critical for the aminoacylation of and multiple structures of PylRS have shown that Y384 is at the same position.(23, 73, 74) With Y384 at its top, the aromatic side chain of 9 might have strong pi-pi stacking interactions with the side chain of Y384 and the two backbone amides of residues 419-421 (not shown in Figure 5). The methoxy oxygen atom of 9 is very close to W417 with a distance of 3.1 AN. It is possible that all para- and meta-substituted phenylalanine derivatives have a similar binding pattern when they interact with N346A/C348A. When a phenylalanine derivative has a small substituent such as azide, halide, nitrile, and nitro moieties, this small substituent that is still considerably larger than an oxygen atom might have a strong steric clash with W417 so that its binding to the active site of N346A/C348A is disfavored. 9 has a flexible O-methyl group that might be located in the deep pocket of N346A/C348A for a favorable interaction. Tyrosine derivatives with larger O-alkyl groups that can potentially interact with the deep pocket with strong hydrophobic interactions might have more favorable interactions with N346A/C348A. The structure of OmeRS also shows that two active site residues at 346 and 348 are very close to the meta position of the phenyl ring of 9. Since these two residues are mutated to alanine residues in N346A/C348A, it is possible that a big hydrophobic pocket forms around the side-chain meta position of phenylalanine when it binds to N346A/C348A. This hydrophobic pocket might contribute to favorable interactions between N346A/C348A and phenylalanine derivatives with small hydrophobic meta-substituents. The incorporation level of one particular phenylalanine derivative is related to the aminoacyl-tRNA synthesis step, the binding of aminoacyl-tRNA to elongation factor Tu, the association of aminoacyl-tRNA to the ribosome, and the final peptidyl transfer reaction. Therefore, there may not be a direct correlation between the incorporation level of a NAA and its binding affinity to N346A/C348A. Assays are necessary to characterize the binding of phenylalanine derivatives to N346A/C348A and their aminoacylation reactions with . In addition, the crystal structures of N346A/C348A complexes with different phenylalanine derivatives will provide additional structural insights. These are directions we intend to pursue in the future.
Figure 5.
The superimposed structures of the PylRS complex with pyrrolysyl-AMP (Pyl-AMP) and the OmeRS complex with 9 (Ome). The PylRS complex with pyrrolysyl-AMP is shown in orange for the protein carbon atoms and pink for the pyrrolysyl-AMP carbon atoms. Four mutated residues in OmeRS and the ligand 9 are shown in cyan for the carbon atoms. Letters in the parentheses indicate four mutated residues in OmeRS.
Conclusion
In summary, we demonstrated genetic incorporation of twelve meta-substituted phenylalanine derivatives with diverse bioorthogonal functional groups into proteins in E. coli using a single PylRS mutant N346A/C348A together with its cognate amber suppressing . This critical development makes it possible to use a single recombinant expression system to synthesize proteins with diverse chemical moieties for parallel studies. Among these twelve NAAs, 15 can be used as a sensitive 19F NMR probe, 17 can potentially serve as a IR probe,(7) and 18 can be potentially used as a distance probe through its ability to quench the intrinsic fluorescence of tryptophan residues in proteins, 13, 19, 20, and 21 can undergo various bioorthogonal reactions for site-specific protein labeling with different biophysical and biochemical probes (see SI Table 1 for other potential applications).(38) Except for 19 that was previously incorporated into proteins using an evolved M. jannaschii pair, genetic incorporation of all other meta-substituted phenylalanine derivatives shown in this study is reported for the first time. In addition, all twelve meta-substituted phenylalanine derivatives resemble phenylalanine in size. The incorporation of these meta-substituted phenylalanine derivatives into proteins even in the protein interiors may not significantly disrupt protein folding as we demonstrated in the sfGFP unfolding studies using a genetically incorporated 15. Since the wild-type pair has been used in S. cerevisiae and mammalian cells for NAA incorporation, it is expected that the pair can be simply transferred to these cellular systems, potentially allowing various bioconjugation reactions to be carried out in these cells. We are currently engaged in transferring the system to eukaryotic cells and the progress will be reported later on.
Experimental Section
NAAs, ligand L1 and dyes
NAAs 1-18 were purchased from Chem Impex Inc. Dye D5 was obtained from Click Chemistry Tools Inc. Ligand L1 and dyes D1-D4 were synthesized according to literature procedures.(43, 51, 75) The synthesis of NAAs 19-21 is described in detail in the supporting information.
Plasmid construction
Construction of pEVOL-pylT-N346A/C348A and pET-sfGFP2TAG was described previously.(27) Plasmid pBAD-sfGFP135TAG that carries a sfGFP gene with an amber mutation at its 135 position was a gift from Dr. Ryan Mehl at Oregon State University. Plasmids pBAD-sfGFP8TAG, pBAD-sfGFP27TAG, and pBAD-sfGFP130TAG that carry sfGFP genes with amber mutations at positions 8, 27, and 130, respectively, were derived from plasmid pBAD-sfGFP, a gift also from Dr. Ryan Mehl. Construction of these plasmids was carried out using a site-directed mutagenesis protocol that was based on Phusion DNA polymerase. In brief, two oligonucleotide primers, one of which covers the mutation site, were used to amplify the whole plasmid of pBAD-sfGFP to give a blunt-end PCR product. This PCR product was phosphorylated by T4 polynucleotide kinase and then underwent self-ligation using T4 DNA ligase. Primers pBAD-sfGFP-F8TAG-F (5′-actggtgttgttcctattcttgttgaacttg-3′) and pBAD-sfGFP-F8TAG-R (5′-ctaaagttcttcacctttagaaaccatggttaattcc-3′) were used for the construction of pBAD-sfGFP8TAG. Primers pBAD-sfGFP-F27TAG-F (5′-tctgttcgtggtgaaggtgaaggtgatg-3′) and pBAD-sfGFP-F27TAG-R (5′-ctatttatgaccattaacatcaccatcaagttc-3′) were used for the construction of pBAD-sfGFP27TAG. Primers pBAD-sfGFP-F130TAG-F (5′-aaagaagatggtaatattcttggtcataaacttg-3′) and pBAD-sfGFP-F130TAG-R (5′-ctaatcaatacctttaagttcaatacgattaac-3′) were used for the construction of pBAD-sfGFP130TAG.
Protein expression of purification
To express sfGFP incorporated with a NAA at its S2 position, E. Coli BL21(DE3) cells were cotransformed with pEVOL-pylT-N346A/C348A and pET-sfGFP2TAG. Cells were recovered in 1 mL LB media for 1 h at 37 °C and then plated on a LB agar plate that contained 34 μg/mL chloramphenicol and 100 μg/mL ampicillin. A single colony was then selected and grown overnight in 10 mL LB media. This overnight culture was used to inoculate 500 mL GMML media supplemented with 34 μg/mL chloramphenicol and 100 μg/mL ampicillin. Cells were grown at 37 °C in an incubating shaker (225 r.p.m.) and protein expression was induced by the addition of 1 mM IPTG, 0.2% arabinose and 2 mM of a NAA (for 19-21, 4 mM was used) when OD600 reached 0.5. After 12 h induction, cells were harvested, resuspended in a lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8.0), and sonicated. The cell lysate was clarified by centrifugation (60 min, 11000 r.p.m., 4 °C). The decanted clear supernatant was incubated with 3 mL Ni2+-NTA resin (Qiagen) (2 h, 4 °C) and washed with 30 mL of the lysis buffer. Protein was finally eluted out using the elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, pH 8.0). Eluted fractions were collected and concentrated. The collected fractions were further purified on an anion exchange column. The buffer was later changed to 10 mM ammonium bicarbonate using an Amicon Ultra-15 Centrifugal Filter Devices (10,000 MWCO, Millipore). The purified proteins were analyzed by 12% SDS-PAGE.
To express sfGFP27-15, E. Coli Top10 cells were cotransformed with pEVOL-pylT-N346A/C348A and pBAD-sfGFP27TAG. Cells were recovered in 1 mL LB media for 1 h at 37 °C and then plated on a LB agar plate that contained 34 μg/mL chloramphenicol and 100 μg/mL ampicillin. A single colony was then selected and grown overnight in 10 mL LB media. This overnight culture was used to inoculate 500 mL LB media supplemented with 34 μg/mL chloramphenicol and 100 μg/mL ampicillin. Cells were grown at 37 °C in an incubating shaker (225 r.p.m.) and the expression of sfGFP27-15 was induced by the addition of 0.2% arabinose and 2 mM 15 when OD600 reached 0.5. The expressed sfGFP27-15 was purified according to the same procedures for other sfGFP variants described above. To express sfGFP8-15, sfGFP130-15, and sfGFP135-15, plasmids pBAD-sfGFP8TAG, pBAD-sfGFP130TAG, and pBAD-sfGFP135TAG were used separately to transform Top10 cells that contained pEVOL-pylT-N346A/C348A. The following expression and purification procedures of sfGFP8-15, sfGFP130-15, and sfGFP135-15 were as same as those for sfGFP27-15.
To express 15N-labeled sfGFP27-15, the Top10 cells transformed with pEVOL-pylT-N346A/C348A and pBAD-sfGFP27TAG were cultured in LB media to OD600 0.7-0.8. The cultured cells were collected and then transferred to GMML media with 15N-labeled ammonium chloride as a sole nitrogen source (15N, 99%, Cambridge Isotope Laboratories Inc.). The expression of 15N-labeled sfGFP27-15 was induced by the addition of 0.2% arabinose and 1 mM 15 for 6 h. 15N-labeled wild-type sfGFP was expressed similarly using Top10 cells transformed with pBAD-sfGFP. The expressed proteins were purified similarly as other sfGFP variants.
ESI-MS analysis of sfGFP variants
ESI-MS experiments were performed using an Applied Biosystems QSTAR Pulsar (Concord, ON, Canada) equipped with a nanoelectrospray ion source. The MS data were acquired in positive ion mode (500-2000 Da) using a spray voltage of +2000 V and flow rate of 700 nL/min. Data analysis and protein signal extraction were performed using the BioAnalyst software (Applied Biosystems). A mass range of m/z 500-2000 was used for spectral deconvolution and the output range was 20,000 to 30,000 Da using a resolution of 0.1 Da and S/N threshold of 20. The final determined molecular weights of sfGFP variants have errors of roughly ± 1 Da.
NMR Measurements
sfGFP27-15 samples for 19F NMR unfolding studies were prepared at a concentration of 0.7 mM in a buffer that contained 12 mM phosphate (pH 7.0), 140 mM NaCl, and 3 mM KCl. In the titration experiment, protein solution was diluted with the same buffer with or without GndCl, and subsequently re-concentrated using a centrifugal concentration device. The concentration of GndCl at each titration point was determined based on 1H NMR. Chemical shifts were referenced against an external standard of trifluoroacetic acid at −75.2 ppm in D2O, which was located in the inner part of a NMR tube with two concentric compartments. 19F spectra were recorded at a temperature of 25 °C on a 400 MHz spectrometer equipped with a Broadband Observe (BBO) probe (Bruker Biospin). The 15N-labeled sfGFP27-15 protein was prepared in the same solution as the unlabeled protein. The HMQC spectrum of 15N-labeled sfGFP27-15 was recorded at 35°C on a 500 MHz NMR spectrometer equipped with a cryoprobe. 19F NMR folding studies of other sfGFP variants were carried out similarly.
Fluorescence equilibrium unfolding analysis
Different sfGFP proteins in PBS buffer were incubated with GndCl (1.0 to 7.0 M) for 12-48 h in room temperature. Their fluorescent intensities at 510 nm were then measured using a FL600 Microplate Fluorescence Reader (excitation wavelength: 450 nm). Midpoint denaturant concentrations of GndCl were determined from fitting the normalized fluorescent intensity (F) data to the equation F = ¼ a + b/(1 + (C/Cm)h), where a, b, Cm and h are adjustable constants, and C is the molarity of the GndCl.
Sonogashira cross coupling
To label sfGFP-13, a stock solution of 10 mM D1 was prepared in DMSO and a stock solution of 80 mM sodium ascorbate was prepared in deionized H2O. To an ice-chilled 0.75-mL centrifuge tube were added sequentially an aliquot of sfGFP-13 (0.69 μL, 2.0 mg/mL), an aliquot of 10 mM Pd(OAc)2·L1 complex solution (0.69 μL, 6.9 nmol), and an aliquot of sodium ascorbate (0.69 μL, 55.2 nmol), and the mixture was stirred at 37 °C for 1 h to obtain an activated reagent cocktail. 8 μL of 10 mL D1 was then added to this activated reagent cocktail. The final mixture was stirred for 2 h. To label sfGFP-20, a stock solution of 10 mM D3 was prepared in DMSO. To an ice-chilled 0.75-mL centrifuge tube were added sequentially an aliquot of D3 (0.69 μL, 6.9 nm), an aliquot of 10 mM Pd(OAc)2·L1 complex solution (0.69 μL, 6.9 nmol), and an aliquot of sodium ascorbate (0.69 μL, 55.2 nmol), and the mixture was stirred at 37 °C for 1 h to obtain an activated reagent cocktail. Separately, to a 0.75-mL centrifuge tube containing 50 μL PBS buffer was added 10 mM D3 (6.21 μL, 62.1 nmol) and 10 μL of sfGFP-20 solutions (2.0 mg/mL in 1x PBS buffer, 0.69 nmol). This mixture of D3 and sfGFP-20 was then added to the previously activated reagent cocktail and stirred at 37 °C for 2 h. The same two reactions were also carried out with wild-type sfGFP. The finally fluorescein-labeled proteins were analyzed by SDS-PAGE (12%) for fluorescence image (BioRad ChemiDoc XRS+) and further Coomassie blue staining.
Oxime formation
sfGFP-19 (100 μL, 2 mg/mL in PBS buffer) was dialyzed against a 10 mM sodium acetate buffer (pH 4) and concentrated to 100 μL. D2 (4 μL, 20 mM in DMSO) was then added and incubated at 25 °C overnight. The same reaction was carried out with wild-type sfGFP. The finally labeled proteins were analyzed by SDS-PAGE (12%) for fluorescence image and further Coomassie blue staining.
Azide-alkyne cycloaddition
For the copper-catalyzed azide-alkyne cycloaddition, to sfGFP-20 (0.5 mg/mL in PBS buffer, 161μL, 2.78 nmol) or sfGFP-21 (2.6 mg/mL in PBS buffer, 31 μL, 2.78 nmol) was added CuSO4 (100 μM final concentration), NiCl2 (1 mM), tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA, stock solution in DMSO, 500 μM) and a corresponding dye D4 or D1 (in DMSO, 50 equiv. of the protein) sequentially, followed by the addition of sodium ascorbate (5 mM). The reaction was performed at 25 °C for 3 h. EDTA (0.5 M) was then added to the reaction mixture to chelate the two metals. The same reaction was carried out with wild-type sfGFP. The copper free cycloaddition reaction between sfGFP-21 (0.5 mg/mL in PBS buffer, 161μL, 2.78 nmol) and D5 (in DMSO, 50 equiv. of the protein) was performed at 25 °C overnight. Same reactions were carried out with wild-type sfGFP. All finally labeled proteins were analyzed by SDS-PAGE (12%) for fluorescence image and further Coomassie blue staining.
Supplementary Material
Acknowledgements
Support from the National Institute of Health (grant 1R01CA161158), the National Science Foundation (grants CHEM-1148684, CHE-0846402 and CHE-0840464), and the Welch Foundation (grant A-1715) is gratefully acknowledged. The authors thank R. Mehl at Oregon State University for providing plasmids pBAD-sfGFP and pBAD-sfGFP135TAG and Y. Rezenom from Laboratory for Biological Mass Spectrometry at Texas A&M University for characterizing proteins with electrospray ionization mass spectrometry.
Footnotes
Authors declare no competing interest.
Associated content
Synthesis of 19-21, all DNA and protein sequences, and additional figures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Roberts MJ, Bentley MD, Harris JM. Chemistry for peptide and protein PEGylation. Adv Drug Deliv Rev. 2002;54:459–476. doi: 10.1016/s0169-409x(02)00022-4. [DOI] [PubMed] [Google Scholar]
- 2.Giepmans BN, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 3.Zhang M, Lin S, Song X, Liu J, Fu Y, Ge X, Fu X, Chang Z, Chen PR. A genetically incorporated crosslinker reveals chaperone cooperation in acid resistance. Nat Chem Biol. 2011;7:671–677. doi: 10.1038/nchembio.644. [DOI] [PubMed] [Google Scholar]
- 4.Chin JW, Schultz PG. In vivo photocrosslinking with unnatural amino Acid mutagenesis. Chembiochem. 2002;3:1135–1137. doi: 10.1002/1439-7633(20021104)3:11<1135::AID-CBIC1135>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 5.Jones DH, Cellitti SE, Hao X, Zhang Q, Jahnz M, Summerer D, Schultz PG, Uno T, Geierstanger BH. Site-specific labeling of proteins with NMR-active unnatural amino acids. J Biomol NMR. 2010;46:89–100. doi: 10.1007/s10858-009-9365-4. [DOI] [PubMed] [Google Scholar]
- 6.Altenbach C, Flitsch SL, Khorana HG, Hubbell WL. Structural studies on transmembrane proteins. 2 Spin labeling of bacteriorhodopsin mutants at unique cysteines. Biochemistry. 1989;28:7806–7812. doi: 10.1021/bi00445a042. [DOI] [PubMed] [Google Scholar]
- 7.Schultz KC, Supekova L, Ryu Y, Xie J, Perera R, Schultz PG. A genetically encoded infrared probe. J Am Chem Soc. 2006;128:13984–13985. doi: 10.1021/ja0636690. [DOI] [PubMed] [Google Scholar]
- 8.Chalker JM, Bernardes GJL, Lin YA, Davis BG. Chemical Modification of Proteins at Cysteine: Opportunities in Chemistry and Biology. Chem-Asian J. 2009;4:630–640. doi: 10.1002/asia.200800427. [DOI] [PubMed] [Google Scholar]
- 9.Hanai R, Wang JC. Protein footprinting by the combined use of reversible and irreversible lysine modifications. Proc Natl Acad Sci U S A. 1994;91:11904–11908. doi: 10.1073/pnas.91.25.11904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L, Schultz PG. Expanding the genetic code. Angew Chem Int Ed Engl. 2004;44:34–66. doi: 10.1002/anie.200460627. [DOI] [PubMed] [Google Scholar]
- 11.Dawson PE, Muir TW, Clark-Lewis I, Kent SB. Synthesis of proteins by native chemical ligation. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 12.Muir TW, Sondhi D, Cole PA. Expressed protein ligation: a general method for protein engineering. Proc Natl Acad Sci U S A. 1998;95:6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen I, Howarth M, Lin W, Ting AY. Site-specific labeling of cell surface proteins with biophysical probes using biotin ligase. Nat Methods. 2005;2:99–104. doi: 10.1038/nmeth735. [DOI] [PubMed] [Google Scholar]
- 14.Carrico IS, Carlson BL, Bertozzi CR. Introducing genetically encoded aldehydes into proteins. Nat Chem Biol. 2007;3:321–322. doi: 10.1038/nchembio878. [DOI] [PubMed] [Google Scholar]
- 15.Gautier A, Juillerat A, Heinis C, Correa IR, Jr., Kindermann M, Beaufils F, Johnsson K. An engineered protein tag for multiprotein labeling in living cells. Chem Biol. 2008;15:128–136. doi: 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 16.Ren H, Xiao F, Zhan K, Kim YP, Xie H, Xia Z, Rao J. A biocompatible condensation reaction for the labeling of terminal cysteine residues on proteins. Angew Chem Int Ed Engl. 2009;48:9658–9662. doi: 10.1002/anie.200903627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilmore JM, Scheck RA, Esser-Kahn AP, Joshi NS, Francis MB. N-terminal protein modification through a biomimetic transamination reaction. Angew Chem Int Ed Engl. 2006;45:5307–5311. doi: 10.1002/anie.200600368. [DOI] [PubMed] [Google Scholar]
- 18.Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 19.Chin JW, Cropp TA, Anderson JC, Mukherji M, Zhang Z, Schultz PG. An expanded eukaryotic genetic code. Science. 2003;301:964–967. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- 20.Wu N, Deiters A, Cropp TA, King D, Schultz PG. A genetically encoded photocaged amino acid. J Am Chem Soc. 2004;126:14306–14307. doi: 10.1021/ja040175z. [DOI] [PubMed] [Google Scholar]
- 21.Neumann H, Peak-Chew SY, Chin JW. Genetically encoding N(epsilon)-acetyllysine in recombinant proteins. Nat Chem Biol. 2008;4:232–234. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]
- 22.Nguyen DP, Lusic H, Neumann H, Kapadnis PB, Deiters A, Chin JW. Genetic encoding and labeling of aliphatic azides and alkynes in recombinant proteins via a pyrrolysyl-tRNA Synthetase/tRNA(CUA) pair and click chemistry. J Am Chem Soc. 2009;131:8720–8721. doi: 10.1021/ja900553w. [DOI] [PubMed] [Google Scholar]
- 23.Yanagisawa T, Ishii R, Fukunaga R, Kobayashi T, Sakamoto K, Yokoyama S. Crystallographic studies on multiple conformational states of active-site loops in pyrrolysyl-tRNA synthetase. J Mol Biol. 2008;378:634–652. doi: 10.1016/j.jmb.2008.02.045. [DOI] [PubMed] [Google Scholar]
- 24.Polycarpo CR, Herring S, Berube A, Wood JL, Soll D, Ambrogelly A. Pyrrolysine analogues as substrates for pyrrolysyl-tRNA synthetase. FEBS Lett. 2006;580:6695–6700. doi: 10.1016/j.febslet.2006.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Young DD, Young TS, Jahnz M, Ahmad I, Spraggon G, Schultz PG. An evolved aminoacyl-tRNA synthetase with atypical polysubstrate specificity. Biochemistry. 2011;50:1894–1900. doi: 10.1021/bi101929e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu W, Brock A, Chen S, Chen S, Schultz PG. Genetic incorporation of unnatural amino acids into proteins in mammalian cells. Nat Methods. 2007;4:239–244. doi: 10.1038/nmeth1016. [DOI] [PubMed] [Google Scholar]
- 27.Wang YS, Fang X, Wallace AL, Wu B, Liu WR. A rationally designed pyrrolysyl-tRNA synthetase mutant with a broad substrate spectrum. J Am Chem Soc. 2012;134:2950–2953. doi: 10.1021/ja211972x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hancock SM, Uprety R, Deiters A, Chin JW. Expanding the genetic code of yeast for incorporation of diverse unnatural amino acids via a pyrrolysyl-tRNA synthetase/tRNA pair. J Am Chem Soc. 2010;132:14819–14824. doi: 10.1021/ja104609m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gautier A, Nguyen DP, Lusic H, An W, Deiters A, Chin JW. Genetically encoded photocontrol of protein localization in mammalian cells. J Am Chem Soc. 2010;132:4086–4088. doi: 10.1021/ja910688s. [DOI] [PubMed] [Google Scholar]
- 30.Greiss S, Chin JW. Expanding the genetic code of an animal. J Am Chem Soc. 2011;133:14196–14199. doi: 10.1021/ja2054034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parrish AR, She X, Xiang Z, Coin I, Shen Z, Briggs SP, Dillin A, Wang L. Expanding the genetic code of Caenorhabditis elegans using bacterial aminoacyl-tRNA synthetase/tRNA pairs. ACS Chem Biol. 2012;7:1292–1302. doi: 10.1021/cb200542j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Q, Wang L. New methods enabling efficient incorporation of unnatural amino acids in yeast. J Am Chem Soc. 2008;130:6066–6067. doi: 10.1021/ja800894n. [DOI] [PubMed] [Google Scholar]
- 33.Pine MJ. Comparative physiological effects of incorporated amino acid analogs in Escherichia coli. Antimicrob Agents Chemother. 1978;13:676–685. doi: 10.1128/aac.13.4.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zenno S, Koike H, Kumar AN, Jayaraman R, Tanokura M, Saigo K. Biochemical characterization of NfsA, the Escherichia coli major nitroreductase exhibiting a high amino acid sequence homology to Frp, a Vibrio harveyi flavin oxidoreductase. J Bacteriol. 1996;178:4508–4514. doi: 10.1128/jb.178.15.4508-4514.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chalker JM, Wood CS, Davis BG. A convenient catalyst for aqueous and protein Suzuki-Miyaura cross-coupling. J Am Chem Soc. 2009;131:16346–16347. doi: 10.1021/ja907150m. [DOI] [PubMed] [Google Scholar]
- 37.Jackson JC, Hammill JT, Mehl RA. Site-specific incorporation of a (19)F-amino acid into proteins as an NMR probe for characterizing protein structure and reactivity. J Am Chem Soc. 2007;129:1160–1166. doi: 10.1021/ja064661t. [DOI] [PubMed] [Google Scholar]
- 38.Tsao ML, Summerer D, Ryu Y, Schultz PG. The genetic incorporation of a distance probe into proteins in Escherichia coli. J Am Chem Soc. 2006;128:4572–4573. doi: 10.1021/ja058262u. [DOI] [PubMed] [Google Scholar]
- 39.Shi P, Wang H, Xi Z, Shi C, Xiong Y, Tian C. Site-specific (1)(9)F NMR chemical shift and side chain relaxation analysis of a membrane protein labeled with an unnatural amino acid. Protein Sci. 2011;20:224–228. doi: 10.1002/pro.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andrews BT, Gosavi S, Finke JM, Onuchic JN, Jennings PA. The dual-basin landscape in GFP folding. Proc Natl Acad Sci U S A. 2008;105:12283–12288. doi: 10.1073/pnas.0804039105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frieden C, Hoeltzli SD, Ropson IJ. NMR and protein folding: equilibrium and stopped-flow studies. Protein Sci. 1993;2:2007–2014. doi: 10.1002/pro.5560021202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang YS, Russell WK, Wang Z, Wan W, Dodd LE, Pai PJ, Russell DH, Liu WR. The de novo engineering of pyrrolysyl-tRNA synthetase for genetic incorporation of L-phenylalanine and its derivatives. Mol Biosyst. 2011;7:714–717. doi: 10.1039/c0mb00217h. [DOI] [PubMed] [Google Scholar]
- 43.Li N, Lim RK, Edwardraja S, Lin Q. Copper-free Sonogashira cross-coupling for functionalization of alkyne-encoded proteins in aqueous medium and in bacterial cells. J Am Chem Soc. 2011;133:15316–15319. doi: 10.1021/ja2066913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hendrickson TL, de Crecy-Lagard V, Schimmel P. Incorporation of nonnatural amino acids into proteins. Annu Rev Biochem. 2004;73:147–176. doi: 10.1146/annurev.biochem.73.012803.092429. [DOI] [PubMed] [Google Scholar]
- 45.Dedkova LM, Fahmi NE, Golovine SY, Hecht SM. Enhanced D-amino acid incorporation into protein by modified ribosomes. J Am Chem Soc. 2003;125:6616–6617. doi: 10.1021/ja035141q. [DOI] [PubMed] [Google Scholar]
- 46.Lin BH, Liu WR, Lin CY, Hsu ST, Yang S, Kuo CC, Hsu CH, Hsieh WF, Chien FS, Chang CS. Single Domain m-Plane ZnO Grown on m-Plane Sapphire by Radio Frequency Magnetron Sputtering. ACS Appl Mater Interfaces. 2012;4:5333–5337. doi: 10.1021/am301271k. [DOI] [PubMed] [Google Scholar]
- 47.Kolb HC, Finn MG, Sharpless KB. Click chemistry: Diverse chemical function from a few good reactions. Angew Chem Int Edit Engl. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 48.Agard NJ, Prescher JA, Bertozzi CR. A strain-promoted [3 + 2] azide-alkyne cycloaddition for covalent modification of biomolecules in living systems. J Am Chem Soc. 2004;126:15046–15047. doi: 10.1021/ja044996f. [DOI] [PubMed] [Google Scholar]
- 49.Wu B, Wang Z, Huang Y, Liu WR. Catalyst-Free and Site-Specific One-Pot Dual-Labeling of a Protein Directed by Two Genetically Incorporated Noncanonical Amino Acids. Chembiochem. 2012;9:1405–1408. doi: 10.1002/cbic.201200281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brustad EM, Lemke EA, Schultz PG, Deniz AA. A general and efficient method for the site-specific dual-labeling of proteins for single molecule fluorescence resonance energy transfer. J Am Chem Soc. 2008;130:17664–17665. doi: 10.1021/ja807430h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wan W, Huang Y, Wang Z, Russell WK, Pai PJ, Russell DH, Liu WR. A facile system for genetic incorporation of two different noncanonical amino acids into one protein in Escherichia coli. Angew Chem Int Ed Engl. 2010;49:3211–3214. doi: 10.1002/anie.201000465. [DOI] [PubMed] [Google Scholar]
- 52.Brustad E, Bushey ML, Lee JW, Groff D, Liu W, Schultz PG. A genetically encoded boronate-containing amino acid. Angew Chem Int Ed Engl. 2008;47:8220–8223. doi: 10.1002/anie.200803240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zeng H, Xie J, Schultz PG. Genetic introduction of a diketone-containing amino acid into proteins. Bioorg Med Chem Lett. 2006;16:5356–5359. doi: 10.1016/j.bmcl.2006.07.094. [DOI] [PubMed] [Google Scholar]
- 54.Liu CC, Schultz PG. Recombinant expression of selectively sulfated proteins in Escherichia coli. Nat Biotechnol. 2006;24:1436–1440. doi: 10.1038/nbt1254. [DOI] [PubMed] [Google Scholar]
- 55.Deiters A, Groff D, Ryu Y, Xie J, Schultz PG. A genetically encoded photocaged tyrosine. Angew Chem Int Ed Engl. 2006;45:2728–2731. doi: 10.1002/anie.200600264. [DOI] [PubMed] [Google Scholar]
- 56.Bose M, Groff D, Xie J, Brustad E, Schultz PG. The incorporation of a photoisomerizable amino acid into proteins in E. coli. J Am Chem Soc. 2006;128:388–389. doi: 10.1021/ja055467u. [DOI] [PubMed] [Google Scholar]
- 57.Deiters A, Schultz PG. In vivo incorporation of an alkyne into proteins in Escherichia coli. Bioorg Med Chem Lett. 2005;15:1521–1524. doi: 10.1016/j.bmcl.2004.12.065. [DOI] [PubMed] [Google Scholar]
- 58.Xie J, Wang L, Wu N, Brock A, Spraggon G, Schultz PG. The site-specific incorporation of p-iodo-L-phenylalanine into proteins for structure determination. Nat Biotechnol. 2004;22:1297–1301. doi: 10.1038/nbt1013. [DOI] [PubMed] [Google Scholar]
- 59.Taskent-Sezgin H, Chung J, Patsalo V, Miyake-Stoner SJ, Miller AM, Brewer SH, Mehl RA, Green DF, Raleigh DP, Carrico I. Interpretation of p-cyanophenylalanine fluorescence in proteins in terms of solvent exposure and contribution of side-chain quenchers: a combined fluorescence, IR and molecular dynamics study. Biochemistry. 2009;48:9040–9046. doi: 10.1021/bi900938z. [DOI] [PubMed] [Google Scholar]
- 60.Chin JW, Santoro SW, Martin AB, King DS, Wang L, Schultz PG. Addition of p-azido-L-phenylalanine to the genetic code of Escherichia coli. J Am Chem Soc. 2002;124:9026–9027. doi: 10.1021/ja027007w. [DOI] [PubMed] [Google Scholar]
- 61.Chin JW, Martin AB, King DS, Wang L, Schultz PG. Addition of a photocrosslinking amino acid to the genetic code of Escherichiacoli. Proc Natl Acad Sci U S A. 2002;99:11020–11024. doi: 10.1073/pnas.172226299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brustad E, Bushey ML, Lee JW, Groff D, Liu W, Schultz PG. A Genetically Encoded Boronate-Containing Amino Acid. Angew Chem Int Edit Engl. 2008;47:8220–8223. doi: 10.1002/anie.200803240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang L, Zhang Z, Brock A, Schultz PG. Addition of the keto functional group to the genetic code of Escherichia coli. Proc Natl Acad Sci U S A. 2003;100:56–61. doi: 10.1073/pnas.0234824100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fleissner MR, Brustad EM, Kalai T, Altenbach C, Cascio D, Peters FB, Hideg K, Peuker S, Schultz PG, Hubbell WL. Site-directed spin labeling of a genetically encoded unnatural amino acid. Proc Natl Acad Sci U S A. 2009;106:21637–21642. doi: 10.1073/pnas.0912009106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Turner JM, Graziano J, Spraggon G, Schultz PG. Structural plasticity of an aminoacyl-tRNA synthetase active site. Proc Natl Acad Sci U S A. 2006;103:6483–6488. doi: 10.1073/pnas.0601756103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tippmann EM, Liu W, Summerer D, Mack AV, Schultz PG. A genetically encoded diazirine photocrosslinker in Escherichia coli. Chembiochem. 2007;8:2210–2214. doi: 10.1002/cbic.200700460. [DOI] [PubMed] [Google Scholar]
- 67.Santoro SW, Wang L, Herberich B, King DS, Schultz PG. An efficient system for the evolution of aminoacyl-tRNA synthetase specificity. Nat Biotechnol. 2002;20:1044–1048. doi: 10.1038/nbt742. [DOI] [PubMed] [Google Scholar]
- 68.Wang L, Brock A, Schultz PG. Adding L-3-(2-Naphthyl)alanine to the genetic code of E. coli. J Am Chem Soc. 2002;124:1836–1837. doi: 10.1021/ja012307j. [DOI] [PubMed] [Google Scholar]
- 69.Zhang Z, Smith BA, Wang L, Brock A, Cho C, Schultz PG. A new strategy for the site-specific modification of proteins in vivo. Biochemistry. 2003;42:6735–6746. doi: 10.1021/bi0300231. [DOI] [PubMed] [Google Scholar]
- 70.Tian F, Tsao ML, Schultz PG. A phage display system with unnatural amino acids. J Am Chem Soc. 2004;126:15962–15963. doi: 10.1021/ja045673m. [DOI] [PubMed] [Google Scholar]
- 71.Kavran JM, Gundllapalli S, O’Donoghue P, Englert M, Soll D, Steitz TA. Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation. Proc Natl Acad Sci U S A. 2007;104:11268–11273. doi: 10.1073/pnas.0704769104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Takimoto JK, Dellas N, Noel JP, Wang L. Stereochemical basis for engineered pyrrolysyl-tRNA synthetase and the efficient in vivo incorporation of structurally divergent non-native amino acids. ACS Chem Biol. 2011;6:733–743. doi: 10.1021/cb200057a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mukai T, Kobayashi T, Hino N, Yanagisawa T, Sakamoto K, Yokoyama S. Adding l-lysine derivatives to the genetic code of mammalian cells with engineered pyrrolysyl-tRNA synthetases. Biochem Biophys Res Commun. 2008;371:818–822. doi: 10.1016/j.bbrc.2008.04.164. [DOI] [PubMed] [Google Scholar]
- 74.Yanagisawa T, Ishii R, Fukunaga R, Kobayashi T, Sakamoto K, Yokoyama S. Multistep engineering of pyrrolysyl-tRNA synthetase to genetically encode N(epsilon)-(o-azidobenzyloxycarbonyl) lysine for site-specific protein modification. Chem Biol. 2008;15:1187–1197. doi: 10.1016/j.chembiol.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 75.Wu B, Wang Z, Huang Y, Liu WR. Catalyst-free and site-specific one-pot dual-labeling of a protein directed by two genetically incorporated noncanonical amino acids. Chembiochem. 2012;13:1405–1408. doi: 10.1002/cbic.201200281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.